Abstract
Spindle and kinetochore-associated complex subunit 3 (SKA3) is a microtubule-binding subcomplex of the outer kinetochore that is required for proper chromosomal segregation and cell division. However, little is known regarding the probable mechanism of SKA3, particularly in terms of prostate cancer (PCA) progression. Multiple databases, including TCGA and GTEx, were utilized to examine the expression of SKA3 in PCA patients and to shed light on the clinical significance and potential mechanism of SKA3 in the onset and progression of PCA. The biological function of SKA3 was evaluated in vitro using RT–qPCR and the CCK8 assay. For statistical analysis, the R 3.6.3 software and its associated packages were utilized. SKA3 was shown to be considerably elevated in PCA patients and was linked to a shorter progress free interval (PFI). Furthermore, we discovered that SKA3 mRNA expression was higher in PCA cells than in normal cells, and inhibition of SKA3 could clearly reduce PCA cell proliferation using the CCK8 assay. Finally, SKA3 could be used as a predictive biomarker in PCA patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40164-022-00337-3.
Keywords: Spindle and kinetochore associated complex subunit 3, Prostate cancer, Biomarker, Enrichment analysis, Targeted therapy
To the editor,
The age-standardized incidence rate and death rate of prostate cancer (PCA) in 2019 were 17.39 and 15.28, respectively [1]. The majority of PCA burden was found in older men [2]. By 2044, the aging population will account for approximately 20.8% of the total population [3]. PCA is an age-related disease whose social conundrum can be compounded by global population aging [4]. Clinical heterogeneity in PCA can be illustrated by regional and clonal genetic variety [5]. With the completion of the cancer genome atlas (TCGA), researchers will have access to a mix of genetic and clinical characteristics to approach precision medicine at several levels. Spindle and kinetochore associated complex subunit 3 (SKA3) is a microtubule-binding subcomplex of the outer kinetochore that is essential for proper chromosome segregation and cell division [6–8]. SKA3 knockdown activates the spindle assembly checkpoint, causing sister chromatid cohesion loss and mitotic arrest during metaphase [9]. Differential expression of SKA3 has been linked to the progression and prognosis of various malignant cancers, including laryngeal squamous cell carcinoma [9] and lung adenocarcinoma [10]. Furthermore, our earlier research found that SKA3 may contribute to poor overall survival, disease-specific survival, and progression-free survival in patients with renal papillary cell carcinoma [11]. There is, however, little published evidence about the putative molecular mechanism of SKA3 carcinogenesis in PCA. Based on several public datasets, we investigated the differential expression of SKA3, as well as its connection with clinicopathological characteristics and prognosis.
Multiple databases, including TCGA and GTEx, were utilized to examine the expression of SKA3 in PCA patients and to shed light on the clinical significance and potential mechanism of SKA3 in the onset and progression of PCA. The biological function of SKA3 was evaluated in vitro using RT–qPCR and the CCK8 assay. For statistical analysis, the R 3.6.3 program and its associated packages were utilized. The extra material contained the full-text article as well as comprehensive methodologies.
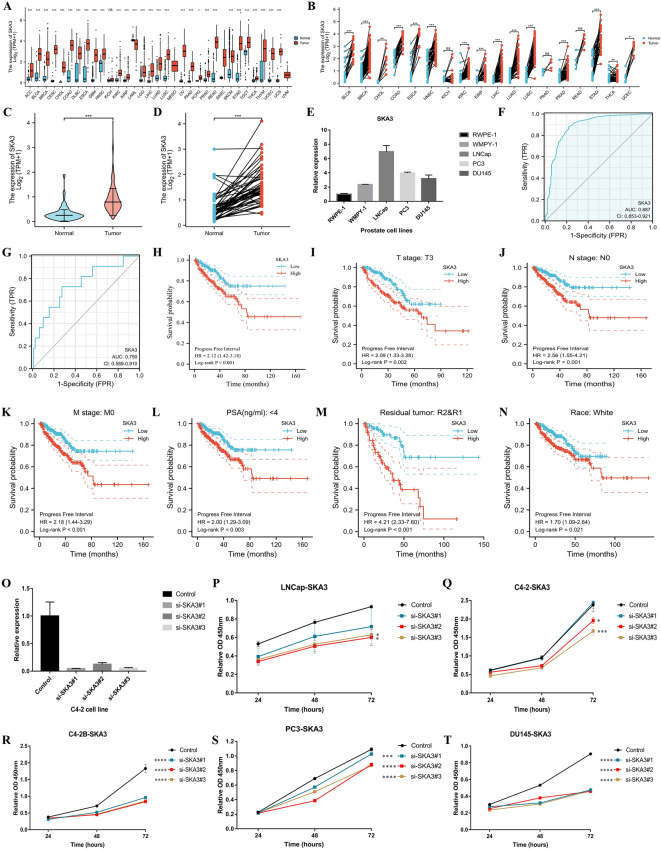
This study included 499 PCA samples and 52 tumor-adjacent tissues from the TCGA database. SKA3 expression was linked to advanced T stage, N1 stage, positive residual tumor, and a higher Gleason score (Table 1). SKA3 mRNA expression was higher in most tumors relative to normal tissues in the pan-cancer analysis of non-paired and paired samples, including PCA (Fig. 1A, B), which was similar with the results of the PCA dataset in the TCGA database (Fig. 1C, D). Furthermore, RT–qPCR revealed that SKA3 was more abundant in PCA cell lines than in normal prostate cell lines (Fig. 1E). The area under the curve (AUC) was 0.887 (95% CI 0.853–0.921), demonstrating that SKA3 may identify tumor from normal patients (Fig. 1F). The AUC in the subgroup analysis of T2–3 versus T4 was 0.750 (95% CI 0.589–0.910). (Fig. 1G). PCA patients with high SKA3 expression had a longer PFI than those with low expression (P0.001, Fig. 1H). Furthermore, patients with higher SKA3 had considerably shorter PFI when compared to their counterparts in the T3 stage, N0 stage, M0 stage, PSA4 ng/ml, positive residual tumor, and white (this signified white population) subgroup analysis (Fig. 1I–N). Using the RT–qPCR test, we discovered that SKA3 expression was downregulated after transfection of the three siRNAs on the proliferation ability of PCA cell lines (Fig. 1O). Furthermore, these siRNAs were able to dramatically inhibit the ability of PCA cells to multiply (Fig. 1P–T), particularly in cells with high levels of malignancy, such as C4-2B, PC3, and DU145, which was consistent with the clinical correlations of SKA3.
Table 1.
The relationships between SKA3 expression and clinicopathological features in prostate cancer patients in the TCGA database
| Characteristic | Low expression of SKA3 (n = 249) | High expression of SKA3 (n = 250) | P value |
|---|---|---|---|
| Age, n (%) | 0.080 | ||
| < = 60 | 122 (54.5%) | 102 (45.5%) | |
| > 60 | 127 (46.2%) | 148 (53.8%) | |
| T stage, n (%) | < 0.001 | ||
| T2 | 124 (65.6%) | 65 (34.4%) | |
| T3 | 118 (40.4%) | 174 (59.6%) | |
| T4 | 2 (18.2%) | 9 (81.8%) | |
| N stage, n (%) | 0.036 | ||
| N0 | 171 (49.3%) | 176 (50.7%) | |
| N1 | 28 (35.4%) | 51 (64.6%) | |
| M stage, n (%) | 1.000 | ||
| M0 | 223 (49%) | 232 (51%) | |
| M1 | 1 (33.3%) | 2 (66.7%) | |
| Race, n (%) | 0.101 | ||
| Asian | 4 (33.3%) | 8 (66.7%) | |
| Black or African American | 35 (61.4%) | 22 (38.6%) | |
| White | 202 (48.7%) | 213 (51.3%) | |
| Residual tumor, n (%) | 0.006 | ||
| R0 | 173 (54.9%) | 142 (45.1%) | |
| R1 | 59 (39.9%) | 89 (60.1%) | |
| R2 | 2 (40%) | 3 (60%) | |
| Zone of origin, n (%) | 0.348 | ||
| Central Zone | 2 (50%) | 2 (50%) | |
| Overlapping / Multiple Zones | 50 (39.7%) | 76 (60.3%) | |
| Peripheral Zone | 69 (50.4%) | 68 (49.6%) | |
| Transition Zone | 3 (37.5%) | 5 (62.5%) | |
| PSA (ng/ml), n (%) | 0.102 | ||
| < 4 | 214 (51.6%) | 201 (48.4%) | |
| >=4 | 9 (33.3%) | 18 (66.7%) | |
| Gleason score, n (%) | < 0.001 | ||
| 6 | 36 (78.3%) | 10 (21.7%) | |
| 7 | 142 (57.5%) | 105 (42.5%) | |
| 8 | 28 (43.8%) | 36 (56.2%) | |
| 9 | 42 (30.4%) | 96 (69.6%) | |
| 10 | 1 (25%) | 3 (75%) | |
| PFI event, n (%) | < 0.001 | ||
| Alive | 218 (53.8%) | 187 (46.2%) | |
| Dead | 31 (33%) | 63 (67%) |
SKA3 spindle and kinetochore-associated complex subunit 3; TCGA the Cancer Genome Atlas; PFI progress free interval
Fig. 1.
The correlation analysis between SKA3 expression and clinical parameters and the effect of SKA3 expression on the proliferation ability of PCA cells. A The differential expressions of SKA3 in pan-cancer level using non-paired samples; B the differential expressions of SKA3 in pan-cancer level using paired samples; C the differential expressions of SKA3 in PCA using non-paired samples; D the differential expressions of SKA3 in PCA using paired samples; E Relative expression of SKA3 among prostate normal and tumor cell lines; F the ROC curve of SKA3 distinguishing tumor from normal; G the ROC curve of SKA3 distinguishing T2–3 from T4; H Kaplan–Meier curve showing progress free survival of high and low expression of SKA3; I Kaplan–Meier curve showing progress free survival of high and low expression of SKA3 in patients with T3 stage; J Kaplan–Meier curve showing progress free survival of high and low expression of SKA3 in patients with N0 stage; K Kaplan-Meier curve showing progress free survival of high and low expression of SKA3 in patients with M0 stage; L Kaplan–Meier curve showing progress free survival of high and low expression of SKA3 in patients with PSA < 4 ng/ml; M Kaplan–Meier curve showing progress free survival of high and low expression of SKA3 in patients with positively residual tumor; N Kaplan–Meier curve showing progress free survival of high and low expression of SKA3 in white patients; O RT–qPCR results of SKA3 siRNAs; P effect of SKA3 siRNAs on LNCap using CCK8 assay; Q effect of SKA3 siRNAs on C4-2 using CCK8 assay; R effect of SKA3 siRNAs on C4-2B using CCK8 assay; S effect of SKA3 siRNAs on PC3 using CCK8 assay; T effect of SKA3 siRNAs on DU145 using CCK8 assay. ROC = receiver operating characteristic curve. R = residual tumor. RT–qPCR = Real-time quantitative polymerase chain reaction
To date, no association between SKA3 and PCA patients has been observed. SKA3 mRNA expression was found to be significantly higher in most malignancies than in normal tissues in our investigation. Furthermore, we discovered that SKA3 mRNA expression was higher in PCA cells than in normal cells, and inhibiting SKA3 may obviously reduce PCA cell growth. Furthermore, those with higher SKA3 mRNA expression had a higher risk of advancement than those with lower SKA3 mRNA expression. Moreover, patients with T3 stage, N0 stage, M0 stage, PSA4 ng/ml, positive residual tumor, or white individuals were more likely to progress if they had high levels of SKA3 mRNA. These findings suggest that increased SKA3 expression may enhance the onset and progression of PCA and has a strong clinical connection. Based on the subgroup analysis, we also postulated a dose impact of SKA3 carcinogenesis.
In conclusion, SKA3 might involve in the progression of PCA and serve as a prognostic biomarker for PCA patients.
Supplementary Information
Additional file 1. The full text of original manuscript containing figures.
Acknowledgements
The results showed here are in whole or part based upon data generated by the Genotype-Tissue Expression (GTEx) Program (https://commonfund.nih.gov/GTEx/) and TCGA Research Network (https://www.cancer.gov/tcga).
Author contributions
DCF proposed the project, conducted data analysis, interpreted the data, and wrote the manuscript; WZZ, XS, DXL, and QX conducted data analysis, interpreted the data; QW and LY, supervised the project, and interpreted the data. All authors read and approved the final manuscript.
Funding
This program was supported by the National Natural Science Foundation of China (Grant Nos. 81974099, 82170785, 81974098, 82170784), National Key Research and Development Program of China (2021YFC2009303), programs from Science and Technology Department of Sichuan Province (Grant Nos. 2021YFH0172), Young Investigator Award of Sichuan University 2017 (Grant No. 2017SCU04A17), Technology Innovation Research and Development Project of Chengdu Science and Technology Bureau (2019-YF05-00296-SN), Sichuan University–Panzhihua science and technology cooperation special fund (2020CDPZH-4). The funders had no role in the study design, data collection or analysis, preparation of the manuscript, or the decision to publish.
Availability of data and materials
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Additional file 1.
Declarations
Ethical approval and consent to participate
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dechao Feng, Weizhen Zhu and Xu Shi contributed equally to this work
Contributor Information
Qiang Wei, Email: weiqiang933@126.com.
Lu Yang, Email: wycleflue@scu.edu.cn.
References
- 1.Lin L, Li Z, Yan L, Liu Y, Yang H, Li H. Global, regional, and national cancer incidence and death for 29 cancer groups in 2019 and trends analysis of the global cancer burden, 1990–2019. J Hematol Oncol. 2021;14:197. doi: 10.1186/s13045-021-01213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zi H, He SH, Leng XY, Xu XF, Huang Q, Weng H, et al. Global, regional, and national burden of kidney, bladder, and prostate cancers and their attributable risk factors, 1990–2019. Mil Med Res. 2021;8:60. doi: 10.1186/s40779-021-00354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gitlin LN, Fuentes P. The Republic of Chile: an upper middle-income country at the crossroads of economic development and aging. Gerontologist. 2012;52:297–305. doi: 10.1093/geront/gns054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng D, Shi X, Zhang F, Xiong Q, Wei Q, Yang L. Mitochondria dysfunction-mediated molecular subtypes and gene prognostic index for prostate cancer patients undergoing radical prostatectomy or radiotherapy. Front Oncol. 2022;12:858479. doi: 10.3389/fonc.2022.858479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng D, Xiong Q, Wei Q, Yang L. Cellular landscape of tumour microenvironment in prostate cancer. Immunology. 2022 doi: 10.1111/imm.13456. [DOI] [PubMed] [Google Scholar]
- 6.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–85. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–52. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–80. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao W, Zhang Y, Luo H, Niu M, Zheng X, Hu W, et al. Targeting SKA3 suppresses the proliferation and chemoresistance of laryngeal squamous cell carcinoma via impairing PLK1-AKT axis-mediated glycolysis. Cell Death Dis. 2020;11:919. doi: 10.1038/s41419-020-03104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu DD, Chen HL, Lou LM, Zhang H, Yang GL. SKA3 promotes lung adenocarcinoma metastasis through the EGFR-PI3K-Akt axis. Biosci Rep. 2020;40:BSR20194335. doi: 10.1042/BSR20194335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng D, Zhang F, Liu L, Xiong Q, Xu H, Wei W, et al. SKA3 serves as a biomarker for poor prognosis in kidney renal papillary cell carcinoma. Int J Gen Med. 2021;14:8591–602. doi: 10.2147/IJGM.S336799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The full text of original manuscript containing figures.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Additional file 1.