Abstract
Noninvasive manipulation of cell signaling is critical in basic neuroscience research and in developing therapies for neurological disorders and psychiatric conditions. Here, the wireless force-induced stimulation of primary neuronal circuits through mechanotransduction mediated by magnetic microdiscs (MMDs) under applied low-intensity and low-frequency alternating magnetic fields (AMFs), is described. MMDs are fabricated by top-down lithography techniques that allow for cost-effective mass production of biocompatible MMDs with high saturation and zero magnetic magnetic moment at remanence. MMDs are utilized as transducers of AMFs into mechanical forces. When MMDs are exposed to primary rat neuronal circuits, their magneto-mechanical actuation triggers the response of specific mechanosensitive ion channels expressed on the cell membranes activating ≈50% of hippocampal and ≈90% of cortical neurons subjected to the treatment. Mechanotransduction is confirmed by the inhibition of mechanosensitive transmembrane channels with Gd3+.
Mechanotransduction mediated by MMDs cause no cytotoxic effect to neuronal cultures. This technology fulfills the requirements of cell-type specificity and weak magnetic fields, two limiting factors in the development of noninvasive neuromodulation therapies and clinical equipment design. Moreover, high efficiency and long-lasting stimulations are successfully achieved. This research represents a fundamental step forward for magneto-mechanical control of neural activity using disc-shaped micromaterials with tailored magnetic properties.
Keywords: alternating magnetic fields, lithography, magnetic nanomaterials, magnetic vortices, mechanotransduction, neurostimulation
1. Introduction
Noninvasive manipulation of cell signaling is critical in basic neuroscience research and in developing therapies for neurological disorders and psychiatric conditions. Noninvasive neuromodulation technologies include transcranial magnetic stimulation (TMS), transcranial current stimulation (tDCS), and the emerging transcranial focused ultrasound stimulation (tFUS). These neuromodulation technologies are classified as “noninvasive” because they facilitate the control of neural activity through the skull and skin.[1] Among them, TMS is the only therapy cleared by the FDA to treat patients suffering from treatment-resistant major depressive disorder,[2] obsessive-compulsive disorder,[3] smoking cessation,[4] and pain associated with migraine headaches preceded by an aura.[5] Much effort is devoted to studying their applications as a potential treatment for a range of other disorders such as post-stroke rehabilitation,[6] epilepsy,[7] Parkinson’s disease,[8] Alzheimer’s,[9] anxiety,[10] schizophrenia,[11] and chronic pain.[12] In general, these noninvasive techniques, whether administered alone or in combination with pharmacological or behavioral therapies, have given rise to positive and enhanced therapeutic outcomes. However, at their current development level, they suffer from a broad radius of action in the order of millimeters to centimeters.[13] This lack of precision consequently results in undesired cell stimulation and ablation of healthy brain tissues.
Another approach for noninvasive, cell-type-specific control of cellular signaling that utilizes external stimuli consists on the use of heat dissipation produced by magnetic nanoparticles (MNPs)[14] and plasmonic nanostructures.[15] Unlike plasmonic nanostructures, which utilizes light as a stimulus and its penetration is limited to a few centimeters into the body, MNPs benefit from the weak magnetic properties and low electrical conductivity of tissue, allowing alternating magnetic fields (AMFs) to reach deep into the body.[16] This advantage makes hysteresis heating of MNPs particularly promising for applications that involve stimulation of brain structures. Both bulk and nanoscale heating of MNPs have been reported to evoke neural activity in vitro and in vivo.[14] Still, this magneto-thermal approach is limited for heat-sensitive neurons and requires transgenes to sensitize neurons to heat. Another limitations of MNPs heat dissipation are the potential off-target heating effects and the need for high-frequency AMFs. The latter makes scaling the electromagnet’s coils extremely challenging, impeding the universal adoption of magnetic hyperthermia in biomedical research and clinical practices.
Alternatively, nanoparticle-assisted mechanotransduction has been proposed to control biological signaling. Zinc sulfite-based nanoparticles have been investigated to convert mechanical forces from ultrasound stimulation into optical energy as a untethered alternative for optogenetics called “sono-optogenetics.”[17] Zinc sulfite-based nanoparticles circulating in the brain vascularate can transduce focused ultrasound into localized 470 nm light, which triggers the reposnse of neurons expressing channelrhodopsin-2. Sono-optogenetics overcomes the invaisive delivery of light required in optogenetics, however, still requires the use of transgenes for sensitizing neurons to light. Mechanotransduction can also be achieved by engineering magnetic nano and microstructures. Recently, a magnetic torque nanoactuator toolkit named “m-Torquer” has been developed for neuromodulation.[18] The nanoactuator toolkit is composed of a rather intricate 500 nm assembly of octahedral iron oxide magnetic nanoparticles and a rotating circular magnet array that favors Brownian over Neel relaxation to produce a torque. The m-Torquer is capable to precisely stimulate neuronal activity in mechanosensitive neurons with spatial accuracy and long working distance. Mechanotransduction can be also realized in magnetic materials with unique spin configurations such as magnetic vortex ground state.[19] Magnetic vortex state –, e.g., a magnetic state with zero net magnetic moments at remanence – can be achieved in low aspect-ratio structures like magnetic microdiscs (MMDs). One tremendous advantage of MMDs with vortex state is that they do not agglomerate when the external magnetic field is off. Therefore, MMDs can be larger than MNPs possessing a higher magnetic moment under the same applied fields.[20]
Due to their low aspect ratio, when AMFs are applied, MMDs rotate to align their magnetic moments with the field direction. The torque generated on the MMDs in response to the AMFs, is essential in controlling cellular activity. Transduction of low-intensity AMFs into mechanical forces by MMDs has been applied to kill cancer cells via the disruption of cell membranes caused by high exerted forces.[21] Additionally, when the actuation forces are regulated, such as in magnetite magnetic nanodiscs, it is possible to use the mechanical forces to control mechano-sensory cells remotely.[22] However, magnetite magnetic nanodiscs present low magnetic moments that limit their efficiency.[22,23]
This work describes the wireless force-induced stimulation of in-vitro primary neurons through mechano-transduction mediated by MMDs under applied low-intensity AMFs. To overcome the limitations of synthesized magnetite nanoparticles, we utilized Nickel-Iron permalloy (Ni80Fe20) MMDs coated with gold. Permalloy microdiscs of 4 μm in diameter and 100 nm in thickness were fabricated by direct writing laser lithography and physical vapor deposition. This technology allows the cost-effective mass production of biocompatible MMDs with the advantage that it can be scaled down into the nanometer range using other top-down lithography methods.[20a,24] The MMDs exhibit a magnetic vortex state configuration. Thus, the large volume of the discs and high magnetization of the permalloy provide the MMDs with a high magnetic moment and zero remanence.[20a] Another advantage of MMDs is their response to small amplitude and low-frequency oscillatory fields. Therefore, MMDs can be used as transducers of low-frequency and low-amplitude AMFs into mechanical forces. When exposed to primary neurons, MMDs magneto-mechanical actuation triggers the response of mechanosensitive ion channels expressed on the cell membranes, allowing for remote control of neural activity (Figure 1). In this work, we corroborated that the observed enhancement of neural signaling mediated by MMDs results from mechano-sensitive ion channels’ force-induced response. Moreover, a percentage around 90% of responsive neurons and a desirable long-lasting post-stimulation neural activity were achieved with this technique. This unique feature of MMDs represents a tremendous potential for neuronal stimulation since it fulfills the requirements of cell-type specificity and weak magnetic fields, limiting factors in the development of noninvasive neuromodulation therapies and the design of clinical equipment.
Figure 1.
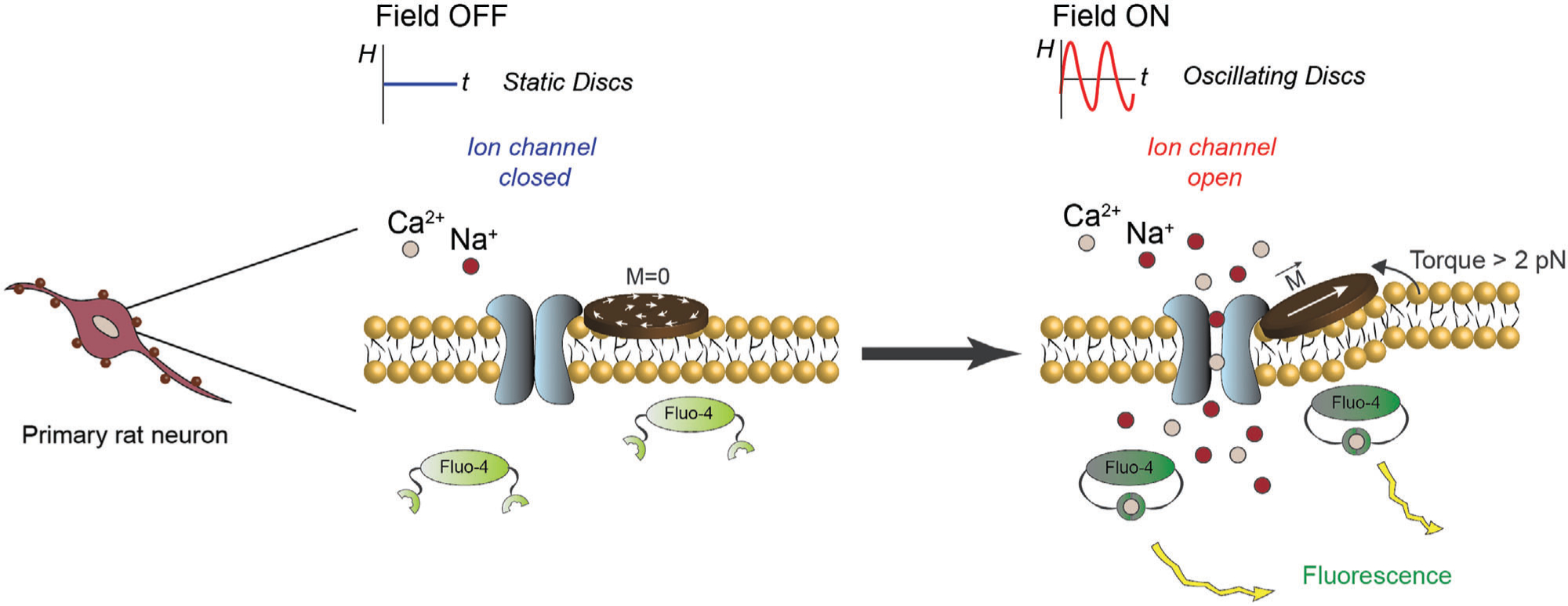
Overview of the experimental scheme. Functionalized MMDs in coculture with primary rat neurons. Upon exposure to AMFs, the torque produced by MMDs triggers the response of mechanosensitive ion channels located in the cell membrane. This response gates calcium ion influx detectable through the fluorescence intensity increase of Fluo-4.
2. Results and Discussion
2.1. MMDs Characterization and Surface Functionalization
Here, we utilize Ni80Fe20 MMDs coated with gold fabricated by direct writing laser lithography. Homogeneity of permalloy MMDs arrays, morphology, and dimensions were verified by Scanning Electron Microscopy (SEM). Figure 2a shows a representative SEM image. The patterning of 4 μm in diameter MNDs is very homogenous as expected from their fabrication technique. Figure 2b shows a representative SEM image of higher magnification with a single MMD. Figure 2c shows the hydrodynamic size distribution of the MMDs in Tyrode characterized by Dynamic Light Scattering (DLS). MMDs display a narrow size distribution with a mean diameter of 3.8 μm and a poly dispersity index (PDI) of 0.042. MMDs size measured by SEM and DLS are consistent with each other for MMDs diameter of 4 μm. It is well known that DLS, which utilized particle diffusion coefficient to infer colloidal particle size via Stokes-Einstein relation,[25] is not reliable for nonspherical particle geometries as the relationship between the diffusion coefficient and dimension can be relatively complex.[26] However, this observation does not apply for 2D particles as MMDs are, where DLS spherical assumption is accurate in determining hydrodynamic diameter.[27]
Figure 2.
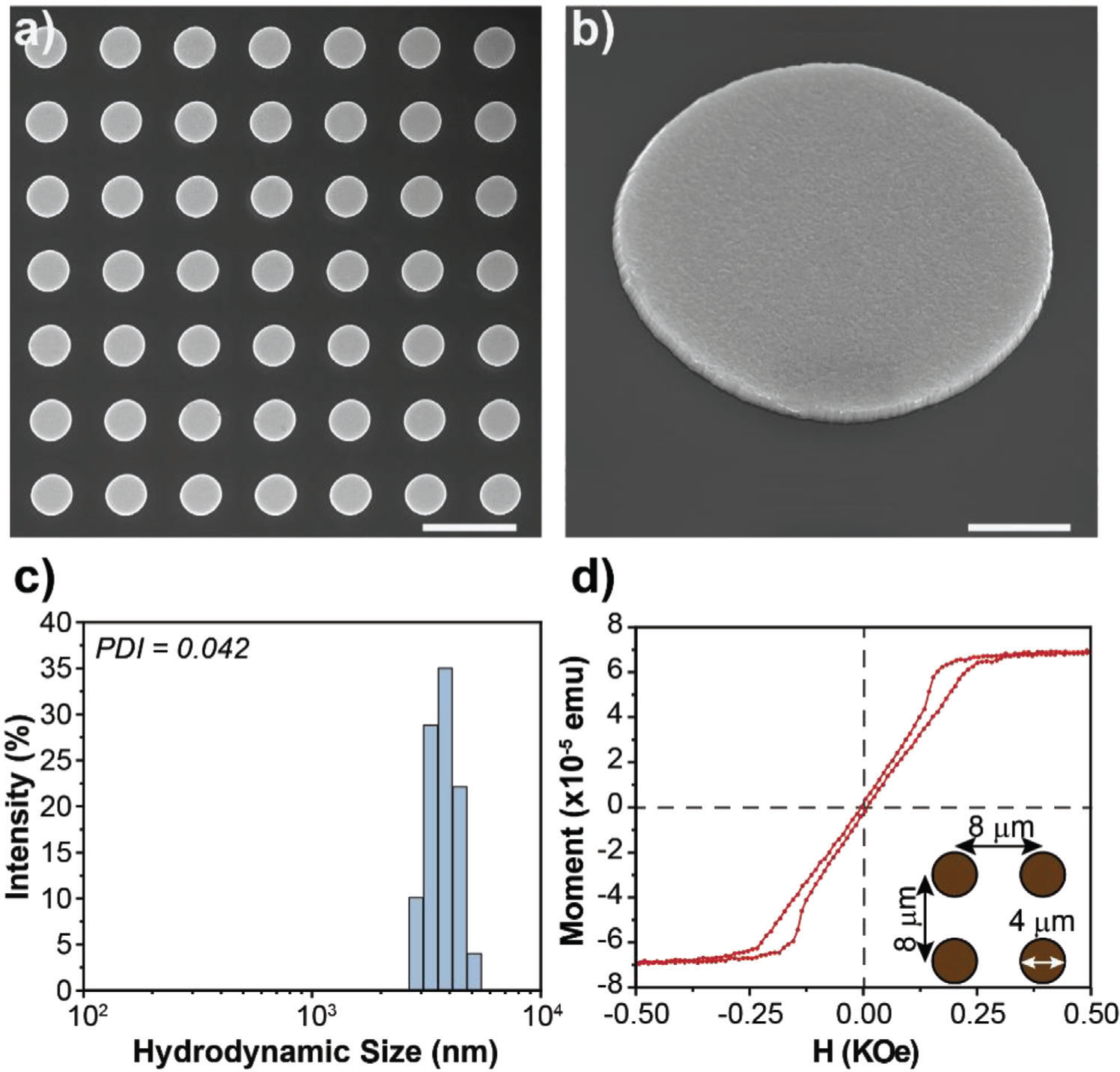
MMDs characterization. a) SEM image of MMDs on the Si wafer. Scale bar = 10 μm, b) SEM image of a tilted single MMD on the Si wafer. Scale bar = 1 μm, c) Hydrodynamic size distribution of MMDs in water after being released from the Si wafer, and d) Hysteresis loop of a square array of MMDs after photoresist liftoff and before release from the Si wafer.
The magnetic characterization of the MMDs is depicted in Figures 2d and Figure S1 (Supporting Information). Figure 2d shows the hysteresis loop of a square array of MMDs with the dimensions shown in the figure’s inset. This sample was cut from a 4 inches wafer after photoresist liftoff. The pinched shape of the hysteresis loop and the lack of magnetic moment at remanence indicates that the MMDs develop a vortex state when the external field is off. The magnetic moment per disc (9.8×10−10 emu disc−1) was estimated from the saturation magnetization and the number of discs (5.6×104) in the sample’s area (3.6 mm2). Figure S1 (Supporting Information) shows the hysteresis loop of the MMDs after being released from the Si wafer. In this case, a silicon chip was coated with 4 μL of MMDs in DI water suspension. The round shape of the loop is caused by the discs’ random distribution and their dipolar interaction. Interestingly, despite the random distribution of the discs on the sample, the remanent magnetic moment remains zero, consistent with the magnetic vortex state. From this measurement, the concentration of MMDs in solution was estimated as 1.6×104 MMDs mL−1. In both cases, the hysteresis loops show that the discs develop a magnetic vortex state and reach saturation at relatively low applied magnetic fields (≈0.5 kOe). The magnetic moment of the MMDs fabricated here (9.8×10−10 emu disc−1) is 1000× higher than that of chemicaly synthesized magnetite nanodisc (6.7×10−13 emu disc−1).[22] Both lithographic and synthesized nanomagnets have a thickness limitation to preserve a vortex state. Interestingly, the lithography method overcomes this issue with the stacking capability, which allows a further enhancement of the magnetic moment of particles. Figure S2a,b (Supporting Information) shows the loss of the vortex spin configuration as permalloy thickness increases from 100 to 200 nm. However, a stacking layer structure of permalloy (100 nm)/titanium (20 nm)/permalloy (100 nm) recovers a magnetization curve that resembles the vortex state profile, but in MMDs with a double magnetic moment (Figure S2c, Supporting Information).
MMDs hysteresis heat dissipation at the AMFs conditions of 0.188 kOe m and 6 Hz was measured for 1 mg of MMDs in water. No increase in the temperature was observed, neither for the MMDs solution nor for water alone.
Although zero-remanence MMDs are highly stable in water, we surface engineered them with polymer brushes to neutralize their charge, enhance their biological fluids’ stability, and avoid nonspecific interactions. Polymer brushes are commonly used in surface chemistry to stabilize suspensions of colloidal particles. When polymer brushes are grafted onto particles surface, the entropy gain and the repulsion between polymer chains, promote steric and/or electrostatic colloidal stability.[28] Poly (ethylene glycol) methyl ether methacrylate (POEGMA) brushes were synthesized from the surface of MMDs by well-established Atom Transfer Radical Polymerization (ATRP) protocols.[28c] ATRP was carried out specifically from the surface of MMDs. Rhodamine B isothiocyanate was used to fluorescently label POEGMA brushes by hydrophobic interactions. Figure 3a shows a representative fluorescent image of MMDs arrays after surface functionalization with POEGMA brushes. The observed fluorescence of the labeled polymer is consistent with the localization of MMDs in the array. After functionalization, MMDs-POEGMA were released from the Si wafer obtaining to partially coated MMDs with POEGMA in solution. MMDs-POEGMA hydrodynamic size was characterized by DLS in Tyrode’s solution. Figure 3b displays a narrow hydrodynamic size distribution of MMDs-POEGMA with a mean diameter of 4.1 μm and PDI of 0.039. The MMDs surface functionalization was also confirmed by ζ-potential measurements, as shown in Figure 3c. Prior functionalization MMDs display a surface charge of −24.4 ± 8 mV. After grafting POEGMA from MMDs surface, the ζ-potential is neutralized to −7.11 ± 3 mV. Together, these results suggest the formation of a nanometer thick POEGMA coating on MMDs.
Figure 3.
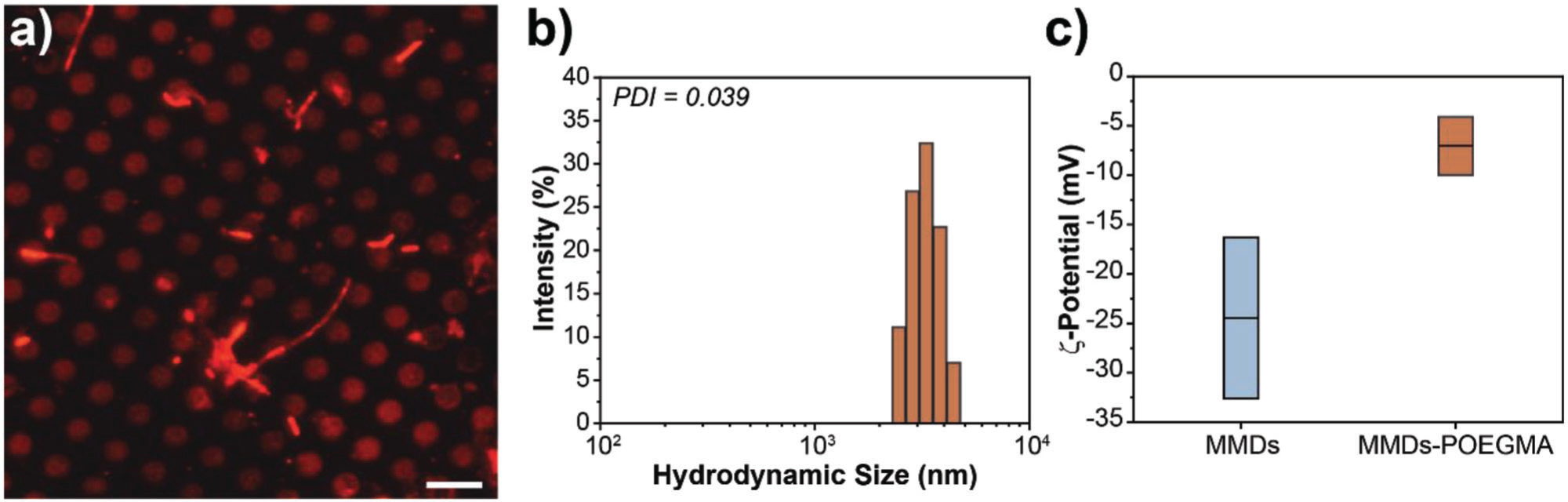
MMDs surface functionalization. a) Fluorescent image of MMDs-POEGMA, where the polymer brushes are fluorescently labeled with Rhodamine B (red). Scale bar = 10 μm, b) Hydrodynamic size distribution of MMDs-POEGMA, c) Zeta-Potential of MMDs before and after functionalization with POEGMA brushes.
2.2. Force-Inducing Neuronal Stimulation Mediated by MMDs
Mechano-sensitive ion channels located in the cell membrane of specific cell populations can gate the influx of ions in response to various mechanical stimuli, including local membrane stretching, cell squeezing, shear stress, cell swelling, deflection, and substrate deformation.[29] To enable cell-type-specific mechanotransduction in neurons through shear stress-mediated by MMDs, while avoiding bulk disruption of tissue, MMDs size and magnetic moment must be tailored to achieve highly efficient neuromodulation in the presence of AMFs on time scales relevant to inter-neuronal communication. The force thresholds for activation of mechanosensitive ion channels or membrane receptors typically range between 0.2 and 1 pN.[30] The mechanical force exerted by MMDs comes from the alignment of the MMDs’ plane with the external magnetic field. The torque magnitude depends on the magnetic moment of disc. For the same material, larger volume will lead to larger magnetic moment. Consequently, to achieve magnetic control of cell mechanoreceptors with low-frequency, low-amplitude AMFs (0.18 – 0.63 kOe, 5 – 10 Hz), the volume of the MMDs must be between 10−10 and 10−22 cm3.[22,31]
MMDs functionalized with POEGMA brushes were used to stimulate neural activity. MMDs were suspended in water and counted using an automatic cell counter. Optimization experiments were done to find the minimum concentration of MMDs needed to evoke neural activity. The optimal concentration for cell stimulation was found to be 10 MMDs per seeded cell, which equates to a total MMDs volume of 1.25×10−11 cm3, consistent with our preliminary estimates. In contrast, previous studies in magneto-mechanical neural stimulation using magnetite nanodiscs[22] required 1000X more particles to detect the stimulation of mechanosensitive ion channels. This observation is in agreement with the reported magnetic moments of magnetite nanodiscs (6.7×10−13 emu disc−1), which is lower than for the MMDs fabricated here.
To confirm the mechanical actuation of MMDs under the applied AMFs of 0.188 kOe and 6 Hz, we first recorded with the bright field the MMDs’ behavior when cocultured with hippocampal neurons and upon exposure to AMFs perpendicular to the plane at a frame rate of 200 ms. A representative video of MMDs’ behavior is shown in the Supporting Information Video S1. The micrometer size and metallic contrast allow us to distinguish MMDs from the neural culture clearly as black dots. We were able to associate MMDs’ vibration with AMFs exposure. When AMFs were turned on, MMDs disappear from the field of view. This observation was attributed to MMDs alignment to the magnetic field (vertically), which position them out of the plane of view. When the AMFs are turned off, MMDs slowly reappear to the field of view in the same positions than before applying the stimulus. These observations confirm that MMDs’ mechanical actuation is due to a response to AMFs.
2.2.1. Force-Inducing Stimulation of the Mechanosensitive Ion Channel eMscL Mediated by MMDs
We first studied MMDs force-inducing neural stimulation in hippocampal neurons. As hippocampal neurons are not associated with overexpression of mechanosensitive ion channels, we transfected the cultures with the mechanosensitive ion channel of large conductance eMscL using Lipofectamine 3000 reagent (Figure S3, Supporting Information). Figure 4a shows calcium images from a representative video of hippocampal neurons expressing eMscL and subjected to 1-minute force-inducing stimulation mediated by MMDs and AMFs. The calcium imaging video was recorded for 2 minutes at a frame rate of 200 ms. The AMFs were turned on at t = 30 seconds and turned off at t = 90 seconds. The calcium images illustrate an increase in the fluorescence intensity of intracellular calcium overtime associated with MMDs mechano-actuation at the cell membrane, which results in the enhancement of transmembrane channels that lead to an influx of ions in the cell. A representative video for hippocampal neurons force-inducing stimulation is shown in the Supporting Information Video S2. The recorded videos were analyzed for intracellular calcium fluorescence over time to quantify cell activity. A threshold relative to five times the standard deviation from the background was set to differentiate active cells from inactive cells (Figure S4, Supporting Information). An automatic code was used to subtract the background and randomize the results presented here. Figure 4b displays the activity of 100 random neurons during AMFs force-inducing stimulation mediated by MMDs. Figure 4c represents the percentage of active neurons over time for the same 100 random neurons. In both figures, the purple shadowed area indicates the exposure to AMFs. Figure 4d summarizes the percentage of responsive neurons quantified from five different samples in each condition. Hippocampal neurons expressing eMscL in the absence of AMFs and MMDs were used as the negative control, while hippocampal neurons in the presence of AMFs and MMDs but not expressing eMscL was used as the negative eMscL control. Representative raster plots and the percentage of active neurons of 100 random neurons for the controls are shown in Figure S5 in the Supporting Information.
Figure 4.
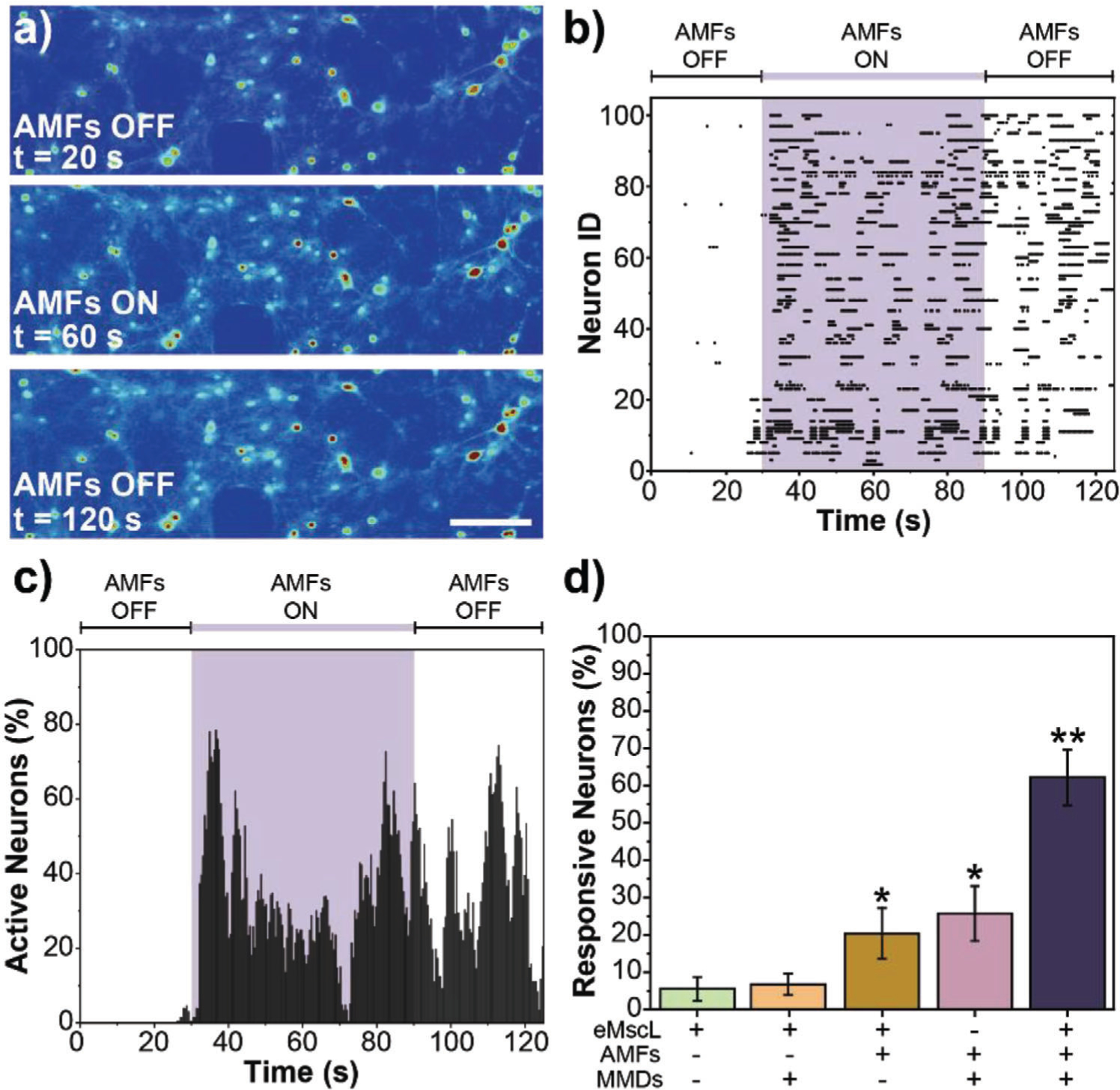
Force-inducing stimulation of hippocampal neurons mediated by MMDs. a) Calcium fluorescence images from a representative video of hippocampal neurons expressing eMscL and subjected to 1-minute force-inducing stimulation mediated by MMDs at the AMFs conditions of 0.188 kOe and 6 Hz. Scale bar = 50 μm, b) Raster plot tracking the activity of 100 random neurons expressing eMscL, cocultured with MMDs and exposed to AMFs, c) Data from the raster plot normalized to the cell population to represent percentage of active neurons as a function of time. In both, (b) and (c), the purple shadowed area indicates the exposure to AMFs, d) Percent response was determined by dividing the number of neurons that were active during AMFs exposure by the total number of neurons in the field of view. Error bars represent standard deviation (n = 5 samples per condition). ANOVA test was performed to determine statistical significance. * Indicates statistical difference with Negative Control (neurons expressing eMscL in the absence of AMFs and MMDs) and ** indicates statistical difference with both Negative Control and eMscL Control (nontransfected neurons in the presence of AMFs and MMDs) (p-value < 0.05).
We were able to evoke neural activity in hippocampal cultures expressing eMscL, cocultured with MMDs, and exposed to low-intensity AMFs. Neural activity was enhanced by 50% within seconds of applying AMFs and continued even after AMFs were turned off. It is also possible to distinguish that more than 40% of the responsive neurons showed synchronized neural activity since the beginning of the stimulation. None of these effects were observed in the control conditions tested. The response of neurons to the force-inducing treatment is ≈35% higher in hippocampal neurons expressing eMscL and in the presence of MMDs and AMFs. These results suggest that the stimulus observed is associated with the mechanical actuation of MMDs on the mechanosensitive ion channel eMscL located in the cell membrane.
2.2.2. Force-Inducing Stimulation of Endogenous Mechanosensitive Ion Channels Mediated by MMDs
We proposed force-inducing neural stimulation mediated by MMDs as a noninvasive neuromodulation approach, we also tested our system, avoiding the use of transgenes, like in eMscL transfection in hippocampal neurons. We studied transgene-free force-inducing neural stimulation mediated by MMDs in primary cortical neurons as they are well known to express different mechanosensitive ion channels endogenously, including TRPV1, TRPV2, TRPV4, Piezo1, TRPC1, TRPM7 TRPP1/2 complex, and indirectly actuated G-protein coupled receptors.[32] Figure 5a shows calcium images from a representative video of cortical neurons subjected to 1-minute force-inducing stimulation mediated by MMDs and AMFs. The calcium imaging video was recorded as described above for hippocampal neurons. The calcium images illustrate an increase in the fluorescence intensity of intracellular calcium overtime associated with MMDs mechano-actuation at the cell membrane. A representative video force-inducing cortical neural stimulation is shown in Supporting Information Video S3. The recorded videos were analyzed following the same protocol described above for hippocampal neurons. Figure 5b shows the activity of 100 random neurons during AMFs force-inducing stimulation mediated by MMDs. Figure 5c represents the percentage of active neurons over time for the same 100 random cortical neurons. In both figures, the purple shadowed area indicates the exposure to AMFs. To confirm that the induced neural activity was an effect of mechanotransduction facilitated by the MMDs response to AMFs, we treated cortical neurons with gadolinium (III) (Gd3+). Gd3+ is a well-studied global inhibitor for mechanosensitive channels that acts by modifying the deformability of the lipid bilayer.[33] The dose of Gd3+ was carefully chosen to avoid altering neural activity by blocking non-mechanosensitive channels.[15b,34] A representative video force-inducing cortical neural stimulation on neurons pretreated with Gd3+ is shown in Supporting Information Video S4. Figure 5d summarizes the percentage of responsive cortical neurons quantified from five different samples in each condition. Cortical neurons in the absence of AMFs and MMDs was used as a negative control. Representative raster plots and percentage of active neurons of 100 random neurons for the controls are shown in Figure S6 in the Supporting Information.
Figure 5.
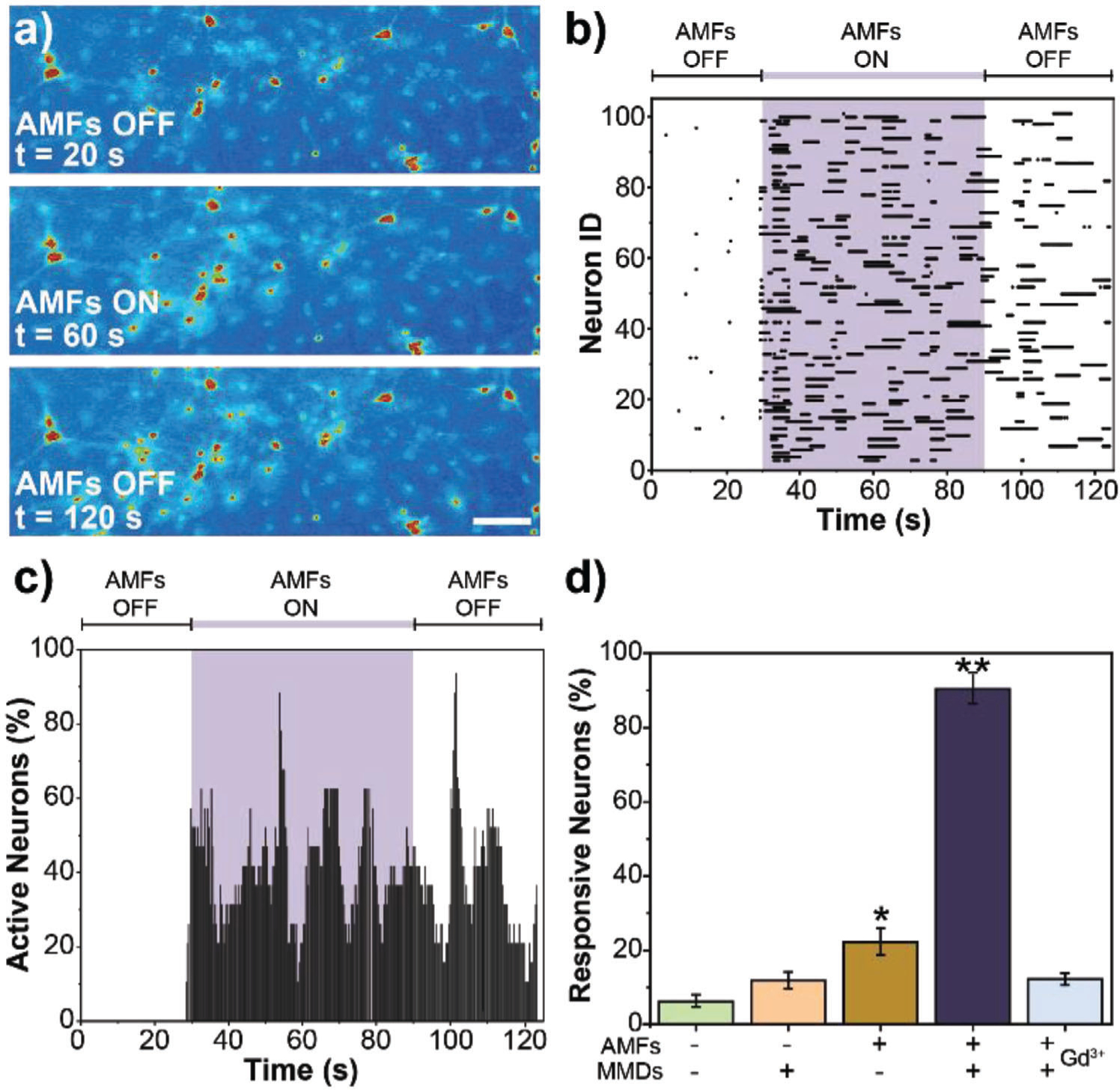
Force-inducing stimulation of cortical neurons mediated by MMDs. a) Calcium fluorescence images from a representative video of cortical neurons subjected to 1-minute force-inducing stimulation mediated by MMDs at the AMFs conditions of 0.188 kOe and 6 Hz. Scale bar = 50 μm, b) Raster plot tracking the activity of 100 random neurons cocultured with MMDs and exposed to AMFs, c) Data from the raster plot normalized to the cell population to represent percentage of active neurons as a function of time. In both, (b) and (c), the purple shadowed area indicates the exposure to AMFs, d) Percent response was determined by dividing the number of neurons that were active during AMFs exposure by the total number of neurons in the field of view. Error bars represent standard deviation (n = 5 samples per condition). ANOVA test was performed to determine statistical significance. * Indicates statistical difference with Negative Control (neurons in the absence of AMFs and MMDs) and ** indicates statistical difference with both Negative Control and AMFs Control (neurons exposed to AMFs with no MMDs) (p-value < 0.05).
We were able to detect neural activity in cortical cultures within seconds of applying AMFs stimulus. Neurons remain active even after completing the 1-minute AMFs stimulus. Almost 90% of cortical neurons were responsive to the treatment. Neural activity was significantly higher in cortical neurons cocultured with MMDs and exposed to AMFs. We observe 30% more evoked activity in cortical neurons than in hippocampal neurons. This observation can be attributed to mechanosensitive ion channels’ endogenous expression in cortical neurons and the low Lipofectamine transfection efficiency of eMscL in hippocampal neurons. In the presence of Gd3+, the evoked activity in cortical neurons cocultured with MMDs and exposed to AMFs was significantly reduced, decreasing more than 80% (Figure 5d; Figure S5d and Video S4, Supporting Information). These results confirmed that mechanosensitive channels are involved in MMDs transduction
2.3. MMDs Colocalization
We investigated the colocalization of MMDs within primary cortical neural networks. Figure 6a shows a representative confocal microscopy image of MMDs within cortical neurons. Cortical neurons were labeled with anti-Tubulin β-III (magenta) to identify neuronal cytoskeleton microtubular networks, with actin-stain 488 phalloidin (cyan) to localize actin stress fibers, with synaptophysin antibody (yellow) as a marker of presynaptic vesicles in mature neurons, and with DAPI (blue) for nuclei identification. MMDs were imaged in the bright field (black), and the color was inverted to facilitate localization in the merged image (white). We also studied MMDs colocalization on the surface of neurons. Figure 6b displays SEM images of cortical neurons exposed to MMDs.
Figure 6.
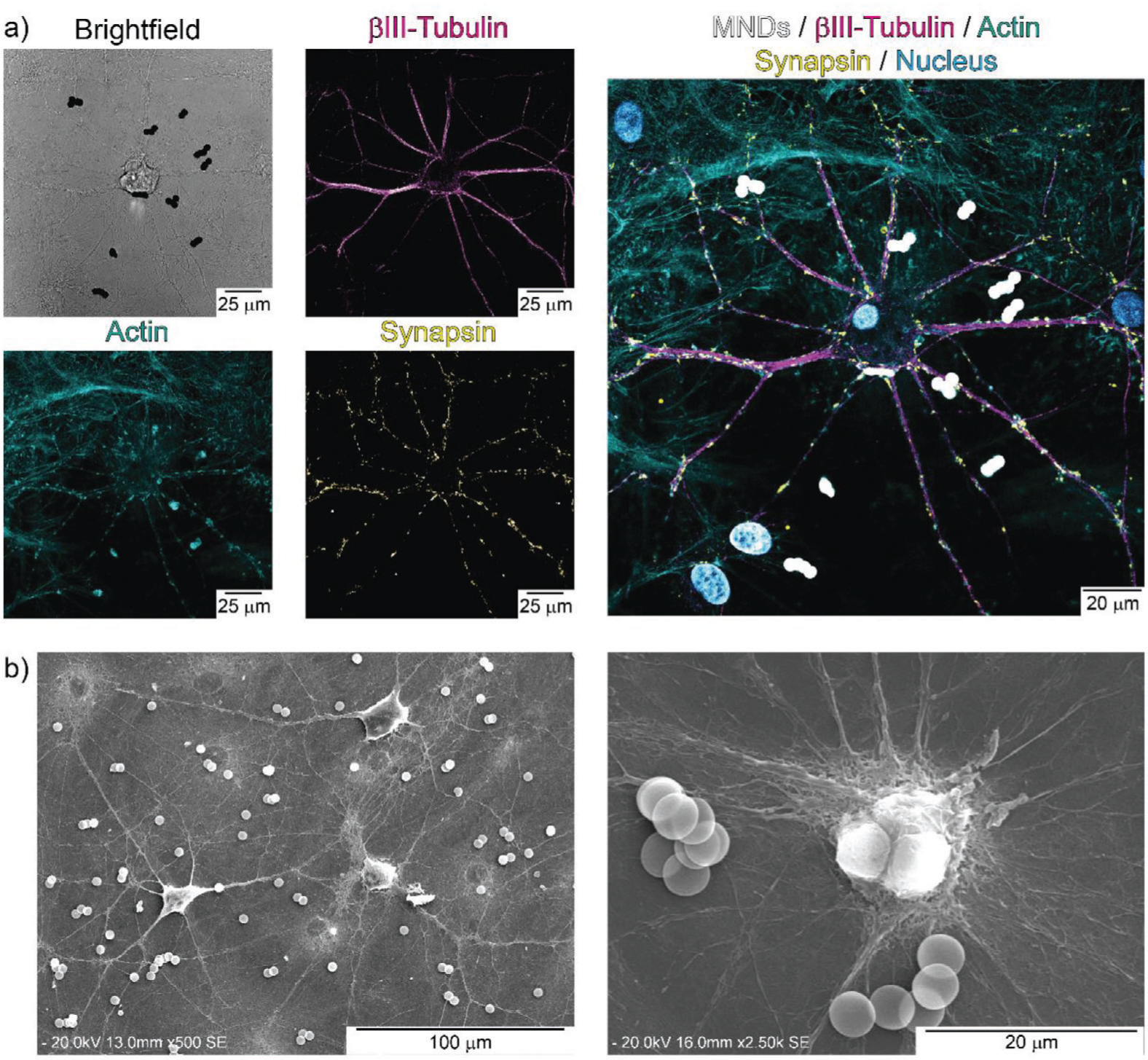
MMDs colocalization within primary cortical neural networks. a) Confocal images of cortical neurons. Cell cytoskeleton microtubules are stained with anti-Tubulin β-III (magenta), actin fibers are stained with Actin-stain 488 Phalloidin (cyan), presynaptic vesicles are labeled with synaptophysin antibody (yellow), nuclei counterstaining was performed with DAPI (blue), MMDs were imaged in bright field (black), and the color was inverted in the merged image (white). b) SEM image of cortical neural networks exposed to MMDs.
All our neural stimulation findings suggest that, indeed, we can evoke neural activity by magnetic mechanotransduction mediated by MMDs and AMFs. Our experiments rely on short exposure times of MMDs with cells to ensure colocalization on the cell membrane and stimulation of mechanosensitive ion channels. MMDs are found to be colocalized in the cell membrane, either around the cell body or the axons, of cortical neurons. Small aggregations of MMDs are observed in both, confocal microscopy, and SEM images. The aggregates are attributed to the post-stimulation treatment of the samples for imaging, which includes exposure to surfactants, vacuum, mechanical shaking, multiple washes with different solvents, among others. Before being manipulated for imaging, MMDs are homogenously distributed in the neuronal cultures and no visible aggregations are detected (Video S1). Future experiments will require specific cell targeting to slow down MMDs internalization and prolongate effective treatment period.
2.4. Cell Viability
Cell viability of cortical neurons exposed to MMDs and AMFs was studied through MTT assay during 2 days at a concentration of 10 MMDs per seeded cell. The cytotoxic effect of MMDs and AMFs were measured independently and coupled as in force-inducing stimulation. Figure 7 shows that the independent and combined exposure of MMDs and AMFs caused minimum to noncytotoxic effects in cortical neurons at the doses required for neurostimulation. No cytotoxic effects were found even after 1- and 2-days post-stimulation.
Figure 7.
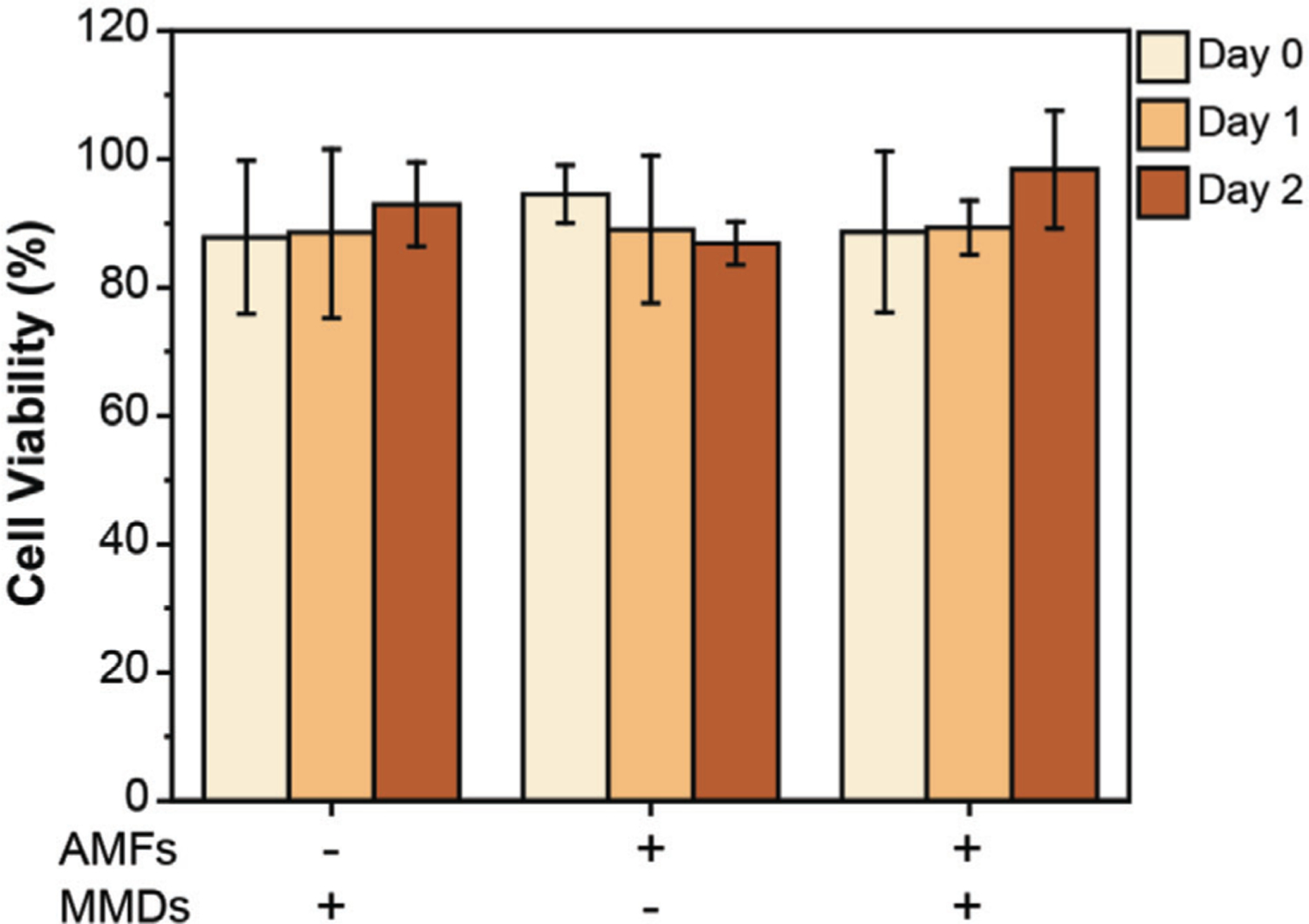
Cell viability of primary cortical neurons over 2 days. Cell viability was performed by MTT assay. Intact primary cortical neurons were used as control. Cell viability was measured immediately after stimulation (Day 0), 1-day (Day 1) and 2-days (Day 2) post-stimulation. ANOVA test was performed to determine statistical significance. No statistical significance was found between samples (p-value < 0.05).
3. Conclusion
Biocompatible gold coated Ni80Fe20 MMDs fabricated by direct writing laser lithography allow the cost-effective mass production of nanomaterials with a high magnetic moment of 9.8×10−10 emu disc−1, and null remanence. Lithography techniques facilitate the production of nano- and micro-magnets with a high magnetic moment, either using pure magnetic materials and alloys or depositing stacking structures that preserve the main features of the vortex state, which is impossible to achieve by conventional chemical synthesis. The surface functionalization of MMDs with POEGMA brushes passivates the surface charge of MMDs in biological fluids from ≈−24 to ≈−7 mV. MMDs act as transducers of low-frequency and low-amplitude AMFs into mechanical forces. When cocultured with cells expressing mechanosensitive ion channels, either artificially or endogenously, the magneto-mechanical actuation of MMDs triggers the response of mechanosensitive ion channels allowing for remote control of neural activity, providing highly efficient and long-lasting stimulations. Activation of endogenously expressed mechanosensitive ion channels was ≈35% higher than those artificially expressed, corroborating the lack of efficiency of commercially available transfection technologies and the primacy of developing transgene-free approaches. MMDs and weak AMFs cause a noncytotoxic effect to cells.
The nanotechnology described in this work may facilitate the wireless force-induced stimulation of biological signaling for basic neuroscience research. The technology’s scalability will allow further investigations on the development of noninvasive therapies for neurological disorders and psychiatric conditions. Future research will include surface engineering to bypass the blood-brain barrier, specifically targeting neural populations, studying MMDs on specific mechanosensitive ion channels endogenously expressed in neurons, and investigating specific disease models.
4. Experimental Section
MMDs Fabrication and Characterization:
Permalloy (Ni80Fe20) MMDs were fabricated using top-down lithography techniques. A four inches silicon (Si) wafer was spin-coated with photoresist. Circular shapes were exposed by direct laser writing (Heidelberg MLA150) and developed to obtain templates of circular holes. Then, a multilayer structure was deposited by electron beam evaporation in high vacuum. Aluminum (20 nm)/Gold (5 nm)/Ni80Fe20 (100 nm)/Gold (5 nm) layers were consecutively evaporated on the resist template. A liftoff process removed the resist leaving MMDs attached to the Si wafer.
The morphology and size of MMDs before releasing them from the Si wafer were characterized by Scanning Electron Microscopy (SEM, Hitachi SU1510) after photoresists liftoff. MMDs were released from the substrate by a selective chemical etching of the Al sacrificial layer, using bath ultrasonication in a 1 m potassium hydroxide (KOH, Sigma Aldrich) solution. After extraction, MMDs were washed 5 times with Millipore water by pulling them with a neodymium magnet and resuspending in Millipore water by sonication. Magnetic properties of released MMDs were characterized using Vibrating Sample Magnetometer (VSM from VersaLab Quantum Design). Magnetization loops were recorded at room temperature (300 K) with the magnetic field parallel to the sample. Two samples were measured, MMDs before releasing from the Si wafer –, i.e., in an ordered square array, as shown in Figure 2d and MMDs from the water solution dispersed (and dried) on the surface of a piece of a Si wafer. For the first sample, a piece of 1.9×1.9 mm2 was cut from the Si wafer right after the photoresist liftoff. This sample was used to confirm the vortex state’s development at remanence (Figure 2d) and estimate the magnetic moment per disc. The second sample was used to verify that the removal process of the MMDs does not affect their magnetic properties. Besides, this data was used to estimate the concentration of MMDs in the solution.
MMDs Surface Functionalization:
The surface of the MMDs was functionalized with poly (ethylene glycol) methyl ether methacrylate (POEGMA) polymer brushes grafted from MMDs surface through atom transfer radical polymerization (ATRP). First, the initiator 2-(2-bromoisobutyryloxy) undecyl thiol was covalently immobilized on the MMDs’ gold-coated surface by immersing the Si wafer with MMDs in a 1 M initiator solution in ethanol under N2 gas flow for 24 h.[35] The Si wafer was rinsed with ethanol to remove the initiator excess. Then, the wafer containing MMDs-initiator was immersed in a previously degassed mixture containing 2.8 mmol ethylene glycol methyl ether methacrylate oligomer (Mn = 300 Da, Sigma Aldrich), 1.9 mmol 2,2’-bipyridyl (Sigma Aldrich), 2 mmol copper (II) chloride (CuCl2, Sigma Aldrich) and 5 mL of dimethylformamide/water (3:2). The mixture was allowed to react overnight at room temperature under N2 gas. The functionalized MMDs wafer was then washed with dimethylformamide/water 3 times, acetone 3 times, ethanol 3 times, and water. MMDs were labeled with Rhodamine B isothiocyanate (Sigma Aldrich) for visualization. Finally, MMDs were released from the substrate with 1 m KOH following the protocol described above. MMDs-POEGMA functionalization was characterized through dynamic light scattering in Tyrode’s solution (DLS, Malvern Zetasizer Nano ZS) and by fluorescence microscopy (Inverted Inflorescent Leica Microscope DM16000 B).
Isolation and Maintenance of Primary Rat Cortical and Hippocampal Neurons:
The UTSA Institutional Animal Care and Use Committee approved all experiments in this study (approval no. MU-RA007). Hippocampal and cortex neurons were extracted from neonatal rats, dissociated, and plated[36] on 35 mm collagen-coated glass-bottom dishes (MatTek). Primary neurons were maintained in Neurobasal media supplemented with 2% B27 and 1% Glutamax-I (ThermoFisher Scientific) at 37 °C and 5% CO2. Glial inhibition was performed after 3 days. Hippocampal neurons were transfected on day 8 with the mechanosensitive ion channel of large conductance eMscL (Addgene) using Lipofectamine 3000.[37] Cells were labeled with the calcium indicator Fluo-4 (ThermoFisher Scientific) before stimulation experiments. Stimulation and control experiments were performed on healthy cultures with an evident expression between days 8 and 12.
Force-Inducing Neuronal Stimulation:
MMDs were incubated with cells for 10 minutes before experimentation at a concentration of 10 MMDs per seeded cell. Exposure to the alternating magnetic field took place in a custom sample holder designed to hold a 35 mm petri dish with 1 mL of Tyrode’s solution fitting into a 50 mm copper varnished wire solenoid driven by a DC-300 amplifier with an input signal of 5 V. A fluorescence stereomicroscope (Leica M205 FCA) equipped with 2× and 5× Plan Apo objectives, and a sCMOS camera (Leica DFC9000) were used to record cell activity during stimulation. All videos were recorded at a frame rate of 200 ms. To block mechanosensitive ion channels, before applying the AMFs stimulation, neurons were treated with Gadolinium (III) (Gd3, Sigma Aldrich) at a final concentration of 20 μM in Tyrode.
Analysis of Calcium Imaging:
The analysis was performed using functions in ImageJ to subtract background and measure fluorescence intensity (Figure S2, Supporting Information). The time resolution of the fluorescence microscopy used for calcium imaging is insufficient to be interpreted as resolving individual spikes. Instead, it might be taken to show time-integrated levels of neural activity. An automatic code in MATLAB was generated to analyze the fluorescence intensity traces of each neuron in each video (Supplementary Note). Briefly, a threshold relative to five times the standard deviation from the background was set to differentiate active cells from inactive cells. For each condition 5 different samples were analyzed. Each sample contained between 300 and 500 neurons. The results presented here correspond to 100 neurons randomly (and automatically) selected from all the neurons analyzed within the 5 samples. The number of neurons exhibiting activity was divided by the total number of cells to determine percent responsiveness. Statistical differences between control trials and force-induced activity mediated by MMDs were assessed by ANOVA test.
Immunohistochemistry:
For the fluorescent staining of cell cytoskeleton microtubules, actin stress fibers, presynaptic vesicles, and cell nucleus, the samples were rinsed twice with PBS and fixed with 4% paraformaldehyde in PBS. After washing 3 times with washing solution, i.e., PBS containing 0.05% Tween-20 (BP337, Fisher Bioreagents), cells were permeabilized with 0.25% Triton X-100 (T8787, Sigma Aldrich) for 10 min followed by 3 washes. Blocking solution, 1% bovine serum albumin (0332, VWR) in PBS, was applied for 1 h. Anti-Tubulin β-III Antibody (clone TU-20, Alexa Fluor555 Conjugate, CBL412A5 from Sigma Aldrich), actin-stain 488 phalloidin (PHDG1-a, Cytoskeleton, Inc.) and Mouse Monoclonal Synaptophysin Antibody (Alexa Fluor647 Conjugate, NBP147483AF647 from Novus Biologicals) were diluted to a working concentration in blocking solution and incubated overnight at 4 °C. After washing 3 times, cell nucleus counterstaining was performed by incubating with DAPI for 5 min (Sigma Aldrich). The samples were washed, and PBS was added to the petri dish before visualization in a Leica TCS SP8 Confocal Microscope.
Sample Preparation for Scanning Electron Microscopy:
Samples were rinsed twice with PBS and fixed with 4% paraformaldehyde in PBS. Cells were gradually dehydrated in a serial change of ethanol in Milli-Q water for 5 min each (0%, 5%, 10%, 25%, 35%, 50%, 65%, 75%, 90%, 100%). Glass bottom was carefully removed from the petri dish before critical point drying (Leica EM CPD300). The samples were mounted on aluminum pin stubs using double-sided carbon tape and gold-coated (PELCO SC-7 Sputter Coater) before imaging in a Hitachi SU1510 Scanning Electron Microscope.
Cell Viability:
Cell viability was determined by a standard colorimetric MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma Aldrich) assay. Primary cortical neurons were seeded in a 96-well plate precoated with Matrigel (Corning Matrigel Matrix, 356 234) at a density of 7000 cells per well. Cortical neurons were exposed to Neurobasal complete media containing MMDs at a concentration of 10 discs per cell. After coculturing with MMDs for 10 minutes, cells were exposed to AMFs for 1 minute. Immediately after stimulation (Day 0), and 1- and 2-days post-stimulation, viability was assessed relative to control samples (intact cortical neurons). 20 μL of MTT compound, prepared at a concentration of 5 mg mL−1 in phosphate buffer saline, was added to each well and incubated for 1 h. The supernatant was aspirated, and 200 μL of dimethyl sulfoxide (DMSO) was added. A plate reader was used to measure absorbance at 570 nm.
Supplementary Material
Acknowledgements
Part of this research was funded by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 734801, the Spanish AEI grant No PID2019-104604RB, and the Basque Country grant No IT1162-19. This project was funded in-part by The University of Texas at San Antonio, Office of the Vice President for Research, Economic Development, and Knowledge Enterprise. This work was supported partially by the National Science Foundation under a CAREER award to Gabriela Romero (CBET – 2044713). Part of the MMDs fabrication process was done at the San Diego Nanotechnology Infrastructure (SDNI) of UCSD, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which was supported by the US National Science Foundation under grant ECCS-1542148. The collaborative UCSD-UTSA research on synthesis and magnetism of nanostructured MMD was supported by NSF under Grant Nos. DMR 1805585 and DMR 1804414. C.C. is deeply grateful to the MARC*U-Star program at the University of Texas at San Antonio that supported her with the undergraduate research training for this work (MARC Grant: NIH GM007717). R.M. thanks for technical and human support provided by the Laser Facility and Magnetic Measurements units of SGIker UPV/EHU.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Claudia Collier, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, TX 78249, USA.
Nicolas Muzzio, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, TX 78249, USA.
Rohini Thevi Guntnur, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, TX 78249, USA.
Amanda Gomez, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, TX 78249, USA.
Carolina Redondo, Department of Physical Chemistry, University of the Basque Country UPV/EHU, Leioa 48940, Spain.
Raquel Zurbano, Department of Physical Chemistry, University of the Basque Country UPV/EHU, Leioa 48940, Spain.
Ivan K. Schuller, Center for Advanced Nanoscience and Department of Physics, University of California San Diego, La Jolla, CA 92093, USA
Carlos Monton, General Atomics, PO Box 85608, San Diego, CA 92186, USA.
Rafael Morales, Department of Physical Chemistry & BCMaterials, University of the Basque Country UPV/EHU, Leioa 48940, Spain; IKERBASQUE, Basque Foundation for Science, Bilbao 48011, Spain.
Gabriela Romero, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, TX 78249, USA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].a) Kubanek J, Neurosurg. Focus 2018, 44, 14; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) di Biase L, Falato E, Di Lazzaro V, Front. Neurol 2019, 10, 549; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wassermann EM, Zimmermann T, Pharmacol. Ther 2012, 133, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Papakostas GI, lonescu DF, Mol. Psychiatry 2015, 20, 1142; [DOI] [PubMed] [Google Scholar]; b) George MS, Taylor JJ, Short B, in Handbook of Clinical Neurology, (Eds: Lozano AM, Hallett M), Vol. 116, Elsevier, Amsterdam: 2013, p. 399; [DOI] [PubMed] [Google Scholar]; c) Levkovitz Y, Harel EV, Roth Y, Braw Y, Most D, Katz LN, Sheer A, Gersner R, Zangen A, Brain Stimul. 2009, 2, 188. [DOI] [PubMed] [Google Scholar]
- [3].a) Trevizol AP, Shiozawa P, Cook IA, Sato IA, Kaku CB, Guimarães FBS, Sachdev P, Sarkhel S, Cordeiro Q, J. ECT 2016, 32; [DOI] [PubMed] [Google Scholar]; b) Berlim MT, Neufeld NH, Van den Eynde F, J. Psychiatr. Res 2013, 47, 999. [DOI] [PubMed] [Google Scholar]
- [4].Dinur-Klein L, Dannon P, Hadar A, Rosenberg O, Roth Y, Kotler M, Zangen A, Biol. Psychiatry 2014, 76, 742. [DOI] [PubMed] [Google Scholar]
- [5].Coppola G, Di Lorenzo C, Parisi V, Lisicki M, Serrao M, Pierelli F, J. Headache Pain 2019, 20, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Bolognini N, Vallar G, Casati C, Latif LA, El-Nazer R,Williams J, Banco E, Macea DD, Tesio L, Chessa C, Fregni F, Neurorehabilitation Neural Repair 2011, 25, 819; [DOI] [PubMed] [Google Scholar]; b) Biou E, Cassoudesalle H, Cogné M, Sibon I, De Gabory I, Dehail P, Aupy J, Glize B, Ann. Phys. Rehab. Med 2019, 62, 104; [DOI] [PubMed] [Google Scholar]; c) Georgiou AM, Lada E, Kambanaros M, Aphasiology 2020, 34, 540; [Google Scholar]; d) Baek H, Sariev A, Lee S, Dong SY, Royer S, Kim H, IEEE Trans. Neural Syst. Rehabil. Eng 2020, 28, 2073. [DOI] [PubMed] [Google Scholar]
- [7].a) Yang D, Wang Q, Xu C, Fang F, Fan J, Li L, Du Q, Zhang R, Wang Y, Lin Y, Huang Z, Wang H, Chen C, Xu Q, Wang Y, Zhang Y, Zhang Z, Zhao X, Zhao X, Li T, Liu C, Niu Y, Zhou Q, Zhou Q, Duan Y, Liu X, Yu T, Xue Q, Li J, Dai X, Han J, Ren C, Xu H, Li N, Zhang J, Xu N, Yang K, Wang Y, Brain Stimul. 2020, 13, 109; [DOI] [PubMed] [Google Scholar]; b) Regner GG, Pereira P, Leffa DT, de Oliveira C, Vercelino R, Fregni F, Torres ILS, Front. Neurosci 2018, 12, 189; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Baumer FM, Pfeifer K, Fogarty A, Pena-Solorzano D, Rolle CE, Wallace JL, Rotenberg A, Fisher RS, J. Clin. Neurophysiol 2020, 37, 170; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bystritsky A, Korb A, Stern J, Cohen M, J. Ther. Ultrasound 2015, 3, O27; [Google Scholar]; e) Ranjan M, Boutet A, Bhatia S, Wilfong A, Hader W, Lee MR, Rezai AR, Adelson PD, Expert Rev. Neurother 2019, 19, 937. [DOI] [PubMed] [Google Scholar]
- [8].a) Song I-U, Na S-H, Im JJ, Jeong H, Chung S-W, Chung Y-A, J. Neuroimaging 2020, 30, 161; [DOI] [PubMed] [Google Scholar]; b) Madrid J, Benninger DH, J. Neurosci. Methods 2020, 108957; [DOI] [PubMed] [Google Scholar]; c) Martínez-Fernández R, Rodríguez-Rojas R, del Álamo M, Hernández-Fernández F, Pineda-Pardo JA, Dileone M, Alonso-Frech F, Foffani G, Obeso I, Gasca-Salas C, de Luis-Pastor E, Vela L, Obeso JA, Lancet Neurol. 2018, 17, 54; [DOI] [PubMed] [Google Scholar]; d) Zaaroor M, Sinai A, Goldsher D, Eran A, Nassar M, Schlesinger I, J. Neurosurg 2017, 128, 202. [DOI] [PubMed] [Google Scholar]
- [9].a) Mayo-Yáñez M, Corrás T, Méndez-Iglesias R, Curr. Psychiatr. Rev 2018, 14, 211; [Google Scholar]; b) Marron EM, Viejo-Sobera R, Quintana M, Redolar-Ripoll D, Rodríguez D, Garolera M, BMC Res. Notes 2018, 11, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Lin Y, Zhang C, Wang Y, Brain Stimul.: Basic, Transl., Clin. Res. Neuromodulation 2019, 12, 403; [Google Scholar]; b) Heeren A, Billieux J, Philippot P, De Raedt R, Baeken C, de Timary P, Maurage P, Vanderhasselt MA, Soc. Cognit. Affective Neurosci 2017, 12, 251; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Costanzo F, Menghini D, Maritato A, Castiglioni MC, Mereu A, Varuzza C, Zanna V, Vicari S, Front. Behav. Neurosci 2018, 12, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jeon D-W, Jung D-U, Kim S-J, Shim J-C, Moon J-J, Seo Y-S, Jung S-S, Seo B-J, Kim J-E, Oh M, Kim Y-N, Schizophr. Res 2018, 197, 378. [DOI] [PubMed] [Google Scholar]
- [12].a) Pinto CB, Teixeira Costa B, Duarte D, Fregni F, j. ECT 2018, 34, 36; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stamenkovic DM, Mladenovic K, Rancic N, Cvijanovic V, Maric N, Neskovic V, Zeba S, Karanikolas M, Ilic TV, Front. Pharmacol 2020, 11, 125; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) O’Neill F, Sacco P, Bowden E, Asher R, Burnside G, Cox T, Nurmikko T, J. Pain Res 2018, 11, 3117; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tremblay S, Rogasch NC, Premoli I, Blumberger DM, Casarotto S, Chen R, Di Lazzaro V, Farzan F, Ferrarelli F, Fitzgerald PB, Hui J, Ilmoniemi RJ, Kimiskidis VK, Kugiumtzis D, Lioumis P, Pascual-Leone A, Pellicciari MC, Rajji T, Thut G, Zomorrodi R, Ziemann U, Daskalakis ZJ, Clin. Neurophysiol 2019, 130, 802. [DOI] [PubMed] [Google Scholar]
- [13].a) Ghanouni P, Pauly KB, Elias WJ, Henderson J, Sheehan J, Monteith S, Wintermark M, Am.J. Roentgenol 2015, 205, 150; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Legon W, Bansal P, Tyshynsky R, Ai L, Mueller JK, Sci. Rep 2018, 8, 10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Chen R, Romero G, Christiansen MG, Mohr A, Anikeeva P, Science 2015, 347, 1477; [DOI] [PubMed] [Google Scholar]; b) Huang H, Delikanli S, Zeng H, Ferkey DM, Pralle A, Nat. Nanotechnol 2010, 5, 602. [DOI] [PubMed] [Google Scholar]
- [15].a) Carvalho-de-Souza JL, Treger JS, Dang B, Kent SBH, Pepperberg DR, Bezanilla F, Neuron 2015, 86, 207; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yoo S, Kim R, Park J-H, Nam Y, ACS Nano 2016, 10, 4274. [DOI] [PubMed] [Google Scholar]
- [16].Duyn JH, Neuroimage 2018, 168, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Wu X, Zhu X, Chong P, Liu J, Andre LN, Ong KS, Brinson K, Mahdi AI, Li J, Fenno LE, Wang H, Hong G, Proc. Natl. Acad. Sci. USA 2019, 116, 26332; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Peng D, Jiang Y, Huang B, Du Y, Zhao J, Zhang X, Ma R, Golovynskyi S, Chen B, Wang F, Adv. Mater 2020, 32, 1907747. [DOI] [PubMed] [Google Scholar]
- [18].Lee J.-u., Shin W, Lim Y, Kim J, Kim WR, Kim H, Lee J-H, Cheon J, Nat. Mater 2021, 20, 1029. [DOI] [PubMed] [Google Scholar]
- [19].a) Mejía-López J, Altbir D, Romero AH, Batlle X, Roshchin IV, Li C-P, Schuller IK, J. Appl. Phys 2006, 100, 104319; [Google Scholar]; b) Dumas RK, Liu K, Li C-P, Roshchin IV, Schuller IK, Appl. Phys. Lett 2007, 91, 202501; [Google Scholar]; c) Mejía-López J, Altbir D, Landeros P, Escrig J, Romero AH, Roshchin IV, Li CP, Fitzsimmons MR, Batlle X, Schuller IK, Phys. Rev. B 2010, 81, 184417. [Google Scholar]
- [20].a) Mora B, Perez-Valle A, Redondo C, Boyano MD, Morales R, ACS Appl. Mater. Interfaces 2018, 10,8165; [DOI] [PubMed] [Google Scholar]; b) Morales R, Flores AN, Vargas NM, Giuliani J, Schuller IK, Monton C, ACS Appl. Nano Mater. 2020, 3, 4037; [Google Scholar]; c) Maziewski A, Zablotskii V, Kisielewski M, Phys. Rev. B 2006, 73, 134415. [Google Scholar]
- [21].Kim D-H, Rozhkova EA, Ulasov IV, Bader SD, Rajh T, Lesniak MS, Novosad V, Nat. Mater 2010, 9, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gregurec D, Senko AW, Chuvilin A, Reddy PD, Sankararaman A, Rosenfeld D, Chiang P-H, Garcia F, Tafel I, Varnavides G, Ciocan E, Anikeeva P, ACS Nano 2020, 14, 8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].a) Yang Y, Li M, Wu Y, Zong B, Ding J, RSC Adv. 2016, 6, 25444; [Google Scholar]; b) Yang Y, Liu X, Lv Y, Herng TS, Xu X, Xia W, Zhang T, Fang J, Xiao W, Ding J, Adv. Funct. Mater 2015, 25, 812; [Google Scholar]; c) Chen L, Yang X, Chen J, Liu J, Wu H, Zhan H, Liang C, Wu M, Inorg. Chem 2010, 49, 8411. [DOI] [PubMed] [Google Scholar]
- [24].Peixoto L, Magalhães R, Navas D, Moraes S, Redondo C, Morales R, Araújo JP, Sousa CT, Appl. Phys. Rev 2020, 7, 011310. [Google Scholar]
- [25].a) Einstein A, Ann. Phys 1905, 322, 549; [Google Scholar]; b) Sutherland W, J Sci. 1905, 9, 781. [Google Scholar]
- [26].Badaire S, Poulin P, Maugey M, Zakri C, Langmuir 2004, 20, 10367. [DOI] [PubMed] [Google Scholar]
- [27].a) Lotya M, Rakovich A, Donegan JF, Coleman JN, Nanotechnology 2013, 24, 265703; [DOI] [PubMed] [Google Scholar]; b) Arenas-Guerrero P, Delgado ÁV, Donovan KJ, Scott K, Bellini T, Mantegazza F, Jiménez ML, Sci. Rep 2018, 8, 9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].a) Llarena I, Romero G, Ziolo RF, Moya SE, Nanotechnology 2009, 21, 055605; [DOI] [PubMed] [Google Scholar]; b) Romero G, Estrela-Lopis I, Castro-Hartmann P, Rojas E, Llarena I, Sanz D, Donath E, Moya S, Soft Matter 2011, 7, 6883; [Google Scholar]; c) Guntnur RT, Muzzio N, Morales M, Romero G, Soft Matter 2021, 17,2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].a) Delmas P, Coste B, Cell 2013, 155, 278; [DOI] [PubMed] [Google Scholar]; b) Wu J, Lewis AH, Grandl J, Trends Biochem. Sci. 2017, 42, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].a) Hughes S, McBain S, Dobson J, El Haj AJ, J. R. Soc., Interface 2008, 5, 855; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Seo D, Southard KM, Kim J.-w., Lee HJ, Farlow J, Lee J.-u., Litt DB, Haas T, Alivisatos AP, Cheon J, Gartner ZJ, Jun Y.-w., Cell 2016, 165, 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].a) Howard J, Hudspeth AJ, Neuron 1988, 1, 189; [DOI] [PubMed] [Google Scholar]; b) Yang Y, Liu X, Lv Y, Herng TS, Xu X, Xia W, Zhang T, Fang J, Xiao W, Ding J, Adv. Funct. Mater 2015, 25, 812. [Google Scholar]
- [32].a) Christensen AP, Corey DP, Nat. Rev. Neurosci 2007, 8, 510; [DOI] [PubMed] [Google Scholar]; b) Ranade SS, Syeda R, Patapoutian A, Neuron 2015, 87, 1162; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Matyas F, Sreenivasan V, Marbach F, Wacongne C, Barsy B, Mateo C, Aronoff R, Petersen CCH, Science 2010, 330, 1240. [DOI] [PubMed] [Google Scholar]
- [33].Cheng Y, Yao H, Lin H, Lu J, Li R, Wang K, Chem.-Biol. Interact 1999, 121, 267. [DOI] [PubMed] [Google Scholar]
- [34].Hamill OP, McBride DW Jr., Pharmacol. Rev 1996, 48, 231. [PubMed] [Google Scholar]
- [35].a) Ohno K, Akashi T, Huang Y, Tsujii Y, Macromolecules 2010, 43, 8805; [Google Scholar]; b) Bao Z, Bruening ML, Baker GL, J. Am. Chem. Soc 2006, 128, 9056. [DOI] [PubMed] [Google Scholar]
- [36].a) Nunez J, J. Visualized Exp 2008, 19, 895; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pacifici M, Peruzzi F, J. Visualized Exp 2012, 63, 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Soloperto A, Boccaccio A, Contestabile A, Moroni M, Hallinan GI, Palazzolo G, Chad J, Deinhardt K, Carugo D, Difato F, J. Cell Sci 2018, 131, jcs210393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


