ABSTRACT
Cellular senescence is a stable form of cell cycle arrest associated with proinflammatory responses. Senescent cells can be cleared by the immune system as a part of normal tissue homeostasis. However, senescent cells can also accumulate in aged and diseased tissues, contributing to inflammation and disease progression. The mechanisms mediating the impaired immune-mediated clearance of senescent cells are poorly understood. Here, we report that senescent cells upregulate the immune checkpoint molecule PD-L1, the ligand for PD-1 on immune cells, which drives immune cell inactivation. The induction of PD-L1 in senescence is dependent on the proinflammatory program. Furthermore, the secreted factors released by senescent cells are sufficient to upregulate PD-L1 in nonsenescent control cells, mediated by the JAK-STAT pathway. In addition, we show that prolongevity intervention rapamycin downregulates PD-L1 in senescent cells. Last, we found that PD-L1 is upregulated in several tissues in naturally aged mice and in the lungs of idiopathic pulmonary fibrosis patients. Together, our results report that senescence and aging are associated with upregulation of a major immune checkpoint molecule, PD-L1. Targeting PD-L1 may offer new therapeutic opportunities in treating senescence and age-associated diseases.
KEYWORDS: PD-L1, SASP, aging, senescence
INTRODUCTION
Cellular senescence restricts proliferation of damaged cells and hence has a beneficial role in restraining tumorigenesis (1). However, senescence can also be detrimental in aging and diseases, mainly due to its proinflammatory features (2). Senescent cells secrete a large array of proinflammatory factors that trigger inflammation and alter tissue microenvironment (2). Strategies to selectively kill senescent cells or to inhibit the proinflammatory program of senescence are important biomedical objectives (3, 4).
Senescent cells in physiological conditions are removed by the immune system: the proinflammatory cytokines secreted by senescent cells recruit immune cells and activate them, leading to immune-mediated clearance (2). Counterintuitively, senescent cells can accumulate in aging and diseases without being removed by the immune system, thereby contributing to inflammation and disease progression (1, 4). The altered interaction between senescence and the immune cells is a major unaddressed area in senescence and aging research.
A similar scenario is seen in cancer, in which cancer cells induce immune checkpoint, resulting in impaired immunosurveillance of cancer (5). Several cancer types show upregulation of PD-L1 in cancer cells, which through binding to PD-1 on immune cells induces inactivation and/or exhaustion of immune cells (6–8). Inhibition of PD-L1/PD-1 binding is a central target in cancer immunotherapy that has transformed treatment of several cancers (7, 9). Whether senescent cells employ similar mechanisms to inhibit immune cell activation has not been explored. In this study, we report that PD-L1 is upregulated in senescence and aging. This finding has implications for understanding how senescent cells accumulate in diseased tissues and suggests new approaches to combat this pathological accumulation.
RESULTS
Senescent cells upregulate PD-L1.
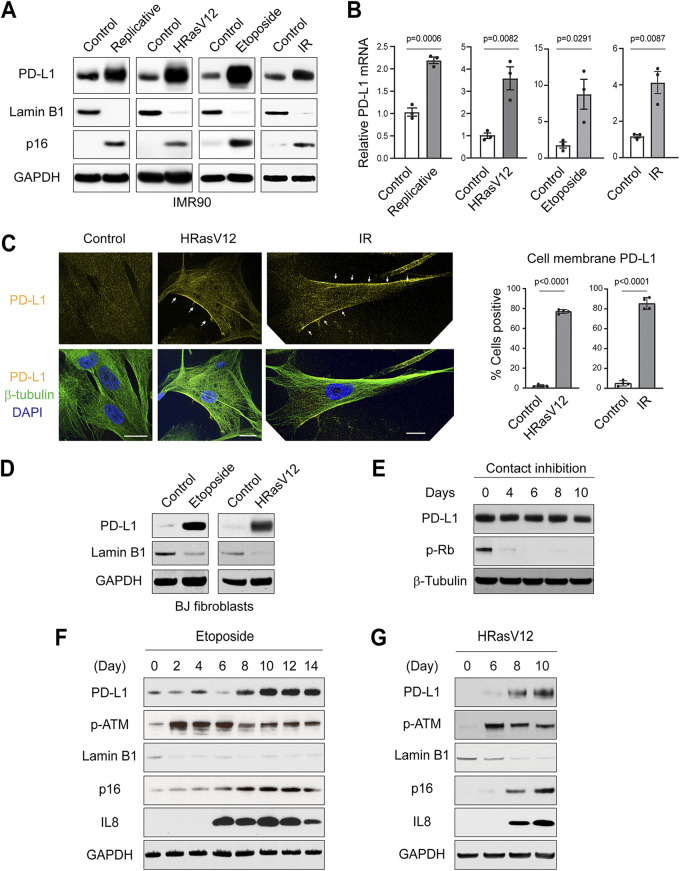
We employed IMR90 primary human diploid lung fibroblasts, cultured in physiological 3% oxygen, as a model system to study senescence. Low-passage-number proliferating cells (population doubling less than 35) were used as controls. Senescence was induced by replication exhaustion, activated oncogene HRasV12, DNA-damaging agent etoposide, or ionizing irradiation (IR), following our previously established conditions (10–13). Induction of senescence was confirmed by loss of lamin B1 and gain of p16ink4a (here referred to as p16) (Fig. 1A). Importantly, PD-L1 is upregulated at the protein level in all forms of senescence, detected by immunoblotting (Fig. 1A). Likewise, the mRNA levels of PD-L1 are significantly increased in these forms of senescence, measured by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 1B). We subsequently observed PD-L1 under confocal microscopy. While PD-L1 in control cells displayed a diffuse pattern, its overall intensities were increased in senescent cells and exhibited prominent localization at or near the cell membrane (Fig. 1C, left, and quantified in Fig. 1C, right).
FIG 1.
Upregulation of PD-L1 in senescent IMR90 cells. (A) Primary IMR90 cells were left untreated or induced to senescence by various means. Replicative senescent cells were harvested 2 weeks after the cells reached cell cycle arrest. HRasV12 was expressed via retroviral vector and harvested 1 week postinfection. Etoposide (50 μM) was used to treat cells for 24 h, and the cells were harvested 1 week after the treatment. Ionizing irradiation (20 Gy)-treated cells were harvested 2 weeks after treatment. Verification of senescence is described under Materials and Methods. The cell lysates were analyzed by immunoblotting with indicated antibodies. Loss of lamin B1 and gain of p16 were used as markers for senescence. (B) Cells as in panel A were analyzed by reverse transcription-quantitative PCR (RT-qPCR) for PD-L1. The results were normalized to lamin A/C and presented as mean values with standard error of the mean (SEM; n = 3). The P values were calculated from two-tailed Student’s t test. (C) IMR90 cells were left untreated or induced to senescence by HRasV12 (1 week) or IR (2 weeks). The cells were fixed and stained with antibodies as indicated, followed by confocal microscopy imaging. The images were acquired under identical settings, under a 63× objective, and representative images are shown. β-Tubulin was used to label the cells. Bars, 20 μm. Arrows indicate PD-L1 at or in close proximity to the cell membrane. (Right) Quantification of cell membrane-localized PD-L1. The data are from four randomly selected fields with over 200 cells, quantified under a 10× objective. The results shown are mean values with standard deviation (SD). The P values were calculated from two-tailed Student’s t test. (D) Primary BJ fibroblasts were treated with etoposide (40 μM) and harvested at day 14 or were infected with HRasV12 and harvested at day 7. The lysates were analyzed by immunoblotting. (E) Low-passage-number proliferating IMR90 cells were left untreated or induced to quiescence by contact inhibition. Cells reaching 100% confluence were considered the start of contact inhibition and were harvested at indicated days. Lysates were subjected to immunoblotting. Phospho-Rb was used as a proliferation marker. (F, G) IMR90 cells were subjected to etoposide treatment (50 μM) (F) or HRasV12 infection (G). The cells were harvested at indicated time points and analyzed by immunoblotting. DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IR, ionizing irradiation.
In addition to IMR90 cells, we examined BJ primary human diploid foreskin fibroblasts and also found PD-L1 to be upregulated in senescence (Fig. 1D). In contrast to senescence, quiescence of IMR90 cells induced by contact inhibition failed to upregulate PD-L1 (Fig. 1E). The induction of PD-L1 appears to be a later event in senescence, since the induction of DNA damage response (measured by phosphorylation of ATM) precedes the upregulation of PD-L1 (Fig. 1F and G). Rather, the induction of PD-L1 follows or correlates with the expression of proinflammatory gene interleukin-8 (IL-8) (Fig. 1F and G). Taken together, these results indicate that PD-L1 is upregulated as a relatively late event of senescence.
SASP program is necessary to induce PD-L1.
The kinetics of PD-L1 induction in senescence (Fig. 1F and G) suggest a possible connection to the senescence-associated secretory phenotype (SASP), characterized by the secretion of a large variety of proinflammatory cytokines, chemokines, growth factors, and proteases (2). The SASP program is induced by multiple mechanisms (2, 14). We and others have reported that the innate immunity cytosolic DNA-sensing cGAS-STING pathway plays an important role in activating the transcription of SASP genes (11, 15–17). To establish the causal relationship between SASP and PD-L1, we induced senescence following our established conditions (11, 12) (Fig. 1), inhibited the SASP program by both genetic and pharmacological means, and evaluated the corresponding changes of PD-L1.
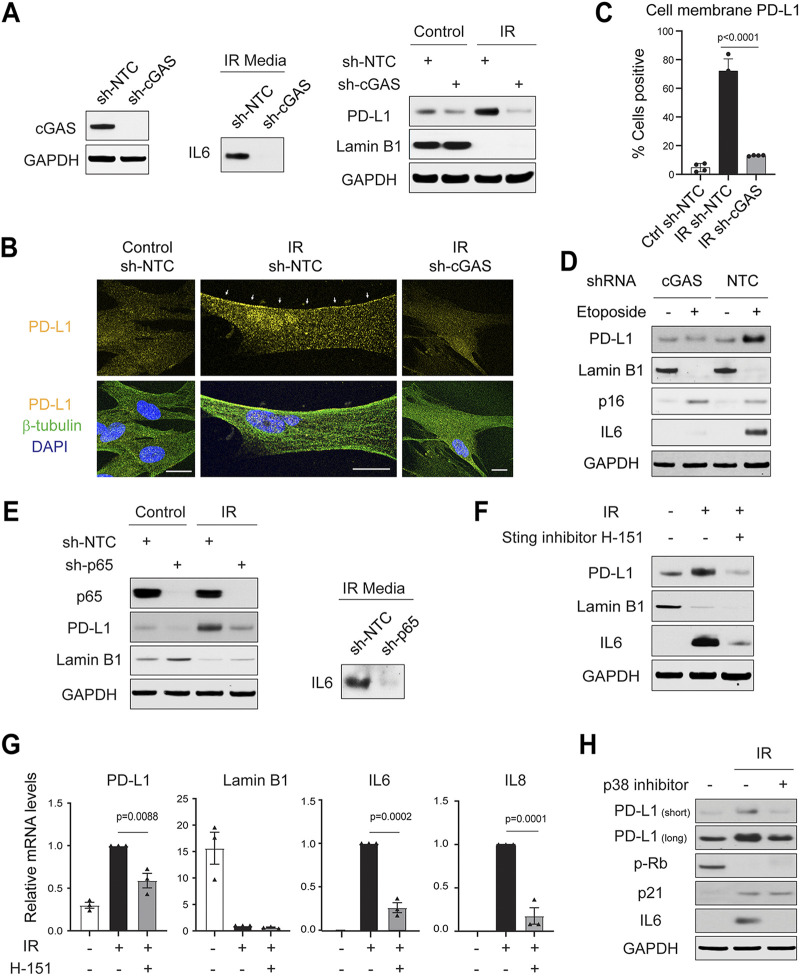
First, we suppressed the SASP by disrupting cGAS, using our previously published short hairpin RNA (shRNA) (11). This resulted in impaired secretion of IL-6 in the conditioned media of IR-induced senescent cells (Fig. 2A, left and middle). Importantly, PD-L1 showed impaired upregulation in cGAS-deficient cells (Fig. 2A, right), accompanied by reduced accumulation at or near the cell membrane (Fig. 2B and C). Likewise, the upregulation of PD-L1 is impaired in etoposide-induced senescence in the absence of cGAS (Fig. 2D).
FIG 2.
The senescence-associated secretory phenotype (SASP) program is required for the induction of PD-L1 in senescence. (A) IMR90 cells were subjected to short hairpin RNA (shRNA)-mediated gene inactivation with a nontargeting control (NTC) hairpin or hairpin targeting cGAS. (Left) Immunoblotting showing cGAS was successfully inactivated by shRNA. (Middle, right) Cells were subjected to ionizing irradiation (IR, 20 Gy) and analyzed at day 14. The conditional media were analyzed for interleukin-6 (IL-6) (middle), and cell lysates were analyzed for PD-L1 and lamin B1. (B) Cells as in panel A were subjected to immunofluorescence analyses of PD-L1, imaged under confocal microscopy. The images were acquired under identical settings, and representative images are shown. β-Tubulin was used to label the cells. Bars, 20 μm. Arrows indicate PD-L1 at or near the cell membrane. (C) Quantification of cell membrane-localized PD-L1. The data are from four randomly selected fields with over 200 cells. The results shown are mean values with SD. The P values were calculated from two-tailed Student’s t test. (D) Related to panel A, IMR90 cells stably expressing sh-NTC or sh-cGAS were subjected to etoposide treatment (50 μM) for 24 h. The cells were then cultured until day 10 and harvested for immunoblotting analyses. (E) IMR90 cells were subjected to shRNA-mediated gene inactivation of NF-κB p65 subunit and were treated with IR (20 Gy) for 2 weeks. Cell lysates were probed with indicated antibodies. (F, G) IMR90 cells were treated with IR (20 Gy) for 10 days and then treated with STING inhibitor H-151 (1 μM) for 4 days. The cells were analyzed by immunoblotting (F) or RT-qPCR analyses normalized to lamin A/C (G). The results shown are mean values with SEM (n = 3) The P values were calculated from two-tailed Student’s t test. (H) IMR90 cells were treated with p38 MAPK inhibitor SB203580 (20 μM) 4 days after IR, and the drug was replenished every other day. Cell lysates were subjected to immunoblotting with indicated antibodies.
Second, we suppressed the SASP by inactivation of NF-κB, a key transcription factor for SASP genes (18). shRNA-mediated inactivation of NF-κB p65 subunit resulted in reduced IL-6 secretion and, notably, impaired induction of PD-L1 (Fig. 2E).
Third, we suppressed the SASP using STING inhibitor H-151 (19). This led to reduced expression of the SASP genes and inhibited PD-L1 upregulation both at the protein level (Fig. 2F) and at the mRNA level (Fig. 2G).
Last, we suppressed the SASP by inhibition of p38 MAPK, another important mediator of the SASP program (20). Addition of a p38 inhibitor, SB203580, inhibited the induction of IL-6 in senescent cells and, importantly, reduced PD-L1 upregulation (Fig. 2H). Collectively, these results suggest that the SASP program is required for the upregulation of PD-L1 in senescence.
SASP program upregulates PD-L1 through the JAK-STAT pathway.
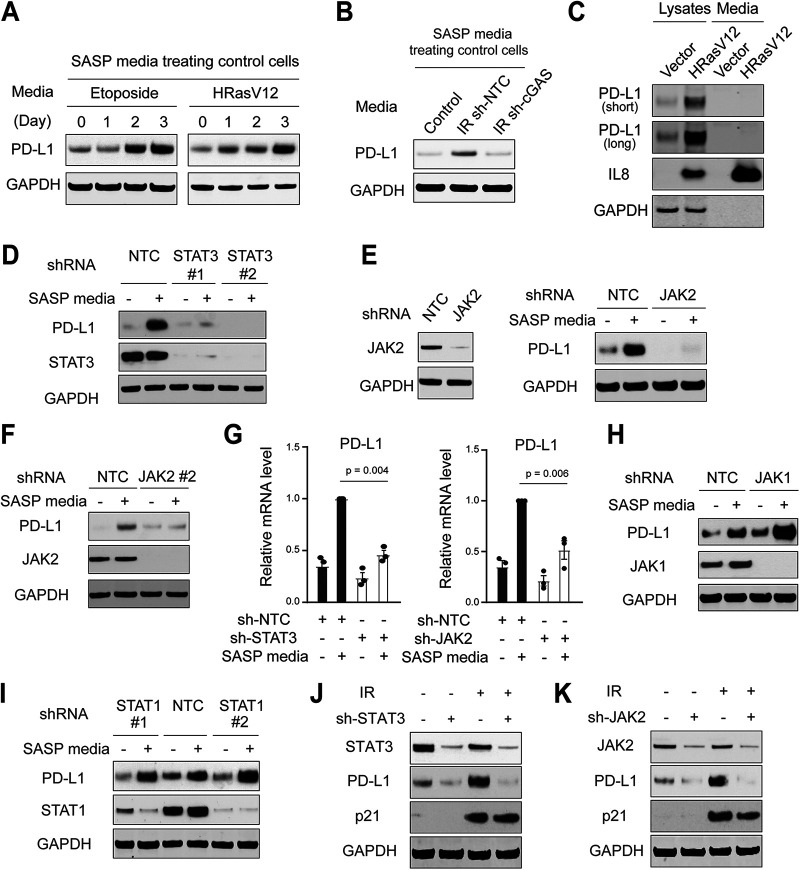
Having established the necessity for the SASP program to induce PD-L1, we subsequently asked whether the SASP program is sufficient to upregulate PD-L1. We collected conditioned media from senescent cells or control cells and used these media to culture low-passage-number control cells (Fig. 3A). The conditioned media from both etoposide-induced senescence and HRasV12-induced senescence upregulated PD-L1 in control cells (Fig. 3A). Further, the conditioned media from wild-type senescent cells, but not from cGAS-deficient senescent cells (as established in Fig. 2A), induced upregulation of PD-L1 in control cells (Fig. 3B). Therefore, these results indicate that the SASP program is sufficient to upregulate PD-L1.
FIG 3.
The SASP upregulates PD-L1 through the JAK-STAT pathway. (A) The conditioned media from etoposide or HRasV12-induced senescent IMR90 cells were harvested and incubated with low-passage-number proliferating IMR90 cells. The protein lysates were analyzed for PD-L1. (B) The conditioned media were harvested from control cells, or IR-induced senescent cells with sh-NTC or cGAS knockdown, as described in the legend to Fig. 2A. These conditioned media were used to incubate low-passage-number proliferating IMR90 cells for 24 h. The cell lysates were probed for PD-L1. (C) IMR90 cells were induced to senescence with HRasV12 for 1 week. The cell lysates and the media were harvested and probed for indicated antibodies. (D) Low-passage-number proliferating IMR90 cells were stably inactivated with shRNA encoding NTC or two independent STAT3 hairpins. The cells were then incubated with conditioned media from HRasV12-induced senescent IMR90 cells for 24 h. The cell lysates were then analyzed with indicate antibodies. (E, F) Similar to panel D, low-passage-number proliferating IMR90 cells were stably inactivated with shRNA encoding NTC or two independent JAK2 hairpins. The cells were then incubated with conditioned media from HRasV12-induced senescent IMR90 cells for 24 h. The cell lysates were probed with indicated antibodies. (G) Cells as in panels D to F were analyzed by RT-qPCR. The results were normalized to lamin A/C and were presented as mean values with SEM (n = 3). The P values were calculated from two-tailed Student’s t test. (H, I) Low-passage-number proliferating IMR90 cells stably expressing shRNA against NTC, JAK1, or STAT1 hairpins were incubated with conditioned media from HRasV12-induced senescent IMR90 cells for 24 h. The cell lysates were probed for indicated antibodies. (J, K) IMR90 cells with indicated hairpins were subjected to IR (20 Gy) and cultured for 14 days. The cell lysates were analyzed as indicated.
We next investigated the mechanisms by which the SASP program induces PD-L1. We first asked whether PD-L1 is present in the conditioned media of senescent cells; if so, the uptake of PD-L1 would explain the increased PD-L1 in control cells. In fact, several cancer cells secrete PD-L1 to the extracellular environment to inhibit immune cell activation (21, 22). We therefore analyzed the cell lysates and conditioned media for PD-L1 (Fig. 3C). While the conditioned media of senescent cells are highly enriched in IL-8, we did not detect PD-L1 there (Fig. 3C), indicating that PD-L1 is not secreted to the conditioned media in our senescence model.
We next asked whether the SASP program upregulates PD-L1 in control cells through signal transduction. The conditioned media in senescent cells harbor a variety of cytokines that can activate the JAK-STAT pathway (2). PD-L1 has been reported to be activated at the transcriptional level by several transcription factors, such as STAT1, STAT3, NF-κB, and Myc (8, 23). We thus hypothesized that the SASP program activates PD-L1 through the JAK-STAT pathway.
We inactivated STAT3 by two independent shRNA hairpins in control cells and cultured the cells with the conditioned media from HRasV12-induced senescence (Fig. 3D). While PD-L1 was upregulated in nontargeting control (NTC) cells, its upregulation was suppressed by STAT3 hairpins (Fig. 3D). Likewise, shRNA against JAK2 inhibited PD-L1 upregulation by the SASP program (Fig. 3E and F). The requirement for JAK2-STAT3 in upregulating PD-L1 was also seen at the mRNA level (Fig. 3G). Inactivation of STAT1 or JAK1 showed minimal effect on PD-L1 induced by the SASP program (Fig. 3H and I), indicating that STAT3 and JAK2 are the predominant isoforms activating PD-L1 in IMR90 cells. The dependency of different JAK and STAT isoforms appears to be cell type specific, as seen in several cancer cell lines that depend on STAT1 to activate PD-L1 transcription (23–25).
We subsequently induced senescence of the STAT3 or JAK2-deficient cells. While PD-L1 is upregulated in IR-induced control senescent cells, this effect is impaired in cells expressing STAT3 or JAK2 shRNA hairpins (Fig. 3J and K). Hence, the JAK-STAT pathway plays an essential role in upregulating PD-L1 in senescence. Further studies are needed to demonstrate a direct effect of STAT1/3 on activating the transcription of PD-L1 gene at the promoter region.
Rapamycin downregulates PD-L1 in senescence.
In the context of cancer immunotherapy, PD-L1/PD-1 checkpoint inhibitors have shown promising clinical outcomes in treating various types of cancer (7–9). However, PD-L1/PD-1 checkpoint inhibitors are known to have side effects (26–28), thus limiting their broad applications to treat senescence/age-associated diseases. We reason that compounds that downregulate PD-L1 in senescent cells will promote immune-mediated clearance of senescent cells, which will be a new approach to intervene in diseases of senescence/aging.
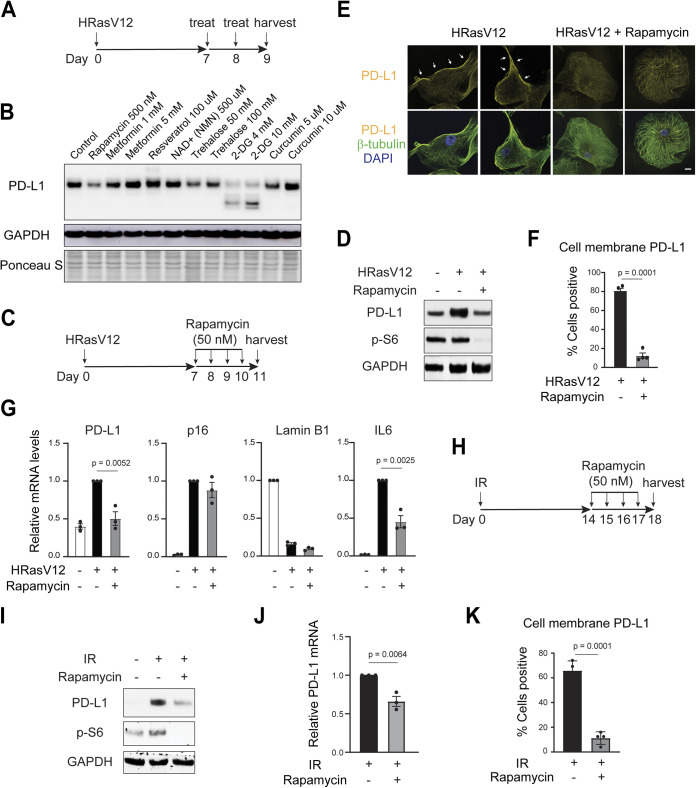
We thus performed a small-scale experiment testing several compounds implicated in antiaging or modulation of PD-L1, in senescent IMR90 cells induced by HRasV12 (scheme shown in Fig. 4A). Among these compounds, rapamycin, metformin, resveratrol, and NAD+ boosting compound nicotinamide mononucleotide have been reported to promote longevity and/or healthy aging (29–33). Trehalose stimulates autophagy and lysosome activities (34). 2-Deoxy-gluocose (2-DG) and curcumin have been shown to modulate PD-L1 in cancer cells (35, 36). Indeed, 2-DG blocked glycosylation of PD-L1 and reduced its stability in our senescent model (Fig. 4B). Interestingly, rapamycin, an mTOR inhibitor that extends the life span of mice, downregulated PD-L1 in senescent cells (Fig. 4B). We confirmed the effect of rapamycin on downregulating PD-L1 in already-established senescent cells (Fig. 4C and D). Further, rapamycin reduced the cell membrane-localized PD-L1 evaluated by immunofluorescence (Fig. 4E and F). In addition, rapamycin suppressed the increased mRNA level of PD-L1 in senescent cells (Fig. 4G) while having minimal effect on lamin B1 and p16 (Fig. 4G).
FIG 4.
Rapamycin downregulates PD-L1 of senescent cells. (A) Scheme of experimental plan. Low-passage-number IMR90 cells were infected with retroviral vector encoding HRasV12 to induce senescence. On days 7 and 8, the cells were treated with compounds and were harvested on day 9 for immunoblotting analysis of PD-L1. (B) Effects of compounds on PD-L1 analyzed by immunoblotting. The doses of compounds are indicated in the figure. (C, D) Rapamycin downregulates PD-L1 in senescent IMR90. (C) Scheme of experimental design. Rapamycin at 50 nM was used. (D) Immunoblotting of PD-L1. Phospho-S6 was shown to indicate that rapamycin successfully inhibited mTOR activity. (E) Cells were prepared as in panel C, fixed, and stained with antibodies as indicated, followed by confocal microscopy imaging. The images were acquired under identical settings, and representative images are shown. β-Tubulin was used to label the cells. Bars, 20 μm. Arrows indicate PD-L1 at or near the cell membrane. (F) Quantification of cell membrane-localized PD-L1. The data are from four randomly selected fields with over 200 cells. The results shown are the mean values with SD. The P values were calculated from two-tailed Student’s t test. (G) Cells as in panel C were analyzed by RT-qPCR with indicated primers. The results were normalized to lamin A/C and were presented as mean values with SEM (n = 3). The P values were calculated from two-tailed Student’s t test. (H) Scheme of experimental design. IMR90 cells were challenged with ionizing irradiation (IR, 20 Gy). On day 14, 50 nM rapamycin was added to the media and was replenished daily. The cells were then harvested at day 18 for analyses. (I, J) Cells treated as in panel H were analyzed for PD-L1 by immunoblotting (I) or by RT-qPCR (J). RT-qPCR results were normalized to lamin A/C and were presented as mean values with SEM (n = 3). The P values were calculated from two-tailed Student’s t test. (K) Quantification of cell membrane-localized PD-L1. The data are from four randomly selected fields with over 200 cells. The results shown are mean values with SD. The P values were calculated from two-tailed Student’s t test.
Rapamycin has been reported to inhibit the SASP program of senescence (37, 38). Consistent with previous reports, we observed reduced IL-6 induction upon rapamycin treatment (Fig. 4G). The effect on the SASP program suggests a possible mechanism by which rapamycin downregulates PD-L1 in senescent cells. In addition to HRasV12-induced senescence, rapamycin also downregulates PD-L1 in IR-induced senescence (Fig. 4H to K). Notably, in our experimental system, senescence was established first, followed by short-term rapamycin treatment (schemes shown in Fig. 4A, C, and H). The ability of rapamycin to reduce PD-L1 in already-established senescent cells illustrates its therapeutic potential in targeting diseases associated with senescence. Future studies are needed to examine the efficacy of rapamycin in facilitating immune-mediated clearance of senescent cells in vivo.
PD-L1 is upregulated in natural aging.
Aging is associated with low-grade but prolonged chronic inflammation in the absence of infection, a conditioned termed as “inflammaging” (39). In addition to senescence and its SASP program, the NLRP3 inflammasome and possibly the cGAS-STING pathway are implicated in generating proinflammatory cytokines in aged tissues (11, 39, 40). Because we found that proinflammatory cytokines upregulate PD-L1 (Fig. 3), we hypothesized that PD-L1 is upregulated in naturally aged tissues.
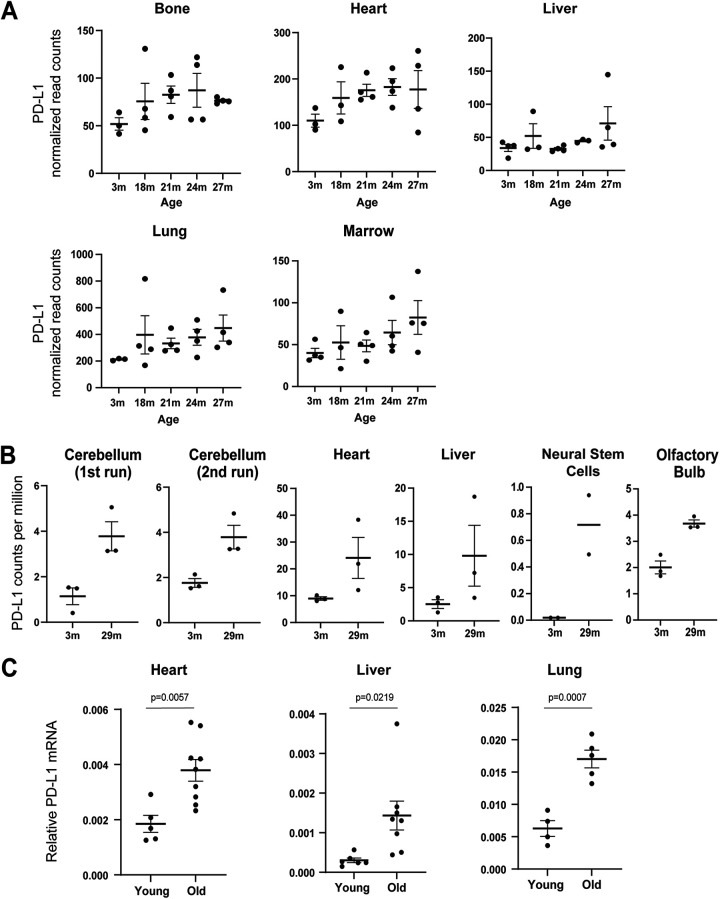
To begin, we performed “data mining” with published resources investigating the transcriptomes of naturally aged mice. First, we explored the Tabula Muris Senis database, a resource for transcriptome sequencing (RNA-seq) of organs across the mouse life span (41). The RNA levels of PD-L1 are generally upregulated in aged bone, heart, liver, lung, and marrow, compared to young controls (Fig. 5A). This effect is not obvious in other tissues from this resource.
FIG 5.
Upregulation of PD-L1 in aging. (A) PD-L1 (CD274) RNA levels in mouse tissues from the Tabula Muris Senis database. The normalized read counts (linear) were used to generate the results. The mean values with SEM are shown. (B) PD-L1 (CD274) RNA levels from Benayoun et al. (42). The transcriptome sequencing (RNA-seq) data were downloaded, and the number of cpm was used. The mean values with SEM are shown. (C) Mice that were 2 to 3 months or 24 to 27 months old were euthanized, and tissues were harvested. The RNA from these tissues were extracted and subjected to RT-qPCR analyses for PD-L1 (CD274) gene. The results were normalized to GAPDH. The mean values with SEM are shown. The P values were calculated from two-tailed Student’s t test.
Second, we explored the RNA-seq data from the work of Benayoun et al., in which heart, liver, cerebellum, olfactory bulb, and neural stem cells were analyzed for their transcriptomes (42). We found that PD-L1 is upregulated in the aged samples of these tissues compared to young controls (Fig. 5B). Due to the limited sample sizes of the two published database, statistical analyses were not shown. These results prompted us to perform experimental examination of PD-L1 in natural aging.
We therefore obtained wild-type mice of the C57BL/6J background, fed ad libitum on a regular diet, comparing young (2 to 3 months) and old (24 to 27 months) tissues. RT-qPCR analyses of PD-L1 revealed statistically significant upregulation in aged heart, liver, and lung (Fig. 5B). PD-L1 protein levels were not examined due to limitation of reliable antibodies to detect mouse PD-L1 protein by immunoblotting. Nevertheless, our results, together with other published studies, collectively indicate that PD-L1 is upregulated in several aged tissues of mice, at least at the RNA level.
PD-L1 is upregulated in the lungs of idiopathic pulmonary fibrosis patients.
The lung is a paradigmatic organ that displays features of senescence in disease and aging (43, 44). Idiopathic pulmonary fibrosis is a form of interstitial lung disease (ILD) characterized by interstitial remodeling, hyperplasia of basal cells (primary stem cell population), abnormal type 2 (AT2) cells (surfactant secreting cells), compromised lung function, and accumulation of fibrogenic senescent cells that promote the progression of the disease (45–47). Clearance of senescent cells in mice and human patients ameliorates disease severity of pulmonary fibrosis (47, 48).
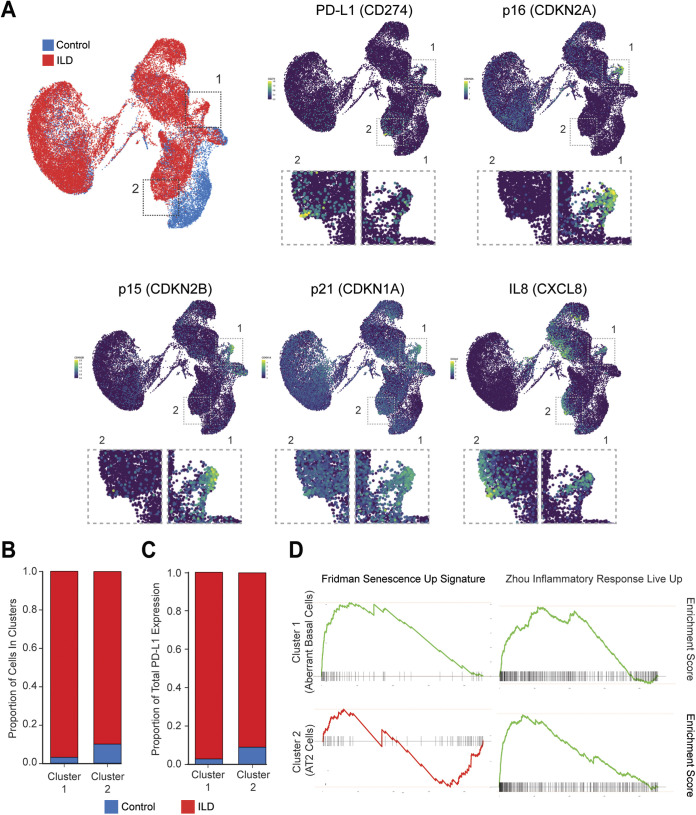
We analyzed a single-cell RNA-seq database of human patients with ILD to explore the expression profile of PD-L1 (49–51). In this database, the expression of 20 fibrotic lungs (red) and 10 nonfibrotic control lungs (blue) is presented in a Uniform Manifold Approximation and Projection (UMAP) plot (49) (Fig. 6A, top left). We found that PD-L1 is substantially upregulated in two clusters: the cells in cluster 1 are predominantly present in ILD patients, and the cells in cluster 2 are qualitatively enriched in ILD samples (Fig. 6A, comparing PD-L1 with top left plot). We subsequently asked whether the PD-L1 high cells in ILD correlate with senescence or inflammatory features and found that the cells in cluster 1 are strongly positive for canonical senescence markers p16, p15, and p21 and that the cells in both clusters 1 and 2 are positive for IL-8 (Fig. 6A).
FIG 6.
Upregulation of PD-L1 in interstitial lung disease (ILD). (A) UMAP expression plots of PD-L1 (CD274), p16 (CDKN2A), p15 (CDKN2B), p21 (CDKN1A), and IL-8 (CXCL8) in merged healthy and ILD human lungs. The data were reanalyzed from Habermann et al. (49). Higher-magnification inset panels below each gene expression are shown to highlight PD-L1 high cells. (B) Bar plot showing relative cell number contributions of healthy control donors versus ILD patients to either cluster 1 or 2. (C) Bar plot showing relative contribution to the expression of PD-L1 from healthy control donors versus ILD patients in cluster 1 or 2. (D) GSEA of the cluster 1 (aberrant basal cells) and cluster 2 (AT2 cells) using MSigDB signatures demonstrating that both clusters 1 and 2 are enriched in inflammatory responses (Zhou_Inflammatory_Response_Live_Up), while only cluster 1 is enriched in senescence signatures (Friedman_Senescent_Up).
We then characterized PD-L1 high cells in the two clusters in greater detail. We found that the cells in both clusters 1 and 2 are predominantly found in ILD patients (Fig. 6B) and that PD-L1 is upregulated in ILD patient samples (Fig. 6C). We then went on further to explore the features of the two clusters high in PD-L1. The single-cell RNA-seq study had identified cluster 1 as a previously unappreciated KRT5−/KRT17+ basaloid population (aberrant basal cells) that expresses p21 and is specifically found in ILD samples (49) (Fig. 6B). We performed Gene Set Enrichment Analysis (GSEA) comparing this population with the rest of the lungs and found that cluster 1 is strongly enriched in senescence signatures (P = 0.001, normalized enrichment score [NES] = 5.27), as well as inflammatory response (P = 0.001, NES = 5.25) (Fig. 6D). For cluster 2, we found that the PD-L1 high cells are derived from AT2 cells, based on their gene expression profile. GSEA revealed that while cluster 2 is not enriched in senescence signature (P = 0.005, NES = −2.56), it is enriched in inflammatory response (P = 0.001, normalized enrichment score [NES] = 5.90) (Fig. 6D). This is consistent with our in vitro observation that inflammatory cytokines alone can induce PD-L1 in control cells (Fig. 2). We also noted that IL-8 high cells are not always positive for PD-L1 (Fig. 6A), suggesting that additional factors are required for the full induction of PD-L1 in the lungs.
Taken together, our data suggest that PD-L1 is upregulated in the lungs of human pulmonary fibrosis patients and that PD-L1 upregulation is linked to senescence and proinflammatory gene expression. These results prompt further studies of targeting PD-L1/PD1 to treat pulmonary fibrosis.
DISCUSSION
In summary, we discovered that PD-L1 is upregulated in senescence and aging. The SASP program of senescence is both necessary and sufficient to induce PD-L1 expression in vitro. While this study focuses on proinflammatory cytokines of the SASP program, interferons are another category of factors secreted by senescent cells (52) and have been reported to induce PD-L1 (24). The paracrine effect of SASP on nonsenescent cells in upregulating PD-L1 may create an overall immunosuppressive environment that inhibits immune-mediated clearance of damaged cells and promotes tumorigenesis. This can help to explain the increased cancer incidences in the elderly population. Hence, targeting PD-L1 of senescent cells may hold promise in restoring tissue homeostasis and in preventing or treating diseases associated with senescence and aging.
In an attempt to find compounds that inhibit PD-L1, we discovered that rapamycin could downregulate PD-L1 in already-established senescent cells. Of note, rapamycin promotes both the health span and the life span of mice (29, 30). Further, transient rapamycin treatment has been reported to increase life span and health span in middle-aged mice (53). These prolongevity effects of rapamycin phenocopy several key features of senescent cell clearance in mice (4, 54, 55). Currently, clinical trials of rapamycin and other mTOR inhibitors in antagonizing dog and human aging are ongoing (56). Short-term rapamycin treatment improved the cardiovascular health of companion dogs (57). Further, topical rapamycin administration decreased p16-positive senescent cells in human skin (58). Future studies are needed to investigate whether part of the antiaging effects of rapamycin in vivo are a result of promoting immune-mediated clearance of senescence. It should be noted that rapamycin and rapalogues are potent immunosuppressants under high doses. The proper doses and frequencies of intervention using rapamycin should be carefully investigated for antiaging purposes.
PD-L1 in cancer cells is regulated by multiple mechanisms, including at the transcriptional, posttranscriptional, and posttranslational levels (23, 59). While we found that the conditioned media of senescence upregulates PD-L1 mRNA level through the JAK-STAT pathway, we do not exclude other alternative mechanisms. For example, IL-6, through JAK1, has been shown to regulate PD-L1 glycosylation and its stability (60). In addition, cancer-associated inflammation, especially tumor necrosis factor (TNF), was reported to modulate PD-L1 ubiquitination and its stability at the protein level (36). Further, it should be noted that rapamycin may downregulate PD-L1 in addition to our proposed mechanism in suppressing the SASP, as rapamycin modulates a variety of cellular events, including autophagy, a reported negative regulator of PD-L1 (61–63).
The interaction between senescent cells and the immune system is incompletely understood. While immune cells play important roles in removing senescent cells (64, 65), recent studies reported that senescent cells also impose inhibitory mechanisms to block immune cell activation. Senescent cells express nonclassical major histocompatibility complex (MHC) molecule HLA-E that binds to the inhibitory receptor NKG2A to inhibit NK and highly differentiated CD8+ T-cells (66). In addition, the NKG2D receptor of immune cells undergoes matrix metalloproteinase (MMP)-dependent shedding of NKG2D ligands, as a consequence of the SASP program of senescence (67). T-cells are a major immune cell type that can kill senescent cells. Mice lacking T-cells exhibited impaired ability to remove senescent cells (68). Conversely, engineered CAR-T-cells that target urokinase plasminogen activator surface receptor (uPAR) of senescent cells reversed senescence-associated pathologies in mice (69). Our findings show that senescent cells upregulate a major T-cell checkpoint molecule PD-L1. While PD-L1 in cancer has been extensively investigated, our study is the first to report the upregulation of PD-L1 in senescence.
Hence, the presence of senescent cells in tissues appears to be a balance of both immune-mediated clearance versus immune tolerance. Under acute senescence-inducing conditions, such as by activated oncogenes, neoantigens derived from mutated proteins of senescent cells are likely to be present. Together with the SASP program that recruits and activates the immune cells, senescent cells can be effectively cleared by the immune system. Under chronic conditions, such as in natural aging, the short of neoantigens together with compromised immune cell functions favors the accumulation of senescent cells, in which the upregulation of immune checkpoint molecules of senescence is involved. In the context of cancer therapy, anti-PD-L1/PD1 immunotherapy works preferentially in PD-L1 high cancers. Consistent with this notion, a recent study found that boosting the SASP program of ovarian cancer improves the response to anti-PD1 therapy (70). Further studies are needed to decipher the specific strategies to modulate immune checkpoints of senescence to intervene in diseases. We hope our findings can spark future studies to explore the therapeutic potential of targeting PD-L1/PD-1 in in vivo models of senescence and aging.
MATERIALS AND METHODS
Cell culture and treatment.
Primary IMR90 and BJ fibroblasts were described previously and were authenticated by genome-wide sequencing analyses (10, 11). The cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) and were intermittently cultured with plasmocin (Invivogen) and tested for mycoplasma using a MycoAlert PLUS mycoplasma detection kit (Lonza). The cells were cultured under physiological oxygen (3%) and were used within population doubling of 35, except for replicative senescence experiments in which the cells were cultured until replication exhaustion (around population doubling 80). For etoposide-induced senescence, IMR90 cells at approximately 60 to 70% confluence were treated with 50 μM etoposide for 24 h and harvested at the days indicated in the figure legends. For BJ cell senescence, the cells at approximately 60 to 70% confluence were treated with 40 μM etoposide for 24 h and harvested at the days indicated in the figure legends. For ionizing irradiation (IR), the cells were irradiated with X-Rad 320 (Precision X-Ray Irradiator) at 20 Gy and harvested as described in the figure legends. For HRasV12-induced senescence, retroviral vector encoding HRasV12 was used to infect cells. Following hygromycin selection, the cells were cultured for 7 or more days as described in the figure legends. These conditions reproducibly induced senescence, confirmed by cell cycle arrest measured by lack of cyclin A and phosphorylated Rb, EdU less than 5% positivity, and senescence associated (SA)-β-galactosidase over 95% positivity, as described in our previous studies (10–13).
Mice.
Young and aged mice were acquired from the National Institute on Aging. C57BL/6 background mice were used. Both sexes were included. The mice were fed ad libitum on a regular diet and handled following institutional regulations and guidelines. The mice were euthanized with CO2 followed by cervical dislocation. Tissues were harvested posteuthanasia and analyzed as indicated.
Reagents and antibodies.
All reagents were purchased from Sigma, unless otherwise stated. STING inhibitor H-151 and p38 inhibitor SB203580 were purchased from Selleck Chemicals. Antibodies used include PD-L1 (Cell Signaling Technology 13684), phospho-S6 (Cell Signaling Technology 2215), STAT1 (Cell Signaling Technology 9172), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Cell Signaling Technology 5174), IL-8 (Abcam ab18672), JAK2 (Cell Signaling Technology 3230), STAT3 (Cell Signaling Technology 9139), p21 (Santa Cruz Biotechnology sc-271532), lamin B1 (Abcam ab16048), p16 (BD Biosciences G175-405), p-ATM (Abcam ab81292), cGAS (Cell Signaling Technology 15102), IL-6 (Cell Signaling Technology 12153), NF-κB p65 (Cell Signaling Technology 8242), JAK1 (Cell Signaling Technology 3344), phospho-Rb (Signaling Technology 9308), and β-tubulin (Sigma T8328).
Retrovirus and lentivirus.
Retroviral WZL-HRasV12 construct was transfected to phoenix packaging cell line, and production of virus for stable expression was performed as previously described (11). Lentiviral pLKO vectors encoding shRNA were transfected with packaging plasmids to HEK293T cells, as described previously (11). Viral supernatant was filtered through a 0.45-μm filter and mixed with trypsinized recipient cells. The infected cells were then selected with puromycin or hydromycin.
pLKO-based shRNA constructs were purchased from Sigma. The following shRNA sequences were used: p65 (TRCN0000014687: CCTGAGGCTATAACTCGCCTA), STAT3 (TRCN0000329886: GCAAAGAATCACATGCCACTT and TRCN0000020843: GCAAAGAATCACATGCCACTT), JAK2 (TRCN0000003181: GCAGAATTAGCAAACCTTATA and TRCN0000003178: CCCTGACCCTAAATAATACAT), STAT1 (TRCN0000280021: CTGGAAGATTTACAAGATGAA and TRCN0000004265: CCCTGAAGTATCTGTATCCAA), and JAK1 (TRCN0000295869:GACAGTCACAAGACTTGTGAA). Nontargeting control and cGAS were described previously (11).
Immunoblotting.
Western blotting was described previously (11), with slight modifications. The cells were lysed in buffer containing 20 mM Tris, pH 7.5, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, supplemented with 1:100 Halt protease and phosphatase inhibitor cocktail (Thermo) and Benzonase (Novagen) at 12.5 U/mL. The lysates were rotated at 4°C for 30 min and boiled at 95°C in the presence of 1% SDS. The resulting supernatants were subjected to electrophoresis using NuPAGE Bis-Tris precast gels (Thermo). After transferring to nitrocellulose membrane, 5% milk in Tris-buffered saline (TBS) was used to block the membrane at room temperature for 1 h. Primary antibodies were diluted in 5% bovine serum albumin (BSA) in TBS supplemented with 0.1% Tween 20 (TBST) and incubated at 4°C overnight. The membrane was washed three times with TBST, each for 10 min, followed by incubation of secondary antibodies at room temperature for 1 h in 5% milk in TBST. The membrane was washed again three times and imaged by film or by an Odyssey imager (Licor Odyssey CLx 2000).
Immunofluorescence.
Immunofluorescence was performed as described previously (11). Briefly, the cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. The cells were washed twice with PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After washing two times, the cells were blocked with 10% BSA in PBS for 1 h at room temperature. The cells were then incubated with primary antibodies in 5% BSA in PBS supplemented with 0.1% Tween 20 (PBST) overnight at 4°C. The next day, the cells were washed four times with PBST, each for 10 min, followed by incubation with Alexa Fluor-conjugated secondary antibody (Thermo) in 5% BSA/PBST for 1 h at room temperature. The cells were then washed four times with PBST, incubated with 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 10 min, and washed twice with PBS. The slides were mounted with ProLong Diamond (Thermo) and imaged with a Leica TCS SP8 fluorescent confocal microscope. Quantification of % positive cells was done under identical microscopy settings between samples. Over 200 cells from 4 randomly selected fields were analyzed.
RT-qPCR.
mRNA from cells were extracted with RNeasy mini kit (Qiagen), with a DNase I (Qiagen) digestion step to minimize genomic DNA contamination. Reverse transcription (RT) was done using a high-capacity RNA-to-cDNA kit (Thermo), and then quantitative PCR (qPCR) was performed using a qPCR machine (Bio-Rad CFX 384 Real-Time System). The results were normalized to lamin A/C for human cells. The following primers were used for RT-qPCR of human cells: PD-L1, TGGTGGTGCCGACTACAAGC and GGGTAGCCCTCAGCCTGACA; lamin B1, CTCGTCGCATGCTGACAGAC and GATCCCTTATTTCCGCCATCT; p16, CCAACGCACCGAATAGTTACG and CCATCATCATGACCTGGATCG; IL-6, CACCGGGAACGAAAGAGAAG and TCATAGCTGGGCTCCTGGAG; and lamin A/C, AGCTGAAAGCGCGCAATACC and GGCCTCCTTGGAGTTCAGCA.
For mouse liver and lung, RNeasy mini kit (Qiagen) was used to extract the RNA. For mouse heart, RNeasy Plus Universal mini kit (Qiagen) was used. The results were normalized to GAPDH for mouse tissues. The following primers were used for RT-qPCR of mouse cells: PD-L1, GGACGCAGGCGTTTACTGCT and AGTGGCTGGATCCACGGAAA; and GAPDH, TGCATCCTGCACCACCAACT and TGGTCATGAGCCCTTCCACA.
Conditioned media.
Control or senescent cells were cultured in regular media for 2 to 3 days before use. The media were then collected and filtered with a 0.45-μm polyvinylidene difluoride (PVDF) filter (Millipore) to remove cells and debris. The resulting supernatant was used for cell treatment or for immunoblotting. The amounts of media used for immunoblotting were quantified based on the protein concentrations of cell lysates, and media corresponding to equal amounts of total cellular proteins were loaded into protein gels.
Data mining from published resources.
For the Tabula Muris Senis database (41), the results were downloaded from the online server, under the bulk RNA-seq section. The normalized read counts (linear) for the CD274 gene were obtained, under corresponding age cohorts in each tissue. The data for male mice were used, because PD-L1 has been reported to be regulated by female hormone cycles (71). For data sets from Benayoun et al. (42), gene expression counts generated using subread 1.4.5-p1 (72) were converted to logarithmic cpm values using EdgeR (version 3.26.8). The expression levels were then filtered to retain counts above a cpm threshold of 0.125. The CD274 gene expression levels (linear) were plotted.
Single-cell RNA-seq analyses and GSEA.
Expression plots were generated using the publicly available data set at http://www.ipfcellatlas.com/. Processed expression values were accessed at GEO (GSE135893) and further analyzed using Seurat (73). Wilcoxon rank sum test was obtained using the R package (74). Fgsea was used to perform GSEA (75, 76) on the entire C2 MSigDB signature set (77). The AT2 cluster was isolated and then subclustered to identify the PD-L1+ subpopulation. Identities were then transferred back onto the full data set prior to GSEA analysis.
Statistical analyses.
Unpaired two-tailed Student’s t test was used for comparison between two groups. One-way analysis of variance (ANOVA) coupled with Tukey’s post hoc test was used for comparisons over two groups. Significance was considered when P value was less than 0.05.
ACKNOWLEDGMENTS
This project was conceived via discussion with Mien-Chie Hung and was initiated in the Shelley Berger laboratory. We thank Kanad Ghosh and Maria Grazia Vizioli for piloting the conditions for this project; Yu Wang, Yuting Tan, and Raymond Lin for technical assistance; and the microscopy core facility of Center for Regenerative Medicine at Massachusetts General Hospital for assistance with confocal microscopy. The single-cell RNA-seq in pulmonary fibrosis idea was inspired by Raghu R. Chivukula. We thank Shuoshuo Wang and Richard T. Lee for advice on heart RNA extraction, as well as Richard T. Lee and Jin Hao for critical reading of our manuscript.
Z.D. is supported by Harvard Stem Cell Institute, Glenn Foundation for Medical Research and an American Federation for Aging Research (AFAR) Grant for Junior Faculty, and by National Institutes of Health (NIH) awards R00AG053406 and R35GM137889. Z.D. and J.R. are also supported by NIH award UG3CA268117, and P.S. is supported by NIH award ZIA AG000679.
Z.D. conceived the project. A.O. and Z.D. performed experiments. A.P.H. and P.D.A. contributed part of the aged mouse tissues. B.L. and J.R. contributed single-cell RNA-seq pulmonary fibrosis data. P.S. contributed RNA-seq data in natural aging of mice. Z.D., A.O., and B.L. wrote the paper. All authors discussed the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.He S, Sharpless NE. 2017. Senescence in health and disease. Cell 169:1000–1011. 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL. 2013. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest 123:966–972. 10.1172/JCI64098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Deursen JM. 2019. Senolytic therapies for healthy longevity. Science 364:636–637. 10.1126/science.aaw1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs BG, Gluscevic M, Baker DJ, Laberge RM, Marquess D, Dananberg J, van Deursen JM. 2017. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov 16:718–735. 10.1038/nrd.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardoll DM. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12:252–264. 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okazaki T, Honjo T. 2007. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol 19:813–824. 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 7.Zou W, Wolchok JD, Chen L. 2016. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med 8:328rv324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun C, Mezzadra R, Schumacher TN. 2018. Regulation and function of the PD-L1 checkpoint. Immunity 48:434–452. 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei SC, Duffy CR, Allison JP. 2018. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 8:1069–1086. 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 10.Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, Catanzaro JM, Ricketts MD, Lamark T, Adam SA, Marmorstein R, Zong WX, Johansen T, Goldman RD, Adams PD, Berger SL. 2015. Autophagy mediates degradation of nuclear lamina. Nature 527:105–109. 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, Capell BC, Xu C, Xu M, Kieckhaefer JE, Jiang T, Shoshkes-Carmel M, Tanim K, Barber GN, Seykora JT, Millar SE, Kaestner KH, Garcia BA, Adams PD, Berger SL. 2017. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550:402–406. 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vizioli MG, Liu T, Miller KN, Robertson NA, Gilroy K, Lagnado AB, Perez-Garcia A, Kiourtis C, Dasgupta N, Lei X, Kruger PJ, Nixon C, Clark W, Jurk D, Bird TG, Passos JF, Berger SL, Dou Z, Adams PD. 2020. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev 34:428–445. 10.1101/gad.331272.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu C, Wang L, Fozouni P, Evjen G, Chandra V, Jiang J, Lu C, Nicastri M, Bretz C, Winkler JD, Amaravadi R, Garcia BA, Adams PD, Ott M, Tong W, Johansen T, Dou Z, Berger SL. 2020. SIRT1 is downregulated by autophagy in senescence and ageing. Nat Cell Biol 22:1170–1179. 10.1038/s41556-020-00579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, Gil J, Hara E, Krizhanovsky V, Jurk D, Maier AB, Narita M, Niedernhofer L, Passos JF, Robbins PD, Schmitt CA, Sedivy J, Vougas K, von Zglinicki T, Zhou D, Serrano M, Demaria M. 2019. Cellular senescence: defining a path forward. Cell 179:813–827. 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Gluck S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, Bridgeman A, Rehwinkel J, Zender L, Ablasser A. 2017. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 19:1061–1070. 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. 2017. cGAS is essential for cellular senescence. Proc Natl Acad Sci USA 114:E4612–E4620. 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller KN, Victorelli SG, Salmonowicz H, Dasgupta N, Liu T, Passos JF, Adams PD. 2021. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184:5506–5526. 10.1016/j.cell.2021.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, Kogan SC, Lowe SW. 2011. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev 25:2125–2136. 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, Heymann M, van der Goot FG, Turcatti G, Behrendt R, Ablasser A. 2018. Targeting STING with covalent small-molecule inhibitors. Nature 559:269–273. 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 20.Freund A, Patil CK, Campisi J. 2011. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J 30:1536–1548. 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. 2018. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560:382–386. 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R. 2019. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 177:414–427.e13. 10.1016/j.cell.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. 2018. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 37:4639–4661. 10.1038/s41388-018-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix BH, Lieskovan S, Damoiseaux R, Lo RS, Ribas A. 2019. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep 29:3766. 10.1016/j.celrep.2019.11.113. [DOI] [PubMed] [Google Scholar]
- 25.Moon JW, Kong SK, Kim BS, Kim HJ, Lim H, Noh K, Kim Y, Choi JW, Lee JH, Kim YS. 2017. IFNgamma induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep 7:17810. 10.1038/s41598-017-18132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmer L, Goldinger SM, Hofmann L, Loquai C, Ugurel S, Thomas I, Schmidgen MI, Gutzmer R, Utikal JS, Goppner D, Hassel JC, Meier F, Tietze JK, Forschner A, Weishaupt C, Leverkus M, Wahl R, Dietrich U, Garbe C, Kirchberger MC, Eigentler T, Berking C, Gesierich A, Krackhardt AM, Schadendorf D, Schuler G, Dummer R, Heinzerling LM. 2016. Neurological, respiratory, musculoskeletal, cardiac and ocular side-effects of anti-PD-1 therapy. Eur J Cancer 60:210–225. 10.1016/j.ejca.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann L, Forschner A, Loquai C, Goldinger SM, Zimmer L, Ugurel S, Schmidgen MI, Gutzmer R, Utikal JS, Goppner D, Hassel JC, Meier F, Tietze JK, Thomas I, Weishaupt C, Leverkus M, Wahl R, Dietrich U, Garbe C, Kirchberger MC, Eigentler T, Berking C, Gesierich A, Krackhardt AM, Schadendorf D, Schuler G, Dummer R, Heinzerling LM. 2016. Cutaneous, gastrointestinal, hepatic, endocrine, and renal side-effects of anti-PD-1 therapy. Eur J Cancer 60:190–209. 10.1016/j.ejca.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 28.Chamoto K, Hatae R, Honjo T. 2020. Current issues and perspectives in PD-1 blockade cancer immunotherapy. Int J Clin Oncol 25:790–800. 10.1007/s10147-019-01588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, Woodward MA, Miller RA. 2012. Rapamycin slows aging in mice. Aging Cell 11:675–682. 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460:392–395. 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, de Cabo R. 2013. Metformin improves healthspan and lifespan in mice. Nat Commun 4:2192. 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. 2008. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab 8:157–168. 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y, Redpath P, Migaud ME, Apte RS, Uchida K, Yoshino J, Imai SI. 2016. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab 24:795–806. 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. 2007. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 282:5641–5652. 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 35.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, Hung MC. 2016. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 7:12632. 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, Kim T, Chang WC, Hsu JL, Yamaguchi H, Ding Q, Wang Y, Yang Y, Chen CH, Sahin AA, Yu D, Hortobagyi GN, Hung MC. 2016. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell 30:925–939. 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, Curran SC, Davalos AR, Wilson-Edell KA, Liu S, Limbad C, Demaria M, Li P, Hubbard GB, Ikeno Y, Javors M, Desprez PY, Benz CC, Kapahi P, Nelson PS, Campisi J. 2015. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol 17:1049–1061. 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ, Raguz S, Acosta JC, Innes AJ, Banito A, Georgilis A, Montoya A, Wolter K, Dharmalingam G, Faull P, Carroll T, Martinez-Barbera JP, Cutillas P, Reisinger F, Heikenwalder M, Miller RA, Withers D, Zender L, Thomas GJ, Gil J. 2015. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol 17:1205–1217. 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franceschi C, Campisi J. 2014. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci 69:S4–S9. 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 40.Youm YH, Grant RW, McCabe LR, Albarado DC, Nguyen KY, Ravussin A, Pistell P, Newman S, Carter R, Laque A, Munzberg H, Rosen CJ, Ingram DK, Salbaum JM, Dixit VD. 2013. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab 18:519–532. 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almanzar N, Antony J, Baghel AS, Bakerman I, Bansal I, Barres BA, Beachy PA, Berdnik D, Bilen B, Brownfield D, Cain C, Chan CKF, Chen MB, Clarke MF, Conley SD, Darmanis S, Demers A, Demir K, de Morree A, Divita T, du Bois H, Ebadi H, Espinoza FH, Fish M, Gan Q, George BM, Gillich A, Gòmez-Sjöberg R, Green F, Genetiano G, Gu X, Gulati GS, Hahn O, Haney MS, Hang Y, Harris L, He M, Hosseinzadeh S, Huang A, Huang KC, Iram T, Isobe T, Ives F, Jones RC, Kao KS, Karkanias J, Karnam G, Keller A, Kershner AM, Khoury N; Tabula Muris Consortium . 2020. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583:590–595. 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benayoun BA, Pollina EA, Singh PP, Mahmoudi S, Harel I, Casey KM, Dulken BW, Kundaje A, Brunet A. 2019. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res 29:697–709. 10.1101/gr.240093.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campisi J. 2016. Cellular senescence and lung function during aging. Yin and yang. Ann Am Thorac Soc 13:S402–S406. 10.1513/AnnalsATS.201609-703AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parikh P, Wicher S, Khandalavala K, Pabelick CM, Britt RD, Jr, Prakash YS. 2019. Cellular senescence in the lung across the age spectrum. Am J Physiol Lung Cell Mol Physiol 316:L826–L842. 10.1152/ajplung.00424.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez-Gonzalez F, Faner R, Rojas M, Agusti A, Serrano M, Sellares J. 2021. Cellular senescence in lung fibrosis. Int J Mol Sci 22:7012. 10.3390/ijms22137012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parimon T, Hohmann MS, Yao C. 2021. Cellular senescence: pathogenic mechanisms in lung fibrosis. Int J Mol Sci 22:6214. 10.3390/ijms22126214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schafer MJ, White TA, Iijima K, Haak AJ, Ligresti G, Atkinson EJ, Oberg AL, Birch J, Salmonowicz H, Zhu Y, Mazula DL, Brooks RW, Fuhrmann-Stroissnigg H, Pirtskhalava T, Prakash YS, Tchkonia T, Robbins PD, Aubry MC, Passos JF, Kirkland JL, Tschumperlin DJ, Kita H, LeBrasseur NK. 2017. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun 8:14532. 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Justice JN, Nambiar AM, Tchkonia T, LeBrasseur NK, Pascual R, Hashmi SK, Prata L, Masternak MM, Kritchevsky SB, Musi N, Kirkland JL. 2019. Senolytics in idiopathic pulmonary fibrosis: results from a first-in-human, open-label, pilot study. EBioMedicine 40:554–563. 10.1016/j.ebiom.2018.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung MI, Taylor CJ, Jetter C, Raju L, Roberson J, Ding G, Wood L, Sucre JMS, Richmond BW, Serezani AP, McDonnell WJ, Mallal SB, Bacchetta MJ, Loyd JE, Shaver CM, Ware LB, Bremner R, Walia R, Blackwell TS, Banovich NE, Kropski JA. 2020. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv 6:eaba1972. 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, Duan Q, Arnett HA, Siddiqui A, Washko GR, Homer R, Yan X, Rosas IO, Kaminski N. 2020. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 6:eaba1983. 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen CI, Ren Z, Verma R, Abdala-Valencia H, Nam K, Chi M, Han S, Gonzalez-Gonzalez FJ, Soberanes S, Watanabe S, Williams KJN, Flozak AS, Nicholson TT, Morgan VK, Winter DR, Hinchcliff M, Hrusch CL, Guzy RD, Bonham CA, Sperling AI, Bag R, Hamanaka RB, Mutlu GM, Yeldandi AV, Marshall SA, Shilatifard A, Amaral LAN, Perlman H, Sznajder JI, Argento AC, Gillespie CT, Dematte J, Jain M, Singer BD, Ridge KM, Lam AP, Bharat A, Bhorade SM, Gottardi CJ, Budinger GRS, Misharin AV. 2019. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 199:1517–1536. 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, Le O, Beausejour C, Ambati J, Ambati K, Simon M, Seluanov A, Gorbunova V, Slagboom PE, Helfand SL, Neretti N, Sedivy JM. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566:73–78. 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bitto A, Ito TK, Pineda VV, LeTexier NJ, Huang HZ, Sutlief E, Tung H, Vizzini N, Chen B, Smith K, Meza D, Yajima M, Beyer RP, Kerr KF, Davis DJ, Gillespie CH, Snyder JM, Treuting PM, Kaeberlein M. 2016. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife 5:e16351. 10.7554/eLife.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, Khazaie K, Miller JD, van Deursen JM. 2016. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530:184–189. 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, Onken JL, Johnson KO, Verzosa GC, Langhi LGP, Weigl M, Giorgadze N, LeBrasseur NK, Miller JD, Jurk D, Singh RJ, Allison DB, Ejima K, Hubbard GB, Ikeno Y, Cubro H, Garovic VD, Hou X, Weroha SJ, Robbins PD, Niedernhofer LJ, Khosla S, Tchkonia T, Kirkland JL. 2018. Senolytics improve physical function and increase lifespan in old age. Nat Med 24:1246–1256. 10.1038/s41591-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mannick JB, Del Giudice G, Lattanzi M, Valiante NM, Praestgaard J, Huang B, Lonetto MA, Maecker HT, Kovarik J, Carson S, Glass DJ, Klickstein LB. 2014. mTOR inhibition improves immune function in the elderly. Sci Transl Med 6:268ra179. 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 57.Urfer SR, Kaeberlein TL, Mailheau S, Bergman PJ, Creevy KE, Promislow DEL, Kaeberlein M. 2017. A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. Geroscience 39:117–127. 10.1007/s11357-017-9972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung CL, Lawrence I, Hoffman M, Elgindi D, Nadhan K, Potnis M, Jin A, Sershon C, Binnebose R, Lorenzini A, Sell C. 2019. Topical rapamycin reduces markers of senescence and aging in human skin: an exploratory, prospective, randomized trial. Geroscience 41:861–869. 10.1007/s11357-019-00113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. 2019. Mechanisms controlling PD-L1 expression in cancer. Mol Cell 76:359–370. 10.1016/j.molcel.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, Wang HL, Yang WH, Yen EY, Chang WC, Zha Z, Lim SO, Lai YJ, Liu C, Liu J, Dong Q, Yang Y, Sun L, Wei Y, Nie L, Hsu JL, Li H, Ye Q, Hassan MM, Amin HM, Kaseb AO, Lin X, Wang SC, Hung MC. 2019. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest 129:3324–3338. 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lastwika KJ, Wilson W, 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. 2016. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res 76:227–238. 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 62.Cao X, Zhao Y, Wang J, Dai B, Gentile E, Lin J, Pu X, Ji L, Wu S, Meraz I, Majidi M, Roth JA. 2017. TUSC2 downregulates PD-L1 expression in non-small cell lung cancer (NSCLC). Oncotarget 8:107621–107629. 10.18632/oncotarget.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang J, Wang L, Wang C, Shen J, Su B, Marisetty AL, Fang D, Kassab C, Jeong KJ, Zhao W, Lu Y, Jain AK, Zhou Z, Liang H, Sun SC, Lu C, Xu ZX, Yu Q, Shao S, Chen X, Gao M, Claret FX, Ding Z, Chen J, Chen P, Barton MC, Peng G, Mills GB, Heimberger AB. 2020. Verteporfin inhibits PD-L1 through autophagy and the STAT1-IRF1-TRIM28 signaling axis, exerting antitumor efficacy. Cancer Immunol Res 8:952–965. 10.1158/2326-6066.CIR-19-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prata L, Ovsyannikova IG, Tchkonia T, Kirkland JL. 2018. Senescent cell clearance by the immune system: emerging therapeutic opportunities. Semin Immunol 40:101275. 10.1016/j.smim.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yousefzadeh MJ, Flores RR, Zhu Y, Schmiechen ZC, Brooks RW, Trussoni CE, Cui Y, Angelini L, Lee KA, McGowan SJ, Burrack AL, Wang D, Dong Q, Lu A, Sano T, O'Kelly RD, McGuckian CA, Kato JI, Bank MP, Wade EA, Pillai SPS, Klug J, Ladiges WC, Burd CE, Lewis SE, LaRusso NF, Vo NV, Wang Y, Kelley EE, Huard J, Stromnes IM, Robbins PD, Niedernhofer LJ. 2021. An aged immune system drives senescence and ageing of solid organs. Nature 594:100–105. 10.1038/s41586-021-03547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pereira BI, Devine OP, Vukmanovic-Stejic M, Chambers ES, Subramanian P, Patel N, Virasami A, Sebire NJ, Kinsler V, Valdovinos A, LeSaux CJ, Passos JF, Antoniou A, Rustin MHA, Campisi J, Akbar AN. 2019. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat Commun 10:2387. 10.1038/s41467-019-10335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muñoz DP, Yannone SM, Daemen A, Sun Y, Vakar-Lopez F, Kawahara M, Freund AM, Rodier F, Wu JD, Desprez P-Y, Raulet DH, Nelson PS, van’t Veer LJ, Campisi J, Coppé J-P. 2019. Targetable mechanisms driving immunoevasion of persistent senescent cells link chemotherapy-resistant cancer to aging. JCI Insight 4:e124716. 10.1172/jci.insight.124716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, Iken M, Vucur M, Weiss S, Heikenwalder M, Khan S, Gil J, Bruder D, Manns M, Schirmacher P, Tacke F, Ott M, Luedde T, Longerich T, Kubicka S, Zender L. 2011. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479:547–551. 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 69.Amor C, Feucht J, Leibold J, Ho YJ, Zhu C, Alonso-Curbelo D, Mansilla-Soto J, Boyer JA, Li X, Giavridis T, Kulick A, Houlihan S, Peerschke E, Friedman SL, Ponomarev V, Piersigilli A, Sadelain M, Lowe SW. 2020. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583:127–132. 10.1038/s41586-020-2403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao X, Zhao B, Zhou W, Liu H, Fukumoto T, Gabrilovich D, Zhang R. 2021. Sensitization of ovarian tumor to immune checkpoint blockade by boosting senescence-associated secretory phenotype. iScience 24:102016. 10.1016/j.isci.2020.102016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shuai C, Yang X, Pan H, Han W. 2020. Estrogen receptor downregulates expression of PD-1/PD-L1 and infiltration of CD8+ T cells by inhibiting IL-17 signaling transduction in breast cancer. Front Oncol 10:582863. 10.3389/fonc.2020.582863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 73.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. 2021. Integrated analysis of multimodal single-cell data. Cell 184:3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korsunsky I, Nathan A, Millard N, Raychaudhuri S. 2019. Presto scales Wilcoxon and auROC analyses to millions of observations. bioRxiv. 10.1101/653253. [DOI]
- 75.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. 2021. Fast gene set enrichment analysis. bioRxiv. 10.1101/060012. [DOI]
- 76.Sergushichev AA. 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 10.1101/060012. [DOI]
- 77.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]