Abstract
Purpose
Following retinal detachment (RD) photoreceptors (PRs) sustain hypoxic stress and eventually die. Hypoxia-inducible factor-1α (HIF-1α) plays a central role in cellular adaptation to hypoxia. The purpose of this study is to determine the necessity of HIF-1α on PR cell survival after RD.
Methods
Experimental RD was created in mice by injection of hyaluronic acid (1%) into the subretinal space. Mice with conditional HIF-1α knockout in rods (denoted as HIF-1αΔrod) were used. HIF-1α expression in retinas was measured real-time polymerase chain reaction (RT-PCR) and Western blotting. PR cell death after RD was evaluated using TUNEL assay. Optical coherence tomography (OCT) and histology were used to evaluate retinal layer thicknesses and PR cell densities. A hypoxia signaling pathway PCR array was used to examine the expression of HIF-1α target genes after RD.
Results
HIF-1α protein levels were significantly increased after RD, and depletion of HIF-1α in rods blunted this increase. A compensatory increase of HIF-2α protein was observed in HIF-1αΔrod mice. Conditional knockout (cKO) of HIF-1α in rods did not lead to any morphologic change in attached retinas but resulted in significantly increased PR cell loss after RD. HIF-1α cKO in rods altered the responses to retinal detachment for 25 out of 83 HIF-1α target genes that were highly enriched for genes involved in glycolysis.
Conclusions
Rod-derived HIF-1α plays a key role in the PR response to RD, mediating the transcriptional activity of a battery of genes to promote PR cell survival.
Keywords: retinal detachment (RD), hypoxia, hypoxia-inducible factor-1α (HIF-1α), photoreceptors (PRs), glycolysis
The retina, as a part of the central nervous system, requires intense metabolic and energy support to maintain vision.1–3 In order to meet this nutrient demand while optimizing optics, the human retina has a dual blood supply, with the inner and outer retina supplied by the retinal and choroidal vasculatures, respectively.4 Photoreceptors (PRs), the cells that detect and convert light signals into electrical signals, are located in the outer retina and receive their nutrient and oxygen supply from the choroidal vasculature through the retinal pigment epithelium (RPE). Retinal detachment (RD) is a pathological condition in which the neurosensory retina is separated from the underlying RPE, leading to the disruption of oxygen and nutrient delivery to PRs and eventually PR cell death.5–8 RD is a common feature of many retinal diseases and a leading cause of blindness.9,10
Due to the high metabolic demand of PR cells, periods of disrupted PR-RPE homeostasis might be expected to result in rapid PR cell death. However, clinical observation shows that patients can obtain useful visual recovery, even if there is delay in re-attachment of the neurosensory retina to the RPE.11–13 Previous studies by our group demonstrated that after RD intrinsic prosurvival pathways, including interleukin-6 and autophagy pathways, became activated to counteract the pro-apoptotic pathway and maintain PR cell survival.14,15 However, the upstream activators of these prosurvival pathways in the context of RD are poorly understood. Our previous work demonstrated that placing 661W PR cells in hypoxic conditions increased the protein levels of hypoxia-inducible factor-1α (HIF-1α) and enhanced autophagy.16 RNA silencing of HIF1α expression in hypoxic 661W cells or detached rat retinas reduced expression of the autophagy marker LC3-II and increased PR cell death. Others have shown that stabilizing HIF-1α, through the retro-orbital administration of a prolyl-4-hydroxylases inhibitor, promoted mitophagy, reduced the generation of reactive oxygen species (ROS), and reduced PR cell death after RD.17 These results emphasize the crucial role of HIF-1α in response to detachment-induced hypoxia and suggest that HIF-1α may be a key intrinsic upstream activator of prosurvival pathways in PRs following RD.
Hypoxia-inducible factors (HIFs) are transcription factors that respond to changes in tissue oxygen levels and allow cells to adapt and survive in hypoxic environments.18 HIFs form heterodimers that consist of a highly regulated alpha subunit and a beta subunit, both of which are constitutively expressed on the mRNA level.18 There are 3 HIF-α isoforms, HIF-1α, HIF-2α, and HIF-3α. HIF-1α and HIF-2α are widely expressed in various tissues and share some overlapping functions but also mediate disparate responses.19 HIF-3α is least characterized and can function as an inhibitor of HIF-1α.20 Under normoxic conditions, HIF-1α is hydroxylated by prolyl hydroxylases (PHDs), which use oxygen as a substrate. Hydroxylated HIF-1α binds to von Hippel-Lindau protein (pVHL), which mediates polyubiquitylation of HIF-1α and subsequent proteasomal degradation.21,22 Under hypoxic conditions, PHD activity is inhibited by the shortage of oxygen, hindering protein hydroxylation and preventing degradation of HIF-1α protein. Subsequently, HIF-1α accumulates, enters the nucleus, heterodimerizes with HIF-1β, binds to a conserved hypoxia responsive element, and activates transcription of a battery of genes crucial for adaptation to hypoxia.23
As HIF-1α is a master regulator of the hypoxic response, it can activate hundreds of genes within any given cell type, and the target genes vary from one cell type to another.24,25 The HIF-1α target genes in PRs following RD are essentially unknown. In this study, we generated transgenic mice with conditional knockout of HIF-1α in rod PRs (HIF1αΔrod) to investigate the necessity of HIF-1α for PR protection during acute RD and to identify HIF-1α target genes in rod PRs. We found that lack of HIF-1α in rods resulted in more PR cell death following RD and differentially altered the expression of a number of genes potentially affecting the prosurvival pathways in rod PRs.
Materials and Methods
Animals
All animal experiments were conducted in adherence to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the University of Michigan Committee on the Use and Care of Animals. Transgenic mice with conditional knockout of the Hif1a gene in rod PRs were used. HIF1αflox strain26 (strain #007561) and rhodopsin-Cre mouse strain27 (strain #015850) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). HIF1αflox mouse strain was crossed with rhodopsin-Cre mouse strain to generate HIF1αflox/flox(Rho)Cre+ mice (denoted as HIF-1αΔrod). Rho-Cre+ mice were used as controls. Mice were genotyped by TransNetX using the genotyping assay provided by the Jackson Laboratory (https://www.jax.org/Protocol?stockNumber=007561&protocolID=28602). All mice were bred and housed in the vivarium at the University of Michigan Kellogg Eye Center under standard 12-hour light/dark cycle.
Experimental Model of Retinal Detachment
Adult mice at 8 to 12 weeks of age were used in the study. Acute retinal detachment was created as previously described.28 Briefly, after mice were anesthetized, their pupils were dilated with topical administration of tropicamide (1%) and phenylephrine (2.5%). A sclerotomy was made with a 30-gauge needle at approximately 1 mm posterior to the limbus. The tip of a 35-gauge needle attached to a microsyringe (NANOFIL-100 injector system; World Precision Instruments, Sarasota, FL, USA) was passed through the sclerotomy, into the vitreous cavity, through the peripheral retina, and into the subretinal space. Sodium hyaluronate (10 mg/mL, Healon; Abott Medical Optics, Santa Ana, CA, USA) was injected into the subretinal space to detach one-third to one-half of the neurosensory retina. The fellow eyes did not receive a sclerotomy or an injection.
Western Blot Analysis
Western blotting was performed as previously reported.29 Primary antibodies used were: HIF-1α (catalog #MAB1536; R&D Systems Inc., Minneapolis, MN, USA) and HIF-2α (catalog #NB100-122; Novus Biologicals, Centennial, CO, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; catalog #AM4300; Applied Biosystems, Foster City, CA, USA) was used as a loading control.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick-End Labeling Assay and Retinal Histology
For the TUNEL assay, eyes were enucleated and fixed in 4% paraformaldehyde for 2 hours at room temperature (RT). Under a dissecting microscope, the cornea and lens were removed. The remaining posterior eyecup was cryoprotected in series of sucrose gradients (5-10-20%) and embedded in a mixture (in 1:1 ratio) of 20% sucrose and optimal cutting temperature compound (Tissue Tek 4583; Sakura Finetek, Tokyo, Japan). A cryostat was used to cut 10-μm thick retinal sections that cross the optic nerve head. To evaluate cell death, the TUNEL assay (DeadEnd Fluorometric TUNEL System catalog #G3250; Promega, Madison, WI, USA) was performed at 3 days post RD according to the manufacturer's instructions. Nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI). Images were obtained with a Leica DM6000 microscope (Leica Microsystems, Wetzlar, Germany) with fixed detection gains. Imaris software (Oxford Instruments plc., Oxford, UK) was used to quantify the TUNEL positive cells in the outer nuclear layer (ONL) in the detached retinas from the optic nerve head to the tip of the retinal periphery by two researchers in a masked manner. Four biological replicates were obtained in each group. The number of TUNEL positive cells of each replicate was the average number from three noncontinuous retinal sections.
For retinal histology, eyes were enucleated, fixed in 4% paraformaldehyde, and embedded in paraffin. Serial retinal sections (6 μm) that cross the optic nerve head were obtained using a Shandon AS325 microtome (Thermo Fisher Scientific, Waltham, MA, USA). Sections were stained with hematoxylin and eosin. To assess PR cell number in the ONL, the detached portion of the retina was imaged under a light microscopy with 20 times magnification. Images from three noncontinuous sections from each replicate retina were analyzed. The nuclei count in the ONL, the area of the ONL, and the area of inner nuclear layer (INL) within each image were measured using ImageJ software (https://imagej.net). The ratio of the ONL area to the INL area was calculated. The area of the INL was used as a normalization control to account for the possible differences in sectioning and allow for inter-sample comparison. The control and HIF-1αΔrod groups contained 9 and 8 biological replicates, respectively.
Spectral Domain Optical Coherence Tomography
A spectral domain optical coherence tomography (OCT) system (Bioptigen, Inc., Durham, NC, USA) was used to assess retinal thickness in mice. After anesthesia, a volume scan (in 1.6 × 1.6 mm square) of the retina with the optic nerve head at the edge of the scan was performed on the attached and detached retinas. The thickness of the retina in the attached and detached retinas was measured at 500 pixels away from the optic nerve head using ImageJ software. The thickness of the ONL and the thickness from the inner limiting membrane (ILM) to the outer limiting membrane (OLM) were obtained and the ratio of ONL/(ILM-OLM) was calculated. Because the detached retina is no longer orthogonal to the OCT light path, a previously developed adjustment based on the angle of the detached retina to the RPE was made to ensure measurements of the retinal layers at the same distance from the optic nerve head as in the attached retina.29
RNA Isolation, Reverse Transcription, Real-Time PCR, and HIF-1α Target PCR Array Quantification
To determine the effects of Hif1a conditional knockout (cKO) on retinal Hif1a mRNA expression, biological replicates from each group were assayed. Retinal samples were collected at desired time points, snap-frozen in liquid nitrogen, and stored at −80°C. Total RNA were isolated using RNeasy Mini kit (catalog #74104; Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA concentrations were measured with NanoDrop One Spectrophotometer (catalog #701058108; Thermo Fisher Scientific). First-strand cDNA synthesis was catalyzed by SuperScript III Reverse Transcriptionase (catalog #18080093; Thermo Fisher Scientific) using 500 ng of total RNA. Quantitative real time-PCR was performed with SYBR Green PCR Master Mix (catalog #4309155; Applied Biosystems) according to the manufacturer's instructions. The mRNA expression level of ribosomal protein L19 (Rpl19) gene was used as the internal control. Mouse primer pair sequences used are as follows: Hif-1α, 5′- TCATCAGTTGCCACTTCCCC-3′ and 5′-CTCACCGGCATCCAGAAGTT-3′ and Rpl19, 5′-ATGCCAACTCCCGTCAGCAG-3′ and 5′-TCATCCTTCTCATCCAGGTCACC-3′.
The levels of mRNAs corresponding to genes induced by hypoxia or related to hypoxia signaling were assayed using the RT2 Profiler PCR Array for the Mouse Hypoxia Signaling Pathway (PAMM-032ZD; Qiagen, Hilden, Germany) according to the manufacturer's instructions. The genes corresponding to the mRNAs examined in the array are shown in Supplementary Table S1. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of genes exhibiting expression affected by Hif1a cKO were performed using ShinyGO 0.76 (http://bioinformatics.sdstate.edu/go/) and importing the list of 83 HIF pathway genes in the array as the background gene list.
Statistical Analyses
Data were presented as mean values ± standard deviation (SD). Statistical analysis was performed using Prism 9 software (GraphPad Software, Inc., San Diego, CA, USA). The significance of data between two groups and more than two groups was determined by the unpaired Student's t-test and 1-way ANOVA followed by Tukey's test for multiple comparisons, respectively. Significance was accepted at P < 0.05.
Results
Conditional Knockout of HIF-1α in Rod PR Cells
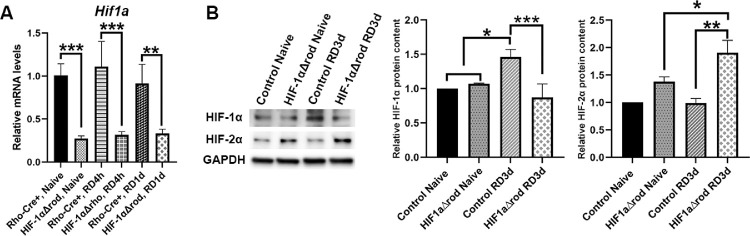
Mice expressing Cre recombinase under the control of rhodopsin promoter were crossed with HIF1αflox mice to generate HIF-1αΔrod mice. Cre recombinase causes excision of exon 2 of the Hif1a gene, thus creating a null allele. To verify cKO of Hif1a in the retina, we first used PCR primers that span the junctions of exon 2 of Hif1a gene in a quantitative RT-PCR assay of Hif1a mRNA using whole retina RNA. As shown in Figure 1A, in naïve mice, the Hif1a mRNA levels were significantly reduced in HIF-1αΔrod mouse retinas relative to Rho-Cre+ controls (72.7% reduction). Conditional knockout of Hif1a in rod PR also significantly decreased the levels of the Hif1a mRNA in detached retinas, both at 4 hours and 1 day post RD (71.6% and 63.7% reductions, respectively).
Figure 1.
HIF-1α ablation in rods significantly reduced HIF-1α expression in the retina and leaded to compensatory increase of HIF-2α protein. (A) Relative Hif1a mRNA levels in naïve attached and detached retinas of Rho-Cre+ and HIF-1αΔrod mice at 4 hours and 1 day post retinal detachment (RD). Hif1a mRNA levels were normalized to the ribosomal protein L19 mRNA levels (Rpl19; N = 4). (B) Western blot analysis of HIF-1α and HIF-2α protein levels in naïve attached and detached retinas of HIF-1αΔrod and control mice at 3 days post RD. Rho-Cre+ mice were used as controls. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as loading controls. Quantification of protein levels based on densitometry of the Western blots are shown in graphs (N = 3). Data are shown as means ± SD. One-way ANOVA and Tukey's test for multiple comparisons were used for statistical analysis. * P < 0.05, **P < 0.01, ***P < 0.001.
To further confirm the effective knockout of HIF-1α, immunoblotting was used to detect HIF-1α protein in retinal whole cell lysates from both naïve and detached retinas (Fig. 1B). Low baseline expression of HIF-1α was detected in the control naïve retinas. RD significantly increased HIF-1α protein levels (1.46-fold increase) at 3 days post RD. Conditional knockout of HIF-1α in rods did not affect the baseline levels of HIF-1α protein in the naïve retinas, but significantly blunted its increase in the detached retinas. In fact, the HIF-1α protein levels remained at baseline in the detached retinas of HIF-1αΔrod mice at 3 days post RD. We also examined the protein levels of the HIF-2α isoform (Fig. 1B). Low levels of HIF-2α protein were detected in the naïve retinas, and RD did not enhance these levels. Conditional knockout of HIF-1α in rods increased HIF-2α protein levels, even in the naïve retinas. RD significantly enhanced the HIF-2α protein expression in HIF-1αΔrod mice compared to either naïve retinas in HIF-1αΔrod mice or detached retinas in control mice.
Depletion of HIF-1α in Rods Leads to Increased PR Cell Loss Following RD
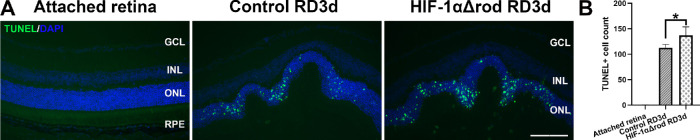
We previously reported that PR apoptosis peaks at 3 days post RD in rodents.28,30,31 Thus, we used TUNEL assay to evaluate the effects of rod-specific deletion of HIF-1α on PR cell apoptosis at 3 days post RD (Fig. 2). Few TUNEL positive cells were detected in the attached retinas. RD sharply increased the number of TUNEL positive cells in the ONL. The number of TUNEL positive cells increased significantly further in HIF-1αΔrod mice compared with the controls.
Figure 2.
Rod-specific HIF-1α ablation resulted in more apoptosis of PR cells after RD. (A) Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) assay was used to detect apoptotic cells in the outer nuclear layer of HIF-1αΔrod and control mice at 3 days post RD. Rho-Cre+ mice were used as controls. Nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI). Green: TUNEL positive cells, blue: DAPI. Scale bar = 100 µm. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. (B) Quantification of TUNEL positive cells in the ONL in the attached and detached retinas from the optic nerve head to the extreme retinal periphery. Four biological replicates were obtained in each group. The number of TUNEL positive cells for each replicate was the average number from three noncontinuous retinal sections. Data are shown as mean ± S.D. * P < 0.05 (unpaired Student's t-test).
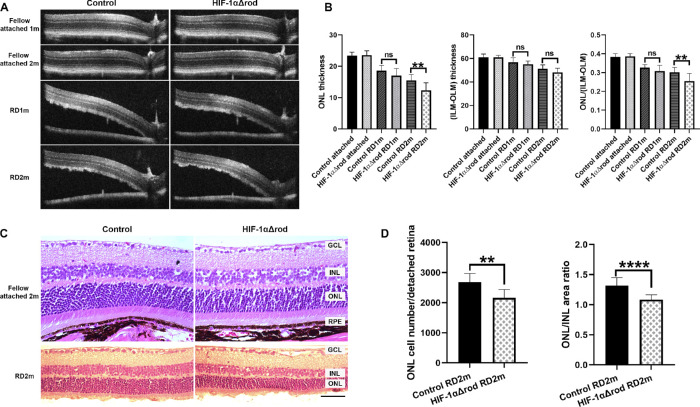
To evaluate the long-term effects of HIF-1α ablation in rods on PR cell survival, we analyzed the thickness of the ONL by OCT in the attached retinas in the fellow eyes and the detached retinas at 1- and 2-months post RD (Figs. 3A, 3B). Without detachment, depletion of HIF-1α in rods did not affect the thickness of ONL compared with the controls, as demonstrated by the similar ONL thicknesses in the attached retinas of control and HIF-1αΔrod mice. The ONL/(ILM-OLM) ratio was calculated to normalize the ONL thickness. As expected, the control mice showed a progressive thinning of the ONL after detachment. At 1 month post RD, the normalized thicknesses of the ONL in the detached retinas of HIF-1αΔrod mice were trending lower compared with the controls, and by 2 months post RD this difference reached statistical significance. Histology of retinal section at 2 months post RD confirmed the observations from OCT. Without detachment, retinal morphology and ONL thickness in HIF-1αΔrod mice remained the same as the control mice (Fig. 3C). Histology confirmed that HIF-1α ablation in rods increased PR cell loss following RD, as revealed by further decreases in absolute ONL cell counts and the ONL/INL area ratios of the detached retinas of HIF-1αΔrod mice, relative to the corresponding controls (see Figs. 3C, 3D).
Figure 3.
Conditional knockout of HIF-1α in rods increased photoreceptor cell loss following RD. (A) Optical coherence tomography (OCT) images of fellow attached and detached retinas at 1 and 2 months post-RD in HIF-1αΔrod and control mice. (B) Graphs show the measurements obtained from A. The thickness of ONL and the thickness from the inner limiting membrane (ILM) to the outer limiting membrane (OLM) were measured at 500 pixels away from the optic nerve head using ImageJ. The ONL/(ILM-OLM) ratio was calculated. (C) Hematoxylin and eosin staining of retinal sections of fellow attached and detached retinas at 2 months post RD in HIF-1αΔrod and control mice. Scale bar = 50 µm. (D) Graphs show the measurements obtained from C. The nuclei count in the ONL per detached retina from the optic nerve head to the tip of retinal periphery was measured using ImageJ. The areas of ONL and INL were also measured and ONL/INL area ratio was calculated. Control group N = 9; HIF-1αΔrod group N = 8. Data are shown as mean ± S.D. ns, not significant. **P < 0.01, ****P < 0.0001 (unpaired Student's t-test).
Expression of HIF-Dependent Genes in the HIF-1αΔrod Mice
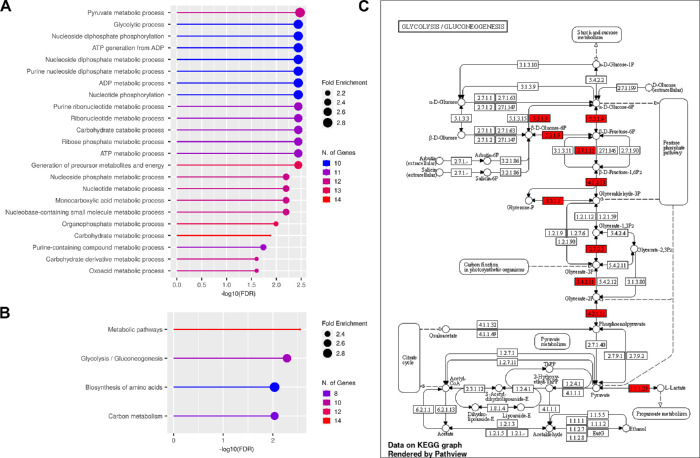
To determine the potential effects of HIF-1α inactivation in rods on target gene expression in attached and detached retinas, we utilized a PCR array examining mRNA levels of genes in the mouse hypoxia signaling pathway (see Supplementary Table S1). A total of 83 genes were analyzed in attached and detached retinas at both the 4 hours and 1 day time points. The results revealed that 25 out of the 83 genes were differentially expressed in the whole retinas of HIF-1αΔrod mice compared with their corresponding controls. GO and KEGG pathway enrichment analyses for the 25 affected genes are shown in Figures 4A and 4B (Supplementary Tables S2, S3). Enriched KEGG categories were for glycolytic production of pyruvate and pathways highly related to or overlapping with glycolysis (Fig. 4C). The 58 genes that did not exhibit altered expression in retinas of HIF-1αΔrod mice included genes in pathways such as: programmed cell death, response to oxidative stress, cell proliferation, cell migration, and blood vessel development (Supplementary Tables S4, S5). However, expression of a key regulator of retinal vessel development, vascular endothelial growth factor A (Vegfa), was affected by HIF-1α cKO in rods.
Figure 4.
The 25 HIF-1α-responsive genes with altered expression following RD in retinas with cKO of HIF-1α in rods were subjected to Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. (A) A lollipop graph for GO pathway enrichment analysis. (B) A lollipop graph for KEGG pathway enrichment analysis. (C) A KEGG glycolysis pathway graph with altered genes by cKO of HIF-1α in rods shown in red.
Discussion
Deprivation of oxygen and nutrients from the underlying choroidal vasculature contributes to immediate and sustained stress to PR cells following RD. The transcription factor HIF-1α is a principal regulator of tissue response to hypoxia and rapidly alters the expressions of a large number of genes. However, the role of HIF-1α on PR cell survival and its downstream regulatory components after RD is largely unknown. In this study, we investigated the consequences of rod-specific HIF-1α knockout on PR cell survival following RD. Our data suggest that lack of functional HIF-1α in rods did not affect retinal morphology or PR cell survival in attached retinas, but accelerated PR cell death after RD. We examined the target gene expressions of the mouse hypoxia signaling pathway after rod-specific HIF-1α inactivation in the retina to explore the possible mechanisms of its protective effects in the detached retina. Deletion of HIF-1α in rods differentially impacted the transcriptional levels of a number of genes in various metabolic pathways centered about glycolysis. Our data strongly support the role of HIF-1α as an upstream regulator of the acute protective response to RD that contributes to PR cell survival through increase anaerobic glycolysis.
Protein levels of HIF-1α and HIF-2α can be regulated at the levels of transcription, translation, and post-translational stability.19 Our data revealed no significant difference of Hif-1α mRNA levels but increased HIF-1α protein levels following RD, suggesting that under hypoxic conditions HIF-1α levels are mainly regulated by oxygen-dependent decrease in degradation.32 HIF-2α shares approximately 50% amino acid sequence identity and similar protein structures with HIF-1α, and is reported to have overlapping functions with HIF-1α.19,33,34 However, these two isoforms also mediate disparate target gene expression, presumably through differential interactions with specific cofactors.35,36 We observed compensatory upregulation of HIF-2α protein levels both in attached and detached retinas when there is depletion of HIF-1α in rods, suggesting that HIF-2α may compensate for part of the functions of HIF-1α, especially after RD. Interestingly, a previous report of a conditional knock out of HIF-1α in cones in both rod-dominant and all-cone retinas found no compensatory increase in HIF-2α expression.37 This discrepancy might be due to the different properties between rods and cones.
OCT measurement showed no significant difference in ONL or retinal thickness in the attached retinas of HIF-1αΔrod mice, consistent with previous reports that the absence of functional HIF-1α or HIF-2α in adult RPE, rod, or cone PR cells does not lead to any morphologic or functional abnormalities in the retina under homeostatic conditions.37–41 However, HIF-1α is instrumental in cellular adaptation to hypoxic stress to promote cell survival.42,43 Our study shows that inactivation of HIF-1α in rods increased PR cell apoptosis and thinning of ONL following RD. Stabilizing HIF-1α through inhibition of prolyl-4-hydroxylases has been shown to promote mitophagy and reduce PR cell death after RD.17 Furthermore, stabilization of HIF-1α protects the retinas from degeneration, inflammation, and reactive oxygen toxicity in a mouse model of retinitis pigmentosa.44 These data suggest that stabilization of HIF during RD may be a strategy for promoting PR survival.
HIF-1α can regulate the expressions of numerous genes in various cell types, and the target genes regulated by HIF-1α varies considerably from one cell type to another.24,25 Our laboratory has previously demonstrated that an increase in autophagy is an important contributor to PR cell survival after RD.14 Although some genes in the HIF pathway array that are involved in the “regulation of autophagy” (Hk2, Hmox1, Ctsa, Vdac1, Bnip3l, and Trp53) were not among the 25 affected genes, the mRNA levels of ankyrin repeat domain 37 (Ankrd37), a target gene through which HIF-1α regulates autophagy,45 was significantly attenuated in the retinas at both 4 hours and 1 day post RD in the of HIF-1αΔrod mice. BCL2/adenovirus E1B 19-kDa-interacting protein 3 (Bnip3) is also a known HIF-1α target gene and plays an important role in autophagy and mitophagy induction.46 The levels of Bnip3 mRNA were upregulated in the detached retinas at 1 day post RD and ablation of HIF-1α in rods significantly blunted this upregulation. Interestingly, no differential expression of the BNIP3-like (Bnip3l) mRNA was apparent in the retinas between HIF-1αΔrod mice and their controls. The levels of immediate early response 3 (Ier3) mRNA, whose encoded protein triggers apoptosis in response to various stimuli,47 were significantly elevated at 1 day post RD in the control mouse retinas; its levels were significantly lower in the HIF-1αΔrod retinas. However, pathway enrichment analysis shows that 33 genes in the GO pathway “programmed cell death” were not in the list of 25 affected genes. Therefore, although the ler3 gene is differentially affected by the cKO of HIF-1α in rods following RD, other apoptosis-related genes are not significantly affected. Nevertheless, the differential expressions of the above-mentioned genes in the detached retinas of HIF-1αΔrod versus control mice indicate that depletion of HIF-1α in rods may affect the balance between autophagy and apoptosis, thus affecting PR cell survival after RD.
Other responses to the hypoxic environment following RD are the modulation of PR metabolism and suppression of protein translation, which serve as mechanisms to conserve cellular energy. We detected increased expression of mRNA for the translation repressor EIF4EBP1 in the detached retina, which was further upregulated after knockout of HIF-1α in rods. Aerobic glycolysis, also known as the Warburg effect, is the main energy source in the retina.48 A number of genes involved in glycolysis are differentially expressed in the retina after depletion of HIF-1α in rods compared with the controls, supporting the previous reports that HIF-1α is the primary regulator of glycolysis.19 These include 8 of the 10 glycolytic enzymes: glucose phosphate isomerase 1 (Gpi1); phosphofructokinase, liver, B-type (Pfkl); phosphofructokinase platelet (Pfkp); aldolase A, fructose-bisphosphate (Aldoa); triosephosphate isomerase 1 (Tpi1); phosphoglycerate kinase 1 (Pgk1); phosphoglycerate mutase 1 (Pgam1); and enolase 1, alpha non-neuron (Eno1). In addition, it includes lactate dehydrogenase A (Ldha), which converts pyruvate produced by glycolysis into lactate; as well as solute carrier family 16 member 3 (Slc16a3), a monocarboxylate transporter that can export the lactate produced from the cell. In addition, a gene encoding a key regulator of glycolysis, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (Pfkfb3), is affected. In addition, nicotinamide phosphoribosyltransferase (Nampt) mRNA level was deceased in detached retinas of HIF-1αΔrod mice. NAMPT is the rate limiting enzyme in the mammalian NAD biosynthesis pathway. Interestingly, glycogen synthase 1 (Gys1), which catalyzes that addition of UDP-glucose to a growing glycogen molecule, is affected. Together these results suggest that HIF-1α is a key regulator of the expression of genes involved with the disposition of glucose in rods in the absence of oxygen.
Hydroxylation of HIF-1α by prolyl hydroxylases (PHDs) leads to ubiquitylation and proteasomal degradation of HIF-1α protein. The protein of Egl-9 family hypoxia inducible factor 1 gene (Egln1) catalyzes the post-translational hydroxylation of HIF-1α, regulating its stability. We found that RD significantly enhanced Egln1 transcriptional levels at 1 day post RD, and conditional knockout of HIF-1α in rods significantly attenuated its increase. The prolyl 4-hydroxylase subunit alpha-1 (P4ha1) gene encodes an enzyme that reduces the proline hydroxylation on HIF-1α, enhancing its stability. Depletion of HIF-1α in rods significantly lowered the mRNA levels of P4ha1 at both 4 hours and 1 day post RD.
Interestingly, cKO of Hif-1α in rods did not significantly alter the expressions of some well-established HIF-1α target genes, such as adrenomedullin (Adm), erythropoietin (Epo), and solute carrier family 2 member 1 (Slc2a1), encoding the facilitated glucose transporter GLUT1, which is the major glucose transporter in PR. In addition, many genes in the GO pathways of programmed cell death, response to oxidative stress, cell proliferation, cell migration, and blood vessel development (with the notable exception of Vegfa) were not affected by cKO of Hif-1α in rods. Given the fact that we observed compensatory increase of HIF-2α protein levels after conditional knockout of Hif-1α in rods, no change in these genes’ expression may reflect the compensatory effects of HIF-2α. Indeed, it is reported that HIF-2α may contribute to the regulation of GLUT1 expression.49
In this work, we used a model of long-lasting detachment, in which dilute hyaluronic acid was injected into the subretinal space. The effect of this substance, versus other material that might allow for more ready diffusion of oxygen from the RPE to the retina, remains to be determined. Based on oxygen diffusion and consumption parameters derived from the cat eye and oxygen diffusion coefficients based upon vitreous fluid, modeling predicted that physical separation caused by retinal detachment reduces oxygen delivery to the outer retina by as much as 70% under light-adapted conditions and by as much as 90% under dark-adapted conditions.5 Thus, physiological retinal detachment severely limits the flow of oxygen to photoreceptors. As such, the 1% hyaluronic acid solution in this model is not expected to greatly exaggerate the human condition, but rather, allows for its replication in the small eye of the mouse. The ability to have a short-term detachment model would also help allow for study of the time-dependence of any of our observations. Finally, our study is limited by the fact that we have only knocked out HIF-1α in rods, and it remains to be determined the effect HIF-1α plays in cone survival.
In conclusion, by using a mouse experimental RD model and cKO of HIF-1α in rods, we demonstrated that HIF-1α plays an important role in PR cell survival after RD. HIF-1α specifically controls a battery of target gene expression in rod PRs, especially those involved in glycolysis. These results provide a foundation to further explore the regulatory pathways that affect PR cell survival after RD and other retinal degenerative diseases and identification of potential new targets for therapeutic intervention to reduce PR death. It is conceivable that expression of many HIF pathway genes not affected by HIF-1α cKO were supported by the compensatory upregulation of HIF-2α expression. Future studies will determine the effect of combined HIF-1α and HIF-2α cKO on photoreceptor survival and gene expression following RD.
Supplementary Material
Acknowledgments
Supported by the National Eye Institute R01-EY-020823 (to S.F.A. and D.N.Z.), Kellogg Eye Center Core Grant P30-EY-007003, Research to Prevent Blindness (to S.F.A.).
Disclosure: B.X. Ross, None; L. Jia, None; D. Kong, None; T. Wang, None; J. Yao, None; H.M. Hager, None; S.F. Abcouwer, None; D.N. Zacks, None
References
- 1. Anderson B Jr, Saltzman HA. Retinal Oxygen Utilization Measured by Hyperbaric Blackout. Arch Ophthalmol. 1964; 72: 792–795. [DOI] [PubMed] [Google Scholar]
- 2. Du J, Linton JD, Hurley JB.. Probing Metabolism in the Intact Retina Using Stable Isotope Tracers. Methods Enzymol. 2015; 561: 149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Country MW. Retinal metabolism: A comparative look at energetics in the retina. Brain Res. 2017; 1672: 50–57. [DOI] [PubMed] [Google Scholar]
- 4. Peet DJ, Kittipassorn T, Wood JP, Chidlow G, Casson RJ.. HIF signalling: The eyes have it. Exp Cell Res. 2017; 356: 136–140. [DOI] [PubMed] [Google Scholar]
- 5. Linsenmeier RA, Padnick-Silver L.. Metabolic dependence of photoreceptors on the choroid in the normal and detached retina. Invest Ophthalmol Vis Sci. 2000; 41: 3117–3123. [PubMed] [Google Scholar]
- 6. Wangsa-Wirawan ND, Linsenmeier RA.. Retinal oxygen: fundamental and clinical aspects. Arch Ophthalmol. 2003; 121: 547–557. [DOI] [PubMed] [Google Scholar]
- 7. Wang S, Linsenmeier RA.. Hyperoxia improves oxygen consumption in the detached feline retina. Invest Ophthalmol Vis Sci. 2007; 48: 1335–1341. [DOI] [PubMed] [Google Scholar]
- 8. Linsenmeier RA, Zhang HF.. Retinal oxygen: from animals to humans. Prog Retin Eye Res. 2017; 58: 115–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chinskey ND, Besirli CG, Zacks DN.. Retinal cell death and current strategies in retinal neuroprotection. Curr Opin Ophthalmol. 2014; 25: 228–233. [DOI] [PubMed] [Google Scholar]
- 10. Huckfeldt RM, Vavvas DG.. Neuroprotection for retinal detachment. Int Ophthalmol Clin. 2013; 53: 105–117. [DOI] [PubMed] [Google Scholar]
- 11. Ross WH. Visual recovery after macula-off retinal detachment. Eye (Lond). 2002; 16: 440–446. [DOI] [PubMed] [Google Scholar]
- 12. Ross WH, Kozy DW.. Visual recovery in macula-off rhegmatogenous retinal detachments. Ophthalmology. 1998; 105: 2149–2153. [DOI] [PubMed] [Google Scholar]
- 13. Ross WH, Stockl FA.. Visual recovery after retinal detachment. Curr Opin Ophthalmol. 2000; 11: 191–194. [DOI] [PubMed] [Google Scholar]
- 14. Besirli CG, Chinskey ND, Zheng QD, Zacks DN.. Autophagy activation in the injured photoreceptor inhibits fas-mediated apoptosis. Invest Ophthalmol Vis Sci. 2011; 52: 4193–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chong DY, Boehlke CS, Zheng QD, Zhang L, Han Y, Zacks DN.. Interleukin-6 as a photoreceptor neuroprotectant in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci. 2008; 49: 3193–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shelby SJ, Angadi PS, Zheng QD, Yao J, Jia L, Zacks DN.. Hypoxia inducible factor 1alpha contributes to regulation of autophagy in retinal detachment. Exp Eye Res. 2015; 137: 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H, Zhu H, Li T, Zhang P, Wang N, Sun X.. Prolyl-4-Hydroxylases Inhibitor Stabilizes HIF-1alpha and Increases Mitophagy to Reduce Cell Death After Experimental Retinal Detachment. Invest Ophthalmol Vis Sci. 2016; 57: 1807–1815. [DOI] [PubMed] [Google Scholar]
- 18. McGettrick AF, O'Neill LAJ. The Role of HIF in Immunity and Inflammation. Cell Metab. 2020; 32: 524–536. [DOI] [PubMed] [Google Scholar]
- 19. Keith B, Johnson RS, Simon MC.. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011; 12: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L.. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Biol Chem. 2002; 277: 32405–32408. [DOI] [PubMed] [Google Scholar]
- 21. Ohh M, Park CW, Ivan M, et al.. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000; 2: 423–427. [DOI] [PubMed] [Google Scholar]
- 22. Jaakkola P, Mole DR, Tian YM, et al.. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001; 292: 468–472. [DOI] [PubMed] [Google Scholar]
- 23. Maxwell PH, Pugh CW, Ratcliffe PJ.. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001; 11: 293–299. [DOI] [PubMed] [Google Scholar]
- 24. Manalo DJ, Rowan A, Lavoie T, et al.. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005; 105: 659–669. [DOI] [PubMed] [Google Scholar]
- 25. Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007; 2007: cm8. [DOI] [PubMed] [Google Scholar]
- 26. Ryan HE, Poloni M, McNulty W, et al.. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000; 60: 4010–4015. [PubMed] [Google Scholar]
- 27. Li S, Chen D, Sauve Y, McCandless J, Chen YJ, Chen CK.. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis. 2005; 41: 73–80. [DOI] [PubMed] [Google Scholar]
- 28. Zacks DN, Hanninen V, Pantcheva M, Ezra E, Grosskreutz C, Miller JW. Caspase activation in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci. 2003; 44: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 29. Ross BX, Choi J, Yao J, Hager HM, Abcouwer SF, Zacks DN.. Loss of High-Mobility Group Box 1 (HMGB1) Protein in Rods Accelerates Rod Photoreceptor Degeneration After Retinal Detachment. Invest Ophthalmol Vis Sci. 2020; 61: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cook B, Lewis GP, Fisher SK, Adler R.. Apoptotic photoreceptor degeneration in experimental retinal detachment. Invest Ophthalmol Vis Sci. 1995; 36: 990–996. [PubMed] [Google Scholar]
- 31. Berglin L, Algvere PV, Seregard S.. Photoreceptor decay over time and apoptosis in experimental retinal detachment. Graefes Arch Clin Exp Ophthalmol. 1997; 235: 306–312. [DOI] [PubMed] [Google Scholar]
- 32. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003; 3: 721–732. [DOI] [PubMed] [Google Scholar]
- 33. Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC.. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003; 23: 9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raval RR, Lau KW, Tran MG, et al.. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005; 25: 5675–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu CJ, Sataur A, Wang L, Chen H, Simon MC.. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol Biol Cell. 2007; 18: 4528–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lau KW, Tian YM, Raval RR, Ratcliffe PJ, Pugh CW.. Target gene selectivity of hypoxia-inducible factor-alpha in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br J Cancer. 2007; 96: 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Samardzija M, Barben M, Todorova V, Klee K, Storti F, Grimm C. Hif1a and Hif2a can be safely inactivated in cone photoreceptors. Sci Rep. 2019; 9: 16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thiersch M, Lange C, Joly S, et al.. Retinal neuroprotection by hypoxic preconditioning is independent of hypoxia-inducible factor-1 alpha expression in photoreceptors. Eur J Neurosci. 2009; 29: 2291–2302. [DOI] [PubMed] [Google Scholar]
- 39. Kurihara T, Westenskow PD, Bravo S, Aguilar E, Friedlander M.. Targeted deletion of Vegfa in adult mice induces vision loss. J Clin Invest. 2012; 122: 4213–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin M, Hu Y, Chen Y, et al.. Impacts of hypoxia-inducible factor-1 knockout in the retinal pigment epithelium on choroidal neovascularization. Invest Ophthalmol Vis Sci. 2012; 53: 6197–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kast B, Schori C, Grimm C.. Hypoxic preconditioning protects photoreceptors against light damage independently of hypoxia inducible transcription factors in rods. Exp Eye Res. 2016; 146: 60–71. [DOI] [PubMed] [Google Scholar]
- 42. Wang GL, Jiang BH, Rue EA, Semenza GL.. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995; 92: 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koh MY, Powis G.. Passing the baton: the HIF switch. Trends Biochem Sci. 2012; 37: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olivares-Gonzalez L, Martinez-Fernandez de la Camara C, Hervas D, Millan JM, Rodrigo R. HIF-1alpha stabilization reduces retinal degeneration in a mouse model of retinitis pigmentosa. FASEB J. 2018; 32: 2438–2451. [DOI] [PubMed] [Google Scholar]
- 45. Deng M, Zhang W, Yuan L, Tan J, Chen Z.. HIF-1a regulates hypoxia-induced autophagy via translocation of ANKRD37 in colon cancer. Exp Cell Res. 2020; 395: 112175. [DOI] [PubMed] [Google Scholar]
- 46. Zhang J, Ney PA.. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009; 16: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arlt A, Schafer H.. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol. 2011; 90: 545–552. [DOI] [PubMed] [Google Scholar]
- 48. Ng SK, Wood JP, Chidlow G, et al.. Cancer-like metabolism of the mammalian retina. Clin Exp Ophthalmol. 2015; 43: 367–376. [DOI] [PubMed] [Google Scholar]
- 49. Chan DA, Sutphin PD, Nguyen P, et al.. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011; 3: 94ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.