ABSTRACT
We report the isolation, identification, and assemblies of three antibiotic-producing soil bacteria (Staphylococcus pasteuri, Peribacillus butanolivorans, and Micrococcus yunnanensis) that inhibit the growth of Neisseria commensals in coculture. With pathogenic Neisseria strains becoming increasingly resistant to antibiotics, bioprospecting for novel antimicrobials using commensal relatives may facilitate discovery of clinically useful drugs.
ANNOUNCEMENT
Antibiotic resistance (AR) in Neisseria gonorrhoeae, the Gram-negative pathogen responsible for the sexually transmitted infection gonorrhea, is a worldwide threat to public health. Resistance to all therapeutics that have been recommended for empirical treatment has emerged (1, 2), and only two drugs, namely, zoliflodacin (currently in phase 3 trials [3, 4]) and gepotidacin (in phase 2 trials [5, 6]), are in development as alternative options. Bioprospecting for antibiotics produced by microbes in soil communities could uncover novel inhibitory compounds against the gonococcus and other important human pathogens (7, 8). This approach can be implemented in undergraduate classrooms as an inquiry-based exercise, which was previously demonstrated by the Small World Initiative (9, 10), Tiny Earth (11, 12), and academic groups (13–16). Developed protocols screen for soil bacteria that produce antibiotics effective against “safe” bacteria (biosafety level 1 [BSL1]), which may also have inhibitory properties against pathogens within the same genus (e.g., ESKAPE pathogens [17, 18]). Here, we expand this methodology to Neisseria, using BSL1 commensals as proxies for pathogens, and identify three soil microbes (WAM01, WAM04, and WAM06) that inhibit commensal Neisseria growth as part of an undergraduate-level classroom exercise in the Thomas H. Gosnell School of Life Sciences at the Rochester Institute of Technology (RIT) (BIOL126-Introductory Biology Laboratory).
Soil samples were collected from Geneseo, New York (USA), and included sediment from an agricultural drainage ditch located at Big Tree Farm (42.798, −77.846) and soil from under an oak tree at the end of Main Street (42.792, −77.815). From these samples, serial dilutions were prepared on 50% tryptic soy agar (TSA), and individual colonies were isolated after 1 week of incubation at room temperature. Commensal Neisseria strains were obtained from the Centers for Disease Control and Prevention (CDC) and Food and Drug Administration (FDA) AR Isolate Bank Neisseria species matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) verification panel, including AR-0944 (Neisseria cinerea), AR-0951 (Neisseria mucosa), and AR-0953 (Neisseria subflava), which were previously characterized and their draft assemblies published (19). Commensal Neisseria strains were plated as a lawn on 50% TSA and were subsequently inoculated with a patch (1 cm by 1 cm) of the soil bacterial strains WAM01, WAM04, and WAM06. Resultant cocultures were incubated at 28°C for 1 week, and the presence or absence of zones of inhibition (ZOIs) was recorded (Table 1 and Fig. 1). WAM06 produced a ZOI against all commensal Neisseria strains tested.
TABLE 1.
Strain attributes and genome assembly overview of the soil bacteria isolated in this studya
| Strain | Isolation site | Species | ZOI with: |
No. of reads | Total assembly length (bp) | No. of contigs | Coverage (×) | N50 (bp) | No. of coding domains | No. of tRNAs | GC content (%) | SRA accession no. | GenBank accession no. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AR-0944 (Neisseria cinerea) | AR-0951 (Neisseria mucosa) | AR-0953 (Neisseria subflava) | |||||||||||||
| WAM01 | Big Tree Farm ditch, Geneseo, NY | Staphylococcus pasteuri | + | − | + | 3,795,320 | 2,563,252 | 34 | 444.20 | 520,728 | 2,541 | 63 | 31.49 | SRR19571305 | JAMWYJ000000000 |
| WAM04 | Main Street oak, Geneseo, NY | Peribacillus butanolivorans | − | − | + | 3,332,040 | 5,994,962 | 213 | 166.74 | 84,680 | 5,885 | 82 | 38.03 | SRR19571306 | JAMWYI000000000 |
| WAM06 | Main Street oak, Geneseo, NY | Micrococcus yunnanensis | + | + | + | 2,289,342 | 2,127,231 | 249 | 322.86 | 23,464 | 2,038 | 41 | 72.5 | SRR19571307 | JAMWYH000000000 |
All assembly statistics are based on contigs of ≥500 bp.
FIG 1.
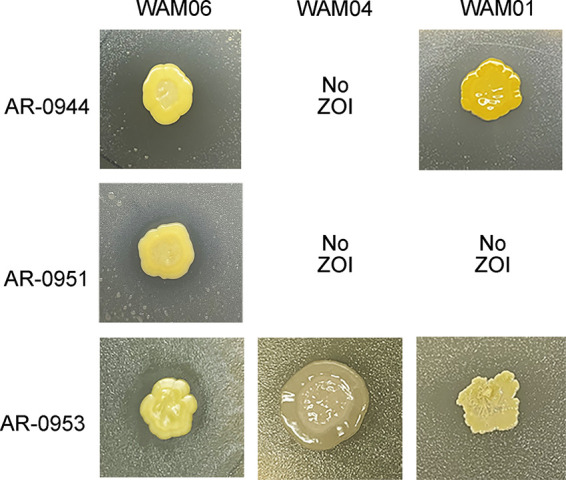
Competitive growth assay on 50% TSA for soil bacteria and commensal Neisseria strains. Micrococcus yunnanensis (WAM06) shows clear ZOIs against all three of the commensal Neisseria species tested. The Peribacillus butanolivorans (WAM04) isolate slightly inhibited the growth of one species, and the Staphylococcus pasteuri isolate inhibited the growth of two.
After incubation for 1 week at room temperature on 50% TSA, DNA was purified from isolates using the Thermo Fisher Scientific PureLink genomic DNA minikit after lysis in Tris-EDTA buffer with 0.5 mg/mL lysozyme and 3 mg/mL proteinase K. The Illumina Nextera XT kit was used to prepare libraries, which were pooled and sequenced using a 600-cycle v3 cartridge (2 × 300 bp) on the Illumina MiSeq platform at the RIT Genomics Core. Default parameters were used for all analyses except where otherwise noted. Paired-end sequencing resulted in an average of 3.13 ± 0.77 million reads, with an average read length of 185.63 ± 50.25 per library. Library quality was assessed using FastQC v0.11.9 (20), and SPAdes v3.14.1 (21) was used for de novo assembly. Assembly statistics were generated with QUAST (http://quast.sourceforge.net/quast), excluding contigs of <500 bp, and are reported in Table 1. Open reading frames (ORFs) were annotated using the GenBank Prokaryotic Genome Annotation Pipeline (PGAP) v5.2 (22) (Table 1), which was also used to assign genera and species, as follows: WAM01, Staphylococcus pasteuri; WAM04, Peribacillus butanolivorans; WAM06, Micrococcus yunnanensis. Further characterization of the anti-Neisseria compounds produced by the bacteria reported here will be reported in a future publication.
Data availability.
The genome assemblies and raw reads are available in GenBank and the SRA, respectively, under the accession numbers listed in Table 1. All code is accessible at https://github.com/wadsworthlab/2022-soil-bacteria.
ACKNOWLEDGMENTS
We acknowledge the generous support for this study provided by the RIT College of Science and the Thomas H. Gosnell School of Life Sciences. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We thank Girish Kumar at the RIT Genomics Core for providing sequencing support.
Contributor Information
Crista B. Wadsworth, Email: cbwsbi@rit.edu.
Vanja Klepac-Ceraj, Wellesley College.
REFERENCES
- 1.Mortimer TD, Grad YH. 2019. Applications of genomics to slow the spread of MDR Neisseria gonorrhoeae. Ann N Y Acad Sci 1435:93–109. doi: 10.1111/nyas.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unemo M, Nicholas RA. 2012. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol 7:1401–1422. doi: 10.2217/fmb.12.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford PA, Miller AA, O'Donnell J, Mueller JP. 2020. Zoliflodacin: an oral spiropyrimidinetrione antibiotic for the treatment of Neisseria gonorrheae, including multi-drug-resistant isolates. ACS Infect Dis 6:1332–1345. doi: 10.1021/acsinfecdis.0c00021. [DOI] [PubMed] [Google Scholar]
- 4.Unemo M, Ahlstrand J, Sánchez-Busó L, Day M, Aanensen D, Golparian D, Jacobsson S, Cole MJ, European Collaborative Group . 2021. High susceptibility to zoliflodacin and conserved target (GyrB) for zoliflodacin among 1209 consecutive clinical Neisseria gonorrhoeae isolates from 25 European countries, 2018. J Antimicrob Chemother 76:1221–1228. doi: 10.1093/jac/dkab024. [DOI] [PubMed] [Google Scholar]
- 5.Scangarella-Oman NE, Hossain M, Dixon PB, Ingraham K, Min S, Tiffany CA, Perry CR, Raychaudhuri A, Dumont EF, Huang J, Hook EW, III, Miller LA. 2018. Microbiological analysis from a phase 2 randomized study in adults evaluating single oral doses of gepotidacin in the treatment of uncomplicated urogenital gonorrhea caused by Neisseria gonorrhoeae. Antimicrob Agents Chemother 62:e01221-18. doi: 10.1128/AAC.01221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor SN, Morris DH, Avery AK, Workowski KA, Batteiger BE, Tiffany CA, Perry CR, Raychaudhuri A, Scangarella-Oman NE, Hossain M, Dumont EF. 2018. Gepotidacin for the treatment of uncomplicated urogenital gonorrhea: a phase 2, randomized, dose-ranging, single-oral dose evaluation. Clin Infect Dis 67:504–512. doi: 10.1093/cid/ciy145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider YK. 2021. Bacterial natural product drug discovery for new antibiotics: strategies for tackling the problem of antibiotic resistance by efficient bioprospecting. Antibiotics 10:842. doi: 10.3390/antibiotics10070842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strobel G, Daisy B. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barral AM, Makhluf H, Broderick NA, Kurt EL. 2016. The Small World Initiative™: an innovative crowdsourcing platform for antibiotics. FASEB J 30:665.13. [Google Scholar]
- 10.Barral AM, Makhluf H, Soneral P, Gasper B. 2014. Small World Initiative: crowdsourcing research of new antibiotics to enhance undergraduate biology teaching (618.41). FASEB J 28:618.41. doi: 10.1096/fasebj.28.1_supplement.618.41. [DOI] [Google Scholar]
- 11.Basalla J, Harris R, Burgess E, Zeedyk N, Wildschutte H. 2020. Expanding Tiny Earth to genomics: a bioinformatics approach for an undergraduate class to characterize antagonistic strains. FEMS Microbiol Lett 367:fnaa018. doi: 10.1093/femsle/fnaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley A, Chevrette MG, Acharya DD, Lozano GL, Garavito M, Heinritz J, Balderrama L, Beebe M, DenHartog ML, Corinaldi K, Engels R, Gutierrez A, Jona O, Putnam JHI, Rhodes B, Tsang T, Hernandez S, Bascom-Slack C, Blum JE, Price PA, Davis D, Klein J, Pultorak J, Sullivan NL, Mouncey NJ, Dorrestein PC, Miller S, Broderick NA, Handelsman J. 2021. Tiny Earth: a big idea for STEM education and antibiotic discovery. mBio 12:e03432-20. doi: 10.1128/mBio.03432-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Groot PWJ, Fernández-Pereira J, Sabariegos R, Clemente-Casares P, Parra-Martínez J, Cid VJ, Moreno DA. 2019. Optimizing Small World Initiative service learning by focusing on antibiotics-producing actinomycetes from soil. FEMS Microbiol Lett 366:fnaa019. doi: 10.1093/femsle/fnaa019. [DOI] [PubMed] [Google Scholar]
- 14.Cavanaugh NT, Parthasarathy A, Wong NH, Steiner KK, Chu J, Adjei J, Hudson AO. 2021. Exiguobacterium sp. is endowed with antibiotic properties against Gram positive and negative bacteria. BMC Res Notes 14:230. doi: 10.1186/s13104-021-05644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeter MN, Gazali SJ, Parthasarathy A, Wadsworth CB, Miranda RR, Thomas BN, Hudson AO. 2021. Isolation, whole-genome sequencing, and annotation of three unclassified antibiotic-producing bacteria, Enterobacter sp. strain RIT 637, Pseudomonas sp. strain RIT 778, and Deinococcus sp. strain RIT 780. Microbiol Resour Announc 10:e00863-21. doi: 10.1128/MRA.00863-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiner KK, Parthasarathy A, Wong NH, Cavanaugh NT, Chu J, Hudson AO. 2020. Isolation and whole-genome sequencing of Pseudomonas sp. RIT 623, a slow-growing bacterium endowed with antibiotic properties. BMC Res Notes 13:370. doi: 10.1186/s13104-020-05216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Oliveira DM, Forde BM, Kidd TJ, Harris PN, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. 2019. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiore MA, Raisman JC, Wong NH, Hudson AO, Wadsworth CB. 2020. Exploration of the Neisseria resistome reveals resistance mechanisms in commensals that may be acquired by N. gonorrhoeae through horizontal gene transfer. Antibiotics 9:656. doi: 10.3390/antibiotics9100656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews S. 2021. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome assemblies and raw reads are available in GenBank and the SRA, respectively, under the accession numbers listed in Table 1. All code is accessible at https://github.com/wadsworthlab/2022-soil-bacteria.


