Abstract
In the last years, tremendous progress has been achieved in the field of gene editing in plants. By the induction of single site-specific double-strand breaks (DSBs), the knockout of genes by non-homologous end joining has become routine in many plant species. Recently, the efficiency of inducing pre-planned mutations by homologous recombination has also been improved considerably. However, very little effort has been undertaken until now to achieve more complex changes in plant genomes by the simultaneous induction of several DSBs. Several reports have been published on the efficient induction of deletions. However, the induction of intrachromosomal inversions and interchromosomal recombination by the use of CRISPR/Cas has only recently been reported. In this review, we want to sum up these results and put them into context with regards to what is known about natural chromosome rearrangements in plants. Moreover, we review the recent progress in CRISPR/Cas-based mammalian chromosomal rearrangements, which might be inspiring for plant biologists. In the long run, the controlled restructuring of plant genomes should enable us to link or break linkage of traits at will, thus defining a new area of plant breeding.
Keywords: Chromosomal rearrangements, CRISPR/Cas, Genome engineering, Inversion, Translocation
Introduction
The variability of the genetic information is a prerequisite for species diversity, and thus for the entire variety of terrestrial life as a result of a sustained evolutionary process. The genetic information is constantly exposed to several exogenous and endogenous factors, leading to different DNA lesions. Most of these lesions can be repaired error free but occasionally, the genetic information is altered during the repair process, resulting in distinct mutations. In addition to simple mutations, such as InDels, large-scale rearrangements of chromosomal fragments within the genome can also occur. Such large-scale changes are referred to as chromosomal rearrangements (CRs), which are defined as “rearrangement of the linear sequence of chromosomes including transposition, duplication, deletion, inversion or translocation of nucleic acid segments” (Gray 2000).
The natural origin of CRs and their consequences
Structural variations within chromosomes can occur naturally and their appearance often goes along with the suppression of meiotic recombination, and thus with evolutionary processes such as species formation and adaptive divergence. The formation of CRs can be mediated by different DNA repair mechanisms. In general, DNA double-strand breaks (DSBs) can be repaired by two main pathways, the non-homologous end joining (NHEJ) pathway and the homologous recombination (HR) pathway (Puchta 2005). Both repair pathways can be divided into several sub-pathways. Whereas HR is based on the utilization of a homologous template, the broken ends within the NHEJ are just re-ligated. Within HR, DSB repair can either be mediated by the single-strand annealing pathway when homologous sequences are present on both ends of the DSB, or by the synthesis-dependent strand annealing (SDSA), or the double-strand break repair (DSBR) pathways, respectively, when the single-stranded 3′ end is able to invade into a donor molecule, resulting in a D-loop structure (reviewed in (Schmidt et al. 2019a). The NHEJ occurs in plants as in other eukaryotes via two different pathways (Salomon and Puchta 1998). In the case of classical NHEJ (cNHEJ), both ends of the DSB are simply reconnected without the use of homologies, resulting in small InDels or correct restoration (Chang et al. 2017). The second NHEJ pathway is based on the usage of small microhomologies (MHs) and the protein polymerase θ, leading to large deletions, insertions or complex combinations of both (Black et al. 2016; Schimmel et al. 2017; Wang and Xu 2017).
Based on these repair pathways, there are several mechanisms as to how CRs can occur (Fig. 1). The occurrence of a single DSB, being repaired according to the DSBR pathway, can already result in large chromosomal changes, whenever a stretch of homology is available, large enough to enable repair via HR (Fig. 1A). This kind of mechanism is called nonallelic homologous recombination (NAHR) and might be responsible for a significant amount of recurrent rearrangements within the genome. Certain areas in the genome, such as low copy repeats and duplications, can promote the formation of CRs leading to the frequent occurrence of rearrangement events in these repetitive regions (Lupski and Stankiewicz 2005).
Fig. 1.
Different mechanisms for the development of CRs. Different DNA molecules and different repair mechanisms can be involved in the development of CRs. (A) If the induction of a DSB occurs in a segment that is homologous to a second DSB on the same chromosome, recombination may occur via a HR-based mechanism called nonallelic homologous recombination (NAHR). The product of this HR-based recombination shown here is an inversion of the DNA stretch between the repeats. (B) If the occurrence of a DSB leads to HR-based strand invasion, an MI process may occur when the invaded strand has homologies to two different donor molecules. Such an intermediate can be dissolved by certain resolvases, which can lead to a translocation between the two invaded donor molecules. (C) The integration of a T-DNA can lead to the formation of a translocation by the independent integration of the two ends of the T-DNA into two DSBs on different chromosomes. As a result, two independent chromosomes can then be linked by a T-DNA. (D) Due to the presence of two DSBs within a chromosome, the area between the two DSBs can be inversely integrated into the genome through NHEJ-based repair
There is also evidence that repetitive DNA complexes play an important role in the evolution of genomes. In a mechanism of concerted evolution, changes in repetitive regions can lead to the formation of novel CRs, and the appearance of such CRs in turn leads to changes in these repetitive regions (Elder and Turner 1995). Dependent on the location of the homology, NAHR can result in deletions, duplications, inversions, translocations and the occurrence of isochromosomes (Barbouti et al. 2004; Park et al. 2016). Research on the evolution of the Triticeae genome has shown recurrent translocation points in several species, where the junction sites are located in genes sharing 90% identity, suggesting that this type of translocation was formed by the NAHR mechanism (Li et al. 2016). A second rearrangement mechanism based on HR can lead to translocations between two intact chromosomes, induced by a lesion on a third chromosome (Fig. 1B). In this multi-invasion (MI)-induced rearrangement, a broken DNA end with two stretches of homology simultaneously invades two independent intact donor molecules, and the subsequent resolution of this intermediate can result in a translocation event (Piazza et al. 2017). A similar case where a third DNA molecule can lead to the formation of a translocation event between two unlinked chromosomes is the integration of a T-DNA in plant cells (Hu et al. 2017). This heterogeneous T-DNA integration can arise when the two ends of a T-DNA independently invade two different DSBs (Fig. 1C). Depending on whether the two DSBs are located on different chromosomes or on the same chromosome, the formation of deletions or translocations can occur.
Two simultaneously occurring DSBs can also lead to the formation of CRs via NHEJ mediated repair (Fig. 1D). In contrast to HR-mediated CRs, the distribution of these types of rearrangements is more random and the same kind of events occur rarely at the same location. However, the role of the NHEJ mechanism in the evolution of CRs has not been clarified so far (Lupski and Stankiewicz 2005). When both DSBs occur on the same chromosome, the fragment in between can be deleted, inverted or duplicated; while the occurrence of simultaneous DSBs on different chromosomes, including homologous chromosomes, can result in translocations or chromosome fusions. Last but not least, transposable elements (TEs) were linked to the formation of CRs and this correlation was initially described by Barbara McClintock (McClintock 1953). In principle, two different mechanisms can be defined for the development of TE-associated CRs. Since TEs are obviously also repetitive sequences, CRs can occur indirectly via HR, as already described (Fig. 1A), or with another mechanism known as alternative transposition. The reaction of alternative transposition is due to the invasion of two TE ends originating from separate elements located on the two sister chromatids (Gray 2000). Both CRs caused directly or indirectly via TEs can result in a wide variety of events such as deletions, duplications, inversions, translocations and dicentric or acentric chromosomes. Thus, the chromosome structure can be altered by NHEJ as well as HR and the occurrence of CRs is favored in the presence of TEs and invading T-DNAs.
Irrespective of the underlying mechanism, such naturally occurring CRs are repeatedly observed in many organisms. In mammals, CRs are mentioned as part of speciation and genome evolution (Navarro and Barton 2003), but mainly these large-scale structural changes are discussed in the context of various genetic diseases and cancer (Abeysinghe et al. 2006; Thompson and Compton 2011). The occurrence of CRs in plants is also linked to speciation and genome evolution, but compared to mammals, CRs in plants are generally associated with their ability to produce genetic isolation between populations and species, because recombination is suppressed in chromosomal heterozygotes (Blanc et al. 2000; Livingstone and Rieseberg 2004; Kirkpatrick and Barton 2006; Fang et al. 2012; Schubert and Vu 2016). Dependent on the type of rearrangement, changes in chromosomes can result in hybrid sterility, centromere shifts, formation of new open reading frames, disruption of already existing genes, removal of chromosomal fragments, alteration of expression profiles, miRNA silencing, as well as the linkage and de-linkage of genes (Allen et al. 2004; Lowry and Willis 2010; Rodríguez-Leal et al. 2017; Schubert 2018). The majority of CRs are more or less randomly distributed across the entire chromosome but some rearrangements occur repeatedly in the same areas of the chromosome, with these “rearrangement hotspots” often being linked to the occurrence of repetitive sequences or heterochromatin–euchromatin borders (Lupski and Stankiewicz 2005; Badaeva et al. 2007; Li et al. 2016; Zapata et al. 2016). In the context of plant breeding, CRs often represent a major obstacle because recombination is inhibited in inverted regions and thus, the transfer of resistance markers from one cultivar to the other is not feasible.
Synthetic generation of CRs
In addition to the targeted modification of selected genes, the targeted modification of the chromosome structure is also an important application in the field of genome engineering. In this field, a new level has been achieved through the development of artificial nucleases, making it possible to target DSBs to almost any sequence in the genome. The general idea behind these synthetically generated nucleases is the combination of a DNA-binding domain with a FokI nuclease domain. This class of nucleases includes zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), and these artificial nucleases provide great possibilities for genome engineering. The recently discovered CRISPR/Cas system takes on the role of key player due to its ease of use for multiplexing approaches, combined with its organism-wide, high cutting efficiencies. The advantage of this system, which has been modified on the basis of the adaptive immune defence system of bacteria and archaea, lies in its simple structure consisting of a Cas protein and a sgRNA, with a quickly and easily adaptable target sequence (Jinek et al. 2012).
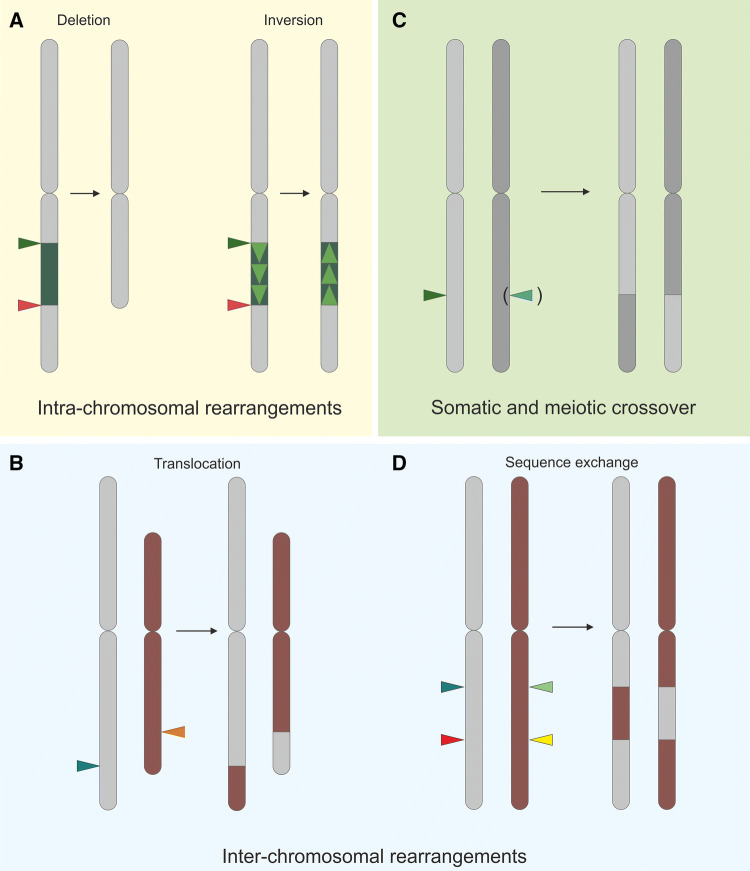
The targeted induction of all sorts of CRs has long been an important part of research in the field of genome engineering. When several DSBs are systematically introduced into the genome, it is possible to modify chromosomes in a directed manner, and thus create completely novel combinations of chromosomal fragments (Fig. 2). If two DSBs are induced on the same chromosome, intra-chromosomal rearrangements may occur by deleting or inverting the area between the two breaks. Inter-chromosomal rearrangements can be achieved by the induction of two or more DSBs on different chromosomes. Depending on the respective tissue, a somatic or meiotic crossover (CO) can be produced by the induction of breaks on one or both homologues.
Fig. 2.
The targeted generation of CRs through the induction of multiple DSBs. Induced intra-chromosomal rearrangements are characterized by break induction on the same chromosome. (A) If the sequence between the two DSBs is removed, a deletion arises. If the excised DNA is reintegrated in reverse orientation, an inversion is induced. (B) If two DSBs are induced on different chromosomes, both chromosome arms can be exchanged in a reciprocal translocation. (C) Furthermore, by inducing (one) or two DSBs at an identical position in homologues, crossovers can be achieved by HR or NHEJ, respectively. (D) By inducing pairs of DSBs on different chromosomes, the sequences between the DSBs can be exchanged between both chromosome arms
Induction of CR in mammals
Because chromosomal abnormalities are often correlated with different cancer types and other diseases in humans, the targeted modification of the chromosome structure is an important challenge in this research field. Initially, the induction of CRs was performed in an undirected manner using radiation or chemical mutagens in mammalian cells, which led to the generation of deletions, inversions, duplications and translocations (Russel 1951; Dutrillaux et al. 1986; Russell et al. 1989; Davisson et al. 1990; Stubbs et al. 1997).
Some of these rearrangements were useful as models for various genetic diseases, such as the duplication of a segment of the mouse Chromosome 16, which is conserved in human Chromosome 21, leading to the imitation of trisomy 21 in mice (Reeves et al. 1995). However, as the location of these rearrangements cannot be predicted when radiation or mutagens are used, the application of this method is limited. To introduce targeted CRs, the site-specific recombination system Cre/loxP was adapted for the use in mouse cells. The use of this method made it possible to specifically induce CRs and generate structural changes that reproduce disease-associated CR in humans (Tsai et al. 1999; Buchholz et al. 2000; Collins et al. 2000). While lox sites need to be integrated into the genome, this strategy is quite time consuming and these sequences remain as scars in the genome.
ZFNs and TALENs have been successfully used in mammals to induce a large number of different CR events (Lee et al. 2010, 2012; Piganeau et al. 2013; Ghezraoui et al. 2014). More recently, CRISPR/Cas was used to induce multiple forms of different chromosomal changes in the genome (Cheong et al. 2018). In addition, this field of research is of particular importance in mammals, as several human cancers and other diseases are associated with the occurrence of translocations and inversions. Through the simultaneous induction of two DSBs, using the CRISPR/Cas system, inversions could be induced very efficiently in human and mouse cell cultures, with an inversely proportional relationship between inversion frequency and inversion size being detected (Canver et al. 2014; Li et al. 2015). Furthermore, a successful transmission of inversion events into the germ line in mice could be achieved. For this purpose, it was possible to inject 120 mouse embryos in the single-cell stage with the two sgRNAs and the Cas9 mRNA at the same time, and the resulting founder mice were examined for inversion formation. In the F0 generation, six mice with an inversion of 1.2 kb were identified in this study, which corresponds to an inversion efficiency of 5%. Furthermore, the same study documented the inheritance of this inversion for the first time. From seven F1 mice tested, two mice with an inversion of the corresponding fragment were identified in the following generation. Overall, the frequencies of induced inversions with a size of 0.7–800 kb were determined, whereby the inversion frequency was relatively constant, up to a size of 6 kb, at about 20% and then gradually decreased to a frequency of 0.7% for the 800-kb inversion (Li et al. 2015). In addition to mouse embryos, inversions with a much lower efficiency could also be successfully induced in zebrafish embryos, whereby transmission of the inversion events into the next generation proved difficult due to the low frequencies (Ota et al. 2014). Additionally, newly formed junctions of induced inversions were investigated in a cell culture of human embryonic kidney cells (Shou et al. 2018). It was shown in this study that a large part of the induced deletions and inversions are the result of an accurate ligation, where no mutations occurred at the junction sites. Nevertheless, the composition of the junction sites in different cells, that are deficient for certain DNA repair factors, was also investigated. In this context, it could be shown that the number of precise repair events in ΔCtIP cells is significantly increased and significantly decreased in ΔXRCC4 and ΔLIG4 cells. This fact suggests that the cNHEJ repair pathway is involved in the development of inversions and deletions in mammalian cells (Guo et al. 2018).
Using the CRISPR/Cas system, it should be possible to reverse pathogenic inversions in the human genome to their original form. Inversions occur, as already mentioned, in connection with different types of cancer, the Hunter syndrome, Emery–Dreifuss muscular dystrophy, as well as with the bleeding disorder hemophilia A (Lakich et al. 1993; Bondeson et al. 1995; Small et al. 1997; Inaki and Liu 2012; Park et al. 2016). An impressive example is the occurrence of an oncogenic inversion on the human Chromosome 2, which leads to the formation of an abnormal fusion protein because of the fusion of the genes EML4 and ALK. Such an inversion is associated with a certain type of lung cancer and this inversion has been successfully induced in mouse and human cells by CRISPR/Cas (Choi and Meyerson 2014). Since this type of lung cancer has an increased sensitivity to ALK inhibitors, the induction of such an inversion can be a valuable tool for the investigation of molecular mechanisms of tumor formation, as well as for the in vivo testing of corresponding drugs (Maddalo et al. 2014).
Besides inversions, translocations also play a role in the development of different leukemia species and sarcomas (Bunting and Nussenzweig 2013). As an example, the very common translocation between the human ETO gene on chromosome 8 and the RUNX1 gene on chromosome 21 is associated with acute myeloid leukemia (Yuan et al. 2001). It has been shown that it is possible to induce this particular translocation, as well as many other translocations, via the use of the CRISPR/Cas system. Thus, it should be possible to reverse cancer associated reciprocal translocations in general (Torres et al. 2014; Lagutina et al. 2015; Torres-Ruiz et al. 2017). Moreover, the number of translocations induced by the CRISPR/Cas system was investigated in different murine cell lines using cell lines deficient in DNA repair factors KU70 and POLQ. These two factors represent the key factors of cNHEJ and MMEJ, respectively. While no changed translocation frequency could be detected in ΔPOLQ cells after DSB induction using Cas9, ΔKU70 cells showed a significantly increased translocation frequency. ku70/polq double mutants showed an increased translocation frequency compared to ku70 mutants, which suggests that these two proteins limit translocation formation in mammalian cells (Wyatt et al. 2016).
Thus, it has been shown for mammals that the CRISPR/Cas system can be used successfully for the targeted induction of a variety of CRs. Using the Cas9 nuclease different naturally occurring inversions and translocations have already been artificially created in vivo. However, application for the targeted revision of oncogenic CRs as part of gene therapy is still pending. Problems in this context are the risk of tumorigenesis, insufficient efficiencies, unexpected outcomes and other safety issues arising during application of CRISPR-Cas9 (Ihry et al. 2018; Kosicki et al. 2018; Sakuma et al. 2018).
Induction of CR in plants
As already discussed in the previous chapter, CRs in plants are associated with evolutionary processes such as adaptation and species formation in a much broader way compared to mammals. An obvious application for the targeted modification of CRs would be the targeted breaking or stabilization of genetic linkages between quantitative trait loci (QTLs), resistance markers or other attractive traits. (Lee et al. 2017; Wellenreuther and Bernatchez 2018). First attempts to induce CR events in plants artificially were made with the use of TEs, site-specific recombinases and meganucleases. TEs were first described in maize by Barbara McClintock, who already recognized their potential to cause genomic instability, leading to the depiction that TEs are directly involved in the formation of gross CRs (McClintock 1987). The Ac/Ds elements were successfully used in the following years to induce different CRs in several plant species like maize, rice and Arabidopsis (Zhang and Peterson 2004; Krishnaswamy et al. 2008; Yu et al. 2012). Since the opportunities to induce CRs using TEs are limited, Ac/Ds elements were additionally combined with the sequence-specific recombination system Cre/Lox to enable the generation of targeted changes in the chromosome structure. Using such sequence-specific recombination systems, it was possible to induce deletions, inversions and translocations in tobacco and Arabidopsis (Qin et al. 1994; Medberry et al. 1995; Osborne et al. 1995; Zhang et al. 2003). Another efficient option to modify the structure of the genome was the induction of a DSB via rare cutting restriction enzymes. A nuclease used in plants for this purpose is the homing endonuclease I-SceI. Initially, induction of a single DSB by I-SceI was used for the increase of gene targeting efficiency by HR (Puchta et al. 1996) as well as for gene knockouts by NHEJ (Salomon and Puchta 1998). When two DSBs were induced simultaneously, the deletion of the fragment in between could be achieved by NHEJ as well as HR (Siebert and Puchta 2002). Moreover, the induction of two DSBs on two different chromosomes in Arabidopsis showed that it is possible to exchange these two chromosome arms, resulting in a reciprocal translocation and this event can be caused by NHEJ and HR (Pacher et al. 2007). A disadvantage of these mentioned procedures is that I-SceI recognition sites and lox sites need to be integrated into the genome, and this can also be quite time-consuming depending on the approach.
This obstacle was overcome as synthetic nucleases were successfully applied in plants (Voytas 2013), and thereby it was possible to induce deletions and inversions in tandemly arrayed genes in Arabidopsis with the use of ZFNs (Qi et al. 2013). Additionally, a transgenic fragment of 4.3 kb was successfully deleted out of the tobacco genome via ZFNs (Petolino et al. 2010). The same kind of CRs was produced using two pairs of TALENs in rice calli, but there was no evidence of successful inheritance of these events into the next generation (Shan et al. 2013). The situation changed with the discovery of the CRISPR/Cas system. Several CRISPR/Cas-based nucleases have been applied for the knockout of various genes by NHEJ, in various plant species. Thus, different kinds of trait improvements have already been achieved in crops [for recent reviews see (Yin et al. 2017; Kumlehn et al. 2018; Langner et al. 2018; Zhang et al. 2018)]. Recently, significant improvements were also achieved by a number of groups to induce pre-planned changes in plant genomes via HR [for a recent review see Huang and Puchta (2019)]. Moreover, by the simultaneous induction of DSBs in several genes, leading to their knockout via NHEJ, a variety of attractive traits can be induced into a plant within a single step [for a recent review see Wolter et al. (2019)]. Therefore, domestication of wild plants to crops could be achieved in a single generation (Lemmon et al. 2018; Li et al. 2018; Zsögön et al. 2018). On the other hand, the induction of several DSBs by Cas9 can also be used for the induction of CRs in plants. In comparison to mammals, applications in plants were mainly focused on the induction of deletions and the transmission of these events into the next generation. Small deletions up to 1 kb can be achieved quite easily in Arabidopsis and other plant species, but interestingly, it was also possible to induce deletions up to 120 kb with low frequencies in Arabidopsis, and stage specific expression of the Cas9 protein promotes the possibility of deletions to enter the germline (Ordon et al. 2017; Durr et al. 2018; Wu et al. 2018). Furthermore, somatic deletion formation with a size of up to 245 kb using CRISPR/Cas was effectively achieved in other plant species like rice, tobacco and Medicago, although the inheritance was only shown for deletions in Medicago and rice (Zhou et al. 2014; Gao et al. 2015; Čermák et al. 2017). Inversion formation following DSB induction, on the other hand, was rarely achieved in plants and inversion sizes were tiny compared to deletions (Gao et al. 2015; Zhang et al. 2017). However, just recently an extensive study about efficient inversion formation in Arabidopsis was conducted, where inversions up to 18 kb were successfully transmitted to the next generation using an egg cell-specific promoter for Cas9 expression (Schmidt et al. 2019b). In addition, the newly formed junctions of the examined inversions and deletions were analyzed to draw conclusions about the involved repair mechanisms. It could be shown that in wild type, the majority of inversions and deletions were formed using an error-free cNHEJ pathway; whereas, only a minor number were formed using MHs. Frequencies of inversions and deletions were enhanced in mutants lacking the protein KU70, a key factor of cNHEJ. In these mutants, both events were generated using a backup NHEJ pathway based on the use of MHs. A further important application for CRISPR/Cas-induced CR in plants will be the induction of heritable chromosomal translocations in plants. Till now, Cas9-induced translocation induction has only been observed as a by-product in somatic cells, at very low frequencies in Arabidopsis (Peterson et al. 2016).
A rather obvious CR desirable for plant breeding is the induction of crossovers between homologous chromosomes. This kind of homologous recombination reaction occurs in a programmed way during meiosis [for a recent review see Wang and Copenhaver (2018)]. Meiotic HR is the origin of genetic diversity and enables the generation of new allelic combinations. The random nature of meiotic recombination diminishes the likelihood for desirable outcomes and the presence of hot- and cold-spots further restricts potential outcomes, due to the inaccessibility of genomic regions [reviewed in Choi (2017)]. Programmable nucleases harbor the potential to make these respective regions accessible to recombination by the specific targeting of DSBs. ZFN, TALENs and the CRISPR/Cas system were harnessed as DNA-binding domains to target SPO11, a subunit of the natural meiotic DSB inducing protein complex, to recombination cold-spots in S. cerevisiae, achieving a certain increase in crossover frequency. However, some regions where natural meiotic DSB induction is suppressed were still inaccessible for targeted DSB formation by SPO11, potentially as a result of regulatory hindrances (Sarno et al. 2017). A major advantage of plants is the late differentiation of germ cells that enables the inheritance of somatically induced mutations, including genomic rearrangements (Das et al. 1990). Thus, targeted CO induction does not have to be exclusively restricted to meiotic cells. Using programmable nucleases for DSB induction, it is possible to bypass meiosis for CO induction. In a ground-breaking study, scientists from the Weizmann institute developed a selection system, using hybrids of two tomato accessions distinguished by SNPs and fruit colour that enables the identification of somatic DSB repair outcomes. By inducing an allele-specific DSB using CRISPR/Cas9, they were able to identify somatic HR events, including gene conversions and one putative crossover event. Due to plant death, inheritable events could only be identified for gene conversions (Filler Hayut et al. 2017). Besides homologous recombination, somatic COs might also be achievable by the use of NHEJ. In this case, DSBs have to induced simultaneously in both homologous chromosomes, which might result in translocations between homologues, in certain frequencies (Fig. 2C).
The impact of these new techniques in the field of plant breeding is remarkable, as it becomes possible to shape chromosomes according to the specific requirement. Besides the direct induction of crossovers in recombination cold regions of the genome or the elimination of adverse traits out of the genome, directed induction of other kinds of CRs is also important to transfer certain traits between cultivars.
CRs contribute to the development of reproductive barriers, because recombination between rearranged areas is suppressed in heterozygotes (Wellenreuther and Bernatchez 2018). This suppression of HR represents a major obstacle in plant breeding because there is no exchange of genetic material between cultivars in parts of the genome that were rearranged during evolution. As an example, in the model plant Arabidopsis thaliana, a large paracentric inversion, the heterochromatic knob hk4S, was identified in the Columbia accession, whilst it is absent in Landsberg, leading to a HR cold-spot in heterozygotes in this area (Drouaud et al. 2006; Giraut et al. 2011; Fransz et al. 2016). These types of rearrangements are leading to several problems in crop breeding because the transfer of certain traits within an inversion from one cultivar to the other is no longer achievable. An accumulation of multiple large CRs might have a drastic effect for the transfer of traits via crossing: the formed hybrids might become sterile because chromosome pairing during meiosis is becoming defective. By inverting or re-inverting certain regions, it is possible to regain accessibility for meiotic HR to these specific chromosome segments. On the other side, if desired, it is also possible to generate new linkage groups to suppress recombination in this area (Fig. 3). As an example of re-inversion to separate linkages, the transfer of the nematode-resistance gene Mi-1, which is associated with an inverted chromosomal segment in tomato, to another cultivar, could be a possibility in future with the aforementioned described techniques. This and many other examples show the potential of targeted modification of chromosome structures for achieving new combinations of alleles out of a pool of species that are not achievable by the use of classical breeding. Therefore, it will be very important to further develop our tools for CRISPR/Cas-induced CRs in plants, in the future.
Fig. 3.
Inversion induction to generate or separate linkage groups. Since the recombination between rearranged areas of the genome is suppressed in heterozygotes, it is possible to generate new linkage groups via the induction of an inversion. When two unlinked genes are located at different locations of the chromosome arm, it is likely that they are separated via CO formation during meiosis. With the induction of an inversion involving both genes, it is possible to link them, and thus ensure that they are inherited together. On the other hand, it should also be possible to reverse inversions that were previously formed and thus eliminate linkage groups caused by this inversion. As a result, it should be possible to separate both genes by a CO event during meiosis
Acknowledgements
We thank Amy Whitbread for critical reading of the manuscript. This work was supported by the European Research Council (Grant number ERC-2016-AdG_741306 CRISBREED).
References
- Abeysinghe SS, Chuzhanova N, Cooper D. Gross deletions and translocations in human genetic disease. In: Volff J-N, editor. Genome and disease. Basel: Karger; 2006. pp. 17–34. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung G-H, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Dedkova OS, Gay G, Pukhalskyi VA, Zelenin AV, Bernard S, Bernard M. Chromosomal rearrangements in wheat: their types and distribution. Genome. 2007;50:907–926. doi: 10.1139/g07-072. [DOI] [PubMed] [Google Scholar]
- Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Höglund M, Johansson B, Hagemeijer A, Park S-S, Mitelman F, Lupski JR, Fioretos T. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black SJ, Kashkina E, Kent T, Pomerantz RT. DNA polymerase θ: a unique multifunctional end-joining machine. Genes (Basel) 2016;7:67. doi: 10.3390/genes7090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson ML, Dahl N, Malmgren H, Kleijer WJ, Tönnesen T, Carlberg BM, Pettersson U. Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum Mol Genet. 1995;4:615–621. doi: 10.1093/hmg/4.4.615. [DOI] [PubMed] [Google Scholar]
- Buchholz F, Refaeli Y, Trumpp A, Bishop JM. Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Rep. 2000;1:133–139. doi: 10.1093/embo-reports/kvd027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29:1196–1217. doi: 10.1105/tpc.16.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong T-C, Blasco RB, Chiarle R. The CRISPR/Cas9 system as a tool to engineer chromosomal translocation in vivo. In: Zhang Y, editor. Chromosome translocation. Advances in experimental medicine and biology. Singapore: Springer; 2018. pp. 39–48. [DOI] [PubMed] [Google Scholar]
- Choi K. Advances towards controlling meiotic recombination for plant breeding. Mol Cells. 2017;40:814–822. doi: 10.14348/molcells.2017.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins EC, Pannell R, Simpson EM, Forster A, Rabbitts TH. Inter-chromosomal recombination of Mll and Af9 genes mediated by cre-loxP in mouse development. EMBO Rep. 2000;1:127–132. doi: 10.1093/embo-reports/kvd021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das OP, Levi-Minzi S, Koury M, Benner M, Messing J. A somatic gene rearrangement contributing to genetic diversity in maize. Proc Natl Acad Sci USA. 1990;87:7809–7813. doi: 10.1073/pnas.87.20.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT, Schmidt C, Akeson EC. Segmental trisomy of murine chromosome 16: a new model system for studying down syndrome. Prog Clin Biol Res. 1990;360:263–280. [PubMed] [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon P-Y, Canaguier A, Bérard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, Quesneville H, Mézard C. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr J, Papareddy R, Nakajima K, Gutierrez-Marcos J. Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci Rep. 2018;8:4443. doi: 10.1038/s41598-018-22667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrillaux B, Sabatier L, Al Achkar W, Muleris M, Aurias A, Couturier J, Dutrillaux AM, Gerbault-Seureau M, Hoffschir F, Lamoliatte E. Radiation induced inversions in human somatic cells. Ann Genet. 1986;29:189–194. [PubMed] [Google Scholar]
- Elder JF, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Fang Z, Pyhäjärvi T, Weber AL, Dawe RK, Glaubitz JC, González JDJS, Ross-Ibarra C, Doebley J, Morrell PL, Ross-Ibarra J. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics. 2012;191:883–894. doi: 10.1534/genetics.112.138578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler Hayut S, Melamed Bessudo C, Levy AA. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat Commun. 2017;8:15605. doi: 10.1038/ncomms15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, Linc G, Lee C-R, Aflitos SA, Lasky JR, Toomajian C, Ali H, Peters J, van Dam P, Ji X, Kuzak M, Gerats T, Schubert I, Schneeberger K, Colot V, Martienssen R, Koornneef M, Nordborg M, Juenger TE, de Jong H, Schranz ME. Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J. 2016;88:159–178. doi: 10.1111/tpj.13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol. 2015;87:99–110. doi: 10.1007/s11103-014-0263-0. [DOI] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud J-B, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Guo T, Feng Y-L, Xiao J-J, Liu Q, Sun X-N, Xiang J-F, Kong N, Liu S-C, Chen G-Q, Wang Y, Dong M-M, Cai Z, Lin H, Cai X-J, Xie A-Y. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 2018;19:170. doi: 10.1186/s13059-018-1518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen Z, Zhuang C, Huang J. Cascade of chromosomal rearrangements caused by a heterogeneous T-DNA integration supports the double-stranded break repair model for T-DNA integration. Plant J. 2017;90:954–965. doi: 10.1111/tpj.13523. [DOI] [PubMed] [Google Scholar]
- Huang T-K, Puchta H. CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep. 2019;38:443–453. doi: 10.1007/s00299-019-02379-0. [DOI] [PubMed] [Google Scholar]
- Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, Kaykas A. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28:550–559. doi: 10.1016/j.tig.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy L, Zhang J, Peterson T. Reversed end Ds element: a novel tool for chromosome engineering in Arabidopsis. Plant Mol Biol. 2008;68:399–411. doi: 10.1007/s11103-008-9377-6. [DOI] [PubMed] [Google Scholar]
- Kumlehn J, Pietralla J, Hensel G, Pacher M, Puchta H. The CRISPR/Cas revolution continues: from efficient gene editing for crop breeding to plant synthetic biology. J Integr Plant Biol. 2018;60:1127–1153. doi: 10.1111/jipb.12734. [DOI] [PubMed] [Google Scholar]
- Lagutina IV, Valentine V, Picchione F, Harwood F, Valentine MB, Villarejo-Balcells B, Carvajal JJ, Grosveld GC. Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease. PLoS Genet. 2015;11:e1004951. doi: 10.1371/journal.pgen.1004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5:236–241. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- Langner T, Kamoun S, Belhaj K. CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol. 2018;56:479–512. doi: 10.1146/annurev-phyto-080417-050158. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim J-S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim J-S. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-R, Wang B, Mojica JP, Mandáková T, Prasad KVSK, Goicoechea JL, Perera N, Hellsten U, Hundley HN, Johnson J, Grimwood J, Barry K, Fairclough S, Jenkins JW, Yu Y, Kudrna D, Zhang J, Talag J, Golser W, Ghattas K, Schranz ME, Wing R, Lysak MA, Schmutz J, Rokhsar DS, Mitchell-Olds T. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat Ecol Evol. 2017;1:119. doi: 10.1038/s41559-017-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, van Eck J, Lippman ZB. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plants. 2018;4:766–770. doi: 10.1038/s41477-018-0259-x. [DOI] [PubMed] [Google Scholar]
- Li J, Shou J, Guo Y, Tang Y, Wu Y, Jia Z, Zhai Y, Chen Z, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol. 2015;7:284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Challa GS, Zhu H, Wei W. Recurrence of chromosome rearrangements and reuse of DNA breakpoints in the evolution of the Triticeae genomes. G3 (Bethesda) 2016;6:3837–3847. doi: 10.1534/g3.116.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol. 2018;36:1160–1163. doi: 10.1038/nbt.4273. [DOI] [PubMed] [Google Scholar]
- Livingstone K, Rieseberg L. Chromosomal evolution and speciation: a recombination-based approach. New Phytol. 2004;161:107–112. doi: 10.1046/j.1469-8137.2003.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han Y-C, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Induction of instability at selected loci in maize. Genetics. 1953;38:579–599. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The discovery and characterization of transposable elements. Genes, Cells and Organisms. Great Books in Experimental Biology. Spokane: Garland Pub; 1987. [Google Scholar]
- Medberry SL, Dale E, Qin M, Ow DW. Intra-chromosomal rearrangements generated by Cre-lox site-specific recombination. Nucleic Acids Res. 1995;23:485–490. doi: 10.1093/nar/23.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Ordon J, Gantner J, Kemna J, Schwalgun L, Reschke M, Streubel J, Boch J, Stuttmann J. Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 2017;89:155–168. doi: 10.1111/tpj.13319. [DOI] [PubMed] [Google Scholar]
- Osborne BI, Wirtz U, Baker B. A system for insertional mutagenesis and chromosomal rearrangement using the Ds transposon and Cre-lox. Plant J. 1995;7:687–701. doi: 10.1046/j.1365-313x.1995.7040687.x. [DOI] [PubMed] [Google Scholar]
- Ota S, Hisano Y, Ikawa Y, Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells. 2014;19:555–564. doi: 10.1111/gtc.12154. [DOI] [PubMed] [Google Scholar]
- Pacher M, Schmidt-Puchta W, Puchta H. Two unlinked double-strand breaks can induce reciprocal exchanges in plant genomes via homologous recombination and nonhomologous end joining. Genetics. 2007;175:21–29. doi: 10.1534/genetics.106.065185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C-Y, Sung JJ, Kim D-W. Genome editing of structural variations: modeling and gene correction. Trends Biotechnol. 2016;34:548–561. doi: 10.1016/j.tibtech.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Peterson BA, Haak DC, Nishimura MT, Teixeira PJPL, James SR, Dangl JL, Nimchuk ZL. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One. 2016;11:e0162169. doi: 10.1371/journal.pone.0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petolino JF, Worden A, Curlee K, Connell J, Strange Moynahan TL, Larsen C, Russell S. Zinc finger nuclease-mediated transgene deletion. Plant Mol Biol. 2010;73:617–628. doi: 10.1007/s11103-010-9641-4. [DOI] [PubMed] [Google Scholar]
- Piazza A, Wright WD, Heyer W-D. Multi-invasions are recombination byproducts that induce chromosomal rearrangements. Cell. 2017;170:760–773. doi: 10.1016/j.cell.2017.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau M, Ghezraoui H, de Cian A, Guittat L, Tomishima M, Perrouault L, René O, Katibah GE, Zhang L, Holmes MC, Doyon Y, Concordet J-P, Giovannangeli C, Jasin M, Brunet E. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot. 2005;56:1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Li X, Zhang Y, Starker CG, Baltes NJ, Zhang F, Sander JD, Reyon D, Joung JK, Voytas DF. Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 (Bethesda) 2013;3:1707–1715. doi: 10.1534/g3.113.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Bayley C, Stockton T, Ow DW. Cre recombinase-mediated site-specific recombination between plant chromosomes. Proc Natl Acad Sci USA. 1994;91:1706–1710. doi: 10.1073/pnas.91.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–480.e8. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Russel WL. X-ray-induced mutations in mice. Cold Spring Harb Symp Quant Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- Russell LB, Hunsicker PR, Cacheiro NL, Bangham JW, Russell WL, Shelby MD. Chlorambucil effectively induces deletion mutations in mouse germ cells. Proc Natl Acad Sci USA. 1989;86:3704–3708. doi: 10.1073/pnas.86.10.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Mochida K, Nakade S, Ezure T, Minagawa S, Yamamoto T. Unexpected heterogeneity derived from Cas9 ribonucleoprotein-introduced clonal cells at the HPRT1 locus. Genes Cells. 2018;23:255–263. doi: 10.1111/gtc.12569. [DOI] [PubMed] [Google Scholar]
- Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno R, Vicq Y, Uematsu N, Luka M, Lapierre C, Carroll D, Bastianelli G, Serero A, Nicolas A. Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 2017;45:e164. doi: 10.1093/nar/gkx739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Kool H, van Schendel R, Tijsterman M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017;36:3634–3649. doi: 10.15252/embj.201796948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Pacher M, Puchta H. DNA break repair in plants and its application for genome engineering. Methods Mol Biol. 2019;1864:237–266. doi: 10.1007/978-1-4939-8778-8_17. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Pacher M, Puchta H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 2019;98:577–589. doi: 10.1111/tpj.14322. [DOI] [PubMed] [Google Scholar]
- Schubert I. What is behind “centromere repositioning”? Chromosoma. 2018;127:229–234. doi: 10.1007/s00412-018-0672-y. [DOI] [PubMed] [Google Scholar]
- Schubert I, Vu GTH. Genome stability and evolution: attempting a holistic view. Trends Plant Sci. 2016;21:749–757. doi: 10.1016/j.tplants.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, Zhang K, Liu J, Voytas DF, Zheng X, Zhang Y, Gao C. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant. 2013;6:1365–1368. doi: 10.1093/mp/sss162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou J, Li J, Liu Y, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell. 2018;71:498–509.e4. doi: 10.1016/j.molcel.2018.06.021. [DOI] [PubMed] [Google Scholar]
- Siebert R, Puchta H. Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell. 2002;14:1121–1131. doi: 10.1105/tpc.001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K, Iber J, Warren ST. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet. 1997;16:96–99. doi: 10.1038/ng0597-96. [DOI] [PubMed] [Google Scholar]
- Stubbs L, Carver EA, Cacheiro NL, Shelby M, Generoso W. Generation and characterization of heritable reciprocal translocations in mice. Methods. 1997;13:397–408. doi: 10.1006/meth.1997.0546. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res. 2011;19:433–444. doi: 10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz R, Martinez-Lage M, Martin MC, Garcia A, Bueno C, Castaño J, Ramirez JC, Menendez P, Cigudosa JC, Rodriguez-Perales S. Efficient recreation of t(11;22) EWSR1-FLI1 + in human stem cells using CRISPR/Cas9. Stem Cell Rep. 2017;8:1408–1420. doi: 10.1016/j.stemcr.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader–Willi syndrome. Hum Mol Genet. 1999;8:1357–1364. doi: 10.1093/hmg/8.8.1357. [DOI] [PubMed] [Google Scholar]
- Voytas DF. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol. 2013;64:327–350. doi: 10.1146/annurev-arplant-042811-105552. [DOI] [PubMed] [Google Scholar]
- Wang Y, Copenhaver GP. Meiotic recombination: mixing it up in plants. Annu Rev Plant Biol. 2018;69:577–609. doi: 10.1146/annurev-arplant-042817-040431. [DOI] [PubMed] [Google Scholar]
- Wang H, Xu X. Microhomology-mediated end joining: new players join the team. Cell Biosci. 2017;7:6. doi: 10.1186/s13578-017-0136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol (Amst) 2018;33:427–440. doi: 10.1016/j.tree.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Wolter F, Schindele P, Puchta H. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 2019;19:176. doi: 10.1186/s12870-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Lucke M, Jang Y-T, Zhu W, Symeonidi E, Wang C, Fitz J, Xi W, Schwab R, Weigel D. An efficient CRISPR vector toolbox for engineering large deletions in Arabidopsis thaliana. Plant Methods. 2018;14:65. doi: 10.1186/s13007-018-0330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Mieczkowski P, Wood RD, Gupta GP, Ramsden DA. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol Cell. 2016;63:662–673. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Gao C, Qiu J-L. Progress and prospects in plant genome editing. Nat Plants. 2017;3:17107. doi: 10.1038/nplants.2017.107. [DOI] [PubMed] [Google Scholar]
- Yu C, Han F, Zhang J, Birchler J, Peterson T. A transgenic system for generation of transposon Ac/Ds-induced chromosome rearrangements in rice. Theor Appl Genet. 2012;125:1449–1462. doi: 10.1007/s00122-012-1925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, Burel SA, Lagasse E, Weissman IL, Akashi K, Zhang DE. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata L, Ding J, Willing E-M, Hartwig B, Bezdan D, Jiao W-B, Patel V, Velikkakam James G, Koornneef M, Ossowski S, Schneeberger K. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc Natl Acad Sci USA. 2016;113:E4052–E4060. doi: 10.1073/pnas.1607532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peterson T. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167:1929–1937. doi: 10.1534/genetics.103.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Raina S, Li H, Li J, Dec E, Ma H, Huang H, Fedoroff NV. Resources for targeted insertional and deletional mutagenesis in Arabidopsis. Plant Mol Biol. 2003;53:133–150. doi: 10.1023/B:PLAN.0000009271.08420.d9. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu C, Weng J, Cheng B, Liu F, Li X, Xie C. Creation of targeted inversion mutations in plants using an RNA-guided endonuclease. Crop J. 2017;5:83–88. [Google Scholar]
- Zhang Y, Massel K, Godwin ID, Gao C. Applications and potential of genome editing in crop improvement. Genome Biol. 2018;19:210. doi: 10.1186/s13059-018-1586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP. De novo domestication of wild tomato using genome editing. Nat Biotechnol. 2018;36:1211–1216. doi: 10.1038/nbt.4272. [DOI] [PubMed] [Google Scholar]