Abstract
A better understanding of the molecular mechanisms that control the UCP1 expression in brown and beige adipocytes is essential for us to modulate adipose cell fate and promote thermogenesis, which may provide a therapeutic view for the treatment of obesity and obesity-related diseases. To systematically identify cis-element(s) that transcriptionally regulates Ucp1, we here took advantage of the high-throughput CRIPSR-Cas9 screening system, and performed an in situ saturating mutagenesis screen, by using a customized sgRNA library targeting the ~ 20 kb genomic region near Ucp1. Through the screening, we have identified several genomic loci that may contain key regulatory element for Ucp1 expression in cultured brown and white adipocytes in vitro, and in inguinal white adipose tissue in vivo. Our study highlights a broadly useful approach for studying cis-regulatory elements in a high-throughput manner.
Electronic supplementary material
The online version of this article (10.1007/s43657-020-00006-7) contains supplementary material, which is available to authorized users.
Keywords: In situ saturating mutagenesis screen, Brown and beige adipocytes, UCP1, sgRNA-9768
Uncoupling protein 1 (UCP1) is a mitochondrial protein, which allows electrons to be released and uncouples oxidative respiration from ATP synthesis, resulting in heat generation (Harms and Seale 2013). UCP1 expression is largely controlled at the transcriptional level, presenting a molecular hallmark of thermogenic adipocytes (Ricquier 2011). Better dissection of the regulatory elements for UCP1 transcription will certainly help to understand the regulation of thermogenesis in adipocytes, and develop treatments to combat obesity. Although the available information on the regulation of UCP1 transcription is rather extensive (Montanari et al. 2017; Bonet and Oliver 1831; Collins et al. 2010; Inagaki et al. 2016), to our knowledge, a systematic genetic screen for cis-element(s) that transcriptionally regulates UCP1 expression has not been performed. Here, we took advantage of the CRISPR technology, in combination with our previously established Ucp1-GFP reporter system (Qiu et al. 2018), and performed an in situ saturating mutagenesis screening, with the aim to identify potential DNA regulatory elements for UCP1 regulation.
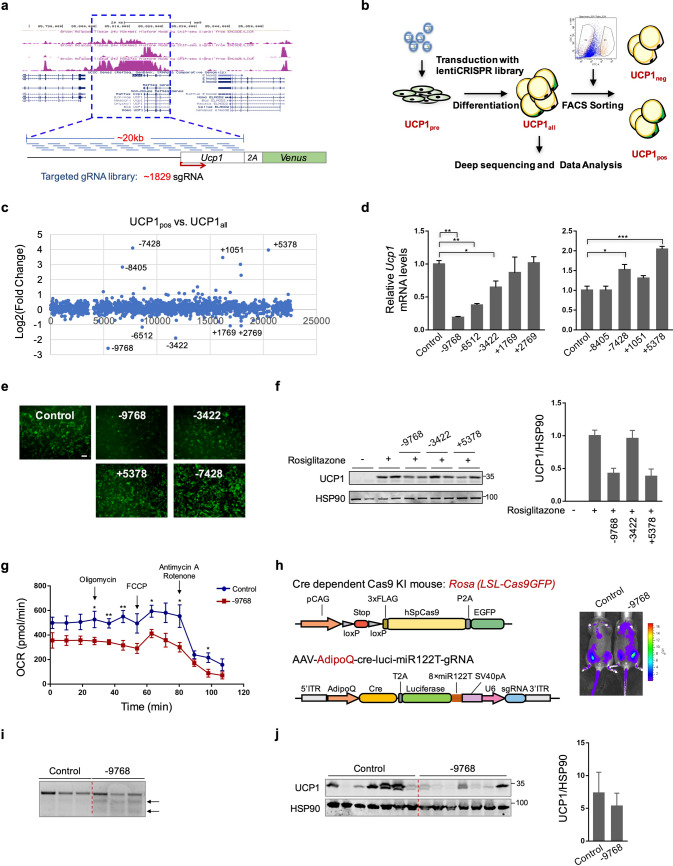
Recent advances in genome engineering technologies allow in situ saturating mutagenesis studies for potential regulatory elements of a specific genomic region in a high-throughput manner (Canver et al. 2015). To elucidate potential DNA regulatory element(s) involved in Ucp1 transcription, we customized an sgRNA library spreading the ~ 20 kb genomic region, covering the 5′ region of the Ucp1 gene as well as all the introns (Fig. 1a). The ~ 20 kb genomic region was intensive for epigenetic signals of enhancers (H3K4me and H3K27ac) or promoters (H3K4me3), according to the ChIP-seq results carried out in brown adipose tissues (ENCODE data) (Fig. 1a). We then performed screenings with this customized sgRNA library using our established screening platform, which was generated with the immortalized Ucp1-GFP primary mouse brown preadipocytes, with GFP signal reflecting the endogenous UCP1 protein level (Qiu et al. 2018; Li et al. 2018) (Fig. 1b). Ucp1-GFP brown preadipocytes (UCP1pre) were transduced with the lentiCRISPR library and differentiated following a standard protocol. Mature brown adipocytes (UCP1all) were then sorted to obtain high GFP + (top 5%) (UCP1pos) and GFP- (bottom 5%) (UCP1neg) populations. Two other cell groups, preadipocytes UCP1pre and presorted UCP1all cells, were also collected. Each group of cells was subjected to genomic DNA extraction and deep sequencing of integrated sgRNAs. Each sgRNA is labeled as a relative targeting locus, with the transcriptional start site (TSS) of Ucp1 designated as 0. Comparisons were primarily carried out between UCP1pos and UCP1all groups to mainly avoid disturbances from cells that may have low UCP1 expression due to unsuccessful differentiation. Three independent screenings were performed, and fold changes of sgRNA counts in UCP1pos vs. UCP1all were averaged between three results. Using a cut-off of log 2(Fold Change [UCP1pos /UCP1all]) ≥ 1 or ≤ - −1, we have identified in total 13 sgRNAs that have significant enrichment in UCP1pos, as well as 5 sgRNAs that have significant depletion in UCP1pos. (Fig. 1c and Table S1). We then validated four of the top-enriched and five of top-depleted sgRNAs individually in brown adipocytes. Consistent with the screening results, cells treated by the sgRNAs − 9768, − 6512, and − 3422 showed significantly downregulated Ucp1 expression, suggesting existing enhancers in these targeted regions, whereas cells treated by sgRNAs -7428 and + 5378 loci resulted in significantly upregulated Ucp1 expression, which indicated the possible repressors in the targeted regions (Fig. 1d and e). No significant difference in Ucp1 expression was observed with sgRNA + 1769, + 2769, − 8405, or + 1051, a reflection of non-specific noise in the screening (Fig. 1d). Further tests using the established primary mouse Ucp1-GFP white adipocytes showed that sgRNA − 9768 and + 5378 treatment led to obvious downregulation of UCP1 expression in the white adipocytes under rosiglitazone stimulation, whereas no significant differences with sgRNA − 3422 and slightly increased UCP1 expression caused by sgRNA − 6512 and − 7428 treatment (Fig. 1f and Fig. S1). Consistently, the oxygen consumption rate (OCR) in sgRNA − 9768-treated cells was significantly lower than that in control cells (Fig. 1g).
Fig. 1.
In situ saturating mutagenesis screening identifies a functional genomic locus that regulates Ucp1 expression. a Schematic view of Ucp1 sgRNA library design for in situ cis-element screen with the Ucp1-GFP reporter cell line. b Workflow of the screening strategy. c Scatterplot displaying enriched or depleted sgRNAs in UCP1pos group as compared to UCP1all group. TSS, transcriptional start site. d Ucp1 mRNA expression analyses in brown adipocytes treated with individual sgRNAs as indicated. n = 4. e Representative images of GFP intensity in brown adipocytes treated with individual sgRNAs. Scale bar = 50 µm. f UCP1 protein expression analyses in white adipocytes under basal level or upon rosiglitazone stimulation, treated with or without indicated sgRNAs. g Oxygen consumption analysis in brown adipocytes under basal level treated with sgRNA -9768 or control vectors. n = 5. h Schematic illustration of the CRISPR-Cas9 KI mice and AAV constructs used in this study (left). In vivo luminescence analysis after 2 weeks’ injection of AAV vectors (right). i Genome editing activity in inguinal white adipose tissues from mice treated with sgRNA − 9768 or control vectors. j UCP1 protein expression analyses in inguinal white adipose tissues from control or sgRNA − 9768-treated mice after exposure to cold (4 °C) for 2 days. n = 7
We then went on to further validate the effect of sgRNA − 9768 in vivo. We delivered sgRNA -9768 to the adipose tissue of adult CRISPR-Cas9 knockin mice (Platt et al. 2014) via in situ injection of adeno-associated virus (AAV) expressing cre-recombinase and sgRNA (Fig. 1h). To specifically target adipose tissue, cre-recombinase was expressed under an adipocyte-specific adipoQ promoter, and eight copies of miR122 targeting sequences were added at the 3′ to limit hepatocyte expression. A firefly luciferase expression cassette was also included in the vector to assist in vivo evaluation of delivery efficiency (Fig. 1h). Three weeks after virus injection, mice were exposed to cold (4 °C) for 2 days to stimulate UCP1 expression in white adipose tissues. Consistent with the results in in vitro cultured cells, we observed clear genome-editing efficiency (Fig. 1i), and a significant downregulation of UCP1 expression in white adipose depots from mice treated with sgRNA − 9768 (Fig. 1j). We also examined possible physiological change after treatment with sgRNA − 9768. Results showed that sgRNA − 9768 treatment did not affect mice body weight or iWAT tissue mass (Fig. S2A and S2B), however, caused decreased UCP1 expression, less multilocular adipocytes, larger adipocyte size, and a decreased trend of the ratio of mitDNA to nuDNA, as well. No significant change in the mRNA levels of other thermogenic genes or lipolysis-related genes was observed, indicating a specific regulation to Ucp1 expression (Fig. S2C–F). Altogether, these results suggest that the − 9768 locus carries functional regulatory elements that are necessary for UCP1 expression in adipocytes.
In this study, we performed an unbiased screening with customized sgRNA library covering potential regulatory regions for UCP1 transcriptional activity, and identified several genomic loci especially the − 9768 locus, that may contain key regulatory elements for Ucp1 expression. However, the exact regulatory sequences and potential transcription factor(s) or epigenetic regulator(s) that bind to these sequences remain to be defined. Our results also showed some inconsistencies in primary mouse brown and white adipocytes. For example, sgRNA − 6512 and + 5378 displayed different functional effects in brown and white adipocytes, suggesting different regulatory mechanisms governing Ucp1 expression in brown and white adipocytes, which warrant further investigation. The most direct physiological change after treatment with sgRNA − 9768 should be the change of body temperature, especially the local temperature of white adipose tissue targeted with sgRNA − 9768, which can be measured by infrared camera. sgRNA targeting known regulatory regions for Ucp1 transcription were included in the designed sgRNA library; however, these sgRNAs were not screened out in this study. This is possibly due to low targeting efficiencies of these sgRNAs, background disturbances from library handling, adipocyte differentiation, etc. Indeed, there exist some limitations in our screening system: (1) as mature adipocytes cannot proliferate, the selection step with the fluorescence activating cell sorting (FACS) is less able to enrich for the desired sgRNAs, compared with screenings in which enhancement of proliferation or acquisition of drug resistance is the desired outcome. The inability for further enrichment in our screening is causing high background disturbances; (2) the lengthy and multi-step adipocyte differentiation procedures are also bringing about a significant amount of variations from differentiation efficiency, which itself leads to variations in expression levels of UCP1, also another big source of background disturbance. Further optimization of the screening strategy may help to reduce false-positive or false-negative results. Nonetheless, by combining a reporter cell line and a CRISPR-mediated genetic screening, our study highlights a useful approach to identify possible regulatory elements for UCP1 transcriptional activity.
Materials and Methods
Animals
All mice were housed under standard conditions at Shanghai Institutes for Biological Sciences (SIBS), China. The animals presented a healthy status and male mice were employed for all experiments.
The colony of Cre-dependent Cas9 knockin mouse (Jackson Laboratory) was maintained by crossing with wild-type C57BL/6 J mice (Shanghai Laboratory Animal Co. Ltd, China). For adipocyte-specific genomic deletions, 7~8 weeks’ old heterozygous Cas9 knockin animals (RosaCas9±) were recorded for body weight. Animals were then randomly divided into groups, and administrated via in situ injection of adeno-associated virus 8 (AAV-8) expressing cre-recombinase and sgRNA targeting specific genomic loci. A firefly luciferase expression cassette was included in the AAV vector to assist in vivo evaluation of delivery efficiency. AAV vectors with cre-recombinase and luciferase cassettes and no sgRNA were used as control viruses.
For in situ AAV virus injection, viruses were dissolved in phosphate-buffered saline (PBS) and mice were injected bilaterally with a dose of 1 ~ 2 × 1012 vg into the inguinal fat pads. After virus injection, animals were fed with normal chow diet (NCD) (Shanghai Laboratory Animal Co. Ltd, P1103F). In vivo bioluminescence imaging of AAV vectors was performed in mice administrated with AAV vectors 2 weeks later. And 3 weeks after virus injection, mice were exposed to cold (4 °C) for 2 days and dissected for further analysis.
Cell Culture and Differentiation
HEK 293 T and NIH 3T3 cell lines (Cell Bank, Type Culture Collection Committee, Chinese Academy of Sciences, Shanghai) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, 8,117,254) containing 10% fetal bovine serum (FBS, Gibco, 16,000,044), and 1% penicillin/streptomycin in a humidified incubator with 5% CO2 at 37 °C. The Ucp1-GFP brown and white adipocytes were used as previously described (Qiu et al. 2018, S1). Briefly, the Ucp1-GFP brown and white adipocytes were isolated and immortalized from the stromal vascular fraction from interscapular brown fat pads or inguinal white fat pads of postnatal day 2 Ucp1-GFP male mice (Qiu et al. 2018). Ucp1-GFP brown and white preadipocytes were cultured at 37 °C in 5% CO2 in primary culture medium (high-glucose Dulbecco’s modified Eagle’s medium (DMEM) with 20% fetal bovine serum (FBS) and 1% penicillin/streptomycin). The differentiation of brown and white preadipocytes were following a standard protocol.
Design and Synthesis of Mouse Lentiviral sgRNA Library
We generated a customized sgRNA library targeting the ~ 20 kb genomic region, including the 5′ region of mouse Ucp1 gene as well as all the introns. Every 20 bp sequence upstream of an NGG PAM sequence on the plus or minus strand was identified as the sequence of a sgRNA. With the removal of sgRNAs with other targets in the genome that match exactly or differ by only 1 base, this customized library is comprised of 1,829 different guides, and they synthesized in CustomeArray (WA, USA).
To produce lentivirus sgRNA library, HEK293T cells in each 15 cm dish were transfected with 14.7 μg pMDLG, 7.9 μg pVSVG, 5.7 μg pREV, and 22.5 μg CRISPR plasmids using polyethylenimine (Polysciences, 23,966–1). Medium was changed 4~6 h after transfection. Lentiviral supernatants were collected at 48 and 72 h post-transfection and centrifuged at 20,000 rpm at 4 °C for 2 h. Viral pellets were then re-suspended in DMEM at 4 °C overnight and titer was calculated using a PCR-based titration kit (Applied Biological Materials Inc, LV900).
In Situ Saturating Mutagenesis Screen for UCP1 Regulators
The Ucp1-GFP brown preadipocytes (UCP1pre) were infected by the lentiviral library at MOI of 2 ~ 3 to achieve an infection efficiency of around ~ 30%. Cells were then treated with 1.5 μg/ml puromycin (Beyotime, ST551) for 2 days to remove non-infected cells. Cells were recovered for one more day after puromycin treatment, and followed by a standard differentiation protocol to obtain mature brown adipocytes (UCP1all). Cells were harvested at day 8 at maturation stage and high GFP + cells (top 5%) or GFP- cells (bottom 5%) were collected by FACS sorting (FACSAria II; BD Biosciences). Control cells, including UCP1all cells (collected before FACS sorting) and infected UCP1pre cells (collected after puromycin treatment), were obtained at indicated time points.
Genomic DNA of cells from different groups was extracted and the sgRNAs were amplified by PCR method using KOD DNA polymerase (TOYOBO, KOD-401). Briefly, in total, 2 μg (200 ng per PCR reaction; 10 separate reactions for each sample) of genomic DNA from each group were used as DNA template; the PCR program was 94 °C 5 min, 98 °C 20 s, 58 °C 30 s, 68 °C 12 s, 32 cycles. Products (158 bp) were gel-purified and quantified. In total, 1.5 μg PCR products from each group were pooled together and sent for deep sequencing (Illumina HiSeq4000 system) by using the pair-ended 150 bp sequencing protocol. PCR primers used for amplification were: 5′-TGAAAGTATTTCGATTTCTTGGCTT-3′, 5′-CGGTGCCACTTTTTCAAGTT-3′. An 8 bp barcode for multiplexing of different biological samples was added at 5′ of each primer.
For data analysis, the sequencing reads of sgRNAs from different samples were first identified by barcode using cutadapt (v1.9) with default parameters. Build-index function of Bowtie [S2] was applied on the sgRNA sequences of sgRNA library to generate Burrows–Wheeler index. The sgRNA sequences were then retrieved and counted by aligning processed reads of each sample to the sgRNA library using Bowtie. Maximum two mismatches were allowed and only the reads with unique alignment were reported. Total reads were normalized to library sequencing depth. sgRNA enrichment score was determined by calculating (1) the ratio of normalized reads in the UCP1pos group as compared to UCP1all group; and (2) log2 transformation. Three independent screenings were performed and the average of three sgRNA enrichment scores were used for identification of interested sgRNAs.
Validation of Individual sgRNAs in Brown and White Adipocytes
The 20 bp sequence of sgRNA targeting specific genomic loci was inserted to lentiCRISPR V2 plasmid and used for lentivirus packaging. The target sequences used are listed in Supplemental Table 1. Lentiviruses carrying CRISPR-sgRNA or empty lentiCRISPR V2 vector as control viruses were packaged. The Ucp1-GFP brown preadipocytes were then infected and selected with 1.5 μg/ml puromycin (Beyotime, ST551) for 2 days to remove non-infected cells. Cells were then recovered for one more day, and subjected to adipocyte differentiation. Brown adipocytes were collected at day 8, and white adipocytes were collected at day 10 for later mRNA or protein expression analyses.
CRISPR Construct and AAV Packaging
The pAAV-GFP plasmid (Cell Biolabs. Inc) was modified, with the original CMV promoter replaced by the adipoQ promoter for adipocyte-specific expression between NotI and EcoRI restriction enzyme sites, the original GFP cassette replaced with the Cre-T2A-Luciferase-sgRNA cassette between EcoRI and XbaI restriction enzyme sites for expression of Cre-recombinase, luciferase, and sgRNA. And an 8 × miR122 targeting sequence at the 3′ region was added after luciferase cassette to limit the AAV expression in the liver tissue. The sgRNA scaffold was inherited from the lentiCRISPR V2 plasmid (Addgene, #52961), and pieces containing Cre-recombinase, T2A-luciferase, 8 × miR122 targeting sequence, and sgRNA were ligated via PCR. The resultant construct was named AAV-AdipoQ-Cre-Luci-8 × miR122T-gRNA construct, which was further used to insert sgRNA sequences targeting specific genomic loci or directly as control vector.
For adipocyte tissue-specific genomic disruption with sgRNA -9768, mouse Ucp1 sgRNA − 9768 (5′-GAATGAAAAAAAAAGGTGAC + AGG-3′) was inserted into the sgRNA scaffold in AAV-AdipoQ-Cre-Luci-8 × miR122T-gRNA construct. And construct was verified by sequencing before using. Genome-editing efficiency by CRISPR targeting in adipose tissue was examined with genomic DNA by T7EI analysis (NEB, E3321). Primers used in T7EI analysis were as follows: 5′-AAAAGAGTCCATGGCCCTGA-3′ and 5′-GATACACAACACAGGCCCAG-3’.
Adeno-associated viruses were generated using packaging plasmids AAV-helper and AAV-8 (Cell Biolabs. Inc) together with AAV-AdipoQ-Cre-Luci-8 × miR122T-gRNA constructs. Viruses were administrated via in situ injection at a dose of 1 ~ 2 × 1012 vg per mouse for adipose tissue-specific CRISPR genomic loci mutation.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from brown adipocytes, white adipocytes, or subcutaneous adipose tissue using Trizol reagent (ThermoFisher, 15,596,018) according to the manufacturer’s instructions. Reverse transcription of isolated RNA was performed using the reverse transcription kit (Takara, RR047A). Quantitative real-time PCR was carried out on the 7900 System (ABI) using SYBR Green supermix (ABI, 4,472,908). Primers used in this study are listed in Table S2.
Western Analysis
Protein from cells or tissues was extracted by the RIPA buffer (Millipore, 20188) and subjected to regular western procedure. The primary antibodies used in the experiments were antibodies to UCP1 (Abcam, ab10983) and HSP90 (Cell Signaling Technology, 4874S).
Cold Challenge Experiment
Heterozygous Cas9 knockin male mice treated with AAV vectors were individually caged with food and water at a cold room (4 °C) for 2 days.
Measurement of Mitochondria Number
Genomic DNA from inguinal white adipose tissues of mice treated with AAV vectors was extracted using the TIANamp Genomic DNA kit (TIANGEN, DP304-03). The presence of amplifiable mitDNA and nuDNA in the extract was assayed through real-time PCR.
Seahorse Assay
The oxygen consumption rate (OCR) was measured using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience). The Ucp1-GFP brown preadipocytes treated with lentiviruses were plated in the XF24 V28 cell culture microplate (Seahorse Bioscience) and subjected to adipocyte differentiation for 8 days. Cells were treated with 2 μM Oligomycin, 1.5 μM FCCP, and 1 μM Rotenone/Antimycin A from the Agilent Seahorse XF Cell Mito Stress Test Kit (Agilent Technologies, 103,015–100) during fixed time intervals.
Histology
Mouse tissues were fixed and embedded in paraffin (for HE staining) or frozen (for UCP1 staining). Sections were stained with hematoxylin and eosin or UCP1 antibody (1:100, Abcam, ab10983) according to standard protocols (Wuhan Servicebio Technology).
Statistics
The unpaired, two-tailed Student’s t test was used for experiments with two groups’ comparison. All data are represented as means with SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author Contributions
YQ and QD designed experiment. YQ performed all experiments. XL performed data analysis for CRIPSR screening. YS and SL assisted with the screening. YW and CT modified the pAAV-GFP plasmid. YQ and QD analyzed the data, prepared figures, and wrote the manuscript. QD supervised the project.
Funding
This work was supported by grants from the National Key R&D Program of China (2017YFA0102800, 2017YFA0103700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030402), and the National Natural Science Foundation of China (31670829, 31971063).
Availability of Data and Materials
All unique materials generated from this study are available from the corresponding author, Qiurong Ding (qrding@sibs.ac.cn).
Compliance with Ethical Standards
Conflict of Interest
All authors declare no conflicts of interest.
Ethics Approval
All mice were housed under standard conditions at the Shanghai Institutes for Biological Sciences (SIBS), China. All animal procedures were performed according to guidelines, and were approved by the Institutional Animal Care and Use Committee of the Shanghai Institutes for Biological Sciences.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional ailiations.
References
- Bonet ML, Oliver P, Palou A. Pharmacological and nutritional agents promoting browning of white adipose tissue. Biochim Biophys Acta. 2013;1831(5):969–985. doi: 10.1016/j.bbalip.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Yehuda-Shnaidman E, Wang H. Positive and negative control of Ucp1 gene transcription and the role of beta-adrenergic signaling networks. Int J Obes (Lond) 2010;34(Suppl 1):S28–33. doi: 10.1038/ijo.2010.180. [DOI] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19(10):1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;17(8):480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li M, Liu X, Yang Y, Wei Y, Chen Y, Qiu Y, Zhou T, Feng Z, Ma D, Fang J, Ying H, Wang H, Musunuru K, Shao Z, Zhao Y, Ding Q. Genetic and chemical screenings identify HDAC3 as a key regulator in hepatic differentiation of human pluripotent stem cells. Stem Cell Rep. 2018;11(1):22–31. doi: 10.1016/j.stemcr.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari T, Poscic N, Colitti M. Factors involved in white-to-brown adipose tissue conversion and in thermogenesis: a review. Obes Rev. 2017;18(5):495–513. doi: 10.1111/obr.12520. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Sun Y, Xu D, Yang Y, Liu X, Wei Y, Chen Y, Feng Z, Li S, Reyad-Ul Ferdous M, Zhao Y, Xu H, Lao Y, Ding Q. Screening of FDA-approved drugs identifies sutent as a modulator of UCP1 expression in brown adipose tissue. EBioMedicine. 2018;37:344–355. doi: 10.1016/j.ebiom.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricquier D. Uncoupling protein 1 of brown adipocytes, the only uncoupler: a historical perspective. Front Endocrinol (Lausanne) 2011;2:85. doi: 10.3389/fendo.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All unique materials generated from this study are available from the corresponding author, Qiurong Ding (qrding@sibs.ac.cn).