Abstract
Background:
Alzheimer’s disease (AD) is a neurodegenerative disease characterized by the accumulation of amyloid-β (Aβ) peptide in the brain.
Objective:
Gain a better insight into alterations in major biochemical pathways underlying AD.
Methods:
We compared metabolomic profiles of hippocampal tissue of 20-month-old female Tg2576 mice expressing the familial AD-associated hAPP695SW transgene with their 20-month-old wild type female littermates.
Results:
The hAPP695SW transgene causes overproduction and accumulation of Aβ in the brain. Out of 180 annotated metabolites, 54 metabolites differed (30 higher and 24 lower in Tg2576 versus wild-type hippocampal tissue) and were linked to the amino acid, nucleic acid, glycerophospholipid, ceramide, and fatty acid metabolism. Our results point to 1) heightened metabolic activity as indicated by higher levels of urea, enhanced fatty acid β-oxidation, and lower fatty acid levels; 2) enhanced redox regulation; and 3) an imbalance of neuro-excitatory and neuro-inhibitory metabolites in hippocampal tissue of aged hAPP695SW transgenic mice.
Conclusion:
Taken together, our results suggest that dysregulation of multiple metabolic pathways associated with a concomitant shift to an excitatory-inhibitory imbalance are contributing mechanisms of AD-related pathology in the Tg2576 mouse.
Keywords: Alzheimer’s disease; excitatory and inhibitory imbalance; fatty acids; hAPP695SW; hippocampus, metabolomics; Tg2576
INTRODUCTION
Alzheimer’s disease (AD) has become a public health crisis, which will be further aggravated by the rise in the number of the aging population. It is estimated that currently, more than 5.5 million Americans, age 65 or older, may have AD and show dementia symptoms [1]. Rodent models of AD, including the Tg2576 murine model of amyloid-β (Aβ) accumulation, are indispensable for exploring mechanisms of pathology and progression of AD, as well as serving as pre-clinical models for testing exploratory treatments and management strategies [2]. The Tg2576 model carries the human 695 splice-variant of amyloid precursor protein (APP) with the double mutation K670M and N671L (hAPP695SW) and expresses this transgene under control of the hamster prion protein gene promoter, which occurs predominantly in neurons [3]. Compared to the endogenous mouse APP, the brains of Tg2576 mice contain around five times higher levels of transgenic mutant human APP [4].
The double mutant APP isoform, first discovered in a Swedish family exhibiting early onset of disease, is frequently referred to as Swedish mutation (APPSW) [5]. The double mutation is located before the amyloid-β peptide (Aβ) region of amyloid-β protein precursor (AβPP), and β-secretase cleavage occurs within the secretory pathway, resulting in increased production and secretion of neurotoxic Aβ peptide. The Tg2576 mouse produces a large amount of Aβ peptides that aggregate extracellularly, forming Aβ plaques and Aβ fibrils that are established in the brain by 11–13 months of age [3]. Aβ plaques are considered as one of AD’s hallmarks and have been linked to age-dependent decline in hippocampal learning and memory. These same impairments are evident in the aged Tg2576 mouse line [6, 7].
The Tg2576 mouse is a well-established and most widely studied preclinical model of AD; however, our knowledge about how the transgene hAPP695SW and the resulting Aβ plaques in aged mice affect metabolic pathways in the brain is limited to three studies that showed increased concentrations of glutamate, creatine, and taurine and decreased concentration of polyunsaturated fatty acid (PUFA)-containing phospholipids and mitochondrial function with age [8–10].
Oxidative stress (OS) plays a key role in the neurotoxicity of Aβ plaques in AD [11, 12]. We have previously reported increased reactive oxygen species (ROS) production and mitochondrial dysfunction [13] as well as reduced dendritic complexity in hippocampal neurons of huAPP695SWE transgene mice compared to wild type (WT) littermates [14].
The goal of the present study was to examine the impact of the hAPP695SW transgene on the hippocampal metabolome in female Tg2576 mice approximately 20 months of age, an age at which cognitive dysfunction can be observed [15]. We hypothesized that the neuropathology of the hAPP695SW transgene is linked to metabolomic abnormalities in brain-relevant functional pathways (i.e., redox homeostasis, fatty acid oxidation, and mitochondrial/peroxisomal function, neurotransmission and signaling, and sphingolipid and ceramide metabolism) in older, female Tg2576 mice.
We selected hippocampal tissue due to its relevance to learning and memory as well as the morphological and biochemical changes we had observed in Tg2576 primary hippocampal neurons described earlier [13, 14]. We focused on female mice because women are affected disproportionately by AD, having a two times higher risk of developing AD compared to men [16–18]. The comprehensive nature of the data, encompassing metabolites, lipids, and various ceramides, the comprehensive analysis of biochemical metabolite classes, biochemical pathways, and brain-relevant functional pathways, all of which had not been done previously in metabolomics studies of AD models, allowed for the identification of biochemical pathways affected by the hAPP695SW transgene. In addition, as a novel approach, we linked metabolite changes with Aβ burden and presence of hAPP695SW transgene to explore how AD related pathology may result in metabolic adaptations in the hippocampus. The observed metabolic changes suggest heightened levels of excitatory neurotransmitters and heightened metabolic activity concomitant with decreased levels of inhibitory neurotransmitters and lowered availability of fatty acids in the female Tg2576 mouse hippocampus.
MATERIALS AND METHODS
Tissue samples
These studies were conducted in accordance with NIH Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the Portland VA Medical Center (IACUC #: 3260–17). Mice used in this study were on a C57BL/6:SJL background, with alternate breeding to WT SJL or C57BL/6 dams for each generation. The female Tg2576 and WT littermates used in these studies were the results of a cross of Tg2576 heterozygous sires with SJL WT dams (Jackson Laboratory, Bar Harbor, ME). Transgene status was assessed using PCR of transgenic human amyloid-β (Aβ) precursor protein (hAβPP, possessing the Swedish mutation – KM670/671NL) from tail sample DNA. Animals were maintained in a climate-controlled environment, on a 12-h lighting schedule. Right brain hemispheres were fixed in 4% formaldehyde for immunohistochemical analysis, while the left hippocampi were dissected and snap-frozen for metabolomic analyses.
Immunohistochemistry (IHC) of Aβ plaque burden
At sacrifice, the right brain hemispheres were surgically removed and fixed in 4% formaldehyde/PBS overnight. Samples were then subjected to increasingly more concentrated solutions of sucrose (0–30%) and stored at −80°C until required. Hemispheres were coronally sectioned in 20 μm thick sections via a cryo-histomat and subsequently processed in Netwell inserts in 6-well plates. Sections of similar depths were incubated in a quenching solution of 30% methanol and 0.3% hydrogen peroxide in 1X TBS to reduce endogenous catalase activity. Sections were then blocked using 10% horse serum, 2% BSA, and 0.5% triton in 1X TBS. Sections were stained using a pan-Aβ antibody (44–136, Invitrogen, Carlsbad, CA) at a concentration of 1:1000 overnight at room temperature. A secondary antibody (anti-rabbit; 1:200; Vector Laboratories, Burlingame, CA) was applied to visualize staining. A DAB counterstain (Sigma Fast 3, 3 Diaminobenzidine Tablet Set, D-4418; Sigma-Aldrich Corp, St. Louis, MO) was applied before sections were mounted on slides and scanned with PrimeHisto XE (Pacific Image Electronics, Torrance, CA). Images collected were quantified using FIJI software. Images were grayscale converted and the threshold was set to zero to remove background staining for WT negative control sections. These settings were applied consistently throughout each staining group by one technician blinded to treatment conditions. Staining in the hippocampus of each sample was measured as the total area stained within the defined region using FIJI software.
Metabolite extraction
Methanol and water (LC-MS-grade) were purchased from EMD Millipore (Burlington, MA, USA). Formic acid (certified ACS reagent) was from Fisher Chemicals (Suwanee, GA, USA). L-Methionine-(methyl-d3), was used as an internal standard (Sigma Aldrich; St. Louis, MO, USA). For metabolomic analysis, hippocampal tissue was analyzed from 9 WT and 12 Tg2576 mice. The extraction protocol was previously reported with some modifications [19]. Briefly, the samples were homogenized using a Precellys™ 24 bead ruptor homogenizer (Bertin Technologies, USA). The whole frozen hippocampus was accurately weighed and placed in a 2 mL homogenization tube prefilled with 1.4 mm ceramic beads. Methanol: ethanol 50:50 v/v containing 1.0 μg/mL of methionine-(methyl-d3) as internal standard was added at 4°C (10 μL of solvent/mg of tissue). The spiked samples were homogenized for 20 s at 5000 rpm three times with 30 s of cool-down in between. Samples were placed at −20°C for 1 h and spun (15,000 g, 10 min, 4°C) to collect the supernatant. The resultant supernatant from each sample was transferred to HPLC vials (Microsolv, Leland, NC, USA) for LC–HRMS/MS analysis. For monitoring platform performance, 20 μl aliquots of each extract were mixed to generate a quality control (QC) pooled sample.
LC–HRMS/MS analysis
Untargeted LC–HRMS/MS analysis was carried out using data dependent acquisitions utilizing an AB SCIEX TripleTOF® 5600 mass spectrometer (AB SCIEX, Concord, Canada) coupled to Shimadzu Nexera UHPLC system as previously described with some modifications [19–21]. Chromatographic separation was performed on an Inertsil Phenyl-3 column (4.6 × 150 mm, 100 Å, 5 μm; GL Sciences, Rolling Hills Estates, CA, USA) held at 50°C. A gradient with two mobile phases was used: Mobile phase A was water (LC-MS grade) with 0.1% v/v formic acid and B, methanol (LC-MS grade) with 0.1% v/v formic acid. After 1 min at 5% B, the linear elution gradient was as follows: 1 min, 5% B; 11 min, 30% B; 20 min, 100% B; 25 min, 100% B; 30 min, 5% B; and 35 min, 5% B. The injection volume was 5 μL with a flow rate of 0.4 mL/min. Samples were randomized before injections. A QC sample was analyzed every five LC runs. The IonSpray voltage was set at 4500 V, and the source temperature was 500°C. Period cycle time was 950 ms; accumulation time 100 ms; m/z scan range 100–1400; and collision energy 35 V with collision energy spread of 15 V. Mass calibration of the TOF analyzer was performed automatically after every fifth LC run.
Metabolomic data processing
LC-HRMS/MS data processing was enabled using Progenesis QI™ software V2.0 (NonLinear Dynamics, UK) and used for peak picking, retention time correction, peak alignment, and metabolite annotations. The metabolite annotation workflow was similar as described by us previously [20–22]. Progenesis supports our in-house IROA library based on the Metabolite Library of Standards consisting of 650 standards (IROA Technologies, Bolton, MA, USA) that allowed L1 annotations. Additional metabolite annotation we received by searching online databases such as METLIN MS/MS library (plugin V1.0.6499.51447), HMDB, and LipidBlast and these annotations are referred to as L2 annotations in Supplementary Tables 1 and 2. Annotation confidence was achieved in accordance with reporting criteria for chemical analysis suggested by the Metabolomics Standards Initiative [23]. Tentative annotations were made based on accurate mass (error < 10 ppm), fragment ion spectral pattern (library score > 50), and isotope pattern (library score > 50). Ions generated from QC samples were retained for annotation and included in the dataset if the coefficient of variation (CV) of their abundance did not exceed 30%. Relative quantities of metabolites were determined by calculating their corresponding peak areas. To account for drift during the metabolomics run, the annotated metabolites were normalized using the support vector regression (SVR) method which is a non-parametric machine learning-based algorithm. SVR was performed using the MetNormalizer package in R (v1.2.5042) software (© 2009–2020 RStudio, PBC) to correct peak intensities across the samples. This algorithm divides each peak intensity in the samples to the predictive peak intensity which was calculated using the SVR model to give normalized abundances of each metabolite [24].
Statistical analysis
Principle component analysis using MetaboAnalyst (v4.0) [25, 26] was used to evaluate overall metabolic alterations of Tg2576 compared to WT mice. Mean decrease accuracy (MDA) values of the random forest (RF) analysis and variable importance in projection (VIP) scores based on partial least squares-discriminant (PLS-DA) model were obtained using the same online platform. To assess individual metabolites changes, fold changes, p-values using parametric t-test and non-parametric Wilcoxon rank-sum tests were obtained using R (v1.2.5042) software and MetaboAnalyst (v4.0). Parametric Pearson correlation coefficients and non-parametric Spearman correlation coefficients were used to correlate individual metabolites with Aβ accumulation in Tg25756 mice. All tests were two-sided. For evaluating the significance of individual metabolites, an FDR-adjusted p ≤ 0.05 was used. For evaluating the significance of biochemical pathways, we did an enrichment analysis of metabolites within the pathway; as cut-off, over 50% of metabolites within the pathway had to differ at p ≤ 0.10. Pathway analysis using MetaboAnalyst (v4.0) was used to evaluate the impact of metabolites alteration on metabolic pathways. Volcano plot, heatmap, diverge plot and correlation heatmap were created using R (V1.3.959) software and bar plots were created using GraphPad Prism 8.4.2. A correlation network using hierarchical clustering was generated using Cytoscape version 3.8.2 [27].
RESULTS
Metabolite Identification
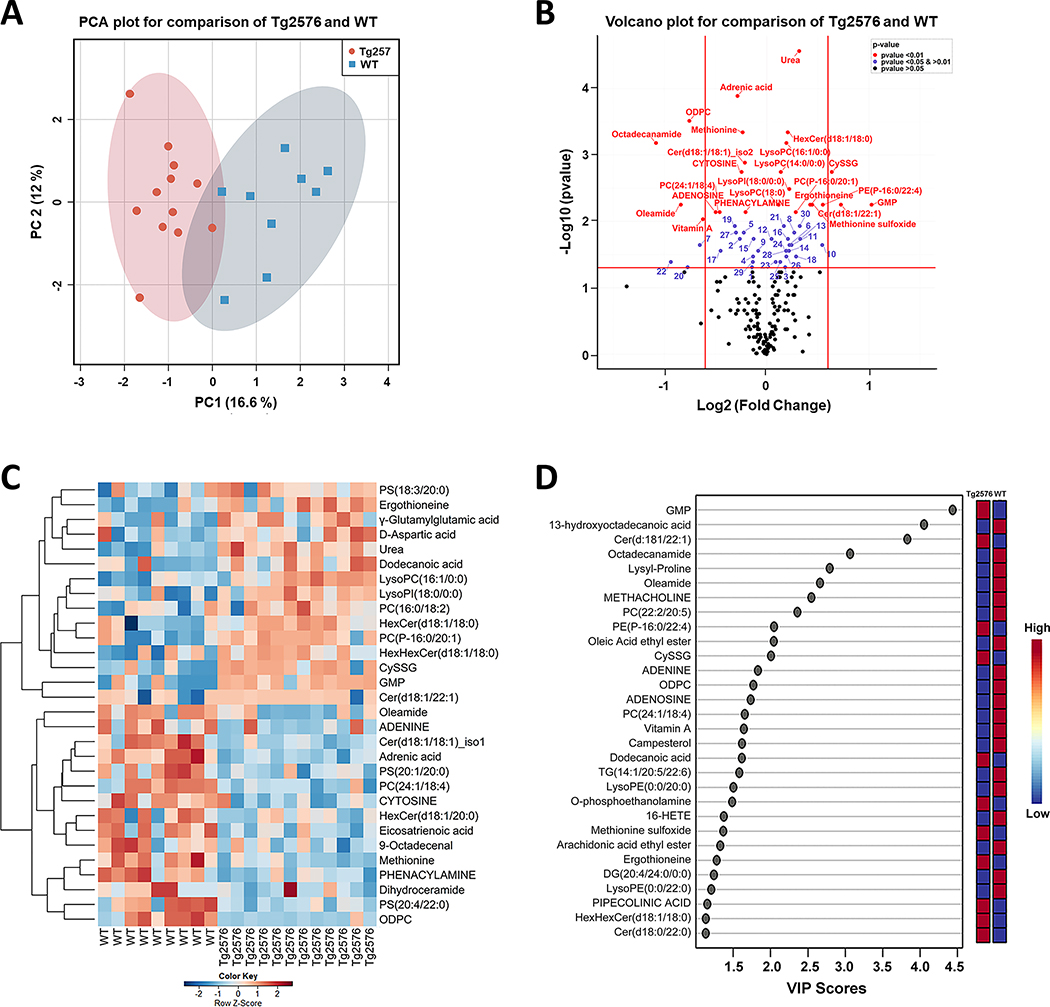
Using the metabolomics data generation and processing workflow shown in Supplementary Figure 1, we annotated 180 metabolites belonging to 23 metabolic classes which include 6 lipid classes (according to Human Metabolome Database (HMDB) and LIPID MAPS classification) with high confidence (Level 1 and 2 annotations, according to the Metabolomics Standards Initiative) (Supplementary Tables 1 and 2) [23, 28, 29]. Metabolites with Level 1 annotations are given in upper case (capitalized) letters. We used the precursor ion peak areas of the annotated metabolites for semi-quantitative assessments of metabolites [20, 28, 30]. For exploratory data analysis, principal component analysis (PCA) was applied to 146 out of the 180 annotated metabolites for which MetaboAnalyst recognized their HMDB IDs [31–33]. PCA results displayed a clear separation between Tg2576 and WT hippocampal tissue metabolomes (Fig. 1A). Using Wilcoxon rank-sum test, 24 (13% at p < 0.01, 13 higher in hAPP695SW hippocampal tissue and 11 lower), 30 (17% at 0.01 < p < 0.05; 17 higher and 13 lower), and 22 (12% at 0.05 < p < 0.10) out of 180 annotated metabolites differed between genotypes; after FDR-adjustment, 12 metabolites remained significant at FDR p < 0.05 (Supplementary Table 3). Differences are graphically represented using a volcano plot as a scatter plot that represents log2 fold change and negative log10 p-values (Fig. 1B) [34]. Comparing Tg2576 versus WT hippocampal tissue, ten metabolites differed at p < 0.05 with a fold change > 1.5. More guanosine monophosphate (GMP), Cer(d18:1/22:1), and cysteineglutathionine disulfide (CySSG) and less octadecanamide, PC(22:2/20:5), oleamide, oleic acid ethyl ester, OCTADECYLPHOSPHOCHOLINE (ODPC), ADENINE, and vitamin A had a lower level in Tg2576 tissue.
Fig. 1.
Integration of metabolomics information of Tg2576 and WT mice hippocampi to evaluate the impact of hAPP695SW transgene on metabolites level. A) PCA 2D score plot shows a clear separation between the hippocampal metabolome of Tg2576 and WT mice. B) Volcano plot summarizes the results of the univariate analysis showing 54 metabolites with a statistically significant difference between genotypes. The x-axis shows cut off values of − 0.6 < log2 (fold change) < 0.6; red, blue, and black dots indicate the metabolites with p-value < 0.01, 0.01 < p-value < 0.05 and p-value > 0.05, respectively. Numbers mark metabolites with p-value < 0.01 (comparing Tg2576 to WT hippocampi): 1: 4-O-Galactopyranosylxylose, 2:5-AMINOPENTANOATE, 3:5’-METHYLTHIOADENOSINE, 4:9-Octadecenal, 5:Eicosatrienoic acid, 6:Acetylcholine, 7:ADENINE, 8:D-Aspartic acid, 9:DESMOSTEROL, 10:Dodecanoic acid, 11:g-Glutamylglutamic acid, 12:GLUTAMIC ACID, 13:GLUTATHIONE, 14:Homoanserine, 15:LEUCINE, 16:LysoPC(18:2), 17:LysoPE(0:0/20:0), 18:N-ACETYL-L-ASPARTIC ACID, 19:Cer(d18:1/16:0), 20:Oleic Acid ethyl ester, 21:PC(16:0/18:2), 22:PC(22:2/20:5), 23:PC(22:6/18:2), 24:PC(P-16:0/22:1), 25: LysoPE(P-16:0/0:0), 26:PS(18:3/20:0), 27:PS(20:4/22:0), 28:TG(18:4/18:3/18:4), 29:Dihydroceramide, 30:HexHexCer(d18:1/18:0). C) Hierarchical clustering heatmap of the 30 most important metabolites responsible for classification based on MDA values found by random forest analysis. The heatmap was colored based on row Z-scores. Positive Z-score values are shown in red while negative Z-score values are shown in blue. D) Variable importance in projection (VIP) scores results using PLS-DA analysis showing the 30 most important discriminatory metabolites. Ten metabolites are in common in both D and E plots.
Level 1 annotations (metabolites with capitalized letter) were obtained from an in-house compound library consisting of >650 authentic standards (MSMLS, IROA Technology, Bolton, USA). Additional annotations (L2) were obtained by querying and comparison with spectral data from METLIN, LipidBlast and HMDB (online versions, August 2019) using Progenesis QI™.
A supervised learning algorithm, RF, was applied to rank order metabolites based on MDA values [35, 36]. The RF consisted of 1000 trees, and the out-of-bag error for classifying metabolites was 0.00 for Tg2576 hippocampal tissue and 0.22 for WT hippocampal tissue. The 30 most important metabolites for classification are shown in Fig. 1C. The top five metabolites based on MDA values and discriminatory power were urea, PC(16:0/20:1), CYTOSINE, ODPC, and HexCer(d18:1/18:0), all of which differed at p < 0.01 in univariate analysis, and ODPC also exceeded 1.5-fold change (Supplementary Table 4). VIP scores based on PLS-DA estimates the importance of each metabolite for discriminating between genotypes (Supplementary Table 5)[26]. The PLS-DA model, validated by Leave-One-Out Cross-Validation (LOOCV) procedure, had accuracy = 0.95, R2 = 0.84, and Q2 = 0.61. The 30 most important metabolites based on VIP scores (scores > 1 are considered important) are shown in Fig. 1D. The five highest VIP-scoring metabolites were GMP, 13-hydroxyoctadecanoic acid, Cer(d18:1/22:1), octadecanamide, and lysyl-proline and differed from the five metabolites with the highest MDA values in RF; GMP was the only metabolite with a > 1.5-fold change, and 13-hydroxyoctadecanoic acid and lysyl-proline did not differ between genotypes at p < 0.05 in univariate analysis.
Correlation of immunohistochemistry result with identified metabolites
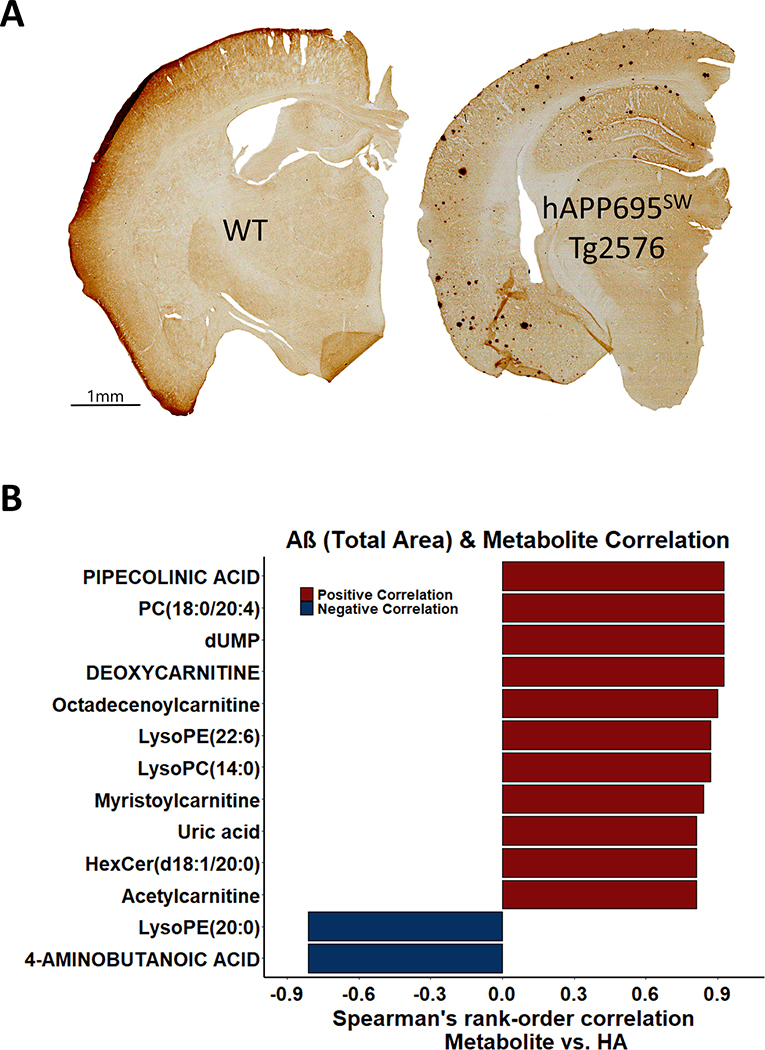
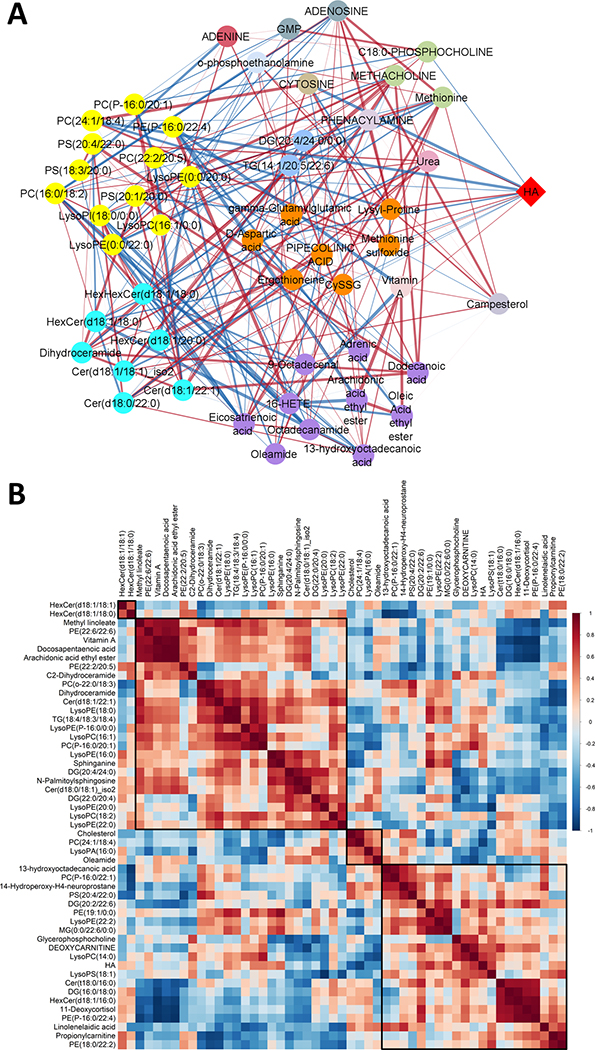
In the pathological examination of the Tg2576 hippocampal tissue, we analyzed the presence of Aβ plaques using IHC. Figure 2A shows pan-Aβ IHC analysis of WT and Tg2576 brain sections. Total plaque area within the Tg2576 hippocampal tissue (HA) was measured (median: 0.51%; range: 0.21–0.58%). To our knowledge, there has not been a comprehensive study examining the relation among total plaque area within the Tg2576 HA and metabolites. Using Spearman’s rank-order correlation test, 13 out of 180 metabolites were correlated at p < 0.05 with HA in Tg2576 hippocampal tissue (Fig. 2B). PIPECOLINIC ACID, uric acid, dUMP, DEOXYCARNITINE, octadecenoylcarnitine, myristoylcarnitine, acetylcarnitine, PC(18:0/20:4), lysoPE(22:6), lysoPC(14:0), and HexCer(d18:1/20:0) were positively correlated and 4-AMINOBUTANOIC ACID (GABA) and lysoPE(20:0) were negatively correlated with HA; however, none of the correlations were significant at FDR p < 0.05. Metabolic changes in Tg2576 hippocampal tissue suggest increased fatty acid transport with Aβ accumulation in Tg2576 mice (4 of 7 non-acylated and acylated carnitines increased with HA). Supplementary Table 6 summarizes the results related to the correlation between metabolites with HA in the hippocampi of Tg2576 mice. The Pearson correlation coefficients were calculated to investigate the linear relationships between metabolites and HA in the hippocampi of Tg2576 mice (Supplementary Table 7). A disproportionate higher number of lipids were positively correlated with HA (64% or 72 of 112 positively correlated; sign test p = 0.003). Figure 3A visualizes a Pearson correlation network analysis between the 50 most important metabolites selected by RF and VIP scores with HA in the hippocampi of Tg2576 mice. Figure 3B shows the Pearson correlation heatmap related to 50 lipids having the same association with HA compared to association among Tg2576 versus WT mice brain tissue. The network analysis using hierarchical clustering reveals a grouping of metabolites based on biochemical metabolite classes and pathways, grouping separately phospholipids, ceramides, fatty acids & modified fatty acids, amino acids, and nucleic acids. Whereas 7 out of 9 fatty acids showed a negative association with both HA and presence hAPP695SW transgene, indicating increased fatty acid oxidation, most phospholipids (21 out of 32) showed a positive association with both HA and presence of hAPP695SW transgene (Fisher’s exact test p = 0.03), supporting the importance of membrane lipids for the assembly of Aβ peptides.
Fig. 2.
The hAPP695SW transgene results in Aβ plaques, the area of which showed a positive correlation with fatty acyl carnitines. A) Immunohistochemistry staining for pan-Aβ of representative coronal sliced brain sections of WT (left) and Tg2576 (right) mice (one brain hemisphere). B) The horizontal bar graph visualizes the association between total Aβ plaque area (HA) (%) and 13 significantly correlated metabolites (at p ≤ 0.05 using Spearman rank correlation coefficients) in hippocampal tissue of Tg2576 mice.
Fig. 3.
A) Pearson correlation network analysis shows correlation of 50 metabolites chosen by RF and PLSDA analysis methods with each other and with total Aβ plaque area (HA) (%). Metabolites are grouped based on HMDB classification. 66 edges show a significant correlation between metabolites and also between metabolites and HA. 26 out of 50 metabolites revealed a negative correlation with HA which CYTOSINE and PC (24:1/18:4) showed a significant negative correlation with HA. Yellow nodes represent glycerophospholipids, vivid cyan represent sphingolipids, purple represent fatty acyls, orange represents carboxylic acids and derivatives, light green represent organonitrogen compounds, light blue represent glycerolipids, grayish cyan represents purine nucleotide and purine nucleoside. The positive and negative correlations are indicated by red and blue edges, respectively. Edge thickness indicates the magnitude of correlation. Metabolites with weak correlations (−0.6 < r < +0.6) are removed. B) Pearson’s correlation coefficient heatmap of 50 metabolites showed the same direction of correlation with HA in the hippocampus of Tg2576 mice and hAPP695SW transgene effect. 30 out of 50 metabolites revealed a positive correlation with HA in the hippocampus of Tg2576 mice and hAPP695SW transgene effect. Positive correlations are shown in red and negative correlations are shown in blue. Darker color indicates a stronger correlation. Metabolites are ordered through hierarchical clustering. The values in the squares are the absolute value of the correlation r.
Biochemical pathway analysis
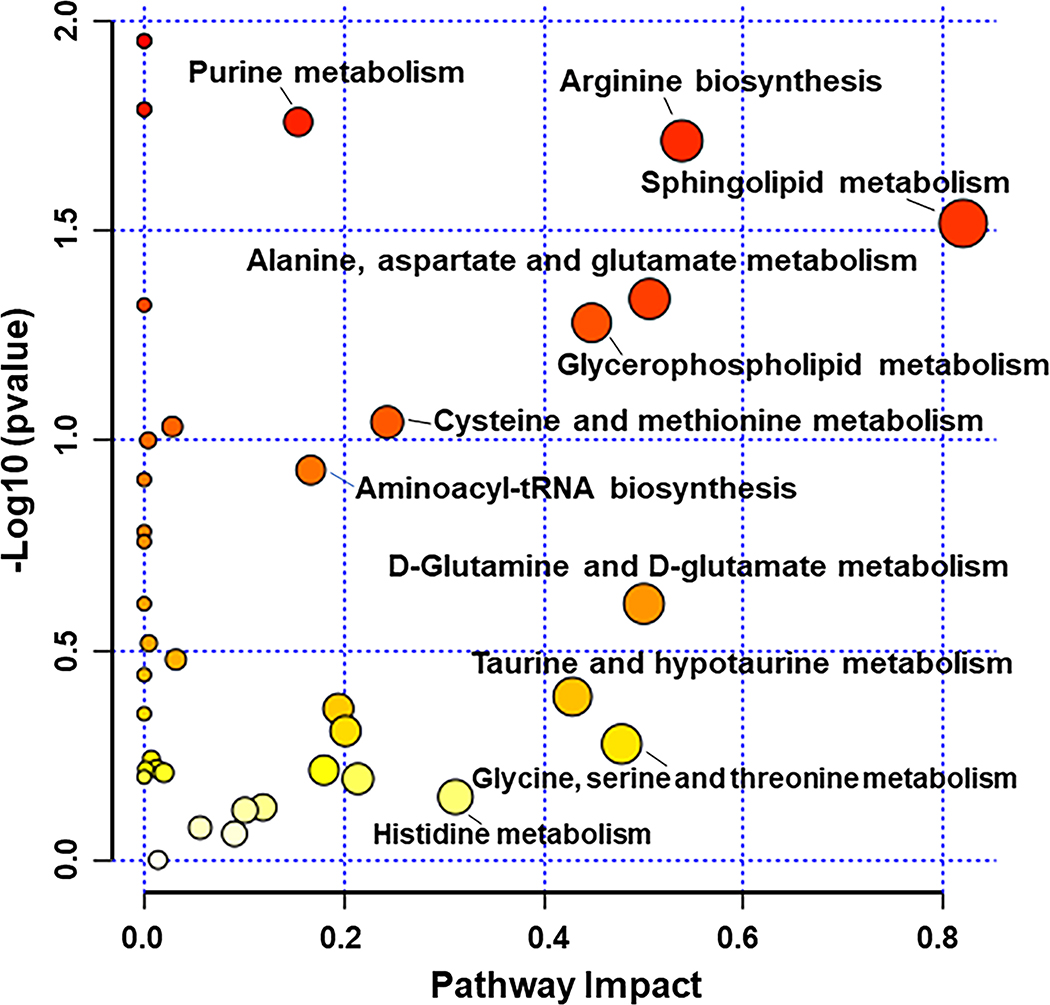
Metabolomic Pathway Analysis (MetPA) was applied to 107 metabolites that had both HMDB IDs and KEGG IDs (Supplementary Table 8). Genotype differences are graphed (Fig. 4) by p-values based on quantitative enrichment analysis and impact values based on pathway topology analysis [36–39]. Sphingolipid metabolism (p = 0.03, impact = 0.82), arginine biosynthesis (p = 0.02, impact = 0.54), metabolism of ALANINE, aspartate, and glutamate (p = 0.05, impact = 0.51), glycerophospholipid metabolism (p = 0.05, impact = 0.45), and purine metabolism (p = 0.02, impact = 0.15), differed at p < 0.05 between Tg2576 and WT hippocampal tissue. However, none of these pathways were FDR p < 0.05, and for none of these pathways the majority of metabolites differed at p < 0.10 in univariate analysis. In the following sections, we describe the different metabolic pathways impacted by hAPP695SW transgene in 20-month-old female mice more comprehensively.
Fig. 4.
Metabolomic pathway analysis (MetPA) for 107 metabolites with annotated HMDB IDs and KEGG IDs. X and Y axes represent pathway impact values and − log10 (p values), respectively. The node color (white to red) is based on the node’s p-value with red indicating significance, and the pathway impact values define the node size.
Amino acid/peptide/amine metabolism
We identified 39 amino acids, peptides, amines, and amino acid metabolites, of which 12 (34%; ten higher and two lower in Tg2576 versus WT hippocampal tissue) differed at p < 0.05 between genotypes (Supplementary Table 9). Metabolite changes in Tg2576 hippocampal tissue suggest 1) increased deamination as indicated by accumulation of urea (p = 0.00003) and 2) upregulation of biochemical pathways linked to the metabolism of acidic (6 of 9 metabolites higher at p < 0.05) and sulfur amino acids (5 of 9 metabolites higher at p < 0.05).
Nucleic acid metabolism
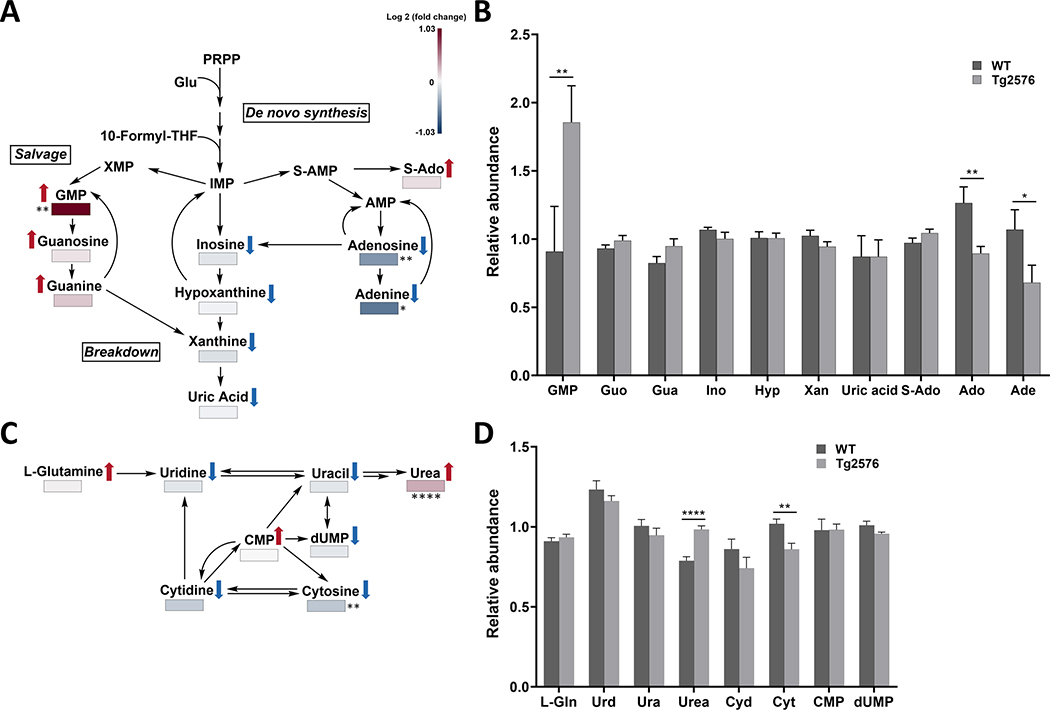
We identified 24 purines and pyrimidines, as well as their precursors and catabolites, of which 8 (33%; four higher and four lower) differed at p < 0.05 between genotypes (Supplementary Table 10). The purine and pyrimidine metabolic pathways as well the relative abundance among metabolites involved in purine and pyrimidine pathways are shown in Fig. 5A and 5C. Excluding precursors and catabolites, five of ten purines (50%; 3 higher and 2 lower) and three of seven pyrimidines (43%; one higher and 2 lower) differed between genotypes at p < 0.10. Metabolite changes in Tg2576 hippocampal tissue suggest a shift in nucleic acid metabolism toward more guanine-derived purines (all three higher), methylated or succinylated nucleic acids (all four higher; also CDP-choline (Supplementary Table 11) was higher), and less other nucleic acids and their catabolites (11 of 12 lower). For the Tg2576 to WT hippocampi ratio level, stronger ratios were observed for nucleotides versus nucleoside and even more versus their respective nitrogenous bases (Fig. 5B, D), indicating lower availability of nucleotide precursors.
Fig. 5.
The hAPP695SW transgene affects purine and pyrimidine metabolism. A, C) Purine and pyrimidine metabolic pathways with metabolites annotated that showed level changes comparing hippocampal extracts of Tg2576 versus WT mice. The metabolites are color-coded from negative log 2 (fold change) to positive log 2 (fold change) on a gradient from blue to red, respectively ( ,
,  the direction of change comparing Tg2576 versus WT mice). B, D) Bar graphs showing changes in relative abundance values (obtained from the mass spectrum) for metabolites linked to the (B) purine pathway and (D) pyrimidine pathway. Data are expressed as mean values ± standard error of the mean. p-values are calculated using Wilcoxon rank-sum test as a non-parametric test (*p < 0.05, **p < 0.01, ***p < 0.001). Double arrows indicate the involvement of other intermediates in the pathway. GMP, Guanosine monophosphate; Guo, Guanosine; Gua, GUANINE; Ino, INOSINE; Hyp, HYPOXANTHINE; Xan, Xanthine; S-Ado, Succinyladenosine; Ado, ADENOSINE; Ade, ADENINE; L-Gln, L-Glutamine; Urd, Uridine; Ura, Uracil; Cyd, CYTIDINE; Cyt, CYTOSINE; CMP, Cytidine monophosphate; dUPM, Deoxyuridine monophosphate
the direction of change comparing Tg2576 versus WT mice). B, D) Bar graphs showing changes in relative abundance values (obtained from the mass spectrum) for metabolites linked to the (B) purine pathway and (D) pyrimidine pathway. Data are expressed as mean values ± standard error of the mean. p-values are calculated using Wilcoxon rank-sum test as a non-parametric test (*p < 0.05, **p < 0.01, ***p < 0.001). Double arrows indicate the involvement of other intermediates in the pathway. GMP, Guanosine monophosphate; Guo, Guanosine; Gua, GUANINE; Ino, INOSINE; Hyp, HYPOXANTHINE; Xan, Xanthine; S-Ado, Succinyladenosine; Ado, ADENOSINE; Ade, ADENINE; L-Gln, L-Glutamine; Urd, Uridine; Ura, Uracil; Cyd, CYTIDINE; Cyt, CYTOSINE; CMP, Cytidine monophosphate; dUPM, Deoxyuridine monophosphate
Metabolites linked to fatty acid oxidation and mitochondrial/peroxisomal function
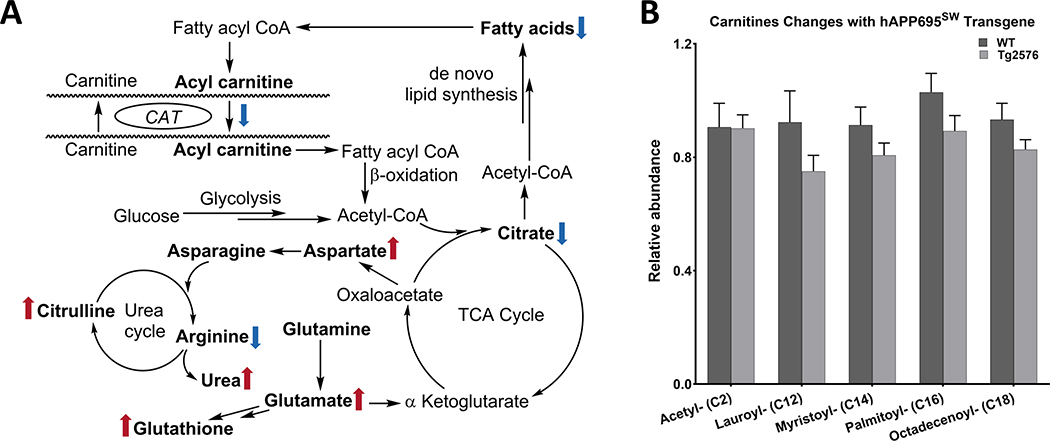
We have previously reported mitochondrial dysfunction in hippocampal neurons isolated from Tg2576 embryos [13]. Here we identified 23 fatty acid (FA) and FA derivatives that are linked to FA oxidation and mitochondrial/peroxisomal function, of which 7 (30%; 1 higher and 6 lower) differed at p < 0.05 between genotypes (Supplementary Table 12). Metabolite changes in Tg2576 hippocampal tissue suggest increased FA oxidation and less FA availability (19 of 23 lower; Supplementary Table 12). This trend was more pronounced when excluding short- and medium-chain FA (16 of 17 lower), which result from incomplete β-oxidation. Specifically, C18-containing FA and derivatives were lower (six of eight (75%) C18-containing FA were lower with p < 0.10). The group of FA derivatives that were most strongly impacted were FA esters, FA amides, and FA aldehydes, of which five of six (83%) were lower at p < 0.10. Also, all fatty acyl carnitines linked to FA metabolism were lower in Tg2576 versus WT hippocampal tissue (Fig. 6A, B); in contrast, propionyl carnitine, which is linked to amino acid catabolism, was higher.
Fig. 6.
Metabolites are linked to fatty acid oxidation and mitochondrial/peroxisomal function. A) The figure represents the pathway in which Fatty acid oxidation, mitochondrial function, and urea cycle are involved ( ,
,  the direction of change comparing Tg2576 versus WT mice). Double arrows indicate the involvement of other intermediates in the pathway. B) The hAPP695SW transgene is linked to increased FA oxidation, as indicated by consistently less medium-chain and long-chain fatty acyl carnitines (obtained from the mass spectrum) in Tg2576 mice.
the direction of change comparing Tg2576 versus WT mice). Double arrows indicate the involvement of other intermediates in the pathway. B) The hAPP695SW transgene is linked to increased FA oxidation, as indicated by consistently less medium-chain and long-chain fatty acyl carnitines (obtained from the mass spectrum) in Tg2576 mice.
Metabolites associated with maintaining redox homeostasis
We examined pathways linked to redox homeostasis and identified 26 metabolites, of which 15 (58%; 11 higher and 4 lower) differed at p < 0.05 between genotypes (Supplementary Table 13). Metabolite changes in Tg2576 hippocampal tissue suggest upregulation of antioxidant pathways, most likely in response to elevated OS. Specifically, all three metabolites associated with the glutathione system and four of five plasmalogens were higher at p < 0.05.
Metabolites linked to neurotransmission and signaling
We identified 33 metabolites that are linked to neurosignaling, of which 12 (36%; seven higher and five lower) differed at p < 0.05 between genotypes (Table 1 and Supplementary Table 11). Metabolite changes in Tg2576 hippocampal tissue point to increased levels of neuro-excitatory metabolites (acetylcholine; GLUTAMIC ACID; N-ACETYL-L-ASPARTIC ACID were higher at p < 0.05; no effect on METHACHOLINE, 5-hydroxy indolic acid, CYSTEINE) and decreased levels neuro-inhibitory metabolites (ADENOSINE; oleamide; 5-AMINOPENTONOATE were lower at p < 0.05; no effect on guanosine, GABA, and TAURINE). Both FA amides, oleamide and octadecanamide, were lower in Tg2576 hippocampal tissue (only oleamide at p < 0.05).
Table 1.
The hAPP695SW transgene results in imbalanced neuro-excitatory and neuro-inhibitory metabolites in Tg2576 versus WT hippocampal tissue
| Neuro-excitatory metabolites | Log 2 (fold change) | p | Neuro-inhibitory metabolites | Log 2 (fold change) | p |
|---|---|---|---|---|---|
|
| |||||
| Acetylcholine | 0.22 | 0.023 | ADENOSINE | −0.5 | 0.0073 |
| GLUTAMIC ACID | 0.048 | 0.018 | Oleamide | −0.84 | 0.0056 |
| N-ACETYL-L-ASPARTIC ACID | 0.29 | 0.034 | 5-AMINOPENTANOATE | −0.26 | 0.018 |
| METHACHOLINE | −0.64 | 0.35 | Guanosine | 0.088 | 0.42 |
| 5-hydroxyindolic acid | −0.0004 | 0.92 | GABA | 0.06 | 0.86 |
| CYSTEINE | 0.18 | 0.42 | TAURINE | 0.08 | 0.19 |
Glycerophospholipid (GPL) metabolism
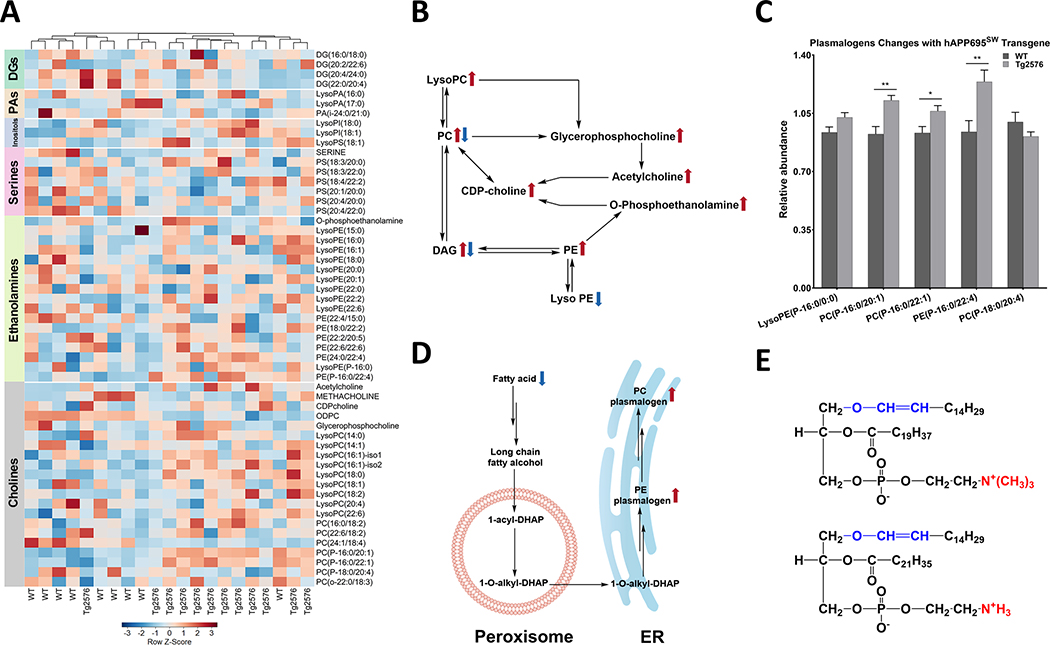
In the present study, we identified 56 metabolites that are linked to glycerophospholipid (GPL) metabolism, of which 18 (32%; 13 higher and 5 lower) differed at p < 0.05 between genotypes (Supplementary Table 14). The heatmap and metabolic pathways of GPLs and diacylglycerols are shown in Fig. 7A and 7B, respectively. Metabolite changes in Tg2576 hippocampal tissue suggest three primary effects (over 50% of metabolites differed) on GPL metabolism: 1) activated peroxisomal plasmalogen synthesis (4 of 5 plasmalogens higher including all C16-containing plasmalogens; Fig. 7C–E), 2) activated choline metabolism (11 of 22 choline-containing metabolites differed at p < 0.05 including 9 higher), and 3) decreased very-long-chain FA availability of Tg2576 versus WT hippocampal tissue (Supplementary Table 14). All seven (lyso)PL that were lower at p < 0.10 contained C20 or longer FA, including three of six lysoPL and three of seven PL containing two C20 or longer FAs. In contrast, six of eight (75%) of C16-containing (lyso)PL were higher at p < 0.10, and seven of 16 (44%) C18-containing (lyso)PL were higher at p < 0.10.
Fig. 7.
Hippocampal glycerophospholipid metabolism is affected in Tg2576 mice. A) Hierarchical clustering analysis of glycosphingolipids is shown as a heatmap which is row Z-score normalized and showing the impact of hAPP695SW transgene. Color bars on the side of the heatmap represent six main classes of metabolites (diacylglycerols (DGs), phosphatidic acids (PAs), inositols, serines, ethanolamines, and cholines). The color-coded gradient is depicted at the bottom. The heatmap is colored based on row Z-scores. Positive Z-score values are shown with red, while negative Z-score values are shown with blue color. B) Glycerophospholipids biosynthesis through the (CDP)-choline pathway. C) Levels of plasmalogens are elevated in the hAPP695SW transgene hippocampi. D) Abbreviated biosynthetic pathway of plasmalogens. E) Structural representation of plasmalogens as ether lipids; representative structures are given: PC(P-16:0/20:1) (upper structure) and PE(P-16:0/22:4). The red color-coded substructure shows the main difference between PC and PE plasmalogens. Metabolites with significant p-value are shown with asterisks which p-values less than 0.001, 0.01, and 0.05 are shown with three (***), two (**), and one asterisks (*), respectively. p-values are calculated using Wilcoxon rank-sum test as a non-parametric test. Double arrows indicate the involvement of other intermediates in the pathway.
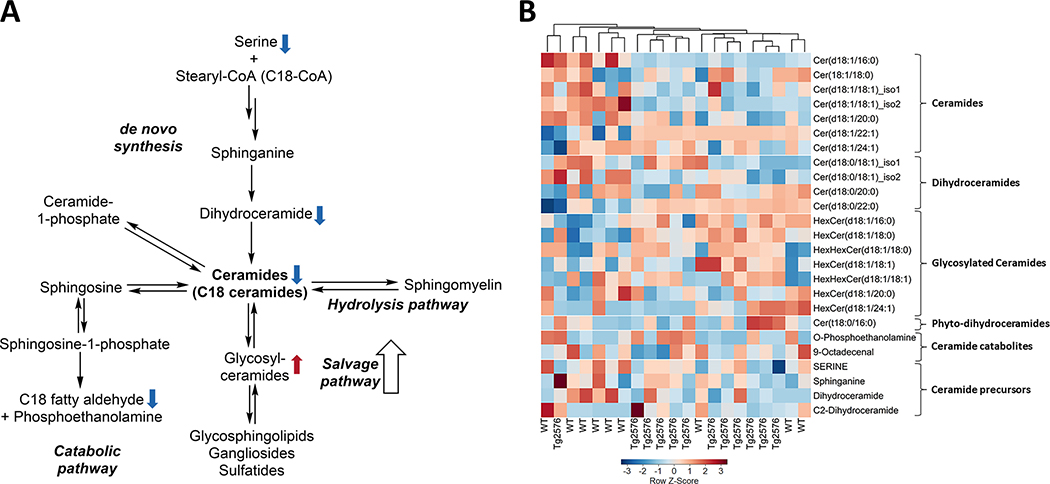
Ceramide metabolism
Our method allowed us to measure ceramides and their metabolites, but not sphingomyelins. We identified 25 metabolites that are linked to ceramide metabolism, of which 7 (28%; three higher and four lower) differed at p < 0.05 between genotypes (Supplementary Table 15). The metabolic pathway, heatmap of ceramides are shown in Fig. 8A and 8B, respectively. Most affected were ceramides and their catabolites; 9 out of 16 metabolites (56%) differed at p < 0.10 between genotypes (Supplementary Table 15). Metabolite changes in Tg2576 hippocampal tissue suggest a shift of ceramide metabolism towards more glycosylated ceramides with a C16 or C18 FA in the acyl position (all 5 higher).
Fig. 8.
Ceramide metabolic pathways and aberrations in Tg2576 hippocampi. A) Fatty acyl ceramides levels were found to be lower in Tg2576 hippocampi whereas glycosylated ceramides were found to be elevated in the Tg2576 hippocampi ( ,
,  the direction of change comparing Tg2576 versus WT mice). B) Hierarchical clustering analysis of ceramides is shown as a heatmap that reveals the impact of hAPP695SW transgene. Values are depicted as Z-scores, with the values color-coded from negative Z-scores to positive Z-scores on a gradient from blue to red, respectively. Double arrows indicate the involvement of other intermediates in the pathway.
the direction of change comparing Tg2576 versus WT mice). B) Hierarchical clustering analysis of ceramides is shown as a heatmap that reveals the impact of hAPP695SW transgene. Values are depicted as Z-scores, with the values color-coded from negative Z-scores to positive Z-scores on a gradient from blue to red, respectively. Double arrows indicate the involvement of other intermediates in the pathway.
DISCUSSION
The present study sought to explore differences between the hippocampal metabolome of Tg2576 mice, a well-established model of AD, and their WT littermates to increase our understanding of the downstream metabolic changes induced by hAPP695SW transgene expression.
The observed metabolite differences in Tg2576 versus WT hippocampal tissue of 20-month-old female mice encompassed several metabolite groups and pathways. The results of this study point to heightened FA metabolism associated with reduced TCA cycle activity, OS, and an excitatory/inhibitory neurosignaling imbalance in Tg2576 hippocampal tissue. The current study supports our earlier work that identified elevated ROS production and neurodegeneration in Tg2576 hippocampal neurons [13, 14].
Metabolite differences, observed in the current study, suggest a heightened metabolic activity, as indicated by depletion of FA and nucleotide precursors. The increase in urea points further toward increased utilization of amino acids as energy precursors. In the brain, ammonia is primarily removed by the glutamate-glutamine cycle; a partial urea cycle primarily removes ammonia via citrulline and arginine. Elevated arginine and polyamine metabolism have been reported for the AD brain [40, 41]. In addition, hypoxia can increase energy needs in the AD brain [41].
We observed in Tg2576 hippocampal tissue less FA and fatty acyl carnitines, the latter transport FA into the mitochondria for FA oxidation. Four of seven carnitines and fatty acyl carnitines increased with Aβ plaque area, indicating increased FA transport and oxidation with Aβ accumulation in Tg2576 mice. Based on our previous research, deficit in mitochondrial bioenergetics is prominent in Tg2576 hippocampal neurons [13], which will further increase energy precursor needs. A study of Tg2576 mouse whole brain metabolome using gas chromatography-mass spectrometry (GC-MS) analysis also observed evidence of altered energy metabolism and mitochondrial dysfunction [9].
Increased FA oxidation increases ROS production, which in turn, activates redox regulatory pathways. Multiple studies show patients diagnosed with AD have higher levels of oxidized biomolecules in frontal and temporal brain sections, including the hippocampus, than normally aging individuals [42]. We observed less methionine and more methionine sulfoxide, the product of methionine oxidation and a potential indicator of OS [43] in Tg2576 versus WT hippocampal tissue, suggestive of a high OS environment within the Tg2576 hippocampus [44, 45]. The increase in ROS production and Aβ activates antioxidant pathways [46–48]. Our previous work reported induction of antioxidant transcription factor Nrf2 expression and target antioxidant response element genes as potential mechanisms for removing ROS [13, 49–52] and improved cognitive function [53–57]. Several metabolites linked to Nrf2 expression differed between Tg2576 versus WT hippocampal tissue. Both FA amides, octadecanamide and oleamide, known to induce Nrf2 expression [58], were lower in Tg2576 versus WT hippocampal tissue, as was vitamin A, known to induce Nrf2 expression and decrease AD risk and outcomes [59]. Four of five annotated plasmalogens, known to be increased by Nrf2 and being less abundant in AD [60–62], were higher in Tg2576 versus WT hippocampal tissue, suggesting a regulated mechanism for removing ROS in Tg2576 hippocampal tissue. Working in conjunction with NRF2-signaling, another mechanism for counteracting ROS is the glutathione system. All three annotated metabolites of the glutathione biosynthesis and degradation pathway (GLUTATHIONE, CySSG, γ-glutamylglutamate) were higher in Tg2576 versus WT hippocampal tissue. In addition, the dipeptide ergothioneine and the non-proteinogenic amino acid homoanserine, both have antioxidant properties [63], were elevated in Tg2576 hippocampi. In addition, Tg2576 hippocampi, cortex and cerebellum and possessed lower levels of common targets of OS including polyunsaturated, very-long chain phospholipids (PC 22:2/20:5 and PC 24:1/18:4) and fatty acids (C20:3 and C22:5) [10, 64].
Glutathione biosynthesis is also linked to excitatory neurosignaling [65]. We observed higher levels of neuro-excitatory metabolites (glutamate, acetylcholine, N-acetyl aspartate) and fewer neuro-inhibitory metabolites (ADENOSINE, oleamide, GABA, 5-AMINOPENTANOATE) in Tg2576 versus WT hippocampal tissue. Similarly, others proposed neuronal hyperexcitability as an early indicator of AD in Tg2576 mice [66]. Glutamate is the primary excitatory neurotransmitter in the brain, binding at metabotropic and ionotropic glutamate receptors including N-methyl D-aspartate (NMDA) receptors [67]. Increased Aβ formation results in NMDA-dependent synaptic depression and spine elimination [67]. We have previously reported reduced spine density in Tg2576 compared to WT mouse hippocampal neurons [13]. The age-related decrease in NMDA receptor binding sites (i.e., NMDA hypofunction) is accelerated with Aβ formation [68]. Suppressed NMDA activity has been linked to excessive release of glutamate in the brain [68], which, in turn, has been linked to excitotoxicity in AD [69].
Suppressed NMDA activity has been linked to excessive release of acetylcholine in the brain [68] and could explain the higher levels of acetylcholine and N-acetyl-aspartate as well as choline- and aspartate-containing metabolites in Tg2576 hippocampal tissue. Similarly, Lalande et al. reported higher levels of taurine in hippocampal tissue of 6- and 11-month-old Tg2576 mice [8]. Suppressed NMDA receptor activity could also explain the higher levels of o-phosphoethanolamine (2-AMINOETHYL DIHYDROGEN PHOSPHATE) in Tg2576 hippocampal tissue [70]. This ties NMDA hypofunction to phospholipid biosynthesis and provides a hypothetical link to the higher levels of choline-containing metabolites that we observed in Tg2576 hippocampal tissue [71].
We observed less neuro-inhibitory metabolites (i.e., ADENOSINE and ADENINE, oleamide, octadecanamide, 5-aminopentonoate, and PHENACYLAMINE) in Tg2576 hippocampal tissues. ADENOSINE deficiency via upregulation of ADENOSINE kinase likely contributes to neuronal death in AD [72]. Adenosine inhibits the release of excitatory acetylcholine and glutamate in the hippocampus via A1 and A2A receptors [73] and can explain the elevated glutamate and acetylcholine in Tg2576 hippocampal tissue. Blockage of those receptors improved cognitive function [73]. The endocannabinoid oleamide may protect against cognitive impairments linked to AD by protecting against excitotoxic insults [74], promoting phagocytosis of Aβ particles [75], and/or inducing choline acetyltransferase activity [76]. Reduced levels of FA amides points to enhanced FA amide hydrolase (FAAH) activity. FAAH plays a critical role in brain function and FAAH inhibitors are promising therapeutics in treatment strategies of AD [77, 78].
We also detected 5-AMINOPENTANOATE, also called 5-aminovaleric acid, which is a lysine degradation product, and which acts as a weak GABA agonist on the receptor level [79]. AD has been linked to dysregulation of GABA (or 4-aminobutanoate), the primary inhibitory neurotransmitter [80]. Whereas GABA levels did not differ (p = 0.87, FDR = 0.91), we did observe in Tg2576 hippocampal tissues elevated levels of the GABA metabolite homoanserine [81]. Moreover, there was a negative correlation between GABA and Aβ plaque area, indicating altered excitation/inhibition balance with Aβ accumulation in Tg2576 mice [82]. Neurotransmitters, such as adenosine, glutamate, GABA, acetylcholine, are expressed by microglia, and microglia-mediated neuroinflammation plays an important role in AD [83, 84]. Phenacylamine is a diet-derived cathinone, which stimulates the release of dopamine and inhibits epinephrine, norepinephrine, and serotonin reuptake, which can result in poor memory [83, 84].
Previous studies reported a link between AD and nucleic acid metabolism, showing the potential of several nucleic acids to serve as early indicators of AD [41]. In the present study, we observed changes in purine and pyrimidine metabolism that are commensurate with previous studies [85, 86]. We observed elevated levels of GMP and other guanosine metabolites (guanosine and GUANINE), whereas ADENOSINE and ADENINE tended to be lower in Tg2576 hippocampal tissue. It is noteworthy that GMP is degraded to guanine by cytosolic 5’nuceotidase II; the inhibition of which induces neuronal death [87]. All pyrimidine metabolism related metabolites tended to be lower in the Tg2576 hippocampal tissue, with CYTOSINE reduction reaching statistical significance.
We also noticed more modified (i.e., methylated, succinylated, CDP-choline) and guanine-derived nucleic acid metabolites in Tg2576 hippocampal tissue. Others reported higher concentrations of methylated nucleic acids in patients with mild-to-moderate AD [88]. Given that DNA methylation alterations are known to play an important role in AD [89], the elevated levels of methylated nucleic acid metabolites in Tg2576 hippocampi indicate a role of OS and oxidative DNA damage in AD.
Disturbances in lipid metabolism in AD have gained more widespread attention in recent years [90–92]. The current study allowed an evaluation of the impact of hAPP695SW expression and Aβ accumulation on glycerophospholipids and sphingolipids. The current study covers mainly phosphatidylcholines (PC), phosphatidylethanolamines (PE), and phosphatidylserine, but did not allow evaluating neutral lipids. The most striking differences, that we observed, were in the levels of LysoPCs and plasmalogens (Fig. 7). LysoPC levels were elevated in Tg2576 hippocampal tissue. Specifically, we observed reduced PUFA content in multiple lysoPLs (specifically those containing choline) in Tg2576 hippocampi. This is in accordance with data showing that Aβ accumulation is associated with OS and neuroinflammation, conditions that trigger enhanced phospholipase A2 activity and as a result, temporal membrane biphasic changes in astrocytes. Previously, increased phospholipase A2 expression was observed in Tg2576 mice, mediating increased neuronal activity and excitotoxic injury [93, 94].
We observed a small number of alkenyl plasmalogens, three choline plasmalogens, and one ethanolamine plasmalogen. The functional characteristics of plasmalogen species are linked to the sn-1 vinyl ether bond and the presence of PUFAs at the sn-2 position. Uniquely compared to other PLs, the biosynthesis of plasmalogens starts in the peroxisomes and is completed in the endoplasmic reticulum. Interestingly, all plasmalogen species were found at higher levels in the Tg2576 than WT hippocampal tissue. We postulate that the observed upregulation of plasmalogens is a mean to counteract an increasingly oxidative environment due to Aβ accumulation [95, 96]. However, others have reported reduced levels of plasmalogens in the AD brain implying enhanced degradation due to susceptibility of the vinyl ether bond to oxidative degradation [97, 98]. Abnormal sphingolipid metabolism has been reported to be associated with brain aging and AD [99]. We observed higher levels of glycosylated ceramides with a C16 or C18 in the acyl position in Tg2576 hippocampal tissue, whereas other ceramides and precursors were lower. Multiple possible mechanisms can be discussed here including enhanced degradation of complex glycosphingolipids and myelin (salvage pathway) and/or defects in the biosynthesis that may lead to elevated levels of glycosylated ceramides. There are two caveats associated with the current study that need to be considered. The current study reflects on the steady-state levels of metabolites, thus no directional information can be drawn. And, secondly, the applied annotation workflow did not allow to delineate different hexose structures. Others have reported that inhibition of glucosylceramide synthesis can decrease aggregation of tau in the hippocampus [100]. Apoptosis is mainly induced by saturated long-chain ceramides (Cer C16:0 and C18:0), whereas very long-chain ceramides (≥C20) may interfere with the pro-apoptotic and ROS-generating effect of saturated long-chain FA [101–103]. Several studies reported a link between accumulation of pro-apoptotic C18:0 ceramides and sphingomyelins in human AD patients, senile plaques, and in normal aging mice [99, 104–107]. Studies in AD model mice that overexpress Aβ but do not show neuronal loss (AβPPSL, AβPPsw, and AβPPV717F) reported no effect of Aβ accumulation on hippocampal ceramide levels [108, 109], which has been explained by the fact that a small rate of hippocampal neuron loss has been observed in these models [110].
There are several strengths and limitations to the current study. The current study covers a wide range of metabolites and lipids, and as such allows evaluating the impact of hAPP695SW transgene and Aβ accumulation on multiple biochemical pathways and thus goes beyond describing disparate individual metabolites in Tg2576 hippocampi. The chosen pathway-centered focus potentially can aid in linking AD-type brain pathologies to broader aspects of AD. The number of animals in the study is small; however, the use of WT littermates as control, the age and sex of animals (i.e., females are at greater risk of AD), and the use of a humanized transgene model (i.e., Tg2576 mouse), which is the most widely studied transgenic AD model [111], make this study an appropriate model for AD. There is limited generalizability with this AD model, as limited neuronal death is observed in the mice and the transgene is indicative of a specific mutation found in Aβ-mediated early-onset AD [5, 107]. Furthermore, there are concerns that the 5-fold APP overexpression in Tg2576 mice results in artificial metabolic phenotypes that are unrelated to human AD pathogenesis; however, the Aβ-afflicted area in the hippocampus was below 1% in all tested Tg2576 mice, making it unlikely to create artificial metabolic phenotypes. Future larger and longitudinal studies in both sexes are warranted to reevaluate our findings in an AD model that overexpresses Aβ species but not APP, such as an APP knock-in model, examine the sequential metabolite changes during different stages of AD, and examine the interplay between different cell types in the hippocampus (e.g., microglia-mediated neuro-inflammation; astrocyte activation). Despite that multiple metabolite groups were covered in our study, all pre-analytical flows have shortcoming and limitations. The applied sample preparation method limits coverage of some metabolite and lipid groups, including cholesterols and triglycerides, which require different extraction methods. Also, during electrospray ionization of complex mixtures ion suppression may occur that hinders detection of some metabolite species, which in this study likely prevented the detection of sphingolipids. Moreover, our metabolomics approach is not suited to examine the impact of the huAPP695SW transgene on neuro-inflammatory pathways, which require proinflammatory biomarkers. However, our method allowed coverage of major metabolite classes and polar lipids, including ceramides of which observations were hitherto absent for the Tg2576 model.
To conclude, in the present study, metabolite differences in Tg2576 versus WT hippocampal tissue of 20-month-old female mice point to heightened metabolic activity, an elevated OS response, and an excitatory and inhibitory imbalance in Tg2576 hippocampal tissue. We observed higher levels of urea, neuro-excitatory, and glutathione metabolites and less FAs, nucleotides, and neuro-inhibitory metabolites (Supplementary Figure 2). We report on disturbances in lipid metabolism that are in accordance with an enhanced OS environment and mitochondrial dysfunction caused by Aβ accumulation. We also discuss several potential pharmacological targets for the observed genotype differences (i.e., phospholipase A2 and FAAH). The current study provides a comprehensive assessment of disturbances in biochemical pathways for a widely used AD rodent model, the Tg2576 mouse.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge NIH grants R01AT008099 (A.S.), S10RR027878 (J.S.), T32 AT002688, and VA Merit Review grant IO1BX003440. The authors acknowledge technical support provided by the Oregon State University’s Mass Spectrometry Center.
Footnotes
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/21-5084r2).
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/jad-000000.
REFERENCES
- [1].Alzheimer’s Disease Fact Sheet, NIH, https://www.nia.nih.gov/health/alzheimers-disease-fact-sheet [Google Scholar]
- [2].Götz J, Bodea L-G, Goedert M (2018) Rodent models for Alzheimer disease. Nat Rev Neurosci 19, 583–598. [DOI] [PubMed] [Google Scholar]
- [3].Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G (1996) Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science 274, 99–103. [DOI] [PubMed] [Google Scholar]
- [4].Sweatt JD (2009) Mechanisms of memory, Academic Press. [Google Scholar]
- [5].Axelman K, Basun H, Winblad B, Lannfelt L (1994) A large Swedish family with Alzheimer’s disease with a codon 670/671 amyloid precursor protein mutation: A clinical and genealogical investigation. Arch Neurol 51, 1193–1197. [DOI] [PubMed] [Google Scholar]
- [6].Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe DJ (1995) The Swedish mutation causes early-onset Alzheimer’s disease by β-secretase cleavage within the secretory pathway. Nat Med 1, 1291–1296. [DOI] [PubMed] [Google Scholar]
- [7].Westerman MA, Cooper-Blacketer D, Mariash A, Kotilinek L, Kawarabayashi T, Younkin LH, Carlson GA, Younkin SG, Ashe KH (2002) The relationship between Aβ and memory in the Tg2576 mouse model of Alzheimer’s disease. J Neurosci 22, 1858–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lalande J, Halley H, Balayssac S, Gilard V, Dejean S, Martino R, Frances B, Lassalle J-M, Malet-Martino M (2014) 1H NMR metabolomic signatures in five brain regions of the AβPPswe Tg2576 mouse model of Alzheimer’s disease at four ages. J Alzheimers Dis 39, 121–143. [DOI] [PubMed] [Google Scholar]
- [9].Trushina E, Nemutlu E, Zhang S, Christensen T, Camp J, Mesa J, Siddiqui A, Tamura Y, Sesaki H, Wengenack TM (2012) Defects in mitochondrial dynamics and metabolomic signatures of evolving energetic stress in mouse models of familial Alzheimer’s disease. PLoS One 7, e32737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bascoul-Colombo C, Guschina IA, Maskrey BH, Good M, O’Donnell VB, Harwood JL (2016) Dietary DHA supplementation causes selective changes in phospholipids from different brain regions in both wild type mice and the Tg2576 mouse model of Alzheimer’s disease. Biochim Biophys Acta 1861, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pike CJ, Cummings BJ, Cotman CW (1992) beta-Amyloid induces neuritic dystrophy in vitro: similarities with Alzheimer pathology. Neuroreport 3, 769–772. [DOI] [PubMed] [Google Scholar]
- [12].Gella A, Durany N (2009) Oxidative stress in Alzheimer disease. Cell Adh Migr 3, 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gray NE, Zweig JA, Matthews DG, Caruso M, Quinn JF, Soumyanath A (2017) Centella asiatica attenuates mitochondrial dysfunction and oxidative stress in Aβ-exposed hippocampal neurons. Oxid Med Cell Longev 2017, 7023091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gray NE, Zweig JA, Murchison C, Caruso M, Matthews DG, Kawamoto C, Harris CJ, Quinn JF, Soumyanath A (2017) Centella asiatica attenuates Aβ-induced neurodegenerative spine loss and dendritic simplification. Neurosci Lett 646, 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Soumyanath A, Zhong Y-P, Henson E, Wadsworth T, Bishop J, Gold BG, Quinn JF (2012) Centella asiatica extract improves behavioral deficits in a mouse model of Alzheimer’s disease: investigation of a possible mechanism of action. Int J Alzheimers Dis 2012, 381974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, Bush WS, Gifford KA, Chibnik LB, Mukherjee S, De Jager PL (2019) Sex differences in the genetic predictors of Alzheimer’s pathology. Brain 142, 2581–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Buckley RF, Mormino EC, Amariglio RE, Properzi MJ, Rabin JS, Lim YY, Papp KV, Jacobs HIL, Burnham S, Hanseeuw BJ, Doré V, Dobson A, Masters CL, Waller M, Rowe CC, Maruff P, Donohue MC, Rentz DM, Kirn D, Hedden T, Chhatwal J, Schultz AP, Johnson KA, Villemagne VL, Sperling RA (2018) Sex, amyloid, and APOE ε4 and risk of cognitive decline in preclinical Alzheimer’s disease: Findings from three well-characterized cohorts. Alzheimers Dement 14, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mazure CM, Swendsen J (2016) Sex differences in Alzheimer’s disease and other dementias. Lancet Neurol 15, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kirkwood JS, Maier C, Stevens JF (2013) Simultaneous, untargeted metabolic profiling of polar and nonpolar metabolites by LC-Q-TOF Mass Spectrometry. Curr Protoc Toxicol Chapter 4, Unit4.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Housley L, Magana AA, Hsu A, Beaver LM, Wong CP, Stevens JF, Choi J, Jiang Y, Bella D, Williams DE (2018) Untargeted metabolomic screen reveals changes in human plasma metabolite profiles following consumption of fresh broccoli sprouts. Mol Nutr Food Res 62, 1700665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Magana AA, Reed RL, Koluda R, Miranda CL, Maier CS, Stevens JF (2020) Vitamin C activates the folate-mediated one-carbon cycle in C2C12 myoblasts. Antioxidants 9, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bading-Taika B, Akinyeke T, Magana AA, Choi J, Ouanesisouk M, Torres ERS, Lione LA, Maier CS, Bobe G, Raber J (2018) Phytochemical characterization of Tabernanthe iboga root bark and its effects on dysfunctional metabolism and cognitive performance in high-fat-fed C57BL/6J mice. J Food Bioact 3, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Viant MR, Kurland IJ, Jones MR, Dunn WB (2017) How close are we to complete annotation of metabolomes? Curr Opin Chem Biol 36, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shen X, Gong X, Cai Y, Guo Y, Tu J, Li H, Zhang T, Wang J, Xue F, Zhu Z-J (2016) Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics 12, 89. [Google Scholar]
- [25].Louppe G, Wehenkel L, Sutera A, Geurts P (2013) Understanding variable importances in forests of randomized trees. Advances in neural information processing systems, pp. 431–439. [Google Scholar]
- [26].Mendez KM, Broadhurst DI, Reinke SN (2020) Migrating from partial least squares discriminant analysis to artificial neural networks: a comparison of functionally equivalent visualisation and feature contribution tools using jupyter notebooks. Metabolomics 16, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR (2007) Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A (2018) HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46, D608–d617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hou Y, He D, Ye L, Wang G, Zheng Q, Hao H (2020) An improved detection and identification strategy for untargeted metabolomics based on UPLC-MS. J Pharm Biomed Anal 191, 113531. [DOI] [PubMed] [Google Scholar]
- [31].Brereton RG (2018) Chemometrics: Data Driven Extraction for Science, John Wiley & Sons. [Google Scholar]
- [32].Khanmohammadi M (2015) Current applications of chemometrics, Nova Publishers. [Google Scholar]
- [33].Shulaev V (2006) Metabolomics technology and bioinformatics. Brief Bioinformatics 7, 128–139. [DOI] [PubMed] [Google Scholar]
- [34].Cui X, Churchill GA (2003) Statistical tests for differential expression in cDNA microarray experiments. Genome Biol 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Breiman L (2001) Random forests. Mach Learn 45, 5–32. [Google Scholar]
- [36].Chong J, Wishart DS, Xia J (2019) Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics 68, e86. [DOI] [PubMed] [Google Scholar]
- [37].Xia J, Wishart DS (2010) MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26, 2342–2344. [DOI] [PubMed] [Google Scholar]
- [38].Xia J, Wishart DS (2010) MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 38, W71–W77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xia J, Wishart DS (2011) Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 6, 743–760. [DOI] [PubMed] [Google Scholar]
- [40].Polis B, Samson AO (2019) A new perspective on Alzheimer’s disease as a brain expression of a complex metabolic disorder. In Alzheimer’s Disease [Internet], Wisniewski T, ed. Codon Publications, Brisbane. [PubMed] [Google Scholar]
- [41].Badhwar A, McFall GP, Sapkota S, Black SE, Chertkow H, Duchesne S, Masellis M, Li L, Dixon RA, Bellec P (2020) A multiomics approach to heterogeneity in Alzheimer’s disease: focused review and roadmap. Brain 143, 1315–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang X, Wang W, Li L, Perry G, Lee H-g, Zhu X (2014) Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim Biophys Acta 1842, 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Suzuki S, Kodera Y, Saito T, Fujimoto K, Momozono A, Hayashi A, Kamata Y, Shichiri M (2016) Methionine sulfoxides in serum proteins as potential clinical biomarkers of oxidative stress. Sci Rep 6, 38299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Arun P, Rittase WB, Wilder DM, Wang Y, Gist ID, Long JB (2018) Defective methionine metabolism in the brain after repeated blast exposures might contribute to increased oxidative stress. Neurochem Int 112, 234–238. [DOI] [PubMed] [Google Scholar]
- [45].Subash S, Essa MM, Al-Asmi A, Al-Adawi S, Vaishnav R, Guillemin GJ (2015) Effect of dietary supplementation of dates in Alzheimer’s disease APPsw/2576 transgenic mice on oxidative stress and antioxidant status. Nutr Neurosci 18, 281–288. [DOI] [PubMed] [Google Scholar]
- [46].Castellani RJ, Lee H-g, Perry G, Smith MA(2006) Antioxidant protection and neurodegenerative disease: The role of amyloid-β and tau. Am J Alzheimers Dis Other Demen 21, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kontush A (2001) Alzheimer’s amyloid-β as a preventive antioxidant for brain lipoproteins. Cell Mol Neurobiol 21, 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ye B, Shen H, Zhang J, Zhu YG, Ransom BR, Chen XC, Ye ZC (2015) Dual pathways mediate β-amyloid stimulated glutathione release from astrocytes. Glia 63, 2208–2219. [DOI] [PubMed] [Google Scholar]
- [49].Gray NE, Harris CJ, Quinn JF, Soumyanath A (2016) Centella asiatica modulates antioxidant and mitochondrial pathways and improves cognitive function in mice. J Ethnopharmacol 180, 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gray NE, Sampath H, Zweig JA, Quinn JF, Soumyanath A (2015) Centella asiatica attenuates amyloid-β-induced oxidative stress and mitochondrial dysfunction. J Alzheimers Dis 45, 933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gray NE, Zweig JA, Caruso M, Zhu JY, Wright KM, Quinn JF, Soumyanath A (2018) Centella asiatica attenuates hippocampal mitochondrial dysfunction and improves memory and executive function in β-amyloid overexpressing mice. Mol Cell Neurosci 93, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Matthews DG, Caruso M, Murchison CF, Zhu JY, Wright KM, Harris CJ, Gray NE, Quinn JF, Soumyanath A (2019) Centella asiatica improves memory and promotes antioxidative signaling in 5XFAD mice. Antioxidants 8, 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kwon D-H, Kim B-S, Chang H, Kim Y-I, Jo SA, Leem Y-H (2013) Exercise ameliorates cognition impairment due to restraint stress-induced oxidative insult and reduced BDNF level. Biochem Biophys Res Commun 434, 245–251. [DOI] [PubMed] [Google Scholar]
- [54].Logan S, Royce GH, Owen D, Farley J, Ranjo-Bishop M, Sonntag WE, Deepa SS (2019) Accelerated decline in cognition in a mouse model of increased oxidative stress. GeroScience 41, 591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nagai T, Yamada K, Kim HC, Kim YS, Noda Y, Imura A, Nabeshima Yi, Nabeshima T (2003) Cognition impairment in the genetic model of aging klotho gene mutant mice: a role of oxidative stress. FASEB J 17, 50–52. [DOI] [PubMed] [Google Scholar]
- [56].Nichol KE, Poon WW, Parachikova AI, Cribbs DH, Glabe CG, Cotman CW (2008) Exercise alters the immune profile in Tg2576 Alzheimer mice toward a response coincident with improved cognitive performance and decreased amyloid. J Neuroinflammation 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tarantini S, Valcarcel-Ares NM, Yabluchanskiy A, Fulop GA, Hertelendy P, Gautam T, Farkas E, Perz A, Rabinovitch PS, Sonntag WE (2018) Treatment with the mitochondrial-targeted antioxidant peptide SS-31 rescues neurovascular coupling responses and cerebrovascular endothelial function and improves cognition in aged mice. Aging Cell 17, e12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Alapafuja SO, Nikas SP, Bharathan IT, Shukla VG, Nasr ML, Bowman AL, Zvonok N, Li J, Shi X, Engen JR (2012) Sulfonyl fluoride inhibitors of fatty acid amide hydrolase. J Med Chem 55, 10074–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Honarvar NM, Saedisomeolia A, Abdolahi M, Shayeganrad A, Sangsari GT, Rad BH, Muench G (2017) Molecular anti-inflammatory mechanisms of retinoids and carotenoids in Alzheimer’s disease: A review of current evidence. J Mol Neurosci 61, 289–304. [DOI] [PubMed] [Google Scholar]
- [60].Wood PL, Locke VA, Herling P, Passaro A, Vigna GB, Volpato S, Valacchi G, Cervellati C, Zuliani G (2016) Targeted lipidomics distinguishes patient subgroups in mild cognitive impairment (MCI) and late onset Alzheimer’s disease (LOAD). BBA Clin 5, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Goodenowe DB, Cook LL, Liu J, Lu Y, Jayasinghe DA, Ahiahonu PW, Heath D, Yamazaki Y, Flax J, Krenitsky KF (2007) Peripheral ethanolamine plasmalogen deficiency: a logical causative factor in Alzheimer’s disease and dementia. J Lipid Res 48, 2485–2498. [DOI] [PubMed] [Google Scholar]
- [62].Uruno A, Matsumaru D, Ryoke R, Saito R, Kadoguchi S, Saigusa D, Saito T, Saido TC, Kawashima R, Yamamoto M (2020) Nrf2 suppresses oxidative stress and inflammation in App knock-in Alzheimer’s disease model mice. Mol Cell Biol 40, e00467–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nakamichi N, Nakayama K, Ishimoto T, Masuo Y, Wakayama T, Sekiguchi H, Sutoh K, Usumi K, Iseki S, Kato Y (2016) Food-derived hydrophilic antioxidant ergothioneine is distributed to the brain and exerts antidepressant effect in mice. Brain Behav 6, e00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ayala A, Muñoz MF, Argüelles S (2014) Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev 2014, 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Baxter PS, Bell KF, Hasel P, Kaindl AM, Fricker M, Thomson D, Cregan SP, Gillingwater TH, Hardingham GE (2015) Synaptic NMDA receptor activity is coupled to the transcriptional control of the glutathione system. Nat Commun 6, 6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kam K, Duffy ÁM, Moretto J, LaFrancois JJ, Scharfman HE (2016) Interictal spikes during sleep are an early defect in the Tg2576 mouse model of β-amyloid neuropathology. Sci Rep 6, 20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liu J, Chang L, Song Y, Li H, Wu Y (2019) The role of NMDA receptors in Alzheimer’s disease. Front Neurosci 13, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Newcomer JW, Farber NB, Olney JW (2000) NMDA receptor function, memory, and brain aging. Dialogues Clin Neurosci 2, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang R, Reddy PH (2017) Role of glutamate and NMDA receptors in Alzheimer’s disease. J Alzheimers Dis 57, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Robert F, Bert L, Stoppini L (2002) Blockade of NMDA-receptors or calcium-channels attenuates the ischaemia-evoked efflux of glutamate and phosphoethanolamine and depression of neuronal activity in rat organotypic hippocampal slice cultures. C R Biol 325, 495–504. [DOI] [PubMed] [Google Scholar]
- [71].Gasull T, Sarri E, DeGregorio-Rocasolano N, Trullas R (2003) NMDA receptor overactivation inhibits phospholipid synthesis by decreasing choline–ethanolamine phosphotransferase activity. J Neurosci 23, 4100–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Boison D (2011) Modulators of nucleoside metabolism in the therapy of brain diseases. Curr Top Med Chem 11, 1068–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rahman A (2009) The role of adenosine in Alzheimer’s disease. Curr Neuropharmacol 7, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Maya-López M, Rubio-López LC, Rodríguez-Alvarez IV, Orduño-Piceno J, Flores-Valdivia Y, Colonnello A, Rangel-López E, Túnez I, Prospéro-García O, Santamaría A (2020) A cannabinoid receptor-mediated mechanism participates in the neuroprotective effects of oleamide against excitotoxic damage in rat brain synaptosomes and cortical slices. Neurotox Res 37, 126–135. [DOI] [PubMed] [Google Scholar]
- [75].Ano Y, Ozawa M, Kutsukake T, Sugiyama S, Uchida K, Yoshida A, Nakayama H (2015) Preventive effects of a fermented dairy product against Alzheimer’s disease and identification of a novel oleamide with enhanced microglial phagocytosis and anti-inflammatory activity. PLoS One 10, e0118512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Heo H-J, Park Y-J, Suh Y-M, Choi S-J, Kim M-J, Cho H-Y, Chang Y-J, Hong B, Kim H-K, Kim E (2003) Effects of oleamide on choline acetyltransferase and cognitive activities. Biosci Biotechnol Biochem 67, 1284–1291. [DOI] [PubMed] [Google Scholar]
- [77].Brunetti L, Laghezza A, Loiodice F, Tortorella P, Piemontese L (2020) Combining fatty acid amide hydrolase (FAAH) inhibition with peroxisome proliferator-activated receptor (PPAR) activation: a new potential multi-target therapeutic strategy for the treatment of Alzheimer’s disease. Neural Regen Res 15, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Oliveira TG, Di Paolo G (2010) Phospholipase D in brain function and Alzheimer’s disease. Biochim Biophys Acta 1801, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Luzzi S, Maggi C, Spagnesi S, Santicioli P, Zilletti L (1985) 5-Aminovaleric acid interactions with GABAA and GABAB receptors in guinea-pig ileum. J Auton Pharmacol 5, 65–69. [DOI] [PubMed] [Google Scholar]
- [80].Giovannetti EA, Fuhrmann M (2019) Unsupervised excitation: GABAergic dysfunctions in Alzheimer’s disease. Brain Res 1707, 216–226. [DOI] [PubMed] [Google Scholar]
- [81].Bauer K, Hallermayer K, Salnikow J, Kleinkauf H, Hamprecht B (1982) Biosynthesis of carnosine and related peptides by glial cells in primary culture. J Biol Chem 257, 3593–3597. [PubMed] [Google Scholar]
- [82].Vyas Y, Montgomery JM, Cheyne JE (2020) Hippocampal deficits in amyloid-β-related rodent models of Alzheimer’s disease. Front Neurosci 14, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Blough BE, Decker AM, Landavazo A, Namjoshi OA, Partilla JS, Baumann MH, Rothman RB (2019) The dopamine, serotonin and norepinephrine releasing activities of a series of methcathinone analogs in male rat brain synaptosomes. Psychopharmacology (Berl) 236, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Leyrer-Jackson JM, Nagy EK, Olive MF (2019) Cognitive deficits and neurotoxicity induced by synthetic cathinones: is there a role for neuroinflammation? Psychopharmacology (Berl) 236, 1079–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ansoleaga B, Jové M, Schlüter A, Garcia-Esparcia P, Moreno J, Pujol A, Pamplona R, Portero-Otín M, Ferrer I (2015) Deregulation of purine metabolism in Alzheimer’s disease. Neurobiol Aging 36, 68–80. [DOI] [PubMed] [Google Scholar]
- [86].Dejakaisaya H, Harutyunyan A, Kwan P, Jones NC (2021) Altered metabolic pathways in a transgenic mouse model suggest mechanistic role of amyloid precursor protein overexpression in Alzheimer’s disease. Metabolomics 17, 1–12. [DOI] [PubMed] [Google Scholar]
- [87].Careddu MG, Allegrini S, Pesi R, Camici M, Garcia-Gil M, Tozzi MG (2008) Knockdown of cytosolic 5′-nucleotidase II (cN-II) reveals that its activity is essential for survival in astrocytoma cells. Biochim Biophys Acta 1783, 1529–1535. [DOI] [PubMed] [Google Scholar]
- [88].Lee SH, Kim I, Chung BC (2007) Increased urinary level of oxidized nucleosides in patients with mild-to-moderate Alzheimer’s disease. Clin Biochem 40, 936–938. [DOI] [PubMed] [Google Scholar]
- [89].Yokoyama AS, Rutledge JC, Medici V (2017) DNA methylation alterations in Alzheimer’s disease. Environ Epigenet 3, dvx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wong MW, Braidy N, Poljak A, Pickford R, Thambisetty M, Sachdev PS (2017) Dysregulation of lipids in Alzheimer’s disease and their role as potential biomarkers. Alzheimers Dement 13, 810–827. [DOI] [PubMed] [Google Scholar]
- [91].Gao H-L, Zhang A-H, Yu J-B, Sun H, Kong L, Wang X-Q, Yan G-l, Liu L, Wang X-J(2018) High-throughput lipidomics characterize key lipid molecules as potential therapeutic targets of Kaixinsan protects against Alzheimer’s disease in APP/PS1 transgenic mice. J Chromatogr B 1092, 286–295. [DOI] [PubMed] [Google Scholar]
- [92].Castellanos DB, Martín-Jiménez CA, Rojas-Rodríguez F, Barreto G, Santos JG (2021) Brain lipidomics as a rising field in neurodegenerative contexts: Perspectives with Machine Learning approaches. Front Neuroendocrinol 61, 100899. [DOI] [PubMed] [Google Scholar]
- [93].Sanchez-Mejia RO, Mucke L (2010) Phospholipase A2 and arachidonic acid in Alzheimer’s disease. Biochim Biophys Acta 1801, 784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hicks JB, Lai Y, Sheng W, Yang X, Zhu D, Sun GY, Lee JC (2008) Amyloid-β peptide induces temporal membrane biphasic changes in astrocytes through cytosolic phospholipase A2. Biochim Biophys Acta 1778, 2512–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Paul S, Lancaster GI, Meikle PJ (2019) Plasmalogens: A potential therapeutic target for neurodegenerative and cardiometabolic disease. Prog Lipid Res 74, 186–195. [DOI] [PubMed] [Google Scholar]
- [96].Kuczynski B, Reo NV (2006) Evidence that plasmalogen is protective against oxidative stress in the rat brain. Neurochem Res 31, 639–656. [DOI] [PubMed] [Google Scholar]
- [97].Gorgas K, Teigler A, Komljenovic D, Just WW (2006) The ether lipid-deficient mouse: tracking down plasmalogen functions. Biochim Biophys Acta 1763, 1511–1526. [DOI] [PubMed] [Google Scholar]
- [98].Su XQ, Wang J, Sinclair AJ (2019) Plasmalogens and Alzheimer’s disease: A review. Lipids Health Dis 18, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP (2004) Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc Natl Acad Sci U S A 101, 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Sardi SP, Viel C, Clarke J, Treleaven CM, Richards AM, Park H, Olszewski MA, Dodge JC, Marshall J, Makino E (2017) Glucosylceramide synthase inhibition alleviates aberrations in synucleinopathy models. Proc Natl Acad Sci U S A 114, 2699–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Stiban J, Perera M (2015) Very long chain ceramides interfere with C16-ceramide-induced channel formation: a plausible mechanism for regulating the initiation of intrinsic apoptosis. Biochim Biophys Acta 1848, 561–567. [DOI] [PubMed] [Google Scholar]
- [102].Zigdon H, Kogot-Levin A, Park J-W, Goldschmidt R, Kelly S, Merrill AH, Scherz A, Pewzner-Jung Y, Saada A, Futerman AH (2013) Ablation of ceramide synthase 2 causes chronic oxidative stress due to disruption of the mitochondrial respiratory chain. J Biol Chem 288, 4947–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Couttas TA, Kain N, Suchowerska AK, Quek L-E, Turner N, Fath T, Garner B, Don AS (2016) Loss of ceramide synthase 2 activity, necessary for myelin biosynthesis, precedes tau pathology in the cortical pathogenesis of Alzheimer’s disease. Neurobiol Aging 43, 89–100. [DOI] [PubMed] [Google Scholar]
- [104].Grösch S, Schiffmann S, Geisslinger G (2012) Chain length-specific properties of ceramides. Prog Lipid Res 51, 50–62. [DOI] [PubMed] [Google Scholar]
- [105].Panchal M, Gaudin M, Lazar AN, Salvati E, Rivals I, Ayciriex S, Dauphinot L, Dargère D, Auzeil N, Masserini M (2014) Ceramides and sphingomyelinases in senile plaques. Neurobiol Dis 65, 193–201. [DOI] [PubMed] [Google Scholar]
- [106].Czubowicz K, Jęśko H, Wencel P, Lukiw WJ, Strosznajder RP (2019) The role of ceramide and sphingosine-1-phosphate in Alzheimer’s disease and other neurodegenerative disorders. Mol Neurobiol 56, 5436–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Koal T, Klavins K, Seppi D, Kemmler G, Humpel C (2015) Sphingomyelin SM (d18: 1/18: 0) is significantly enhanced in cerebrospinal fluid samples dichotomized by pathological amyloid-β 42, tau, and phospho-tau-181 levels. J Alzheimers Dis 44, 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Barrier L, Fauconneau B, Noël A, Ingrand S (2011) Ceramide and related-sphingolipid levels are not altered in disease-associated brain regions of APPSL and APPSL/PS1M146L mouse models of Alzheimer’s disease: relationship with the lack of neurodegeneration? Int J Alzheimers Dis 2011, 920958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Cheng H, Zhou Y, Holtzman DM, Han X (2010) Apolipoprotein E mediates sulfatide depletion in animal models of Alzheimer’s disease. Neurobiol Aging 31, 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Irizarry MC, McNamara M, Fedorchak K, Hsiao K, Hyman BT (1997) APPSw transgenic mice develop age-related Aβ deposits and neuropil abnormalities, but no neuronal loss in CA1. J Neuropathol Exp Neurol 56, 965–973. [DOI] [PubMed] [Google Scholar]
- [111].Elder GA, Gama Sosa MA, De Gasperi R (2010) Transgenic mouse models of Alzheimer’s disease. Mt Sinai J Med 77, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.