ABSTRACT
Acinetobacter baumannii is a formidable opportunistic pathogen that is notoriously difficult to eradicate from hospital settings. This resilience is often attributed to a proclivity for biofilm formation, which facilitates a higher tolerance toward external stress, desiccation, and antimicrobials. Despite this, little is known regarding the mechanisms orchestrating A. baumannii biofilm formation. Here, we performed RNA sequencing (RNA-seq) on biofilm and planktonic populations for the multidrug-resistant isolate AB5075 and identified 438 genes with altered expression. To assess the potential role of genes upregulated within biofilms, we tested the biofilm-forming capacity of their respective mutants from an A. baumannii transposon library. In so doing, we uncovered 24 genes whose disruption led to reduced biofilm formation. One such element, cold shock protein C (cspC), had a highly mucoid colony phenotype, enhanced tolerance to polysaccharide degradation, altered antibiotic tolerance, and diminished adherence to abiotic surfaces. RNA-seq of the cspC mutant revealed 201 genes with altered expression, including the downregulation of pili and fimbria genes and the upregulation of multidrug efflux pumps. Using transcriptional arrest assays, it appears that CspC mediates its effects, at least in part, through RNA chaperone activity, influencing the half-life of several important transcripts. Finally, we show that CspC is required for survival during challenge by the human immune system and is key for A. baumannii dissemination and/or colonization during systemic infection. Collectively, our work identifies a cadre of new biofilm-associated genes within A. baumannii and provides unique insight into the global regulatory network of this emerging human pathogen.
KEYWORDS: A. baumannii, Csp, biofilms
INTRODUCTION
Acinetobacter baumannii is a formidable pathogen that causes >8,000 cases of multidrug-resistant infections annually in the United States and is recognized globally as an urgent health care threat (1, 2). Fundamental to the success of this pathogen is its ability to persist on surfaces for prolonged periods (3–6), its capacity to rapidly spread via health care personnel despite conventional prevention measures (7–9), and its genomic plasticity, driving rapid acquisition of antibiotic resistance (10). Thus, it has been suggested that A. baumannii employs a “persist and resist” strategy rather than relying on classic virulence factors that may otherwise dictate an organism’s potential for virulence (reviewed in reference 11).
Like many bacteria, the resilience of A. baumannii can be attributed to biofilm formation. Biofilms are multicellular aggregates surrounded by exopolymeric substance (EPS), including extracellular DNA, proteins, and polysaccharides (12–14), which enhance tolerance toward external stress, such as desiccation (15). To date, only a handful of contributing factors and conditions for A. baumannii biofilm formation have been identified (reviewed in references 16 and 17). In particular, pilus production plays a pivotal role in the first step of biofilm initiation: the irreversible attachment of bacteria to a surface. Two highly conserved A. baumannii attachment factors have been identified to date, the chaperone-usher pili (Csu) and outer membrane protein A (OmpA) (16, 18, 19). OmpA has been shown to facilitate attachment to abiotic surfaces as well as epithelial cells (19). Comparatively, the Csu assembly system (CsuA/B, CsuA-E) has a more critical role mediating attachment to abiotic surfaces via hydrophobic interaction with structurally variable substrates, including plastic and glass (18, 20). Transcriptional regulation of the csu operon is governed, in part, by the biofilm formation-regulating two-component system BfmRS (21). During sub-MIC antibiotic treatment, BfmRS also enhances the expression of another major biofilm determinant, the major capsular biosynthesis gene cluster (K locus) (22).
To identify novel factors that drive A. baumannii biofilm formation, here we used a transcriptomic approach, comparing gene expression in biofilms to that of planktonic cells. We then identified genes for which transcription was enriched within biofilm, obtained transposon mutants for these genes, and screened them for changes in biofilm integrity. In so doing, we identified 24 genes whose disruption resulted in a notable decrease in biofilm mass compared to the wild type. One such factor was cold shock protein C (CspC), the disruption of which resulted in a mucoidal, but nonmucoviscous, colony phenotype. Cold shock proteins (Csps) are important for cell aggregation (23), extracellular polysaccharide production, and membrane fluidity in other organisms (24). Furthermore, Csps are multifunctional proteins that act pleiotropically as transcriptional regulators and/or RNA chaperones (25–27). Although Csps often play important roles in bacterial stress response, they have not been previously implicated in the regulation of A. baumannii biofilm formation. In this work, we reveal that disruption of cspC leads to enhanced tolerance to polysaccharide degradation, altered antibiotic resistance profiles, and diminished abiotic surface adherence. The cspC mutant also exhibited diminished survival in a murine model of infection. Collectively, these findings establish CspC as a novel regulator of A. baumannii biofilm formation and disease causation.
RESULTS
Identification of A. baumannii biofilm-associated genes.
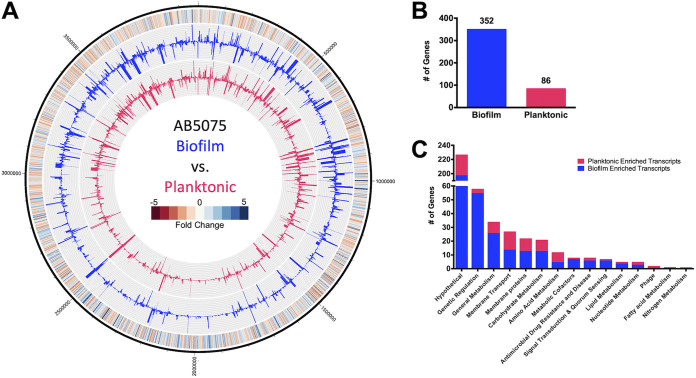
To inform on factors potentially important to biofilm formation, global gene expression within AB5075 wild-type biofilm populations was compared to that of planktonic counterparts (Fig. 1A, see Table S2 in the supplemental material). Of the genes differentially expressed by a magnitude of ≥2-fold, 352 genes were expressed higher in biofilm cell populations (Fig. 1B), while just 86 genes were enriched ≥2-fold within planktonic cells.
FIG 1.
A. baumannii biofilms exhibit differential expression patterns compared to planktonic cell populations. (A) The genomic map depicts changes in planktonic (inner circle, pink) and biofilm (outer circle, blue) transcriptomes, reported as TPM expression values. The outermost circle is a heat map demonstrating the fold change in expression, where red and blue indicate higher expression in the biofilm and planktonic cells, respectively. (B and C) The numbers of genes that were preferentially expressed ≥2-fold in biofilm (blue) or planktonic (pink) cell populations were tallied (B) and further sorted by function based on KEGG ontology (C).
When sorted ontologically, most biofilm-associated transcripts were categorized within the genetic regulation group, fulfilling roles pertaining to DNA replication, transcriptional regulation, and translation (Fig. 1C). Of the 58 genes within this cluster, 27 are annotated as transcription factors. Furthermore, an overwhelming 25 of these transcription factors were enriched in the biofilm population. The upregulation of a cadre of genes involved in metabolic pathways was also apparent within biofilms. The most drastically enriched transcripts were those responsible for energy metabolism, and specifically, the sulfur metabolism pathway (28). This included genes encoding for subunits of the taurine transporter permease (tauA, tauB, and tauC), taurine dioxygenase (tauD), sulfate transporter permeases (cysT, cysW), and thiosulfate-binding protein (ABUW_RS05010), each of which showed ≥7.69-fold enrichment. Several additional genes involved in the sulfur metabolism pathway were also upregulated by ≥2-fold in biofilms, including sulfate transporter permease (ABUW_RS04990), thiosulfate-binding protein (ABUW_RS01375), and sulfate adenylyl transferase subunits (cysN and cysD).
Six genes identified as contributors to antimicrobial drug resistance and disease were also upregulated in biofilm. This group included ABUW_RS09485 and ABUW_RS09480, which encode a putative efflux pump and its accompanying membrane fusion protein component, respectively, which are homologous to the Escherichia coli efflux pump, EmrAB. Other ontologies with altered expression in biofilms included additional membrane transporters (e.g., – ABUW_RS11790, ABUW_RS11795, ABUW_RS18795) and putative membrane proteins (e.g., – ABUW_RS10540, ABUW_RS17095, ABUW_RS18900), carbohydrate metabolism (e.g., – ABUW_RS08155, leuA, ABUW_RS10665), and amino acid metabolism (e.g., – yncA, ABUW_RS13045, mmsB). Thus, it is clear that the biofilm has unique regulatory networks activated compared to that of planktonic cells, which reflect distinct metabolic needs and response to the environment.
To validate RNA sequencing (RNA-seq) findings, a selection of genes were assayed by real-time quantitative reverse transcription PCR (RT-qPCR), with fold changes in expression proving comparable to that of RNA-seq findings (Fig. S1).
Physiological impact of biofilm-associated genes on biofilm integrity.
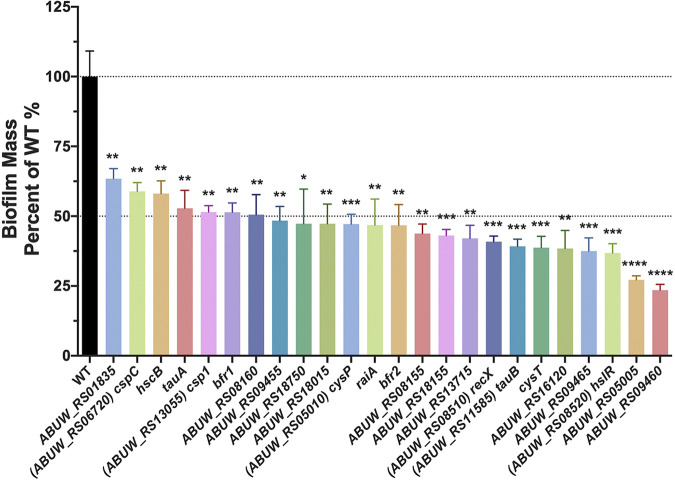
Preferential expression within biofilm populations does not necessary indicate that a given gene product has a role in biofilm formation. Thus, to explore genes with a tangible role in biofilm formation, we first narrowed the list of 352 genes with biofilm-enriched transcripts to those meeting the following criteria: confident read coverage (>85% uniquely mapped reads), relatively strong expression (transcripts per million [TPM], ≥115 in biofilm), and greatest fold increase (>3-fold) (Table 1). Of the genes fitting these criteria, the most highly expressed gene was ssrS (62,175.62 TPM), which produces the well-conserved 6S RNA (29), while the most upregulated gene within biofilms compared to planktonic populations was ABUW_RS05005 (+60.17-fold), which encodes an uncharacterized alpha/beta hydrolase fold protein. We next assessed whether the 40 genes within these parameters have an impact on biofilm formation by performing classic crystal violet biofilm assays on transposon mutants for each them (from the University of Washington Acinetobacter baumannii AB5075 transposon mutant library [30]). In so doing, we uncovered 24 genes whose disruption led to reduced biofilm formation (Fig. 2). Of these, 17 mutant strains showed >50% reduction in biofilm. The largest changes were observed for ABUW_RS09460 and ABUW_RS05005, with >72% reduction in biofilm mass for their respective mutant strains. Although both ABUW_RS09460 and ABUW_RS05005 mutants show comparable reduction in biofilm mass, ABUW_RS09460 was only modestly upregulated (3.07-fold), whereas ABUW_RS05005 was markedly upregulated (60.17-fold) in biofilm. Significant changes in biofilm formation were also observed for cspC and hscB mutants (>40% reduction), with cspC being the highest expressed gene (1,324.94 TPM), while hscB was one of the lowest expressed genes (167.95 TPM) within biofilms investigated in these assays. Altogether, neither fold change in expression nor magnitude of expression exclusively correlate with physiological impact on biofilm formation, but rather, each feature must be considered holistically.
TABLE 1.
Prioritized genes exhibiting preferential expression within biofilms
| Gene | Expression (TPM)a | Fold changeb |
|---|---|---|
| ssrS | 62,175.62 | 3.15 |
| ffs | 4,153.04 | 3.38 |
| leuA | 1,513.08 | 3.55 |
| cspC (ABUW_RS06720) | 1,324.94 | 3.16 |
| hslR (ABUW_RS08520) | 1,059.73 | 3.24 |
| ABUW_RS08155 | 1037.86 | 3.58 |
| ABUW_RS02640 | 737.36 | 3.18 |
| ABUW_RS09455 | 717.76 | 3.53 |
| cysD | 565.84 | 3.95 |
| ABUW_RS01835 | 554.93 | 3.92 |
| bfr_2 | 532.31 | 3.36 |
| ABUW_RS08160 | 511.43 | 4.02 |
| cysP (ABUW_RS05010) | 440.27 | 51.8 |
| iscA | 370.19 | 3.93 |
| ABUW_RS20285 | 330.21 | 3.71 |
| csp1 (ABUW_RS13055) | 325.75 | 15.48 |
| ABUW_RS09460 | 322.14 | 3.07 |
| raiA | 318.52 | 3.65 |
| ABUW_RS05005 | 293.8 | 60.17 |
| tauA | 242.59 | 16 |
| ABUW_RS18750 | 211.35 | 3.98 |
| tauB (ABUW_RS11585) | 207.38 | 15.64 |
| ABUW_RS09465 | 184.01 | 3.96 |
| ABUW_RS04445 | 175.35 | 7.93 |
| hscB | 167.95 | 3.53 |
| ABUW_RS16120 | 165.53 | 3.66 |
| ABUW_RS17360 | 163.51 | 3.24 |
| ABUW_RS20445 | 160.93 | 17.11 |
| ABUW_RS10540 | 153.08 | 12.13 |
| ABUW_RS01965 | 152.46 | 3.17 |
| recX (ABUW_RS08510) | 145.88 | 4.62 |
| tauC | 138.39 | 11.13 |
| ABUW_RS18155 | 136.84 | 5.31 |
| ABUW_RS07450 | 133.65 | 9.93 |
| ABUW_RS09695 | 131.04 | 3.56 |
| ABUW_RS13715 | 128.98 | 3.94 |
| ABUW_RS18015 | 122.17 | 4.61 |
| cysT | 121.76 | 30.66 |
| tauD | 118.66 | 7.69 |
| bfr_1 | 115.41 | 3.43 |
Expression is reported as the TPM of the biofilm sample measured by RNA-seq.
Fold change is reported as the expression of biofilm relative to the planktonic sample.
FIG 2.
Physiological impact of gene disruption for those preferentially expressed in biofilms. Crystal violet biofilm assays were performed with transposon mutants of select genes preferentially expressed within biofilms. Alterations in biofilm mass are reported as a percentage of the wild type. Assays were performed in biological triplicate with 3 technical replicates each. Error bars represent the ± standard error of the mean SEM; Student’s t test was used to determine statistical significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To assess whether the observed changes in biofilm production may be due to a growth defect in the mutant strains, we measured cell density over time in liquid culture compared to the wild type (Fig. S2A). In so doing, we noted that the vast majority of mutants behaved just like the wild type, with only a handful of strains exhibiting growth defects. These included the transposon mutant for csp1, which grew in a similar manner to the parent for the first 3 h but then essentially plateaued in growth, reaching a maximum cell density (optical density at 600 nm [OD600]) of 0.883 ± 0.014 at 8.5 h (WT OD600, 1.133 ± 0.006 at 11.5 h). Another strain with a notable growth defect was the tauA mutant, which exhibited a less pronounced exponential growth phase, reaching a cell density of 0.602 ± 0.011 by 4 h (WT OD600 = 0.769 ± 0.018 by 4 h). The remaining mutant strains grew at rates comparable to those of the wild type and reached similar, if not higher, maximum cell densities. This indicates that the observed reduction in biofilm formation is unlikely to be an artifact of poor growth, with the exception of the csp1 and tauA mutant strains.
Disruption of cspC results in a highly mucoid colony phenotype.
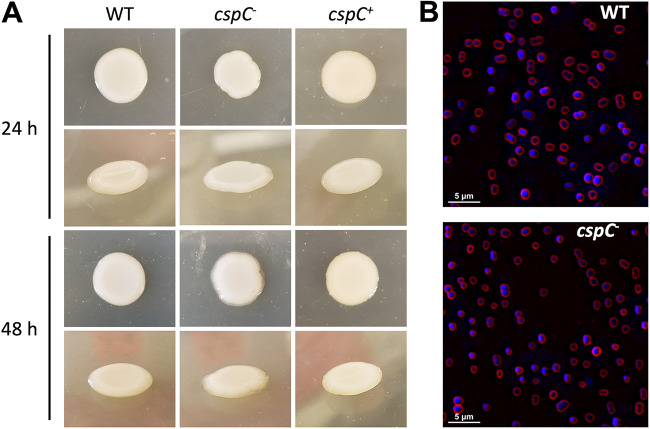
During our screen, we noted that one particular mutant, cspC::tn (+3.2-fold transcription in biofilm), produced a highly mucoid phenotype when grown on LB agar (Fig. 3A). These colonies resembled mucus but did not produce viscous, sticky strings when lifted by a toothpick. The cspC gene was one of the highest expressed genes within the biofilm (1,324.94 TPM) and encodes a putative cold shock protein, the role of which has not been previously characterized in A. baumannii. Importantly, complementation of cspC in trans eliminated the mucoid colony phenotype. When observing cspC mutant cells by fluorescence microscopy, cell morphology was unaltered with respect to shape, size, and chaining (Fig. 3B), indicating that obvious morphological differences are unlikely to contribute to the phenotype observed.
FIG 3.
cspC mutants display a highly mucoid colony phenotype but unaltered cellular morphology. (A) Wild-type (WT), cspC–, and cspC+ strains were grown on LB agar supplemented with hygromycin at 37°C for 24 h (top) and subsequently left to grow for an additional 24 h at room temperature (bottom). Images are representative of 3 experimental repeats. (B) Fluorescence microscopy was used to visualize cell morphology for the wild-type and cspC mutant strain. Cells harvested from bacterial colonies were stained with FM4-64 and DAPI to visualize cell membranes (red) and DNA (blue), respectively. Images are representative of three biological replicates.
cspC mutants display impaired biofilm formation across the growth cycle and altered polysaccharide production.
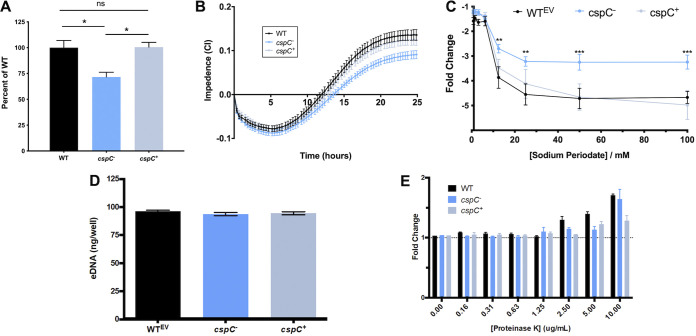
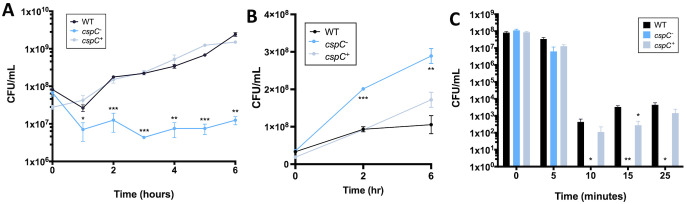
To understand the impact of cspC deletion on A. baumannii cells more deeply, we next performed additional biofilm formation assays. As expected, crystal violet assays generated results similar to those of our screen, with the cspC mutant displaying a significantly impaired ability to form biofilms in comparison to the wild-type and complementing strains (Fig. 4A), with no measurable differences in growth (Fig. S2B). To gain insight into biofilm formation from a kinetic perspective, we used an xCELLigence real-time cell analyzer (RTCA) to track biofilm formation in real time. The RTCA measures biofilm progression based on impedance of electrical signals between electrodes lining each well of a specialized 96-well plate and expresses these measurements as a relative unit cell index (CI). CI measurements are influenced by cell adherence, secretion of extracellular polymeric substance (EPS), and cell spreading (31, 32). When CI values reach their maximum and remain steady, this signifies that the biofilm has entered maturation (31). In agreement with our crystal violet studies, the rate of increase and overall maximum CI value reached were noticeably lessened for the cspC mutant strain compared to the parent and complement strains (Fig. 4B).
FIG 4.
Disruption of cspC results in impaired biofilm formation and enhanced resistance to sodium periodate. (A and B) Comparison of biofilm formation using crystal violet staining following 24 h growth (A) and continuously using an xCELLigence RTCA (B) over a 25-h period. Assays were performed in biological triplicate with 3 technical replicates each. Assays in panel A are corrected for subtle OD600 variations between the wild-type and mutant/complement strains after 24 h of growth under static conditions (Fig. S2). Student’s t test was used to determine statistical significance compared to the wild type. (C) For the biofilm inhibition assay, biofilms were seeded with increasing concentrations of sodium periodate and incubated for 24 h, with the resulting biofilms measured using a crystal violet assay. Data points are from 10 biological replicates and 3 technical replicates. The fold change is reported relative to no-treatment controls for each strain. A two-way analysis of variance (ANOVA) was used to determine statistical significance from the wild type. (D) Extracellular DNA production within the biofilm matrix was measured using a PicoGreen assay kit after 24 h of formation. (E) Biofilms were seeded with increasing concentrations of proteinase K and then measured using a crystal violet assay after 24 h of formation. Change is reported as the fold difference from the no-treatment control. Assays were performed in biological triplicate; Student’s t test determined no statistically significant differences between strains for panels D and E. Error bars are shown as the ±SEM. **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant.
As Csps have demonstrated importance in cell aggregation (23), extracellular polysaccharide production, and membrane fluidity (24), we hypothesized that the altered colony morphology and decreased CI values of the cspC mutant may be due to changes in EPS production. Indeed, upon testing, we noted that cspC mutant biofilms showed increased tolerance to polysaccharide degradation by sodium meta-periodate compared to the wild type and complement strains (Fig. 4C). Conversely, the cspC mutant biofilm contained an amount of environmental DNA (eDNA) comparable to that of the wild type (Fig. 4D). Additionally, when we performed biofilm profiling assays in the presence of proteinase K, no change in biofilm integrity was observed (Fig. 4E), suggesting that protein and eDNA components are not contributing to the biofilm defect observed upon cspC disruption.
cspC is induced under cold shock conditions but does not impact cold shock survival.
CspC bears the same domain architecture as classic Csps, which are nucleic acid-binding proteins that are induced during temperature downshifts (26). To explore whether this is true of cspC, its transcription was measured during cold stress. Upon analysis, we observed that cspC transcript levels were moderately increased by approximately 1.5-fold compared to expression at 37°C (Fig. S3A). When the cspC mutant was challenged with cold stress for 1 h, however, we did not observe any impact on cell viability (Fig. S3B). Furthermore, the cspC mutant also did not exhibit impaired survival during sustained cold stress (Fig. S3C). It is noteworthy that the cspC transcript is relatively abundant at 37°C, under both biofilm (1,324.94 TPM) and planktonic conditions (408.71 TPM). Active transcription under varied conditions, coupled with the apparent lack of impact during cold stress, suggests that the role of CspC may extend beyond cold adaptation in A. baumannii.
Transcriptional regulation by CspC.
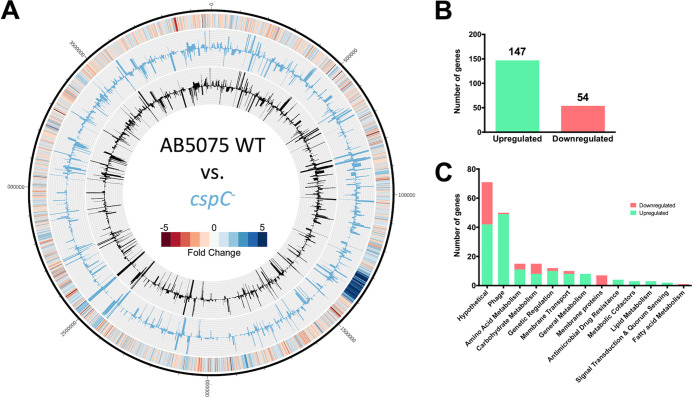
Csps are multifunctional proteins that can wield impacts at the level of transcription, posttranscription, and translation (reviewed in references 26 and 27). They exert their pleiotropic effects as either a transcriptional regulator or RNA chaperone (25). To inform on the mechanism(s) governing the observed physiological changes upon cspC disruption in A. baumannii, we performed RNA-seq with our mutant and wild-type strains (Fig. 5A, Table S3). In so doing, we observed 147 genes upregulated and 54 genes downregulated by ≥2-fold upon cspC disruption (Fig. 5B). When sorted ontologically, it is apparent that a large portion of upregulated transcripts pertained to phage-related genes within the same chromosomal locus (n = 49) (Fig. 5C). Of these, 31 fall within the region predicted to encode Acinetobacter phage Bϕ-B1251, and the remaining 17 are within Ab105-1ϕ. Recent work has identified these regions as encoding active Siphoviridae and Myoviridae family phages, respectively (33).
FIG 5.
A. baumannii cspC disruption leads to global transcriptional changes. (A) The genomic map depicts transcription profiles of the wild type and the cspC mutant reported as TPM expression values. Inner histograms display RNA-seq expression values of the wild type (black) and cspC mutant (blue) reported as TPM. The outermost circle is a heat map illustrating the fold change in expression upon cspC disruption relative to the wild type, where blue and red indicate the fold -increase and -decrease of expression, respectively. (B and C) The numbers of genes upregulated ≥2-fold (green) or downregulated (red) were tallied (B) and parsed by function based on KEGG ontology (C).
A notable trend of opposing regulation of membrane transporter proteins compared to nontransporter membrane proteins was also apparent in our data set. All 8 differentially expressed membrane protein genes, with no transport function, were downregulated, including type 1 pilus subunits csuAB (6.19-fold) and csuC (2.7-fold) and a fimbrial subunit, fimA (3.28-fold), each of which is important for attachment of A. baumannii to abiotic surfaces (18, 20, 34). Of the differentially expressed membrane transporters, 8 of 10 showed increased transcription, including adeFG (≥4.78-fold), which encode components of multidrug efflux pumps. Also upregulated were the accompanying component adeH (3.29-fold) and additional efflux pump genes, adeIJ (≥2.08-fold) and abeS (ABUW_RS06550, 2.43-fold).
Other ontologies with enhanced expression in the cspC mutant included amino acid metabolism (e.g., pepD, glyA), regulatory factors (e.g., rpoH, adeL, ABUW_RS06565), and carbohydrate metabolism (e.g., – gapN, atoA, atoD). Collectively, it is clear that cspC has a global impact on the transcriptional homeostasis of A. baumannii and acts to regulate factors with many and varied roles within the cell.
To validate the RNA-seq findings, a selection of genes were assayed by real-time quantitative reverse transcription PCR (RT-qPCR). Trends in differential expression within the cspC mutant relative to the wild type were comparable to that of RNA-seq findings (Fig. S4).
CspC influences the antimicrobial resistance profile of A. baumannii.
Previous work employing a spontaneous mutation screen identified cspC as one of several genes able to restore, at least in part, antibiotic resistance in an AB5075 bfmRS-null strain (35). Considering this, and the upregulation of multidrug efflux pump components observed in our study, we hypothesized that the cspC mutant would display phenotypic antibiotic tolerance. Accordingly, we conducted an MIC screening panel for the cspC mutant using a variety of antimicrobial agents. In so doing, we noted that disruption of cspC resulted in markedly increased tolerance to ciprofloxacin, chloramphenicol, and streptomycin (Table 2) yet also resulted in marginally increased sensitivity to gentamicin, kanamycin, neomycin, and fosfomycin.
TABLE 2.
Antibiotic susceptibility of the AB5075 wild type and the cspC mutant straina
| Strain | MIC (μg/mL) for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| CIP | CM | STR | GM | KAN | NEO | FOS | OX | |
| AB5075 wild type | 45 | 84.38 | 1,070 | 2,000 | 2,250 | 253.13 | 562.5 | 1,130 |
| AB5075 cspC::tn | 80 | 150 | 3,380 | 1,500 | 1,690 | 189.84 | 316.41 | 1,130 |
CIP, ciprofloxacin; CM, chloramphenicol; STR, streptomycin; GM, gentamicin; KAN, kanamycin; NEO, neomycin; FOS, fosfomycin; OX, oxacillin.
CspC regulates mRNA stability of transcriptional regulators adeL and ABUW_RS06565.
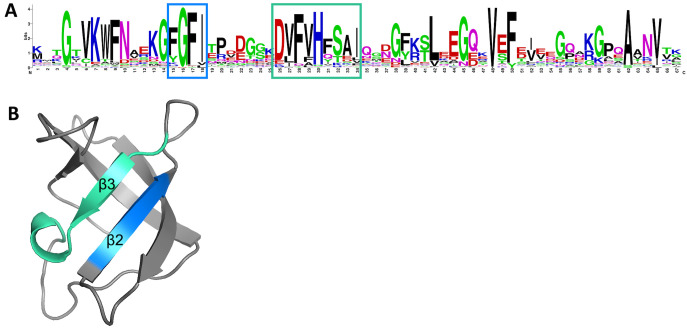
When bacteria experience temperature downshifts, single-stranded DNA (ssDNA) and RNA secondary structures are consequently stabilized. This leads to inhibited transcription and/or translation and RNA degradation, which adversely effects cellular function. Csps possess a conserved nucleic acid-binding domain that allows them to bind single-stranded DNA and RNA and rescue undesirable secondary structures. Specifically, this nucleic acid-binding domain typically comprises a ribonucleoprotein (RNP)-1 and RNP-2 motif, which facilitates interaction with nucleic acids and has chaperone activity (26). When interrogating the amino acid sequence of CspC, a conserved cold shock domain signature was apparent (Fig. S5). This domain contains both an RNP-1 and an RNP-2 motif (Fig. 6A). Structural prediction of CspC revealed that these motifs are both within the β2 and β3 strands of the antiparallel β-barrel structure (Fig. 6B). This arrangement makes the RNP motifs spatially available for binding to ssDNA and RNA, as is the hallmark characteristic of Csps (36, 37).
FIG 6.
CspC contains conserved RNP-1 and RNP-2 motifs. (A) A sequence logo generated using the UniProtKB/SWISS-PROT sequences for the cold shock domain profile of AB5075 CspC detected using ScanProsite. Putative RNP-1 (blue) and RNP-2 (green) motifs are boxed. (B) A 3-D representation of the A. baumannii CspC protein structure is shown. Key residues within the β2 and β3 sheets are colored corresponding with the RNP motif residues indicated in panel A.
In order to determine if CspC functions as an RNA chaperone, we assessed the impact of cspC mutation on the rate of decay for putative target mRNAs. To do so, cells were grown to the exponential phase and treated with rifampin to inhibit transcription. RNA was then isolated from wild type and cspC mutant strains at consecutive time points post-transcriptional arrest. Using RT-qPCR, we assessed RNA transcript levels at each time point and determined the decay rate by plotting the change in transcript abundance relative to transcript level immediately prior to rifampin treatment. The close arrangement of efflux pump-encoding genes adeFGH indicates that these genes are likely within an operon, and similar conclusions can be drawn for adeIJK. Further supporting this, RNA-seq read alignments (data not shown) revealed transcript readthrough between the individual genes of each efflux pump. Considering this, we selected adeG and adeJ, located in the middle of their respective operons, for mRNA half-life studies. The half-life of adeJ mRNA was slightly longer than that of adeG (Fig. S6, Table 3) in the wild-type strain; however, half-lives for both transcripts were comparable between the parental and cspC-deficient strains. This suggests that CspC has no impact on the stability of adeJ and adeG mRNA. Importantly, the half-life of our control gene, recA, was found to be 4.674 min in the wild type, which is comparable to the previously determined half-life of 4.5 min (38), indicating that the experimental approach used here is reproducible.
TABLE 3.
Mean mRNA half-lives of CspC-regulated transcripts as determined by RNA decay assaysa
| Gene | Wild type |
cspC::tn |
||||
|---|---|---|---|---|---|---|
| Half-life (min) | Decay constant (k) | R2 | Half-life (min)a | Decay constant (k) | R2 | |
| adeJ | 5.326 | 0.1301 ± 0.015 | 0.954 | 5.206 | 0.1331 ± 0.020 | 0.928 |
| adeG | 4.773 | 0.1452 ± 0.016 | 0.926 | 4.236 | 0.1636 ± 0.024 | 0.884 |
| adeL | 1.280 | 0.5417 ± 0.020 | 0.999 | 2.540 | 0.2728 ± 0.022 | 0.969 |
| ABUW_RS06565 | 1.753 | 0.396 ± 0.025 | 0.990 | 2.565 | 0.270 ± 0.016 | 0.993 |
| recA | 4.674 | 0.1483 ± 0.002 | 0.999 | 4.890 | 0.1418 ± 0.031 | 0.880 |
The decay constant (k) mean was used to calculate the half-life using t1/2 = ln(2)/k, where t1/2 represents the mRNA half-life. The decay constant is reported as the mean ± the standard error.
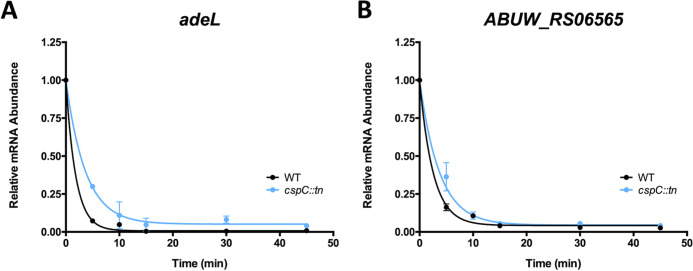
Considering that the half-lives of the major efflux pump transcripts were unaffected, we next considered whether CspC could be acting as an mRNA chaperone for transcriptional regulators. Transcription of adeFGH is repressed by the LysR-type transcriptional regulator adeL (upregulated +2.8-fold in the cspC mutant; Table S3, Fig. S4), located immediately upstream of the adeFGH operon (39). When assessed experimentally, we found that the half-life of adeL was increased in our mutant from 1.28 min in the parent to 2.54 min in the cspC– strain (Fig. 7A, Table 3). We also investigated the half-life of ABUW_RS06565, an uncharacterized GntR family transcriptional regulator, which was upregulated 2.98-fold in the cspC mutant. ABUW_RS06565 was the only transcriptional regulator with altered transcription, aside from adeL, that was not part of the collectively altered Bϕ-B1251 and Ab105-1ϕ phage loci genes. The half-life of ABUW_RS06565 rose from 1.75 min in the wild-type strain to 2.57 min in the mutant—a 1.5-fold increase in the cspC mutant compared to the parent (Fig. 7B, Table 3). This indicates that the mRNAs of transcriptional regulators adeL and ABUW_RS06565 are, in fact, more stable upon cspC disruption. It is noteworthy that, although the average mRNA half-life varies depending on bacterial species and growth condition, it is typically between 2 and 5 min (40–43). Considering this scale, the extended half-lives of adeL and ABUW_RS06565 RNA in the cspC mutant are likely substantial.
FIG 7.
CspC mutation results in extended mRNA half-life for transcriptional regulators adeL and ABUW_RS06565. (A and B) Exponentially growing wild-type and cspC mutant strains were treated with 250 μg/mL rifampin to arrest transcription, and changes in transcript abundance were measured by RT-qPCR for adeL (A) and ABUW_RS06565 (B). Values represent the mean fold change in transcript abundance relative to that immediately prior to rifampin treatment (t = 0). Data are from triplicate cultures ± SD. The lines shown are the exponential, one-phase decay curve, represented as R(t) = R0e−kt, which were used to calculate mRNA half lives.
CspC is required to resist challenge by components of the human immune system.
Biofilm production contributes to bacterial survival against host immune responses (44), and previous studies have found that a majority of A. baumannii bloodstream isolates are capable of producing robust biofilms (45). Furthermore, altering the balance of attachment, growth, and dispersal of a biofilm results in negative impacts on the capacity to cause bloodstream infections (46). We thus speculated that the loss of CspC would impact A. baumannii survival in human blood. Accordingly, we grew the wild-type, mutant, and complement strains in whole human blood for 6 h. Upon analysis, we noted that AB5075 cell viability declined 67.6% from 0 to 1 h, followed by an increase in viability as time progressed (Fig. 8A). In contrast, the cspC mutant strain fared markedly worse, demonstrating a far more severe decline in cell viability from 0 to 1 h (86.6%), with cell viability counts remaining relatively stagnant as time progressed. As expected, the cspC complement strain had a survival capacity similar to that of the wild type. These data indicate a clear requirement for CspC for survival during A. baumannii engagement with human blood.
FIG 8.
The cspC mutant has abrogated survival in human blood and during oxidative stress. (A to C) Cell viability of the cspC mutant was assessed in whole human blood (A) and human serum (B) and in the presence of 2 mM hydrogen peroxide (C). Assays were performed in biological triplicate. Data are presented as the mean, and error bars represent the ± SEM. For certain points, the error is so small as to be unplottable. Student’s t test was used to determine statistical difference from the wild type at each time point. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
As human blood is largely composed of serum, which is also known to kill A. baumannii, we next explored whether this was the reason for the survival defect displayed by our mutant strain. Accordingly, our wild-type, cspC mutant, and complemented strains were separately inoculated into human serum, in a manner akin to our whole-human blood studies. When these data were analyzed, we noted no killing of any of our strains over the 6-h test period (Fig. 8B). Moreover, we noted increased survival of the mutant strain compared to the wild-type and complemented strains, albeit at relatively modest levels (~2 fold). As such, it appears that killing by human serum is not causative for the survival defect observed for our cspC mutant strain in whole human blood.
Bacterial clearance in human blood is mediated by several factors, including cell-mediated immunity. A primary killing mechanism of leukocytes during engagement with bacterial pathogens is through reactive oxygen species (ROS). It is known that clearance of A. baumannii is dependent on host reduction of NADPH oxidase and subsequent accumulation of ROS (47, 48). Considering this, we next performed a hydrogen peroxide killing assay (Fig. 8C). Upon analysis, we noted that the viability of all strains declined after 10 min of exposure to 2 mM hydrogen peroxide, which is a clear indication that the cells are experiencing oxidative stress. At the conclusion of these studies (25 min) we observed a comparable number of viable cells for the wild type and complement strains. Conversely, the cspC mutant showed no recovery beyond 10 min of exposure, which indicates that CspC has a major role in bacterial survival for A. baumannii in the presence of ROS.
CspC is required for survival during A. baumannii systemic infection.
Given our findings regarding the ability of the cspC mutant to survive challenge by components of the human immune system and ROS, we next considered whether its survival during in vivo infection would be impaired. Accordingly, using a murine model of sepsis infection, we compared the pathogenic potential of the cspC mutant to wild-type AB5075. When assessing mortality, we observed no apparent differences between the strains (data not shown); however, when comparing dissemination of infection, bacterial load was substantially reduced for the cspC mutant strain in all organs evaluated (Fig. 9). The most significant reduction observed was for that of the liver, which showed a 43-fold reduction in bacterial load compared to the wild type. Although not quite as striking, reduced dissemination of the cspC mutant to the other organs was equally remarkable (spleen, 12.16-fold; brain, 6.58-fold; lungs, 5.62-fold; kidneys, 4.32-fold, heart, 3.14-fold). These findings support CspC as a novel regulator of dissemination and/or colonization, proving critical for A. baumannii survival when challenged by the host innate immune response.
FIG 9.
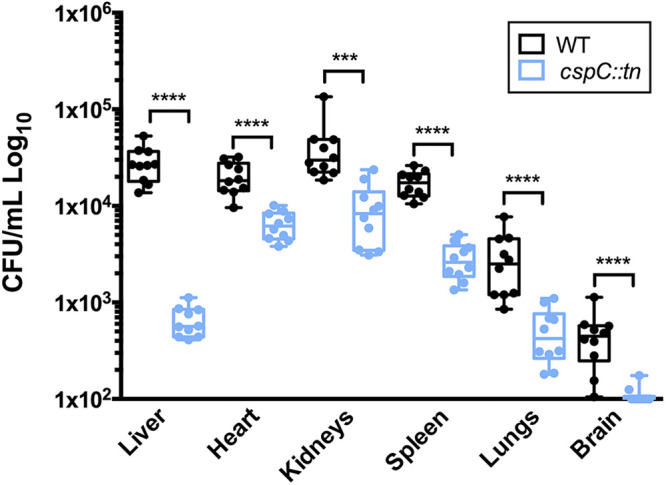
cspC is required for A. baumannii survival in a murine model of sepsis. Cohorts of BL/6 mice (n = 10) were inoculated by retroorbital injection with 2.5 × 107 CFU of wild-type (black) and cspC::tn mutant strains (blue). At 6 h postinfection, mice were euthanized, and organs were collected to determine bacterial load. The bounds of each box represent the 25th to 75th percentiles, whiskers represent the 5th to 95th percentiles, the center line denotes the median, and individual data points are plotted as circles. Statistical significance was determined using a Mann-Whitney nonparametric test. ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
In this study we investigated the global transcriptional regulation occurring within biofilms formed by A. baumannii AB5075. RNA-sequencing revealed a distinct regulatory response within the biofilm, with 10% of all known transcription factors activated in biofilms and 1% deactivated, suggesting a distinct global regulatory response within this population. The most upregulated gene within biofilms was ABUW_RS05005 (+60.17-fold), which encodes an uncharacterized alpha/beta hydrolase fold protein coexpressed with the cys sulfate transporters (cysP, cysT, cysW, cysA). Flanking genes cysP and cysT were also significantly upregulated within the biofilm (+51.8-fold and +30.66-fold, respectively). Microbiosis (0.1 to 0.3% O2) has been shown to induce sulfur metabolism gene transcription (49), and anaerobic regions are known to occur within biofilms (50, 51); thus, conditions may be prompting a metabolic shift toward sulfur metabolism in our studies. ssrS was the most highly expressed gene within biofilms (62,175.62 TPM), which produces the well-conserved, 6S RNA (29). Studies of E. coli have demonstrated that 6S RNA accumulates during the stationary phase and represses transcription, in turn, enabling cell survival amid nutrient-limiting conditions (52, 53). It is quite possible that nutrient scarcity within the biofilm’s dense bacterial community may induce 6S RNA transcription to reorder gene expression circuits to circumvent nutrient limitation.
Among the enriched transcripts within biofilms, we identified 24 that had a significant physiological impact on biofilm integrity. Mutation of the most highly expressed of these (cspC) resulted in pleiotropic impacts on the cell. CspC is one of four known Csp genes in AB5075 and one of two upregulated during biofilm growth—the other being csp1 (+15.48-fold). In the biofilm, CspC influences extracellular polysaccharide production, as demonstrated by enhanced tolerance of the mutant to sodium meta-periodate, which cleaves PNAG, a component required for A. baumannii biofilm formation (54, 55). Conversely, extracellular protein and eDNA matrix components are unaffected by CspC, although this is perhaps unsurprising, as A. baumannii clinical isolate biofilms are primarily composed of polysaccharides (56). Increased tolerance to PNAG degradation may indicate overproduction and/or structural variation of this matrix component. If the former were true, we would anticipate enhanced biofilm production, but this was not the case. We suggest that structural variation, which would render PNAG resistant to sodium meta-periodate, may be impairing biofilm integrity in the cspC mutant.
How exactly the cspC mutant is able to resist the action of sodium meta-periodate is unclear. In general terms, altered colony morphology and abrogated biofilm formation as consequences of polysaccharide modification have been well described for a variety of bacteria (reviewed in reference 14). When looking for more specific answers, we found that expression of the first three genes of the PNAG-producing pgaABCD operon was unchanged between parent and mutant. The expression of pgaD, however, is diminished −2.2-fold in the mutant strain, although it just missed our coverage cutoff to be considered significant. It is noteworthy that a study of E. coli proposed that PgaD (and its counterparts in other Gram-negative bacteria) assists PgaC in the polymerization of β-1,6-N-acetylglucosamine subunits (55). Importantly, sodium meta-periodate cleaves exactly these polymeric β-1,6-GlcNAc linkages. Thus, one can envisage that a putative decrease in pgaD expression might alter linkages in PNAG sufficiently to render it less sensitive to the action of this degradative enzyme.
In previous work, spontaneous mutation of cspC bypassed antibiotic hypersensitivity and cell morphology defects in a bfmRS mutant strain, which was attributed to reduced transcription of the well-known capsule factor, the K locus (35). No transcriptional change for cspC was evident in bfmRS mutant strains (35), and in our work, bfmRS transcription was unaffected by cspC disruption. Similarly, no measurable changes in expression for any genes within the K locus were noted within our cspC mutant RNA-seq data (Table S4), and no detectable change in the amount of capsular polysaccharide produced was observed for the mutant strain (Fig. S7). As such, this would tend to indicate that CspC may act independently from the capsule biosynthesis factors within the K locus. Instead, our findings suggest that CspC may mediate biofilm formation through regulation of the csu pilus assembly system and fimA fimbrial subunit transcription. Both of these factors have demonstrated roles in the attachment of A. baumannii to abiotic surfaces and were downregulated in the cspC mutant strain (18, 20, 34). In particular, Csu pili are adhesive organelles that belong to the archaic chaperone-usher pilus class, which facilitate strong adherence to hydrophobic plastics, including polypropylene and polyethylene, which are widely used in medical equipment (20) and are indistinguishable from the materials used in this study. In support of this notion, we performed pilot adherence studies with our strains and reveal that the cspC mutant is impaired in its ability to attach to plastic surfaces (Fig. S8). Furthermore, the cspC mutant also exhibited elevated multidrug efflux pump component (adeFGH, adeIJK) transcription. Overexpression of adeIJK has previously been shown to decrease production of CsuAB, CsuC, and FimA, in turn, altering membrane composition and decreasing biofilm formation in A. baumannii (57).
In addition to biofilm abrogation, it is likely that altered expression of efflux pumps in the cspC mutant are also causative for changes in antibiotic sensitivity. Previous studies have shown that adeIJK overexpression mutants exhibit enhanced ciprofloxacin resistance, and adeIJK null strains demonstrate sensitivity to this drug (39, 57–60). Similarly, transcription of adeFGH and adeIJK also correlate with chloramphenicol resistance (39, 57, 58). Thus, it seems likely that the upregulation of adeIJK in the cspC mutant is contributing to ciprofloxacin tolerance and that upregulation of both adeIJK and adeFGH may be contributing to chloramphenicol resistance.
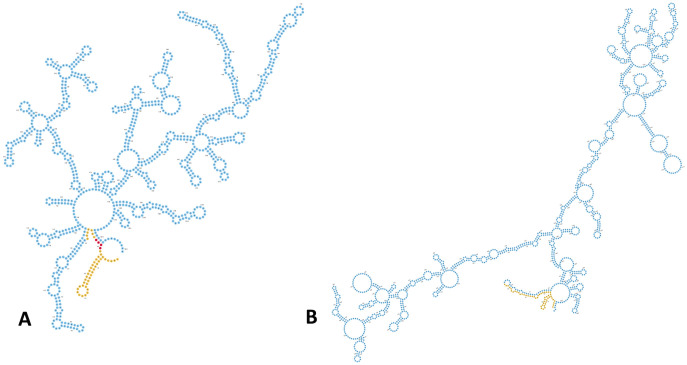
Interestingly, transcription of the adeFGH repressor, adeL (39), was also upregulated in the cspC mutant. It is important to note that the increased half-life of adeL does not necessarily indicate enhanced AdeL repression of adeFGH. Csps can destabilize RNA secondary structures in order to liberate ribosome binding sites (RBSs) and permit translation (reviewed in reference 27); therefore, the absence of CspC could cause enhanced adeL stability, resulting in increased transcription but abrogated translation. In support of this notion, RNA secondary structure prediction revealed a stable hairpin formed in the 5′ untranslated region (UTR) of adeL, immediately upstream of a putative AGGAG ribosomal binding site (Fig. 10). This is of particular interest, as previous studies measuring mRNA half-lives in E. coli determined that an AGGAG motif in 5′ UTRs, specifically within 2 to 8 nucleotides of translational start codons, is frequently found in transcripts with enhanced stability compared to the global mRNA population (61). The ABUW_RS06565 transcript presents a curiosity, as an obvious RBS is not apparent; however, a similar, stable 5′ UTR structure is also evident. Additionally, the ABUW_RS06565 transcript contains an unusual, A/T-rich region beginning at the 5th codon. A/T-rich sequences within RNA, particularly from the 5th to 8th codon, have been shown to significantly enhance translation initiation in E. coli. This suggests that the inaccessibility of this motif due to enhanced RNA stability may also impede translation initiation of this transcription factor upon cspC deletion (62).
FIG 10.
The adeL and ABUW_RS06565 mRNAs contain stable structures proximal to their translation initiation regions. (A and B) The secondary structure for adeL (A) and ABUW_RS06565 (B) mRNA predicted using RNAfold is shown. Nucleotide colors correspond to stable 5′ regions (yellow), putative ribosomal binding site (red), and protein coding sequence (blue). For ABUW_RS06565, a ribosomal binding site was not apparent.
Structural investigation revealed that CspC contains a conserved RNP-1 and RNP-2 nucleic acid binding motif, a hallmark characteristic of Csps. Interestingly, the RNP-2 motif contains an arginine residue at position 34, where most bacteria contain a serine residue. Furthermore, the RNP-1 and RNP-2 motifs are bridged by six amino acids, as opposed to the usual seven. The shortened motif separation, but not arginine residue substitution, is a common occurrence for all four A. baumannii Csps (CspC, ABUW_RS12225, Csp1, ABUW_RS15360) (63), with ABUW_RS15360 having the shortest bridge of four amino acids. The combination of both abridged RNP motifs and residue substitution in RNP-2, may alter the affinity of CspC to nucleic acid ligands.
In summary, this study identified 24 genes with substantial influence on A. baumannii biofilms, many of which remain uncharacterized and have not previously been implicated in this regard. We reveal that one of them, CspC, is critical not only for biofilm formation, but for antibiotic resistance and dissemination and/or colonization within the host. Future work will determine if CspC influences cellular behavior and infection via RNA chaperone activity, as we have suggested, and the importance of the identified and uncharacterized genes to A. baumannii biofilm formation and pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used for this study are listed in Table 4. A. baumannii and Escherichia coli strains were routinely cultured in lysogeny broth (LB) with shaking or on LB agar at 37°C. When appropriate, medium was supplemented with tetracycline (5 μg/mL) and hygromycin (160 μg/mL). A. baumannii biofilm formation was assessed as described previously for microtiter plate biofilms (64) with slight modifications. Briefly, overnight cultures were normalized to an OD600 of 5.0 in phosphate-buffered saline (PBS), before 20 μL was added to 180 μL of fresh LB in 96-well microtiter plates for a final OD600 of 0.5. For assays using strains harboring pMQ557, medium was supplemented with hygromycin.
TABLE 4.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Cloning strain | 85 |
| A. baumannii strains | ||
| AB5075 | Parent strain | 30 |
| cspC::tn | AB5075 with transposon insertion in ABUW_1377 (cspC) | 30 |
| WT | AB5075 containing pMQ557 | This study |
| cspC– | cspC::tn containing pMQ557 | This study |
| cspC+ | cspC::tn containing pMQ557::cspC | This study |
| Plasmids | ||
| pMQ557 | Cloning vector for complementation | Gift, R. Shanks, University of Pittsburgh |
| pLSBT1 | pMQ557::cspC | This study |
Mutant strain confirmation and complement strain construction.
Transposon mutant strains were obtained from the A. baumannii AB5075 transposon mutant library (30). Transposon insertion was confirmed for the cspC transposon mutant by PCR and sequencing using primers OL5087 and OL5088, which flank cspC (all primers are listed in Table S1). A cspC-complement PCR fragment was generated using primers OL5088 and OL5214 and cloned into plasmid pMQ557. In addition to the complete cspC coding region, the complement fragment includes 382 nucleotides 5′ and 97 nucleotides 3′ of cspC to ensure that the native promoter and full-length mRNA (as revealed by RNA-seq) were incorporated. Complementation plasmid pMQ557::cspC was transformed into chemically competent E. coli (DH5α) and confirmed using pMQ557 screening primers OL4163 and OL4164, followed by sequencing. Plasmid containing the correct sequence was transformed into the cspC transposon mutant strain via electroporation (65) to generate the complement strain, cspC+. Given that vector is present in cspC+, the empty pMQ557 vector was also transformed into the wild-type strain and cspC transposon mutant (cspC–). For all assays cspC+, the wild-type, and the cspC mutant strains bearing pMQ557 were used for comparison.
RNA sequencing.
A. baumannii biofilm formation was initiated in 96-well microtiter plates as described above in biological triplicate for 24 h at 37°C in a static incubator. To collect planktonic samples, 75 μL of supernatant was removed from each well and pooled. Planktonic cells were immediately combined with 5 mL of ice-cold PBS and pelleted by refrigerated centrifugation. For biofilm samples, the remaining supernatant was removed, and biofilm-containing wells were washed three times with 200 μL of ice-cold PBS. Ice-cold PBS was added a final time and pipetted vigorously to disrupt biofilm cells. Biofilm cells from different wells were pooled, immediately combined with an additional 5 mL of ice-cold PBS, and pelleted by refrigerated centrifugation.
For collection of wild-type and cspC::tn mutant samples, strains were grown in LB overnight with shaking at 37°C in biological triplicate. Overnight cultures were diluted 1:100 into 5 mL of fresh LB, grown to the exponential phase, and subsequently used to seed new 100-mL cultures at an OD600 0.05. Samples were harvested after 3 h of growth, added to an equal volume of ice-cold PBS, and pelleted by centrifugation at 4°C.
Total RNA was isolated from cell pellets as described previously (66) using an RNeasy kit (Qiagen), with DNA removal performed using a TURBO DNA-free kit (Ambion). DNA removal was confirmed by PCR using OL398 and OL399. Sample quality was assessed using an Agilent 2100 Bioanalyzer system and corresponding RNA 6000 Nano kit (Agilent) to confirm RNA integrity. Samples with a RNA integrity number (RIN) of ≥9.9 were used in this study. Prior to mRNA enrichment, biological triplicates were pooled at equal RNA concentrations. rRNA was then removed using a Ribo-Zero kit for Gram-negative bacteria (Illumina), followed by a MICROBExpress bacterial mRNA enrichment kit (Agilent). Removal efficiency of rRNA was confirmed using an Agilent 2100 Bioanalyzer system and RNA 6000 Nano kit (Agilent). Enriched mRNA samples were then used for RNA sequencing using an Illumina NextSeq instrument. Library preparation and RNA sequencing were performed following TruSeq stranded mRNA kit (Illumina) recommendations, omitting the mRNA enrichment steps. The quality, concentration, and average fragment size of each sample were assessed using an Agilent 2100 Bioanalyzer system and RNA 6000 Nano kit (Agilent) prior to sequencing. The library concentration for pooling of barcoded samples was assessed by RT-qPCR with a KAPA Biosystems library quantification kit as recommended for high sensitivity. Samples were run on an Illumina NextSeq instrument with a corresponding 150-cycle NextSeq midoutput kit v2.5.
RNA-seq bioinformatics.
Data were exported from BaseSpace (Illumina) in fastq format and analyzed using CLC Genomics Workbench v20 (Qiagen Bioinformatics). Reads were imported and failed reads were removed using the Illumina paired importer tool, with quality score parameter options set to Illumina Pipelines 1.8 and later. The total number of reads generated for each sample was at least 15.49 million and up to 22.22 million, resulting in ≥545× read coverage for each sample. Reads corresponding to rRNA were filtered, removed by aligning to known rRNA sequences, and discarded. Samples contained between 0.27% and 0.71% rRNA. The remaining read sequences were aligned using the RNA-seq Analysis tool (v 0.1) with default parameters and defined strand specificity to the A. baumannii AB5075 NCBI reference genome (CP008706.1). Gene expression values were calculated using the Expression Browser tool v1.1, specifying transcripts per million (TPM) as the output. Differential expression values between samples were generated using the Differential Expression in Two Groups tool v1.1 for whole-transcriptome RNA-seq samples. Differential expression is reported as the fold change of expression for biofilm relative to planktonic samples and cspC mutant relative to wild-type samples. Library size normalization was automatically performed using the trimmed mean of M values (TMM) method with the Differential Expression in Two Groups tool (67). Ontology classification of genes was assigned based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) (68). Genomes and differential expression visualizations (Fig. 1A and 5A) were generated using Circos (69).
RT-qPCR transcriptional analysis.
To validate RNA-seq findings, a selection of genes were assayed by real-time quantitative reverse transcription PCR (RT-qPCR). Strains were grown, RNA was harvested, DNA was removed, and sample quality was assessed as described above. One microgram from each sample was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was then performed using gene-specific primers (Table S1) and TB green premix Ex Taq (TaKaRa). Levels of gene expression were normalized to the 16S rRNA gene (OL4498 and OL4449), and the fold change of expression was assessed for biofilm relative to planktonic samples and for the cspC mutant relative to wild-type samples, using the 2–ΔΔCT method (70).
Similarly, transcriptional analysis was used to assess changes in cspC transcript levels under cold stress. A. baumannii wild-type cultures were grown overnight as described above, in biological triplicate, and subcultured into 5 mL of fresh medium. After 3 h of growth at 37°C, new 5-mL cultures were seeded and standardized to an OD600 of 0.05. Bacteria were transitioned to 15°C for 15 min to induce cold shock, and control samples were left at 37°C. Following this, samples were combined with an equal volume of ice-cold PBS and pelleted by centrifugation at 4°C. Total RNA isolation, DNA removal, and reverse transcription were performed as described above. Quantitative, real-time RT-PCR (qRT-PCR) was then performed using cspC-specific primers OL5216 and OL5217 and TB green premix Ex Taq (TaKaRa). Levels of gene expression were normalized to the 16S rRNA gene (OL4498 and OL4449), and the fold change of expression was assessed for cold shocked samples relative to non-cold shocked samples using the 2–ΔΔCT method (70).
Crystal violet and real-time biofilm assays.
A. baumannii biofilm formation was performed as described above in biological triplicate. After 24 h of static growth, biofilms were washed 3 times with PBS and fixed with 100 μL of 100% ethanol. After drying, 200 μL of crystal violet was added and incubated at room temperature for 15 min, and biofilms washed 3 times with PBS. After a second drying step, 100 μL of 100% ethanol was added to solubilize the crystal violet. The absorbance of solubilized crystal violet was measured at OD543 and reported as the percent variance compared to that of the wild-type strain.
An xCELLigence MP real-time cell analyzer (RTCA) (ADCEA Bioscience) instrument was used to monitor biofilm formation over time. The xCELLigence MP RTCA was placed in a 37°C incubator for 1 h prior to experimentation to allow the instrument temperature to equilibrate. Next, 96-well E-plates were loaded with 180 μL of LB, positioned in the RTCA, and measured for background signal. Using the same plate, A. baumannii biofilms were prepared as described above and statically incubated in the RTCA, with reads taken every 15 min for 25 h. The data generated here are from nine biological replicates per strain.
Microscopy.
Cell morphology was assessed by fluorescence microscopy as previously described (71) with minor modifications. Briefly, single 24-h wild-type, cspC::tn mutant, and complemented strain colonies were resuspended in 100 μL of 1× PBS. Cell membranes and DNA were stained with FM4-64 and 4′,6-diamidino-2-phenylindole (DAPI), respectively, at final concentrations of 1 μg/mL. Cell suspensions (5 μL) were spotted onto a glass coverslip on glass-bottom dishes (MatTek) and subsequently covered with a sterile pad of 1% agarose in water. Imaging was completed at room temperature inside a DeltaVision Elite deconvolution fluorescence microscope (GE Applied Precision) environmental chamber. Photos were captured using a CoolSNAP HQ2 camera (Photometrics), and images were acquired by taking 17 z-stacks at 200-nm intervals. All images were deconvolved using softWoRx (GE Applied Precision) imaging software.
Extracellular DNA assays.
Extracellular DNA production of A. baumannii biofilms was analyzed quantitatively as described previously (72). Briefly, A. baumannii biofilm formation was initiated as described above in biological triplicate. After 24 h of growth, supernatant was removed, and biofilms were washed once with 200 μL PBS. eDNA in biofilms was quantified by Quant-iT PicoGreen double-stranded DNA (dsDNA) labeling (Thermo Fisher) and fluorescence measured using a Synergy2 plate reader (BioTek).
Biofilm inhibition by proteinase K and sodium meta-periodate.
Disruption of A. baumannii biofilms was tested using sodium meta-periodate, which cleaves poly-β-1-6-N-acetylglucosamine (PNAG), a component required for A. baumannii biofilm formation (54, 55), and proteinase K, as described previously (73). Briefly, A. baumannii biofilm formation was initiated as described above in biological triplicate. Biofilms were supplemented with a final concentration of 100, 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78, or 0 mM sodium meta-periodate. Alternatively, biofilms were supplemented with a final concentration of 50 μg/mL of proteinase K. Biofilms were allowed to form for 24 h at 37°C and then quantified by crystal violet assay as described above.
Cold-shock recovery and survival.
A. baumannii cultures were grown overnight, in biological triplicate, and subcultured into 5 mL of fresh medium. After 3 h of growth, new cultures were seeded and standardized to an OD600 of 0.05. For cold stress recovery, after 1 h of growth at 37°C, bacteria were transitioned to 15°C for 1 h. Following this cold stress, bacteria were returned to 37°C with shaking. Bacteria were plated on tryptic soy agar (TSA) every hour to determine the CFU/mL as a measure of recovery rate. Alternatively, for cold stress survival, bacteria were immediately placed at 15°C after culture standardization and plated on TSA every hour to determine the CFU/mL for monitoring survival.
Antibiotic susceptibility assays.
Antibiotic sensitivity was assessed by performing MIC assays as previously described (74). Briefly, A. baumannii strains were grown in LB overnight, in biological triplicate, at 37°C with shaking. Overnight cultures were diluted 1:1,000 with fresh LB, and 195 μL was added to 96-well microtiter plates. Subsequently, antibiotics were serial diluted, and 5 μL of each concentration, or solvent (no-treatment control), was added. Antibiotic solvents were as follows: ciprofloxacin, 0.1 M NaOH; gentamicin, 100% EtOH; chloramphenicol, 70% EtOH; streptomycin, kanamycin, neomycin, fosfomycin, and oxacillin, all H2O. Cultures were grown overnight at 37°C with shaking. The MIC is reported as the lowest antibiotic concentration resulting in inhibition of growth compared to the no-treatment control.
CspC protein architecture analysis.
Domain and motif scanning was performed using ScanProsite (75) with the amino acid protein sequence of CspC (GenBank version no. AKA31122.1) as the input. The alignment of 207 UniProtKB/SWISS-PROT sequences of true-positive hits for the detected cold shock domain profile (PS51857) were retrieved, and a sequence logo was generated from this alignment on Prosite. Three-dimensional protein modeling via homology modeling was completed using SWISS-MODEL (76). The SWISS-MODEL template library contained 244 templates matching the CspC amino acid sequence, with the most closely related, and the sequence with the highest global model quality estimate, being CspA of Escherichia coli (77).
Transcriptional arrest and determination of mRNA half-life.
Determination of the RNA half-life was performed as described previously with minor modifications (38). Six biological replicates of A. baumannii cultures were grown overnight and subcultured in 5 mL of fresh medium. After 3 h of growth, new cultures were seeded and standardized to an OD600 of 0.05 in 100 mL of fresh LB. After 3 h of growth to reach the exponential phase, and prior to transcriptional arrest, 5 mL of each culture was collected, immediately combined with 5 mL of ice-cold PBS, and pelleted by refrigerated centrifugation (t = 0). Rifampin at a final concentration of 250 μg/mL was then added to the bacterial cultures. At 5, 10, 15, 30, and 45 min posttreatment, 5 mL of each sample was collected, immediately combined with 5 mL of ice-cold PBS, and pelleted by refrigerated centrifugation. Immediately following each refrigerated centrifugation step, supernatant was removed, and cell pellets were stored at −80°C. Total RNA extraction, confirmation of RNA quality, and RT-qPCR were performed as described above for each sample (the primers used are listed in Table S1). RNA abundance at each time point posttreatment was calculated for each biological replicate and measured in technical triplicate for each time point using 2–ΔΔCT relative to the initial RNA abundance (t = 0). These values were plotted as a function of time and an exponential; one phase decay curve was fitted using GraphPad Prism. The decay curve is represented as R(t) = R0e–kt, where R0 and R(t) are the relative RNA abundance at initial and subsequent time points, respectively (78). The decay rate constant, k, is equal to ln(2)/t1/2, where t1/2 is the mRNA half-life. Accordingly, the half-life was derived from this equation as t1/2 = ln(2)/k.
RNA secondary structure predictions.
RNA secondary structure predictions were generated using RNAfold (ViennaRNA package v2.4.18) using default parameters (79). Full-length mRNA sequences, as revealed by RNA-seq read mapping, were used as input. All structures predicted using RNAfold were inspected and compared. Consensus structures with the lowest minimum free energy were downloaded in Vienna format and used to draw the RNA structure using the forna software (ViennaRNA package v2.4.18) (80).
Human blood and serum survival assays.
Measurement of survival in whole human blood and human serum was performed as previously described (81) with minor modifications. Briefly, A. baumannii cultures were grown overnight, in biological triplicate, and subcultured into 5 mL of fresh LB. After 3 h of growth, 10 mL of cells was centrifuged, washed with PBS, and diluted to an OD600 of 0.5. Cells were then added to 1 mL of deidentified whole human blood or serum (BioIVT) at a final OD600 of 0.05. The initial inoculum of each strain was determined at this time by serial dilution and plating on LB agar. Blood and serum cultures were incubated at 37°C with agitation, and the CFU/mL of each strain was determined, every hour for 6 h, by serial dilution and plating on LB agar.
Oxidative stress assay.
Oxidative stress was assessed using hydrogen peroxide as described previously (82) with minor modifications. Briefly, A. baumannii cultures were grown overnight in biological triplicate and subcultured into 5 mL of fresh medium. After 3 h of growth, new cultures were seeded and standardized to an OD600 of 0.05 in fresh LB. Hydrogen peroxide was then added to the cell suspensions for a final concentration of 2 mM and grown at 37°C with agitation. Cells were collected (500 μL) at each indicated time point and supplemented with catalase (10 μg/mL) to neutralize the effects of hydrogen peroxide. Cells were then serial diluted and plated on LB agar to determine the surviving CFU/mL.
Murine infection model.
The experiments were performed with the prior approval of the University of South Florida Institutional Animal Care and Use Committee. A murine model of dissemination/survival was performed based on previous studies (83, 84). Briefly, 13-week-old, female BL-6 mice were purchased from Charles River Laboratories and allowed to acclimate for 2 weeks prior to the start of experimentation. AB5075 wild-type and csp::tn strains were grown in LB as described above in biological triplicate. Overnight cultures were then subcultured 1:100 in fresh LB and grown for an additional 3 h. Cultures were then pelleted by centrifugation, washed twice in PBS, and standardized to an OD600 of 5.0. From this resuspension, the average number of CFU/mL was calculated for each strain by plating on LB agar. This was repeated on three separate days using two biological replicates per strain, and the average number of CFU/mL across replicates was calculated for each strain. This average number of CFU/mL was used to determine the volume of bacteria needed to obtain a 5-mL inoculum of 2.5 × 108 ([2.5 × 108/average CFU/mL] × 5 mL). On the day of infection, appropriate aliquots from fresh overnight cultures were prepared in the same manner and diluted to 2.5 × 108 CFU/mL, and 100 μL of suspension was administered to 10 mice per strain via retroorbital injection, providing a final inoculum of 2.5 × 107 CFU/mL. Infections were monitored, and the mice were sacrificed 6 h postinfection. At this time, liver, heart, kidneys, spleen, lungs, and brain were harvested and immediately stored at −80°C. Organs were individually homogenized using a bullet blender (Next Advance) in 1.5 mL PBS and serially diluted onto LB agar to determine bacterial burden (CFU/mL). A Mann-Whitney nonparametric test was performed to determine the statistical significance of bacterial burden for each organ between the mutant and wild-type strains.
Ethics statement.
All animal work was performed under the approval of the University of South Florida’s Institutional Animal Care and Use Committee (IACUC).
Data availability.
The experimental data from this study are deposited in the NCBI Gene Expression Omnibus (GEO) database (GEO accession numbers GSE164233 and GSE164290).
ACKNOWLEDGMENTS
This study was supported by grants AI124458 and AI157506 (L.N.S.) from the National Institute of Allergy and Infectious Diseases and GM133617 (P.J.E.) from the National Institute of General Medical Sciences. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We extend our thanks to the USF Genomics Program Genomics Equipment Core for the use of their facilities for RNA sequencing.
Conceptualization: B.R.T., L.N.S.; investigation: B.R.T., G.A.D., R.S.B., N.J.T., J.K.J., P.J.E.; methodology: B.R.T., J.L.A.; formal analysis: B.R.T.; writing, original draft preparation: B.R.T.; writing, review and editing: B.R.T., L.N.S.; funding: L.N.S.
We declare that there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Lindsey N. Shaw, Email: shaw@usf.edu.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.CDC. 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, Antibiotic Resistance Coordination and Strategy Unit, Atlanta, GA. [Google Scholar]
- 2.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 3.Weber DJ, Anderson D, Rutala WA. 2013. The role of the surface environment in healthcare-associated infections. Curr Opin Infect Dis 26:338–344. 10.1097/QCO.0b013e3283630f04. [DOI] [PubMed] [Google Scholar]
- 4.Shamsizadeh Z, Nikaeen M, Nasr Esfahani B, Mirhoseini SH, Hatamzadeh M, Hassanzadeh A. 2017. Detection of antibiotic resistant Acinetobacter baumannii in various hospital environments: potential sources for transmission of Acinetobacter infections. Environ Health Prev Med 22:44. 10.1186/s12199-017-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catalano M, Quelle LS, Jeric PE, Di Martino A, Maimone SM. 1999. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J Hosp Infect 42:27–35. 10.1053/jhin.1998.0535. [DOI] [PubMed] [Google Scholar]
- 6.Thom KA, Johnson JK, Lee MS, Harris AD. 2011. Environmental contamination because of multidrug-resistant Acinetobacter baumannii surrounding colonized or infected patients. Am J Infect Control 39:711–715. 10.1016/j.ajic.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayuga S, Zeana C, Sahni J, Della-Latta P, el-Sadr W, Larson E. 2002. Prevalence and antimicrobial patterns of Acinetobacter baumannii on hands and nares of hospital personnel and patients: the iceberg phenomenon again. Heart Lung 31:382–390. 10.1067/mhl.2002.126103. [DOI] [PubMed] [Google Scholar]
- 8.Huang YC, Su L-H, Wu T-L, Leu H-S, Hsieh W-S, Chang T-M, Lin T-Y. 2002. Outbreak of Acinetobacter baumannii bacteremia in a neonatal intensive care unit: clinical implications and genotyping analysis. Pediatr Infect Dis J 21:1105–1109. 10.1097/00006454-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Morgan DJ, Liang SY, Smith CL, Johnson JK, Harris AD, Furuno JP, Thom KA, Snyder GM, Day HR, Perencevich EN. 2010. Frequent multidrug-resistant Acinetobacter baumannii contamination of gloves, gowns, and hands of healthcare workers. Infect Control Hosp Epidemiol 31:716–721. 10.1086/653201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer LD, Minor KE, Mettlach JA, Rivera ES, Boyd KL, Caprioli RM, Spraggins JM, Dalebroux ZD, Skaar EP. 2020. Modulating isoprenoid biosynthesis increases lipooligosaccharides and restores Acinetobacter baumannii resistance to host and antibiotic stress. Cell Rep 32:108129. 10.1016/j.celrep.2020.108129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int J Artif Organs 34:824–831. 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 13.Fong JNC, Yildiz FH. 2015. Biofilm matrix proteins. Microbiol Spectr 3:3.2.28. 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limoli DH, Jones CJ, Wozniak DJ. 2015. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr 3:3.3.29. 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinal P, Marti S, Vila J. 2012. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect 80:56–60. 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Colquhoun JM, Rather PN. 2020. Insights into mechanisms of biofilm formation in Acinetobacter baumannii and implications for uropathogenesis. Front Cell Infect Microbiol 10:253. 10.3389/fcimb.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eze EC, Chenia HY, El Zowalaty ME. 2018. Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect Drug Resist 11:2277–2299. 10.2147/IDR.S169894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology (Reading) 149:3473–3484. 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 19.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pakharukova N, Tuittila M, Paavilainen S, Malmi H, Parilova O, Teneberg S, Knight SD, Zavialov AV. 2018. Structural basis for Acinetobacter baumannii biofilm formation. Proc Natl Acad Sci USA 115:5558–5563. 10.1073/pnas.1800961115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology (Reading) 154:3398–3409. 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 22.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eshwar AK, Guldimann C, Oevermann A, Tasara T. 2017. Cold-shock domain family proteins (Csps) are involved in regulation of virulence, cellular aggregation, and flagella-based motility in Listeria monocytogenes. Front Cell Infect Microbiol 7:453. 10.3389/fcimb.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White-Ziegler CA, Um S, Pérez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology (Reading) 154:148–166. 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 25.Keto-Timonen R, Hietala N, Palonen E, Hakakorpi A, Lindström M, Korkeala H. 2016. Cold shock proteins: a minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front Microbiol 7:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horn G, Hofweber R, Kremer W, Kalbitzer HR. 2007. Structure and function of bacterial cold shock proteins. Cell Mol Life Sci 64:1457–1470. 10.1007/s00018-007-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phadtare S, Severinov K. 2010. RNA remodeling and gene regulation by cold shock proteins. RNA Biol 7:788–795. 10.4161/rna.7.6.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroger C, MacKenzie KD, Alshabib EY, Kirzinger MWB, Suchan DM, Chao T-C, Akulova V, Miranda-CasoLuengo AA, Monzon VA, Conway T, Sivasankaran SK, Hinton JCD, Hokamp K, Cameron ADS. 2018. The primary transcriptome, small RNAs and regulation of antimicrobial resistance in Acinetobacter baumannii ATCC 17978. Nucleic Acids Res 46:9684–9698. 10.1093/nar/gky603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. 2015. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol 197:2027–2035. 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez D, Hidalgo-Cantabrana C, Rodríguez A, García P, Ruas-Madiedou P. 2016. Monitoring in real time the formation and removal of biofilms from clinical related pathogens using an impedance-based technology. PLoS One 11:e0163966. 10.1371/journal.pone.0163966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atienza JM, Zhu J, Wang X, Xu X, Abassi Y. 2005. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J Biomol Screen 10:795–805. 10.1177/1087057105279635. [DOI] [PubMed] [Google Scholar]
- 33.Loh B, Chen J, Manohar P, Yu Y, Hua X, Leptihn S. 2020. A biological inventory of prophages in A. baumannii genomes reveal distinct distributions in classes, length, and genomic positions. Front Microbiol 11:579802. 10.3389/fmicb.2020.579802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rumbo-Feal S, Gómez MJ, Gayoso C, Álvarez-Fraga L, Cabral MP, Aransay AM, Rodríguez-Ezpeleta N, Fullaondo A, Valle J, Tomás M, Bou G, Poza M. 2013. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One 8:e72968. 10.1371/journal.pone.0072968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geisinger E, Mortman NJ, Vargas-Cuebas G, Tai AK, Isberg RR. 2018. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog 14:e1007030. 10.1371/journal.ppat.1007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murzin AG. 1993. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J 12:861–867. 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theobald DL, Mitton-Fry RM, Wuttke DS. 2003. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 32:115–133. 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ching C, Gozzi K, Heinemann B, Chai Y, Godoy VG. 2017. RNA-mediated cis regulation in Acinetobacter baumannii modulates stress-induced phenotypic variation. J Bacteriol 199. 10.1128/JB.00799-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4389–4393. 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein JA, Lin P-H, Cohen SN, Lin-Chao S. 2004. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci USA 101:2758–2763. 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res 13:216–223. 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts C, Anderson KL, Murphy E, Projan SJ, Mounts W, Hurlburt B, Smeltzer M, Overbeek R, Disz T, Dunman PM. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J Bacteriol 188:2593–2603. 10.1128/JB.188.7.2593-2603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hambraeus G, von Wachenfeldt C, Hederstedt L. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol Genet Genomics 269:706–714. 10.1007/s00438-003-0883-6. [DOI] [PubMed] [Google Scholar]
- 44.Monem S, Furmanek-Blaszk B, Łupkowska A, Kuczyńska-Wiśnik D, Stojowska-Swędrzyńska K, Laskowska E. 2020. Mechanisms protecting Acinetobacter baumannii against multiple stresses triggered by the host immune response, antibiotics and outside-host environment. Int J Mol Sci 21:5498. 10.3390/ijms21155498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozkul C, Hazirolan G. 2021. Oxacillinase gene distribution, antibiotic resistance, and their correlation with biofilm formation in Acinetobacter baumannii bloodstream isolates. Microb Drug Resist 27:637–646. 10.1089/mdr.2020.0130. [DOI] [PubMed] [Google Scholar]
- 46.Marks LR, Davidson BA, Knight PR, Hakansson AP. 2013. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4:e00438-13. 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu H, KuoLee R, Harris G, Chen W. 2009. Role of NADPH phagocyte oxidase in host defense against acute respiratory Acinetobacter baumannii infection in mice. Infect Immun 77:1015–1021. 10.1128/IAI.01029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. 2019. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun 87:e00413-18. 10.1128/IAI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gil-Marques ML, Pachón-Ibáñez ME, Pachón J, Smani Y. 2018. Effect of hypoxia on the pathogenesis of Acinetobacter baumannii and Pseudomonas aeruginosa in vitro and in murine experimental models of infection. Infect Immun 86:e00543-18. 10.1128/IAI.00543-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cendra MDM, Blanco-Cabra N, Pedraz L, Torrents E. 2019. Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms. Sci Rep 9:16284. 10.1038/s41598-019-52726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. 2007. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol 189:4223–4233. 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wassarman KM, Storz G. 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101:613–623. 10.1016/S0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 53.Trotochaud AE, Wassarman KM. 2004. 6S RNA function enhances long-term cell survival. J Bacteriol 186:4978–4985. 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Preston JF, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186:2724–2734. 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, Romeo T. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol 190:3670–3680. 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amin M, Navidifar T, Shooshtari FS, Rashno M, Savari M, Jahangirmehr F, Arshadi M. 2019. Association between biofilm formation, structure, and the expression levels of genes related to biofilm formation and biofilm-specific resistance of Acinetobacter baumannii strains isolated from burn infection in Ahvaz, Iran. Infect Drug Resist 12:3867–3881. 10.2147/IDR.S228981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon E-J, Nait Chabane Y, Goussard S, Snesrud E, Courvalin P, Dé E, Grillot-Courvalin C. 2015. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6:e00309-15. 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52:557–562. 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45:3375–3380. 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leus IV, Adamiak J, Trinh AN, Smith RD, Smith L, Richardson S, Ernst RK, Zgurskaya HI. 2020. Inactivation of AdeABC and AdeIJK efflux pumps elicits specific nonoverlapping transcriptional and phenotypic responses in Acinetobacter baumannii. Mol Microbiol 114:1049–1065. 10.1111/mmi.14594. [DOI] [PubMed] [Google Scholar]
- 61.Esquerré T, Moisan A, Chiapello H, Arike L, Vilu R, Gaspin C, Cocaign-Bousquet M, Girbal L. 2015. Genome-wide investigation of mRNA lifetime determinants in Escherichia coli cells cultured at different growth rates. BMC Genomics 16:275. 10.1186/s12864-015-1482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qing G, Xia B, Inouye M. 2003. Enhancement of translation initiation by A/T-rich sequences downstream of the initiation codon in Escherichia coli. J Mol Microbiol Biotechnol 6:133–144. 10.1159/000077244. [DOI] [PubMed] [Google Scholar]
- 63.Casella LG, Weiss A, Pérez-Rueda E, Antonio Ibarra J, Shaw LN. 2017. Towards the complete proteinaceous regulome of Acinetobacter baumannii. Microb Genom 3:mgen000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cassat JE, Lee CY, Smeltzer MS. 2007. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol 391:127–144. 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacobs AC, Thompson MG, Gebhardt M, Corey BW, Yildirim S, Shuman HA, Zurawski DV. 2014. Genetic manipulation of Acinetobacter baumannii. Curr Protoc Microbiol 35:6G.2.1–11. 10.1002/9780471729259.mc06g02s35. [DOI] [PubMed] [Google Scholar]
- 66.Carroll RK, Weiss A, Shaw LN. 2016. RNA-sequencing of Staphylococcus aureus messenger RNA. Methods Mol Biol 1373:131–141. [DOI] [PubMed] [Google Scholar]
- 67.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 71.Brzozowski RS, Tomlinson BR, Sacco MD, Chen JJ, Ali AN, Chen Y, Shaw LN, Eswara PJ. 2020. Interdependent YpsA- and YfhS-mediated cell division and cell size phenotypes in Bacillus subtilis. mSphere 5:e00655-20. 10.1128/mSphere.00655-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang L, Schramm A, Neu TR, Revsbech NP, Meyer RL. 2013. Extracellular DNA in adhesion and biofilm formation of four environmental isolates: a quantitative study. FEMS Microbiol Ecol 86:394–403. 10.1111/1574-6941.12168. [DOI] [PubMed] [Google Scholar]
- 73.Sager M, Benten WPM, Engelhardt E, Gougoula C, Benga L. 2015. Characterization of biofilm formation in [Pasteurella] pneumotropica and [Actinobacillus] muris isolates of mouse origin. PLoS One 10:e0138778. 10.1371/journal.pone.0138778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fleeman R, LaVoi TM, Santos RG, Morales A, Nefzi A, Welmaker GS, Medina-Franco JL, Giulianotti MA, Houghten RA, Shaw LN. 2015. Combinatorial libraries as a tool for the discovery of novel, broad-spectrum antibacterial agents targeting the ESKAPE pathogens. J Med Chem 58:3340–3355. 10.1021/jm501628s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin H, Jiang W, Inouye M, Heinemann U. 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA 91:5119–5123. 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wada T, Becskei A. 2017. Impact of methods on the measurement of mRNA turnover. Int J Mol Sci 18:2723. 10.3390/ijms18122723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6:26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kerpedjiev P, Hammer S, Hofacker IL. 2015. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics 31:3377–3379. 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, Miller HK, Davenport JE, Riordan JT, Potempa J, Barber DS, Koziel J, Elasri MO, Shaw LN. 2011. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology (Reading) 157:2206–2219. 10.1099/mic.0.049692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. 2016. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palmer LD. 2019. Assessing Acinetobacter baumannii virulence and persistence in a murine model of lung infection. Methods Mol Biol 1946:289–305. 10.1007/978-1-4939-9118-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choby JE, Monteith AJ, Himmel LE, Margaritis P, Shirey-Rice JK, Pruijssers A, Jerome RN, Pulley J, Skaar EP. 2019. A phenome-wide association study uncovers a pathological role of coagulation factor X during Acinetobacter baumannii infection. Infect Immun 87:e00031-19. 10.1128/IAI.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salisbury V, Hedges RW, Datta N. 1972. Two modes of “curing” transmissible bacterial plasmids. J Gen Microbiol 70:443–452. 10.1099/00221287-70-3-443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S8 and Tables S1 and S4. Download iai.00376-22-s0001.pdf, PDF file, 2.5 MB (2.6MB, pdf)
Table S2. Download iai.00376-22-s0002.xlsx, XLSX file, 0.02 MB (26.3KB, xlsx)
Table S3. Download iai.00376-22-s0003.xlsx, XLSX file, 0.02 MB (17.2KB, xlsx)
Data Availability Statement
The experimental data from this study are deposited in the NCBI Gene Expression Omnibus (GEO) database (GEO accession numbers GSE164233 and GSE164290).