Abstract
Vaccines are among the most cost-effective public health interventions for preventing infection-induced morbidity and mortality, yet much remains to be learned regarding the mechanisms by which vaccines protect. Systems immunology combines traditional immunology with modern ‘omic profiling techniques and computational modeling to promote rapid and transformative advances in vaccinology and vaccine discovery. The NIH/NIAID Human Immunology Project Consortium (HIPC) has leveraged systems immunology approaches to identify molecular signatures associated with the immunogenicity of many vaccines. However, comparative analyses have been limited by the distributed nature of some data, potential batch effects across studies, and the absence of multiple relevant studies from non-HIPC groups in ImmPort. To support comparative analyses across different vaccines, we have created the Immune Signatures Data Resource, a compendium of standardized systems vaccinology datasets. This data resource is available through ImmuneSpace, along with code to reproduce the processing and batch normalization starting from the underlying study data in ImmPort and the Gene Expression Omnibus (GEO). The current release comprises 1405 participants from 53 cohorts profiling the response to 24 different vaccines. This novel systems vaccinology data release represents a valuable resource for comparative and meta-analyses that will accelerate our understanding of mechanisms underlying vaccine responses.
Subject terms: Databases, Vaccines
| Measurement(s) | Transcriptomics • Hemagglutination Inhibition Assay • IgG IgM IgA Total Measurement • Virus-neutralizing Antibody • ELISA |
| Technology Type(s) | Microarray • RNA sequencing • Hemagglutination Inhibition Assay • ELISA • Microneutralization Assay • serum neutralization of viral infectivity assay |
| Sample Characteristic - Organism | Homo sapiens |
Background & Summary
Vaccines, one of humanity’s greatest public health achievements, save millions of lives every year by preventing infectious diseases1,2. Despite their widespread use and efficacy, much remains to be learned regarding their molecular mechanisms of action. This is true both for vaccines against pandemic infections such as influenza3, and SARS-coronavirus-24, as well as for infections for which there are currently no authorized or approved vaccines such as HIV5–7. Elucidating the commonalities and differences in the immune responses induced by different vaccines and their association with protective antibody responses will provide deeper insight and a framework for the evidence-based design of better vaccines or vaccination strategies. Recent technologies have provided tools to probe the immune response to vaccination and integrate hierarchical levels of the biological system8. Alluded to as systems vaccinology9, this new application of systems biology tools provides new insights into molecular mechanisms of vaccine-induced immunogenicity and protection10–13.
The National Institute of Allergy and Infectious Diseases (NIAID) established a multi-institutional consortium, Human Immunology Project Consortium (HIPC)14,15, to characterize the immune system in diverse populations in response to a stimulus, such as vaccination, using high-dimensional ‘omic platforms and modern computational tools14. Since the inception of the consortium in 2010, members of HIPC have published >500 articles, including many that describe molecular signatures associated with vaccine-induced protection. These studies include molecular signatures that predict the immunogenicity of vaccination against yellow fever16–19, seasonal influenza in healthy young adults, elderly20–24, and children25, shingles26,27, dengue28,29, malaria30,31, and meta-analyses of common signatures across different vaccines32,33. These molecular signatures resulted from large-scale data analysis using high-throughput systems biology approaches coupled with detailed clinical phenotyping in well-characterized human cohorts.
Predicting immunogenicity from ‘omic signatures remains challenging, prompting methodological innovation to advance the field towards delivering on the promises of precision vaccination34–36. The factors that contribute to robust vaccination responses are highly complex and span multiple biological scales. The vast collection of high-dimensional profiling datasets poses significant challenges for comparative analysis of these studies, including biological variability as well as data challenges such as volume, technical noise, and diverse sample processing pipelines. Data integration of cellular and molecular signatures to predict vaccine responses requires harmonization and normalization of data from multiple sources37. The generation of big data poses simultaneous challenges and opportunities with the potential of contributing to precision medicine. The biological interpretation of the resulting molecular features correlated with robust responses is another key factor. Understanding how effective vaccines stimulate protective immune responses, and how these mechanisms may differ between vaccine types and targeted pathogens remains a substantial challenge for the field. Moreover, the systems vaccinology field has been limited by a lack of a formal framework to standardize immune signatures gathered from diverse studies, creating a bottleneck for comparative analysis. To address these challenges, and in support of advances in systems vaccinology by the HIPC project and the broader scientific community, we present the creation of the Immune Signatures Data Resource, a compendium of systems vaccinology studies that enables standardized comparative analysis to identify molecular signatures that correlate with the outcomes of vaccinations.
The current release of the Immune Signatures Data Resource consists of 4795 transcriptomic samples from 1405 participants curated from 30 ImmPort studies (16 from HIPC-related studies, 14 non-HIPC studies) (Fig. 2, Table 1). The transcriptomic profiling dataset is derived from 53 cohorts of 820 young adults (18–49 years old) and 585 (≥50 years old) older adult samples. The data resource covers 24 vaccines targeting 11 pathogens and 6 vaccine types (Figs. 1b, 4a, Table 2), thus creating a critical mass of data that will serve as a valuable resource for the broader scientific community. Additionally, data assembly and integration of these data set enables derivation of comparable signatures for each study for comparative analysis of the underlying data.
Fig. 2.
Flow chart diagram of the Immune Signatures Data Resource.
Table 1.
Overview of Immune Signatures Data Resource Study Participants Metadata.
| Study Accession | Pathogen (Vaccine Type) | Number of Participants | Number of Samples | Vaccine | Adjuvant | Race | Ethnicity | Cohort | Matrix | Pubmed ID | Geographical Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SDY1373 | Ebola47 (Recombinant Viral Vector) | 13 | 46 | UKE Phase I rVSV ZEBOV | VSV | Not Specified | Not Specified | dose 20 × 10^6 ofu,dose 3 × 10^6 pfu | SDY1373_WholeBlood_highDose_Geo,SDY1373_WholeBlood_lowDose_Geo | 28854372 | Metropolitan France |
| SDY1328 | Hepatitis A/B48 (Inactivated/Recombinant protein) | 164 | 325 | Twinrix | None | White | Not Hispanic or Latino | healthy adults | SDY1328_WholeBlood_HealthyAldults_Geo | 26742691 | Canada |
| SDY1291 | HIV49 (Recombinant Viral Vector) | 10 | 50 | Ad5/HIV | AdV | White, Black, or African American | Not Hispanic or Latino | healthy HIV-1-uninfected adults | SDY1291_PBMC_HealthyHIVUninfected_Geo | 23151505 | US: Washington |
| SDY1119 | Influenza22 (Inactivated) | 72 | 177 | TIV (2011) | None | Not Specified | Not Specified | young and old type 2 diabetes cohorts | SDY1119_PBMC_youngT2D_Geo, SDY1119_PBMC_youngHealthy_Geo,SDY1119_PBMC_oldHealthy_Geo,SDY1119_PBMC_oldT2D_Geo | 26682988 | US: Georgia |
| SDY1276 | Influenza50 (Inactivated) | 218 | 828 | TIV (2008) | None | Not Specified | Not Specified | Validation Cohort; Females 2008-2009 trivalent influenza vaccine,Discovery Cohort; Males 2008?2009 trivalent influenza vaccine | SDY1276_WholeBlood_Validation_Geo,SDY1276_WholeBlood_Discovery_Geo | 21357945 | US: Texas |
| SDY180 | Influenza51 (Inactivated) | 12 | 102 | TIV (2009) | None | Asian,Whit e,Black or African American | Not Hispanic or Latino | Study group 2 2009-2010 Fluzone,Study group 1 2009-2010 Fluzone | SDY180_WholeBlood_Grp2Fluzone_G eo,SDY180_WholeBlood_Grp1Fluzone_Geo | 23601689 | US: Texas |
| SDY212 | Influenza52 (Inactivated) | 90 | 90 | TIV (2008) | None | Oth er,Wh ite,As ian,American I,ndian or Alaska Native | Not Hispanic or L atino,Hispanic or Latino | Cohort_1,Cohort_2 | SDY212_WholeBlood_Young_Geo,SDY212_PBMC_Young_geo,SDY212_WholeBlood_Older_Geo,SDY212_PBMC_Older_Geo | 23591775 | US: California |
| SDY224 | Influenza53 (Inactivated) | 5 | 55 | TIV (2010) | None | White,Black or African American,American Indian or Alaska Native | Not Hispanic or Latino,Hispanic or Latino | TIV 2010 | SDY224_PBMC_TIV2010_ImmPort | 23900141 | US: New York |
| SDY269 | Influenza23 (Inactivated) | 28 | 80 | TIV (2008) | None | White,Asian,Black or African American | Not Hispanic or Latino,Hispanic or Latino | TIV Group 2008 | SDY269_PBMC_TIV_Geo | 21743478 | US: Georgia |
| SDY270 | Influenza23 (Inactivated) | 28 | 83 | TIV (2009) | None | White,Black or African American,Asian | Not Hispanic or Latino,Hispanic or Latino | TIV Group 2009 | SDY270_PBMC_TIVGroup_Geo | 21743478 | US: Georgia |
| SDY400 | Influenza21 (Inactivated) | 30 | 120 | TIV (2012) | None | White,Asian,Black or African American,Other | Not Hispanic or Latino,Hispanic or Latino | Young adults 21-30 years old,Older adults > = 65 years old | SDY400_PBMC_Young_Geo,SDY400_PBMC_Older_Geo | 32060136 | US: Connecticut |
| SDY404 | Influenza25 (Inactivated) | 39 | 156 | TIV (2011) | None | White,Unknown,Other,Asian,Black or African American | Not Hispanic or Latino,Hispanic or Latino | Young adults 21-30 years old,Older adults > = 65 years old | SDY404_PBMC_Young_Geo,SDY404_PBMC_Older_Geo | 25596819 | US: Connecticut |
| SDY520 | Influenza21 (Inactivated) | 24 | 94 | TIV (2013) | None | White,Asian,Black or African American | Not Hispanic or Latino,Hispanic or Latino | Young adults 21-30 years old,Older adults > = 65 years old | SDY520_WholeBlood_Young_geo,SDY520_WholeBlood_Older_Geo | 32060136 | US: Connecticut |
| SDY56 | Influenza22 (Inactivated) | 63 | 288 | TIV (2010) | None | White,Asian,Black or African American | Not Hispanic or Latino,Hispanic or Latino | Healthy adults 25-40 years old receiving TIV flu vaccine,Healthy adults >65 years old receiving TIV flu vaccine | SDY56_PBMC_Young,SDY56_PBMC_Older | 26682988 | US: Georgia |
| SDY61 | Influenza23 (Inactivated) | 9 | 27 | TIV (2007) | None | White | Not Hispanic or Latino,Hispanic or Latino | TIV Group 2007 | SDY61_PBMC_TIVGrp | 21743478 | US: Georgia |
| SDY63 | Influenza25 (Inactivated) | 19 | 72 | TIV (2010) | None | White,Asian,Other,Black or African American | Not Hispanic or Latino | Young adults 21-30 years old,Older adults > = 65 years old | SDY63_PBMC_Young_Geo,SDY63_PBMC_Older_Geo | 25596819 | US: Connecticut |
| SDY640 | Influenza21 (Inactivated) | 20 | 79 | TIV (2014) | None | White,Asian,Unknown | Not Hispanic or Latino,Hispanic or Latino | Young adults 21-30 years old,Older adults > = 65 years old | SDY640_WholeBlood_Young_Geo,SDY640_WholeBlood_Older_Geo | 32060136 | US: Connecticut |
| SDY80 | Influenza54 (Inactivated) | 61 | 286 | TIV (2009) + pH1N1 | None | White,Asian,Other,Black or African American | Other,Hispanic or Latino | Cohort2 | SDY80_PBMC_Cohort2_geo | 24725414 | US: Maryland |
| SDY269 | Influenza23 (Live attenuated) | 28 | 83 | LAIV (2008) | LAIV | White,Black or African American,Asian | Not Hispanic or Latino,Hispanic or Latino | LAIV group 2008 | SDY269_PBMC_LAIV_Geo | 21743478 | US: Georgia |
| SDY1293 | Malaria55 (Recombinant protein) | 44 | 165 | RTS,S/AS01 or RTS,S/AS02 | AS01/AS02 | Not Specified | Not Specified | adjuvanted RTS,S malaria vaccine cohort | SDY1293_PBMC_Vaccinated_geo | 20078211 | US: Maryland |
| SDY1260 | Meningococcus33 (Conjugate) | 17 | 51 | MCV4 | None | Not Specified | Not Specified | MCV4 | SDY1260_PBMC_MCV4_Geo | 24336226 | US: Georgia |
| SDY1325 | Meningococcus56 (Conjugate) | 5 | 10 | MenACWY-CRM | None | Not Specified | Not Specified | Intramuscular MenACWY-CRM | SDY1325_WholeBlood_IntramuscularCRM_Geo | 28137280 | England |
| SDY1260 | Meningococcus33 (Polysaccharide) | 13 | 39 | MPSV4 | None | Not Specified | Not Specified | MPSV4 | SDY1260_PBMC_MPSV4_Geo | 24336226 | US: Georgia |
| SDY1325 | Meningococcus56 (Polysaccharide) | 5 | 10 | MenACWY-PS | None | Not Specified | Not Specified | Intramuscular MenACWY-PS | SDY1325_WholeBlood_IntramuscularPS_Geo | 28137280 | England |
| SDY180 | Pneumococcus51 (Polysaccharide) | 12 | 101 | Pneumovax23 | None | White,Black or African American,Asian | Not Hispanic or Latino,Hispanic or Latino | Study group 2 Pneunomax23,Study group 1 Pneunomax23 | SDY180_WholeBlood_Grp2Pneunomax23_Geo,SDY180_WholeBlood_Grp1Pneunomax23_Geo | 23601689 | US: Texas |
| SDY1370 | Smallpox57 (Live virus) | 4 | 24 | DryVax | Vaccinia | Unknown | Not Specified | DryVax | SDY1370_PBMC_dryvax_geo | 21921208 | US: Massachusetts |
| SDY1370 | Smallpox57 (Live virus) | 4 | 24 | LC16m8 | Vaccinia | Unknown | Not Specified | LC16m8 | SDY1370_PBMC_lc16m8_geo | 21921208 | US: Massachusetts |
| SDY1364 | Tuberculosis58 (Recombinant Viral Vector) | 12 | 36 | MVA85A | Vaccinia | Not Specified | Not Specified | MVA85A intramuscular | SDY1364_PBMC_IntraMuscular_Geo | 23844129 | England |
| SDY984 | Varicella Zoster27 (Live attenuated) | 72 | 288 | Zostavax | VZV | White,Black or African American,Unknown,Asian | Not Hispanic or Latino,Hispanic or Latino | young,elderly | SDY984_PBMC_Young_Geo,SDY984_PBMC_Elderly_Geo | 28502771 | US: Georgia, US: Colorado |
| SDY1264 | Yellow Fever19 (Live attenuated) | 25 | 87 | YF17D | YF17D | Not Specified | Not Specified | Trial2,Trial1 | SDY1264_PBMC_Trial2_Geo,SDY1264_PBMC_Trial1_Geo | 19029902 | US: Georgia |
| SDY1289 | Yellow Fever18 (Live attenuated) | 25 | 117 | YF17D | YF17D | Not Specified | Not Specified | in vivo vaccination study Montreal adult cohort,in vivo vaccination study Lausanne adult cohort | SDY1289_WholeBlood_MontrealCohort_Geo,SDY1289_WholeBlood_LausanneCohort_Geo | 19047440 | Canada, Switzerland, US: Georgia |
| SDY1294 | Yellow Fever59 (Live attenuated) | 21 | 109 | YF17D | YF17D | Asian | Not Hispanic or Latino | Chinese cohort | SDY1294_PBMC_ChineseCohort_Geo | 28687661 | China |
| SDY1529 | Yellow Fever18 (Live attenuated) | 36 | 180 | YF17D | YF17D | Black or African American | Not Hispanic or Latino | healthy adults | SDY1529_WholeBlood_HealthyAdults_PreVax_Geo,SDY1529_WholeBlood_HealthyAdults_PostVax_Geo | 19047440 | Uganda |
Fig. 1.
HIPC Immune Signatures Data Resource pipeline and study demographics. (a) Systems vaccinology datasets from existing HIPC studies, as well as published systems vaccinology papers and databases, were submitted to the ImmPort database. ImmuneSpace captures these datasets to create a combined compendium dataset. Quality control assessments of these data include array quality checks for microarray studies, batch correction, imputations for missing age and sex/y-chromosome presence information, and normalization per study. The combined virtual study included transcriptional profiles and antibody response measurements from 1405 participants across 53 cohorts, profiling the response to 24 different vaccines. Note that Hepatitis A/B (Twinrix) cohort also received Diphtheria/Tetanus toxoid (Td) and Cholera inactivated vaccine at the same time (Dukoral). (b) Demographic data included biological sex, race, vaccine, and number of participants.
Fig. 4.
Immune Signatures Transcriptomics Overview for young and old datasets. (a) Number of samples available for each data type, including transcriptomics (TX), hemagglutination inhibition assay (HAI), neutralizing antibody assay (NAB), and ELISA assays (ELISA). (b) Bar plot depicting the number of samples at each time point. The colors within each bar indicate the breakdown for each unique combination of pathogen and vaccine type. Day -7 and day 0 correspond to times pre-vaccination. (c) Box plot depicting the participant’s age distribution for each unique combination of pathogen and vaccine type. Note that Hepatitis A/B (Twinrix) cohort also received Diphtheria/Tetanus toxoid (Td) and Cholera inactivated vaccine at the same time (Dukoral). (d) Each area-proportional Euler diagram represents the total number of participants with corresponding data types.
Table 2.
Overview of Transcriptomics Datasets Included in the Resource.
| Study Accession | Pathogen (Vaccine type) | Sample type | featureSetName | featureSetName2 | featureSetVendor | Time post last vaccination | GEO Accession |
|---|---|---|---|---|---|---|---|
| SDY1373 | Ebola (Recombinant Viral Vector) | Whole blood | SDY1373_customAnno | RNA-seq | NA | 0, 1, 3, 7 | GSE97590 |
| SDY1328 | Hepatitis A/B (Inactivated/Recombinant protein) | Whole blood | Affy_HumanRSTAcustom | RNA-seq | Affymetrix | 0, 7 | GSE65834 |
| SDY1291 | HIV (Recombinant Viral Vector) | PBMC | Affy_HumanExonST_1_0_v2 | Affy_HumanExonST_1_0_v2 | Affymetrix | 0, 0.25, 1, 3, 7 | GSE22768 |
| SDY1119 | Influenza (Inactivated) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 3, 7 | GSE74817 |
| SDY1276 | Influenza (Inactivated) | Whole blood | HumanHT-12_v3_2018 | HumanHT-12_2018 | Illumina | 0, 1, 3, 14 | GSE48024/GSE48018 |
| SDY180 | Influenza (Inactivated) | Whole blood | HumanHT-12_v3_2018 | HumanHT-12_2018 | Illumina | −7, 0, 0.5, 1, 3, 7, 10, 14, 21, 28 | GSE48762 |
| SDY212 | Influenza (Inactivated) | Whole blood | HumanHT-12_v3_2018 | HumanHT-12_2018 | Illumina | 0 | GSE41080 |
| SDY224 | Influenza (Inactivated) | PBMC | SDY224_CustomAnno | RNA-seq | NA | 0, 1, 2,3, 4, 5, 6, 7, 8, 9, 10 | GSE45735 |
| SDY269 | Influenza (Inactivated) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 3, 7 | GSE29615/GSE29617/GSE29614 |
| SDY270 | Influenza (Inactivated) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 3, 7 | GSE29617/GSE29614 |
| SDY400 | Influenza (Inactivated) | PBMC | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 2, 4, 7, 28 | GSE59743/GSE95584 |
| SDY404 | Influenza (Inactivated) | PBMC | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 2, 4, 7, 28 | GSE59654 |
| SDY520 | Influenza (Inactivated) | Whole blood | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 2, 7, 28 | GSE101709 |
| SDY56 | Influenza (Inactivated) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 1, 3, 7, 14 | GSE74817 |
| SDY61 | Influenza (Inactivated) | PBMC | hgu133plus2 | hgu133plus2 | Affymetrix | 0, 3, 7 | GSE29617/GSE29614 |
| SDY63 | Influenza (Inactivated) | PBMC | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 4, 7, 28 | GSE59635 |
| SDY640 | Influenza (Inactivated) | Whole blood | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 2, 7, 28 | GSE101710 |
| SDY80 | Influenza (Inactivated) | PBMC | HuGene-1_0-st-v1 | HuGene-1_0-st-v1 | Affymetrix | −7, 0, 1, 7, 70 | GSE47353 |
| SDY269 | Influenza (Live attenuated) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 3, 7 | GSE29615/GSE29617/GSE29614 |
| SDY1293 | Malaria (Recombinant protein) | PBMC | hgu133plus2 | hgu133plus2 | Affymetrix | 0, 1, 3, 14 | GSE18323 |
| SDY1260 | Meningococcus (Conjugate) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 3, 7 | GSE52245 |
| SDY1325 | Meningococcus (Conjugate) | Whole blood | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 7 | GSE92884 |
| SDY1260 | Meningococcus (Polysaccharide) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 3, 7 | GSE52245 |
| SDY1325 | Meningococcus (Polysaccharide) | Whole blood | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 7 | GSE92884 |
| SDY180 | Pneumococcus (Polysaccharide) | Whole blood | HumanHT-12_v3_2018 | HumanHT-12_2018 | Illumina | −7, 0, 0.5, 1, 3, 7, 10, 14, 21, 28 | GSE48762 |
| SDY1370 | Smallpox (Live virus) | PBMC | HEEBOHumanSetV1_2019 | HEEBOHumanSetV1_2019 | Stanford Functional Genomics Facility | 0, 3, 7, 10, 13, 21 | GSE22121 |
| SDY1370 | Smallpox (Live virus) | PBMC | HEEBOHumanSetV1_2019 | HEEBOHumanSetV1_2019 | Stanford Functional Genomics Facility | 0, 3, 7, 10, 13, 21 | GSE22121 |
| SDY1364 | Tuberculosis (Recombinant Viral Vector) | PBMC | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 2, 7 | GSE40719 |
| SDY984 | Varicella Zoster (Live attenuated) | PBMC | HGU133_plus_PM | HGU133_plus_PM | Affymetrix | 0, 1, 3, 7 | GSE79396 |
| SDY1264 | Yellow Fever (Live attenuated) | PBMC | hgu133plus2 | hgu133plus2 | Affymetrix | 0, 1, 3, 7, 21 | GSE13485 |
| SDY1289 | Yellow Fever (Live attenuated) | Whole blood | IlluminaHumanRef8_v2 | IlluminaHumanRef8_v2 | Illumina | 0, 3, 7, 10, 14, 28, 60 | GSE13699 |
| SDY1294 | Yellow Fever (Live attenuated) | PBMC | AffyPrimeView_2016 | AffyPrimeView_2016 | Affymetrix | 0, 0.166666666666667, 1, 2, 3, 5, 7, 14, 28 | GSE82152 |
| SDY1529 | Yellow Fever (Live attenuated) | Whole blood | HumanHT-12_v4_2018 | HumanHT-12_2018 | Illumina | 0, 3, 7, 14, 84 | GSE125921/GSE136163 |
Methods
Database background information and structure
Compatibility with immport and immunespace, the central databases of the human immunology project consortium
Given the exponential growth of the number of datasets of multiple modalities, an urgent need emerged for data sharing across the broader scientific community. The HIPC implements the NIH Data Sharing policy to promote the principles of Findability, Accessibility, Interoperability, and Reusability (FAIR) via ImmPort, created under the National Institute of Allergy and Infectious Diseases Division of Allergy, Immunology, and Transplantation (NIAID-DAIT). ImmPort (ImmPort.org) is an open repository of participant-level large-scale human immunology data designed to aid scientists with data standards and guidelines for efficient secondary analyses38,39. ImmPort facilitates data sharing of immunology studies creating a centralized knowledge base and resources, and serves as a central data repository for HIPC. ImmuneSpace14,33 extends ImmPort, providing access to additional data (e.g., standardized gene expression matrices) and web-based R tools for data accession, analysis, and reporting. Studies in the Immune Signatures Data Resource are archived through the Shared Data Portal on ImmPort and ImmuneSpace repositories and may be updated over time. To provide a consistent data source for reproducible results, we also archived a static copy of the data as a “virtual study” in ImmuneSpace (Figs. 1a and 2).
Identification of vaccine study cohorts with transcriptomic profiles
Through a literature search conducted from July 2017 to January 2020 with terms including “Vaccine [AND] signatures”, “Vaccine [AND gene expression”, “Vaccine [AND] immune response [AND] gene expression”, we identified target publications containing transcriptomics profiling datasets and vaccination responses. We found 16 HIPC-funded vaccinology studies in ImmPort with transcriptomics datasets generated with matching immune response outcomes and surveyed HIPC centers of their publications. We excluded non-human study cohorts, cohorts with B cell and T cell transcriptomics since most studies are PBMC or whole blood-derived, studies other than with intramuscular mode of vaccine route, studies with subjects beyond our target age range (<18), and those studies that lack vaccine stimulation. Notably, we have supplemented the HIPC data previously available in ImmPort by curating and submitting 14 additional human vaccination studies to ImmPort. For studies that were not in ImmPort/ImmuneSpace, we located the underlying data by surveying public transcriptome databases (e.g., Gene Expression Omnibus (GEO)) or reaching out to study authors to request data access, allowing us to submit to ImmPort on their behalf. These datasets were then made available via ImmuneSpace to be processed for standardization, preprocessing checks, and normalization. The standard analytical pipeline enables reproducibility and comparability of future studies to be correlated with publicly available immune response measurement. This process created the virtual study for the HIPC named the Immune Signatures Data Resource (Figs. 1a, 2).
Gene expression data processing pipeline
Data were read directly from ImmuneSpace using ImmuneSpaceR functions and subsequently preprocessed, quality controlled, and integrated using the following pipeline:
Quality control of microarray experiments
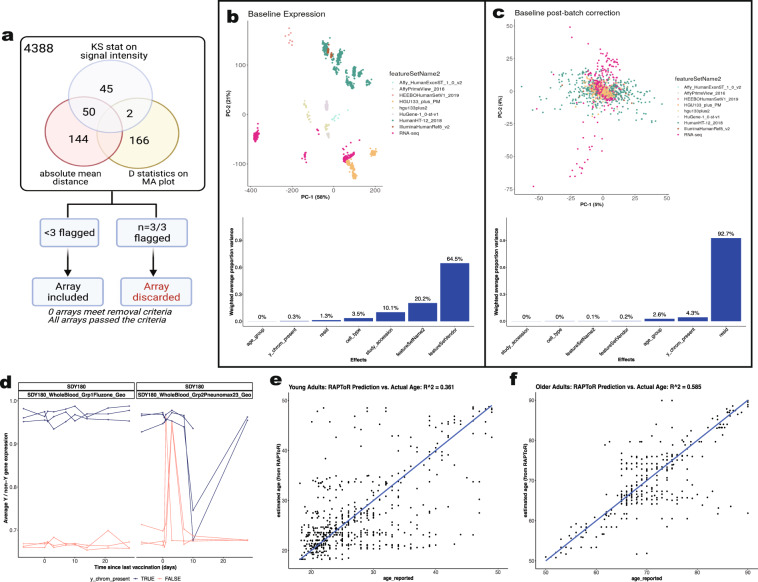
The ArrayQualityMetrics R package40 was used for quality control and assurance of all microarray experiments (Fig. 3a). Outlier detection was based on the following statistics: i) Mean absolute difference of M-values (log-ratios) of each pair of arrays, ii) the Kolmogorov-Smirnov statistic Ka between each array’s signal intensity distribution and the distribution of the pooled data and, iii) the Hoeffding’s statistic Da on the joint distribution of A (average) and M values for each array. Using pre-specified criteria within an established public microarray data reuse pipeline40, we flagged for removal arrays that failed all three quality control statistics.
Fig. 3.
Quality control assessments of transcriptomics data. (a) Sample quality assessments of gene expression datasets using Array Quality metrics. Array quality metrics package was employed to assess quality of microarray datasets by checking the following criteria: (a) absolute mean difference between arrays to check the probe and median intensity across all arrays, (b) Kolmogorov-Smirnov statistics to check the signal intensity distribution of arrays, comparing each probe versus distribution of test statistics for all other probes, (c) Hoeffding’s D-statistics for arrays. Arrays were excluded if they fail all three criteria above. (b,c) Principal component analysis (Top) and Principal Variation component Analysis (PVCA) of baseline expression data per study before (B) and after batch correction (C). (d) Biological sex imputation based on expression of Y-chromosome genes. We used 13 Y-chromosome-associated genes to cluster samples into 2 groups assuming biological male or female. (e,f) Age imputation based on transcriptomic profiles for studies without reported ages (SDY1260, SDY1264, SDY1293, SDY1294, SDY1364, SDY1370, SDY1373, SDY984) via the RAPToR R package44. Virtual studies were split into young (age < 50, E) and older (age > = 50, F) for two separate predictive models.
Preprocessing
Raw probe intensity data for Affymetrix studies were background-corrected and summarized using the RMA algorithm41 while the function read.ilmn (limma R package) was used to read and background correct Illumina raw probe intensities. To integrate RNA-seq and microarray data, raw counts for RNA-seq data were transformed using the variance stabilizing transformation (VST). VST yields expression values that are normalized across samples and by library size and approximately homoskedastic. After a proper log-2 transformation they can be analyzed as microarray data, using linear models in the limma framework. Expression data within each study were quantile normalized and log-transformed separately for each cohort/sample type.
Annotation
We annotated the manufacturing IDs (probes from microarray/Illumina) to their corresponding gene alias. Gene aliases were mapped to the recent gene symbols from the HUGO Gene Nomenclature Committee42 [accessed Dec 23, 2020]. For the rare case where a gene alias mapped to more than one gene symbol, the mapping was resolved by the following: i) If a gene alias mapped to itself as a symbol, as well as other symbols, then it was mapped to itself; ii) if the gene alias mapped to multiple symbols that did not include itself, then the gene alias was dropped from the study. As a result, the raw gene expression matrix was reduced to 10086 HUGO gene aliases with known unique mapping.
Gene-based expression profiles
Expression data were summarized at the probe level (for microarray data) and gene-alias level (RNA-seq) to the canonical Gene-Symbol level. The probes/gene-aliases were summarized by selecting the probe or gene-alias with the highest average expression (mean of probes across all samples, take the highest mean) across all samples within the matrix (cohort and sample type).
Cross-Study normalization
One of the main assumptions in expression analysis is that differences in gene expression across conditions occur in a relatively small number of processes. As such, the distribution across conditions should be similar, and departures of these assumptions are corrected, for example, using quantile normalization. This procedure usually creates a target distribution using all samples available, but we observed dissimilar distributions in our collection stemming from various platforms used. Such differences lead to extensive distributions and introduce artifacts in the data (Fig. 3b,c). The target distribution was obtained from samples using Affymetrix platforms, resulting in a well-defined distribution, and each sample in our collection was quantile normalized to this target distribution. Before cross-study normalization, there were 35,725 representative gene symbols present. There were 25,639 genes removed after normalization, as these genes were not present in all the studies. This yielded a final expression matrix of 4795 samples from 1405 participants representing 10,086 genes (Fig. 2).
Determining and adjusting for technical confounders
We studied the primary sources of variation in the data, including the study effect (which also encompasses the impact of different expression platforms (RNA-seq, Affymetrix arrays, Illumina arrays, etc.), sample types (Whole blood, PBMC), as well as demographics. We conducted Principal Component Analysis (PCA) to visualize such associations in a bidimensional space of principal components (PCs) and applied Principal Variance Component Analysis (PVCA)43 to quantify the amount of variability attributed to different experimental conditions. This approach models the multivariate distribution of the PCs computed for the PCA as a function of experimental factors and estimates the total variance explained by each factor via mixed-effect models. Since many studies included only one vaccine, temporal variations due to vaccine response were confounded with the study effect. The assessment of the primary technical sources of variation was carried out using only the pre-vaccination data, not affected by the targeted pathogen and vaccine type used in the different studies. Of note, all studies enrolled healthy volunteers, and the first biosample was obtained pre-vaccination. The targeted pathogen and vaccine type should not affect these baseline data.
Platform, study, and sample types were identified as significant sources of variation in the gene expression matrix. The effect of those three variables was estimated by modeling gene expression at baseline (at which no vaccine or timepoint effect exists) with a linear model using the limma framework, including feature set vendor (Platform/Affy), study (batch factors), and sample type, Y-chromosome genes presence, as covariates. Study and cell-type effects were estimated using a linear model with age, Y-chromosome genes presence (biological sex), study, sample type (Whole Blood/PBMC), study, and platform as additive effects. From here, the study, platform, and cell-type effects were eliminated from the entirety of the expression matrix. There were three studies (SDY1276, SDY1264, SDY180) that contained multiple cohorts and were treated as separate studies.
Biological sex imputation
Imputation of biological sex, as defined by the presence of a Y-chromosome, was carried out based on the gene expression profiles of 13 Y-chromosome genes. Within each study, a multidimensional scaling was first applied to the Y-chromosome gene expression profiles. K-means clustering was then used to cluster samples into two groups. Participants in the cluster with higher mean expression values were considered male (i.e., the Y-chromosome was present) while those in the cluster with lower expression were considered female (i.e., the Y-chromosome was absent). The consistency of the Y-chromosome presence assignment across time points was verified (Fig. 3d). In the (few) cases where imputation was not in agreement across all time points, the reported sex was used and if no sex was reported, imputation followed a majority rule principle.
Age imputation
Age imputation for studies without reported ages (SDY1260, SDY1264, SDY1293, SDY1294, SDY1364, SDY1370, SDY1373, SDY984) employed the RAPToR R v1.1.5 package44. The RAPToR algorithm takes in a reference set of gene expression time series with reported ages and generates a near-continuous, high-temporal resolution from the interpolated reference dataset. Transcriptomic profiles of participants without reported ages were compared to the reference dataset via a correlation profile, providing age estimates for the sample. Finally, random subsets of genes from the subject’s transcriptomic profile were bootstrapped to ascertain a confidence interval for the imputed age. We generated the reference dataset using the transcriptomic profiles of 21 studies in our resource for which age was reported. The studies were split into younger (age <50) and older (age ≥50) cohorts, thus two different models were generated, and only baseline transcriptomic profiles were used in the reference dataset. As RAPToR also enables phenotypic data to be incorporated into the interpolation model, each possible combination of phenotypic features was tested. These phenotypic features included the top variables found during our PVCA tests as well as demographic information such as reported age, cohort and matrix type, Y chromosome imputation, study accession, feature set vendor and platform names, and cell types. For each combination, RAPToR predicted the age of participants in the 21 studies with known age, and the goodness of fit was evaluated by the coefficient of determination (R2) and confirmed via RMSE. The best model for the younger and older cohorts was then used to impute ages for the 7 studies without reported age (Fig. 3e,f)
Immune response datasets processing pipeline
To identify the molecular signatures that correlate with vaccine immunogenicity, we included immune response readouts in the creation of this data resource. For studies that were missing vaccine response endpoints in their public data deposition, we contacted study authors and requested available antibody response measures to vaccine antigens. Once shared, these data were submitted to ImmPort and linked to the relevant studies. These readouts include neutralizing antibody titers (Nab), hemagglutination inhibition assay (HAI) results for influenza studies, and Immunoglobulin IgG ELISA assay results. In participants for whom the humoral immune response was measured with multiple assays, the preference was given to HAI for influenza or Nab for non-influenza studies, then IgG ELISA datasets. The antibody measures were normalized within each study by estimating the fold-change differences between the post-vaccination time-point (generally between day 28 or day 30) compared to the baseline measurement. For influenza studies where the vaccine included multiple strains, the fold changes between the post-vaccination versus baseline were calculated for each strain, and the maximum fold change (MFC) over the strains was selected33. Due to the variability in baseline antibody (Ab) levels and immune memory such as influenza vaccines, we also estimated the maximum residual after baseline adjustment (maxRBA) method by calculating the maximum residual across all vaccine strains to adjust for variable baseline Ab levels using the R package titer20. A total of 30 studies with 1405 participants and 4795 samples have both transcriptomics and immune response readout data available (Fig. 2). This dataset enables researchers to carry out comparative analyses using immunogenicity data as well as prediction of the quality of response across multiple vaccines.
Data Records
The Immune Signatures Data Resource is available online for download by the research community from this website45: 10.6084/m9.figshare.17096978. The data is hosted on ImmuneSpace and can be accessed in full detail via the R package ImmuneSpaceR (https://rglab.github.io/ImmuneSpaceR/). The resource is available for use by the scientific community and can be downloaded from a research data repository IS2 https://www.ImmuneSpace.org/is2.url. A summary of datasets17,18,20–22,24,26,32,46–58, with their corresponding study ID, accession numbers and DOI, is provided in Table 3.
Table 3.
Studies with corresponding Immune Response Data.
| Study Accession | Pathogen Vaccine Type | Number of Participants | Number of Samples | Assay | Digital Object Identifier (DOI) |
|---|---|---|---|---|---|
| SDY1328 | Hepatitis A/B (Inactivated/ Recombinant protein) | 160 | 320 | ELISA | 10.21430/M3ID8ZC1AT |
| SDY1119 | Influenza (Inactivated) | 72 | 177 | HAI | 10.21430/M3ZU72TO6V |
| SDY1276 | Influenza (Inactivated) | 214 | 816 | HAI, NAb | 10.21430/M3J92GN8I3 |
| SDY180 | Influenza (Inactivated) | 12 | 102 | HAI, NAb | 10.21430/M3I44H8R17 |
| SDY212 | Influenza (Inactivated) | 88 | 88 | HAI | 10.21430/M37NGTHMDS |
| SDY224 | Influenza (Inactivated) | 5 | 55 | HAI | 10.21430/M37KMO7JLW |
| SDY269 | Influenza (Inactivated) | 28 | 80 | HAI | 10.21430/M3CDX6TL4I |
| SDY270 | Influenza (Inactivated) | 28 | 83 | HAI | 10.21430/M3H9N1SFLO |
| SDY400 | Influenza (Inactivated) | 30 | 120 | HAI | 10.21430/M3U7GDOFIT |
| SDY404 | Influenza (Inactivated) | 39 | 156 | HAI | 10.21430/M3GWQRC8DT |
| SDY520 | Influenza (Inactivated) | 24 | 94 | HAI | 10.21430/M3KVVHM735 |
| SDY56 | Influenza (Inactivated) | 30 | 148 | HAI | 10.21430/M3X9SKF8RQ |
| SDY61 | Influenza (Inactivated) | 9 | 27 | HAI | 10.21430/M3FH0SA2W0 |
| SDY63 | Influenza (Inactivated) | 19 | 72 | HAI | 10.21430/M38WXGBDTS |
| SDY640 | Influenza (Inactivated) | 20 | 79 | HAI | 10.21430/M3A6GYD5L0 |
| SDY67 | Influenza (Inactivated) | 159 | 477 | HAI | 10.21430/M3OYWCJHO1 |
| SDY80 | Influenza (Inactivated) | 60 | 281 | NAb | 10.21430/M3STAI2V6T |
| SDY269 | Influenza (Live attenuated) | 28 | 83 | HAI | 10.21430/M3CDX6TL4I |
| SDY1260 | Meningococcus (Conjugate) | 17 | 51 | ELISA | 10.21430/M3F47KSLLP |
| SDY1325 | Meningococcus (Conjugate) | 4 | 8 | NAb | 10.21430/M3Q1ZBWOG2 |
| SDY1260 | Meningococcus (Polysaccharide) | 13 | 39 | ELISA | 10.21430/M3F47KSLLP |
| SDY1325 | Meningococcus (Polysaccharide) | 5 | 10 | NAb | 10.21430/M3Q1ZBWOG2 |
| SDY180 | Pneumococcus (Polysaccharide) | 6 | 54 | NAb | 10.21430/M3I44H8R17 |
| SDY1370 | Smallpox (Live virus) | 4 | 24 | ELISA | 10.21430/M3QHF445NF |
| SDY1364 | Tuberculosis (Recombinant Viral Vector) | 12 | 36 | ELISA | 10.21430/M3NJTLGRT4 |
| SDY984 | Varicella Zoster (Live attenuated) | 35 | 140 | ELISA | 10.21430/M36N1BYFT5 |
| SDY1264 | Yellow Fever (Live attenuated) | 25 | 87 | NAb | 10.21430/M3XTBR8F18 |
| SDY1289 | Yellow Fever (Live attenuated) | 14 | 84 | NAb | 10.21430/M37CO9E6FQ |
| SDY1294 | Yellow Fever (Live attenuated) | 21 | 109 | NAb | 10.21430/M3LT8WVHVH |
| SDY1529 | Yellow Fever (Live attenuated) | 36 | 180 | NAb | 10.21430/M36X4BH892 |
Technical Validation
Quality control and assurance
For global quality control across all public microarray data, we used a well-established pipeline available through the ArrayQualitymetrics R package40. Using pre-specified criteria established in the existing public microarray data reuse pipeline59, arrays that failed 3 out of 3 calculated quality control statistics were flagged for removal (see Methods). Consistent with standard practice to perform such quality control analysis prior to downstream analysis and dataset submission to the Gene Expression Omnibus, none of the samples were outliers by all three statistics (Fig. 3a). As expected for data from published peer-reviewed studies, all the identified studies passed the quality assurance method using the Arrayqualitymetrics method.
Y-chromosomal presence and age imputation
A few studies were missing information for sex and for age. To achieve data completeness, we included the biological sex imputation based on the imputed presence of the Y-chromosome using gene expression, as well as imputation of age when the variable was missing or defined by a broad range of values. Age imputation employed the RAPToR tool using 21 studies with reported age to define the best predictive model for the younger (age <50 years) and older (age ≥ 50 years) cohorts separately. The model with the lowest root mean square error (RMSE) from the young cohort was generated by taking into account the model (X ~ age_reported + matrix) with a coefficient of determination of R2 = 0.367 (Fig. 3e), while the old cohort yielded a prediction with R2 of 0.536 for their highest performing model (Fig. 3f).
Definition of vaccination studies transcriptomic cohort
Data preprocessing in ImmuneSpace yielded a total of 30 studies and 59 cohorts, with 1482 participants and 5413 samples. After the data was preprocessed and quality control measures were performed, we further assessed the identified cohorts as defined in the flow diagram (Fig. 2). This curation included: i) removing participants that were not relevant to the objective (n = 34); ii) removing samples due to inconsistencies with time design determination (n = 178); iii) removing participants with no baseline expression data (n = 42). Some studies, such as SDY1368 and SDY67, were dropped from the normalized data sets as they did not include subjects within our target age range (18–50 years). In summary, we report that the final Immune Signatures Data Resource contains 53 cohorts from 30 studies with 1405 participants and 4795 samples.
Assessment and adjustment of the batch effects
We evaluated the main sources of variation on the gene expression matrix to identify and adjust technical confounders (RNA-seq, Affymetrix arrays, Illumina arrays, etc.), study, and specimen types (e.g., whole blood vs. PBMCs) using the baseline samples. Since all studies enrolled healthy volunteers, and the first sample was taken pre-vaccination, pathogen and vaccine type would not affect the baseline data. Figure 3b clearly demonstrates robust clustering of samples by study, which are also grouped by platform type. The study effect and type of platform used accounted for the vast majority (95%) of variation, followed by specimen types (3.6%). It is thus essential that the data are corrected for these major effects prior to any analytical usage [see Materials and Methods for further details]. The study, platform type, and specimen type-specific effects were estimated using a linear model that also included age and Y-chromosome presence as additive effects using only baseline expression. Once the study, platform, and specimen-type effects were estimated, they were eliminated from the entirety of the expression matrix. Figure 3b shows that those effects can successfully be adjusted from the data, thus leading to a matrix of expression that is free of most technical biases induced by the laboratory and cell-type effects.
Immune signatures transcriptomics and immune response datasets
We report the total number of assay samples collected from the transcriptomic and immune response datasets tallied by targeted pathogen and vaccine type, across multiple systems vaccinology datasets (Fig. 4a). We captured about ~3000 HAI antibody titer results from influenza studies that were measured by the standard HAI assay pre- and at multiple time points post-vaccination, depending on the study. Mean titers were calculated for the reported strains of the virus and were based on the highest dilution reported at day 28–30 post-vaccination. In addition, neutralizing antibody (NAB) titers and IgG ELISA results specific to each pathogen were determined by each study and are summarized (Fig. 4a). The overall transcriptomics dataset comprises multiple time points from 7 days pre-vaccination up to day 180 days post-vaccination (Fig. 4b). While most of the datasets focus on the young adult population (ages 18–50 years old), the data resource also includes studies that profile older adults following hepatitis B, influenza, and varicella vaccination (Fig. 4c) that may be useful for analysis. The Euler diagram describes the dataset overlap of participants with transcriptomics datasets and corresponding to one or more immune response datasets (Fig. 4d).
Heterogeneity of the immune response to vaccination across targeted pathogens and vaccine types was reflected in variation in the longitudinal trajectories of HAI and NAB titer measurements (Fig. 5a,b). HAI and NAB titers generally increased by 14–28 days after vaccination but attenuated at different times for each vaccine (Fig. 5a,b). Change in NAB titers after vaccination were significantly different across the 5 unique combinations of targeted pathogen and vaccine types where these measurements were reported (ANOVA p < 10−10), with significant differences across all 5 groups except between meningococcus and yellow fever vaccines (Fig. 5c). Some influenza vaccination studies reported both HAI and NAB measures of immunogenicity, and there was a significant positive correlation between the vaccination-induced changes in these titers across participants (Spearman’s rho = 0.45, p < 10−10) (Fig. 5d).
Fig. 5.
Immune Response Dataset Overview. (a) The longitudinal trajectory (summarized as a loess curve) of hemagglutinin inhibition assay (HAI) measurements (in log2 scale) by influenza vaccine type and year. (b) The longitudinal trajectory of neutralizing antibody (NAB) titers (in log2 scale) for influenza, meningococcus, pneumococcus, and yellow fever vaccines. (c) Neutralizing antibody titers were plotted for each unique combination of targeted pathogen and vaccine type to compare each participants’ post-vaccination (day 28-30) values versus baseline (day 0). The violin plot shows the variation in magnitude for each unique combination of targeted pathogen and vaccine type. (d) The correlation plot of influenza studies compares the maximum fold change (MFC) across strains for hemagglutinin inhibition assay (HAI) titers versus neutralizing antibody (NAB) titers. Size is proportional to the number of samples analyzed.
Usage Notes
The expression data and accompanying meta-data have been made available with different formats and options to ease usage. Data are available as standard expression sets (eSet) objects, the R/Bioconductor structure unifying expression values, metadata, and gene annotation Both normalized data and batch-adjusted data are available (Table 4). Users interested in a single study or those planning to work exclusively within participants’ changes may opt for the normalized data without batch adjustment. For comparison of time points across studies or developing algorithms that use expression data, batch corrected matrices should be employed. Imputed age values for participants with no reported age were included to facilitate the use of age as a covariate in future analysis. Such analysis can be carried out with the complete data set and can be followed up by a sensitivity analysis using the small cohort with age-reported data. For the use of expression sets with the corresponding immune response per participant, these are available in eSets noted with a response. The selected immune response outcome per study is also summarized in Table 3.
Table 4.
List of data files for the Immune Signatures Data Resource.
| File name | Description |
|---|---|
| all_noNorm_eset.rds | Gene expression matrix of all participants, log2-normalized expression |
| all_noNorm_withResponse_eset.rds | Gene expression matrix of all participants with matched immune response data, log2-normalized expression |
| all_norm_eset.rds | Gene expression matrix of all participants that are cross-study normalized and batch corrected |
| all_norm_withResponse_eset.rds | Gene expression matrix of all participants with matched simmune response dataset, cross-study normalized and batch corrected |
| young_noNorm_eset.rds | Gene expression matrix of participants aged 18–50, log2-normalized |
| young_noNorm_withResponse_eset.rds | Gene expression matrix of participants aged 18–50 with matched immune response data, log2-normalized |
| young_norm_eset.rds | Gene expression matrix of participants aged 18–50, cross-study normalized and batch corrected |
| young_norm_withResponse_eset.rds | Gene expression matrix of participants aged 18–50 with matched immune response data, cross-study normalized and batch corrected |
| old_noNorm_eset.rds | Gene expression matrix of participants aged 60–90, log2-normalized |
| old_noNorm_withResponse_eset.rds | Gene expression matrix of participants aged 60–90 with matched immune response data, log2-normalized expression |
| old_norm_batchCorrectedFromYoung_eset.rds | Gene expression matrix of participants aged 60–90, cross-study normalized and batch corrected using age correction coefficients from young |
| old_norm_batchCorrectedFromYoung_withResponse_eset.rds | Gene expression matrix of participants aged 60–90 with matched immune response data, cross-study normalized and batch corrected using age correction coefficients from young |
| extendedOld_noNorm_eset.rds | Gene expression matrix of participants aged 50–90, log2-normalized expression |
| extendedOld_noNorm_withResponse_eset.rds | Gene expression matrix of participants aged 50–90 with matched immune response data, log2-normalized counts |
| extendedOld_norm_batchCorrectedFromYoung_eset.rds | Gene expression matrix of participants aged 50–90, log2-normalized expression |
| extendedOld_norm_batchCorrectedFromYoung_withResponse_eset.rds | Gene expression matrix of participants aged 50–90 with immune response data, cross-study normalized, and batch corrected using correction coefficients from young |
Acknowledgements
This research was conducted within the Human Immunology Project Consortium (HIPC) and supported by the National Institute of Allergy and Infectious Diseases. This work was supported in part by NIH grants U19AI128949, U19AI118608, U19AI118626, and U19AI089992, U19AI090023, U19AI089992, U19AI128914, U19AI118610, U19AI128913. The HIPC projects are listed at https://www.immuneprofiling.org/hipc/page/showPage?pg=projects. This work was supported in part by the Canadian Institutes of Health Research [funding reference number FDN-154287]
Author contributions
All authors identified the datasets, performed quality control, and assurance and analyzed the datasets. J.D.-A., H.M., S.H.K. and M.S.F. led the writing and organization of the manuscript. H.M., E.H., P.D., together with the ImmuneSpace and ImmPort team, implemented the pipeline for data access and visualization. The HIPC Consortium contributed to the conception and design of the work, as well as the acquisition of data. All authors edited and approved the manuscript.
Code availability
The source codes for the Immune Signatures Data Resource and all data are available in ImmuneSpace (https://www.immunespace.org/is2.url) and in Zenodo60 (10.5281/zenodo.5706261) and FigShare45: (10.6084/m9.figshare.17096978). Pre-processing code and supplementary data in full detail can be found in the ImmuneSignatures2 R package hosted on Github (https://github.com/RGLab/ImmuneSignatures2).
Competing interests
S.H.K. receives consulting fees from Northrop Grumman and Peraton. OL is an inventor on several patents relating to vaccine adjuvants and human in vitro systems predicting vaccine action. R.G. has received consulting income from Illumina, Takeda, and declares ownership in Ozette Technologies and Modulus Therapeutics. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Joann Diray-Arce, Helen E. R. Miller, Evan Henrich, Steven H. Kleinstein, Mayte Suárez-Fariñas.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Joann Diray-Arce, Email: joann.arce@childrens.harvard.edu.
Mayte Suárez-Fariñas, Email: mayte.suarezfarinas@mssm.edu.
The Human Immunology Project Consortium (HIPC):
Alison Deckhut-Augustine, Raphael Gottardo, Elias K. Haddad, David A. Hafler, Eva Harris, Donna Farber, Ofer Levy, Julie McElrath, Ruth R. Montgomery, Bjoern Peters, Adeeb Rahman, Elaine F. Reed, Nadine Rouphael, Ana Fernandez-Sesma, Alessandro Sette, Ken Stuart, Alkis Togias, and John S. Tsang
References
- 1.Piot P, et al. Immunization: vital progress, unfinished agenda. Nature. 2019;575:119–129. doi: 10.1038/s41586-019-1656-7. [DOI] [PubMed] [Google Scholar]
- 2.Pulendran B. Systems vaccinology: probing humanity’s diverse immune systems with vaccines. Proc Natl Acad Sci USA. 2014;111:12300–12306. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fineberg HV. Pandemic preparedness and response–lessons from the H1N1 influenza of 2009. N Engl J Med. 2014;370:1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 4.Fauci AS, Lane HC, Redfield RR. Covid-19 - Navigating the Uncharted. N Engl J Med. 2020 doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauci AS. An HIV Vaccine Is Essential for Ending the HIV/AIDS Pandemic. JAMA. 2017;318:1535–1536. doi: 10.1001/jama.2017.13505. [DOI] [PubMed] [Google Scholar]
- 6.Fauci AS, Folkers GK, Marston HD. Ending the global HIV/AIDS pandemic: the critical role of an HIV vaccine. Clin Infect Dis. 2014;59(Suppl 2):S80–84. doi: 10.1093/cid/ciu420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauci AS, Marston HD. Ending the HIV-AIDS Pandemic–Follow the Science. N Engl J Med. 2015;373:2197–2199. doi: 10.1056/NEJMp1502020. [DOI] [PubMed] [Google Scholar]
- 8.Diercks A, Aderem A. Systems approaches to dissecting immunity. Curr Top Microbiol Immunol. 2013;363:1–19. doi: 10.1007/82_2012_246. [DOI] [PubMed] [Google Scholar]
- 9.Pulendran B, Li S, Nakaya HI. Systems Vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang JS, et al. Improving Vaccine-Induced Immunity: Can Baseline Predict Outcome? Trends Immunol. 2020;41:457–465. doi: 10.1016/j.it.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2012;4:193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakaya HI, Pulendran B. Systems vaccinology: its promise and challenge for HIV vaccine development. Curr Opin HIV AIDS. 2012;7:24–31. doi: 10.1097/COH.0b013e32834dc37b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zak DE, Aderem A. Overcoming limitations in the systems vaccinology approach: a pathway for accelerated HIV vaccine development. Curr Opin HIV AIDS. 2012;7:58–63. doi: 10.1097/COH.0b013e32834ddd31. [DOI] [PubMed] [Google Scholar]
- 14.Brusic V, Gottardo R, Kleinstein SH, Davis MM, committee, H. s. Computational resources for high-dimensional immune analysis from the Human Immunology Project Consortium. Nat Biotechnol. 2014;32:146–148. doi: 10.1038/nbt.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poland GA, Quill H, Togias A. Understanding the human immune system in the 21st century: the Human Immunology Project Consortium. Vaccine. 2013;31:2911–2912. doi: 10.1016/j.vaccine.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 16.Muyanja E, et al. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. J Clin Invest. 2014;124:3147–3158. doi: 10.1172/JCI75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Querec T, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avey S, et al. Seasonal Variability and Shared Molecular Signatures of Inactivated Influenza Vaccination in Young and Older Adults. J Immunol. 2020;204:1661–1673. doi: 10.4049/jimmunol.1900922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakaya HI, et al. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh JZ, et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakar J, et al. Aging-dependent alterations in gene expression and a mitochondrial signature of responsiveness to human influenza vaccination. Aging (Albany NY) 2015;7:38–52. doi: 10.18632/aging.100720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakaya HI, et al. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc Natl Acad Sci USA. 2016;113:1853–1858. doi: 10.1073/pnas.1519690113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, et al. Metabolic Phenotypes of Response to Vaccination in Humans. Cell. 2017;169:862–877 e817. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan, N. L. et al. Breadth and Functionality of Varicella-Zoster Virus Glycoprotein-Specific Antibodies Identified after Zostavax Vaccination in Humans. J Virol92, 10.1128/JVI.00269-18 (2018). [DOI] [PMC free article] [PubMed]
- 28.Michlmayr D, et al. Comprehensive Immunoprofiling of Pediatric Zika Reveals Key Role for Monocytes in the Acute Phase and No Effect of Prior Dengue Virus Infection. Cell Rep. 2020;31:107569. doi: 10.1016/j.celrep.2020.107569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katzelnick LC, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazmin D, et al. Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. Proc Natl Acad Sci USA. 2017;114:2425–2430. doi: 10.1073/pnas.1621489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mpina M, et al. Controlled Human Malaria Infection Leads to Long-Lasting Changes in Innate and Innate-like Lymphocyte Populations. J Immunol. 2017;199:107–118. doi: 10.4049/jimmunol.1601989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Team, H.-C. S. P. & Consortium, H.-I. Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci Immunol2, 10.1126/sciimmunol.aal4656 (2017). [DOI] [PMC free article] [PubMed]
- 34.Azuaje F. Computational models for predicting drug responses in cancer research. Brief Bioinform. 2017;18:820–829. doi: 10.1093/bib/bbw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia S, Li J, Liu Y, Zhu F. Precision immunization: a new trend in human vaccination. Hum Vaccin Immunother. 2020;16:513–522. doi: 10.1080/21645515.2019.1670123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao, A. et al. Predicting the Immunogenicity of T cell epitopes: From HIV to SARS-CoV-2. bioRxiv, 10.1101/2020.05.14.095885 (2020).
- 37.Chaussabel D. Assessment of immune status using blood transcriptomics and potential implications for global health. Semin Immunol. 2015;27:58–66. doi: 10.1016/j.smim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharya S, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya S, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 42.Bruford EA, et al. Guidelines for human gene nomenclature. Nat Genet. 2020;52:754–758. doi: 10.1038/s41588-020-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boedigheimer MJ, et al. Sources of variation in baseline gene expression levels from toxicogenomics study control animals across multiple laboratories. BMC Genomics. 2008;9:285. doi: 10.1186/1471-2164-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulteau, R. & Francesconi, M. Real age prediction from the transcriptome with RAPToR. bioRxiv, 2021.2009.2007.459270, 10.1101/2021.09.07.459270 (2021). [DOI] [PubMed]
- 45.Diray-Arce J, 2021. HIPC-II Immune Signatures Data Resource. figshare. [DOI]
- 46.Rechtien A, et al. Systems Vaccinology Identifies an Early Innate Immune Signature as a Correlate of Antibody Responses to the Ebola Vaccine rVSV-ZEBOV. Cell Rep. 2017;20:2251–2261. doi: 10.1016/j.celrep.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zak DE, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Natl Acad Sci USA. 2012;109:E3503–3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bucasas KL, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henn AD, et al. High-resolution temporal response patterns to influenza vaccine reveal a distinct human plasma cell gene signature. Scientific reports. 2013;3:2327. doi: 10.1038/srep02327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsang JS, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vahey MT, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS,S malaria vaccine. J Infect Dis. 2010;201:580–589. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- 54.O’Connor D, et al. High-dimensional assessment of B-cell responses to quadrivalent meningococcal conjugate and plain polysaccharide vaccine. Genome Med. 2017;9:11. doi: 10.1186/s13073-017-0400-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy JS, et al. Safety and immunogenicity of LC16m8, an attenuated smallpox vaccine in vaccinia-naive adults. J Infect Dis. 2011;204:1395–1402. doi: 10.1093/infdis/jir527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsumiya M, et al. Roles for Treg expansion and HMGB1 signaling through the TLR1-2-6 axis in determining the magnitude of the antigen-specific immune response to MVA85A. PLoS One. 2013;8:e67922. doi: 10.1371/journal.pone.0067922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou J, et al. A Systems Vaccinology Approach Reveals Temporal Transcriptomic Changes of Immune Responses to the Yellow Fever 17D Vaccine. J Immunol. 2017;199:1476–1489. doi: 10.4049/jimmunol.1700083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fourati S, et al. Pre-vaccination inflammation and B-cell signalling predict age-related hyporesponse to hepatitis B vaccination. Nat Commun. 2016;7:10369. doi: 10.1038/ncomms10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah N, et al. A crowdsourcing approach for reusing and meta-analyzing gene expression data. Nat Biotechnol. 2016;34:803–806. doi: 10.1038/nbt.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.2021. RGLab/ImmuneSignatures2: 1.0.6. Zenodo. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
Data Availability Statement
The source codes for the Immune Signatures Data Resource and all data are available in ImmuneSpace (https://www.immunespace.org/is2.url) and in Zenodo60 (10.5281/zenodo.5706261) and FigShare45: (10.6084/m9.figshare.17096978). Pre-processing code and supplementary data in full detail can be found in the ImmuneSignatures2 R package hosted on Github (https://github.com/RGLab/ImmuneSignatures2).