ABSTRACT
Root caries in geriatric patients is a growing problem as more people are maintaining their natural teeth into advanced age. We determined the levels of various bacterial species previously implicated in root caries disease or health using quantitative real-time PCR in a pilot study of 7 patients with 1 to 4 root caries lesions per person. Levels of 12 different species on diseased roots compared to healthy (contralateral control) roots were measured. Four species were found at significantly higher levels on diseased roots (Streptococcus mutans, Veillonella parvula/dispar, Actinomyces naeslundii/viscosus, and Capnocytophaga granulosa) compared across all plaque samples. The level of colonization by these species varied dramatically (up to 1,000-fold) between patients, indicating different patients have different bacteria contributing to root caries disease. Neither of the two species previously reported to correlate with healthy roots (C. granulosa and Delftia acidovorans) showed statistically significant protective roles in our population, although D. acidovorans showed a trend toward higher levels on healthy teeth (P = 0.08). There was a significant positive correlation between higher levels of S. mutans and V. parvula/dispar on the same diseased teeth. In vitro mixed biofilm studies demonstrated that co-culturing S. mutans and V. parvula leads to a 50 to 150% increase in sucrose-dependent biofilm mass compared to S. mutans alone, depending on the growth conditions, while V. parvula alone did not form in vitro biofilms. The presence of V. parvula also decreased the acidification of S. mutans biofilms when grown in artificial saliva and enhanced the health of mixed biofilms.
KEYWORDS: biofilms, metabolism, root caries
INTRODUCTION
Root caries is a degradative disease of teeth where plaque microorganisms on exposed root surfaces generate acids, leading to demineralization of cementum and dentin (1). Proteolysis of the organic components of cementum and dentin (primarily collagen) can also facilitate the progression of this disease (2).
The microbiology of root caries has been an ongoing area of study. As the frequency of elderly patients keeping their natural teeth increases, so does the incidence of root caries in this population, largely due to the increased exposure of roots upon gingival recession. Several studies have indicated that Streptococcus mutans is highly associated with root caries due to its ability to efficiently generate lactic acid from carbohydrates (3, 4). In some studies, both S. mutans and Lactobacillus species were associated with the highest incidence of root caries (5, 6), and some studies showed the highest risk of root caries when both organisms were present on the same tooth (7, 8). Early studies also suggested a role for Actinomyces species in root caries (6); however, other studies have indicated no difference in colonization by Actinomyces species in carious versus healthy tooth roots (9, 10). In fact, in one study, Actinomyces species were detected more frequently in healthy controls rather than carious root lesions (11).
Studies using culture-independent microbiological techniques found elevated levels of previously unappreciated bacterial species such as Olsenella species, Selenomonas species, Veillonella dispar, Prevotella multisaccharivorax, Prevotella denticola, and Propionibacterium acidifaciens in carious root lesions (10, 12), as well as strains potentially protective against root caries disease, Capnocytophaga granulosum and Delftia acidovorans (12).
Given advances in the detection of nonculturable and slow-growing bacteria from the oral cavity using quantitative PCR and deep-sequencing techniques and the lack of consensus from previous studies regarding the role of certain microorganisms in root caries, we compared the microorganisms present in the plaque of carious root lesions in seven geriatric patients to the plaque of healthy contralateral control root surfaces in the same patients using quantitative PCR analysis for the detection of various bacteria previously implicated in the development of root caries or protection from root caries (4, 5, 7–15). To assess whether species found to be protective against enamel caries in some studies also protect against root caries, Streptococcus sanguinis and Veillonella parvula levels were also determined (16, 17). Finally, we also performed microbiome analysis on plaque from matched carious and healthy teeth from 5 of these patients to determine whether any novel bacterial not detected by our PCR analysis of 12 bacterial species were prevalent in our patient population.
We found the bacteria analyzed by quantitative PCR were a good reflection of those found in the complete microbiome analysis. Furthermore, we found a strong correlation between S. mutans and V. parvula/dispar with carious lesions as well as with each other. Unlike a previous study (12), we found a positive correlation between C. granulosa and root caries, rather than a protective role for this species. In vitro biofilm studies were performed to more closely assess the relationship between S. mutans and V. parvula. Finally, we present the data from our quantitative PCR analysis at various levels of analysis (as an aggregated group, on a patient-by-patient basis, and as a tooth-by-tooth comparison) to demonstrate the importance of presenting the data at various levels of analysis to observe differences that are masked when the data are presented solely as an aggregate of measurements across numerous patients.
RESULTS
Analysis of three traditional root caries species.
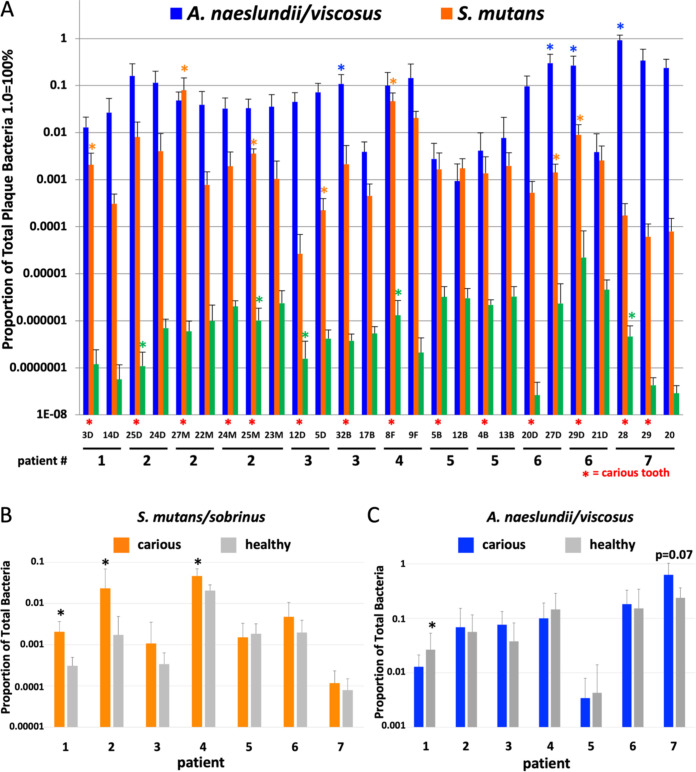
Given previous studies implicating S. mutans, Actinomyces species, and/or Lactobacillus species in root caries, we initially analyzed the levels of these organisms in root caries lesions by using 14 paired carious and noncaries teeth from 7 geriatric root caries patients. Our pairwise analysis demonstrated that in 5 out of 7 cases where significantly higher S. mutans were found on one tooth in a pair, the tooth with higher S. mutans levels was the carious tooth (Fig. 1A). Only four pairs of teeth showed a tooth with a statistically significant difference in Actinomyces naeslundii/viscosus colonization comparing carious to noncarious teeth. In three cases, the tooth with more A. naeslundii/viscosus was carious (Fig. 1A). Tooth 32 in patient 3 and tooth 29 in patient 6 had especially high levels of Actinomyces naeslundii/viscosus compared to their healthy contralateral controls (Fig. 1A). The PCR primer used in this study recognized both A. naeslundii and A. viscosus, but not Actinomyces odontolyticus (see Fig. S1 in the supplemental material). Lactobacillus casei levels were low on all teeth tested (<0.01% of total plaque bacteria; Fig. 1A), and general Lactobacillus species levels were <0.1% of total plaque bacteria on all but one tooth tested (tooth 24 in patient 2; Fig. S2), suggesting Lactobacillus did not contribute to caries in our patients.
FIG 1.
Reverse transcription-quantitative PCR (qRT-PCR) analysis of DNA isolated from plaque from seven root caries patients (n = 6 to 8). Five of seven teeth with increased S. mutans levels relative to their contralateral controls had caries. Three of four teeth with increased Actinomyces levels relative to their contralateral controls had caries. In general, Lactobacillus levels on all teeth were low, but in two cases, increased Lactobacillus casei levels correlated with caries. (A) Analysis of three species by tooth pair (carious and noncarious). (B and C) Analysis of S. mutans/sobrinus (B) or Actinomyces naeslundii/viscosus (C) by combining colonization levels of all carious or healthy teeth per patient. *, P ≤ 0.05 by Student’s t test (A) or Mann-Whitney test for multiple teeth per patient (B and C). Tooth numbers are shown in panel A. Tooth surfaces are represented as follows: D, distal; M, mesial; B, buccal; and F, facial.
We also determined overall colonization trends within each patient by aggregating data from all carious teeth or healthy controls within an individual. These analyses demonstrated that in only 3 out of 7 patients did we find a significantly elevated level of S. mutans on carious teeth (Fig. 1B), and in 0 out of 7 patients, we found elevated levels of A. naeslundii/viscosus on diseased teeth (Fig. 1C; patient 7 showed a trend toward increased A. naeslundii/viscosus on diseased teeth; P = 0.07). However, if one combines data from all patients into just two categories, carious lesion versus noncarious teeth, one finds S. mutans and A. naeslundii/viscosus are then significantly higher on diseased teeth (Fig. 2; P = 0.014 and P = 0.03, respectively). These analyses emphasize the importance of analyzing microbiological data for root caries at various levels of sample grouping.
FIG 2.
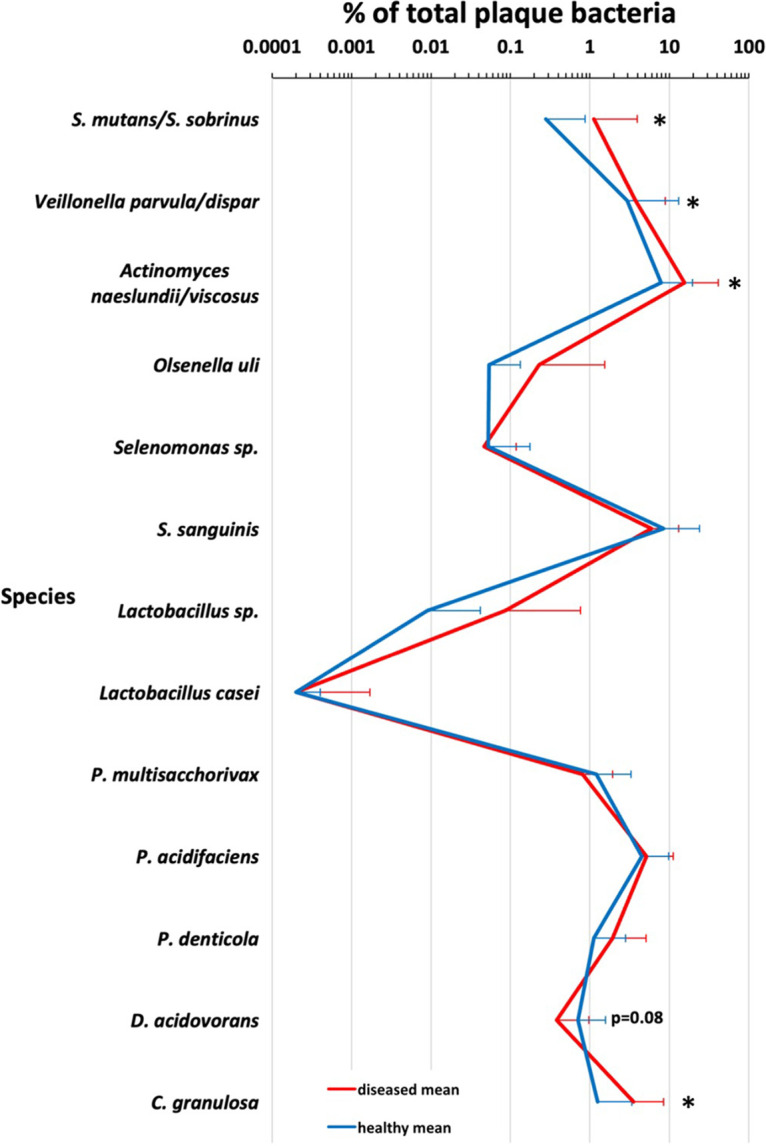
Four species tested showed a correlation between higher levels on teeth and caries, including S. mutans, A. naeslundii/viscosus, V. parvula/dispar, and C. granulosa. One species, D. acidovorans, showed a trend toward association with health (P = 0.08). *, P < 0.05 using Mann-Whitney multivariate logistic regression analysis to account for differences between patients. n = 6 to 10, except general Lactobacillus species, where n = 3 to 7.
Assessment of Olsenella uli and Selenomonas species association with root caries.
We next investigated two more recently appreciated genera identified previously in root caries studies, Selenomonas species and Olsenella species (10). We rarely found statistically higher levels of either bacterium on a carious tooth compared to its matched contralateral healthy control, and levels of each genus were generally low, composing only about 0.01 to 0.1% of the total microbial population (Fig. S2 and S3). Our Selenomonas-specific primer was designed to detect Selenomonas strain CS002 (10); however, whole-microbiome analysis indicated increased levels of Selenomonas noxia in some patients, although, in some cases, high levels of S. noxia were found on healthy teeth (Table S3).
S. sanguinis is not associated with reduced S. mutans root colonization.
Previous studies have suggested elevated levels of S. sanguinis on enamel surfaces lead to reduced S. mutans levels and reduced caries risk (17–19). Thus, we compared the levels of S. mutans and S. sanguinis on healthy and diseased tooth pairs from our seven patients. There was no inverse correlation in the levels of S. mutans and S. sanguinis on root surfaces and no protection from root caries by elevated levels of S. sanguinis (Fig. S4 and Table S2). In fact, in only 3 out of 7 cases where higher levels of S. sanguinis were detected on one tooth in a pair did we detect a concomitant decrease in S. mutans colonization relative to the other tooth, and 4 out of 7 times, the increased levels of S. sanguinis were on the carious tooth rather than the healthy tooth (Fig. S4).
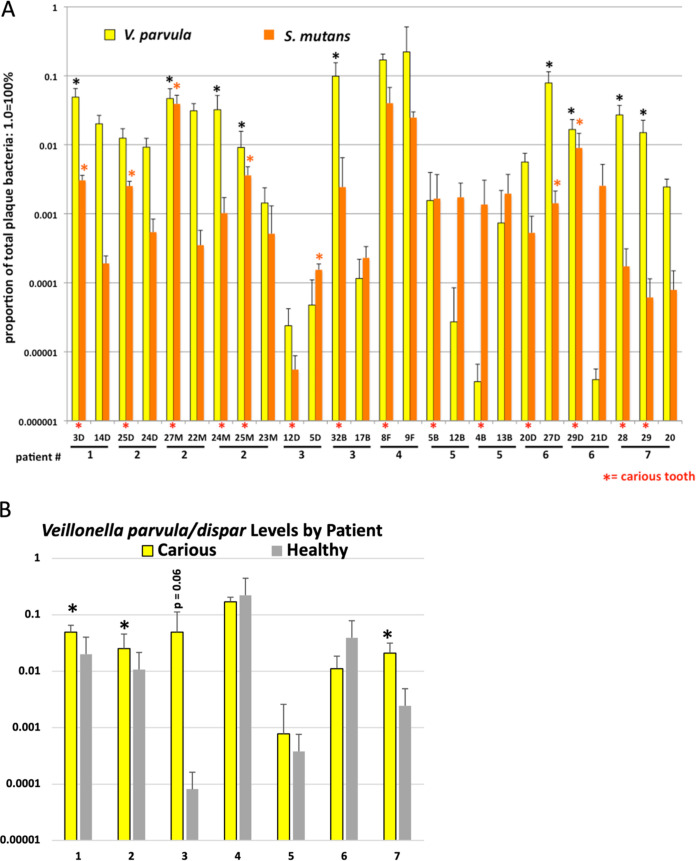
V. parvula/dispar are associated with plaque containing elevated levels of S. mutans and root caries.
There are conflicting reports in the literature regarding the role of Veillonella species in S. mutans-mediated caries. In one study, coinoculation with Veillonella alcalescens led to protection from caries (20), while in another study, Veillonella alcalescens coinoculation increased enamel degradation by S. mutans in an artificial mouth model (21). An association has also been found with elevated Veillonella species in early childhood caries (19, 22), and V. dispar was associated with higher levels of root caries in one study (12). Thus, we sought to determine whether our patient pool had any association between Veillonella species and root caries or root health. Dramatically, V. parvula/dispar levels in our population were the most highly correlated with increased root caries risk. In 8 out of 9 times that one tooth in the pair had statistically higher levels of V. parvula/dispar, that tooth was carious (Fig. 3A). This correlation between V. parvula/dispar and caries was maintained in 3 out of 7 patients when all data from each patient were combined, with a strong trend toward caries in a fourth patient (patient 3, P = 0.06) (Fig. 3B). There were also higher levels of V. parvula/dispar when all patient data were combined comparing all carious teeth to all healthy teeth (Fig. 2). Finally, since V. parvula/dispar can metabolize lactic acid released by S. mutans, we determined whether there was a positive correlation between S. mutans and V. parvula/dispar levels. In three patients (patients 2, 5, and 6), we found a strong or very strong positive correlation between V. parvula/dispar and S. mutans colonization levels (Table 1) as well as when we aggregate data from all seven patients on carious teeth (Table 1). The seemingly large negative correlation between V. parvula/dispar and S. mutans in patient 4 is not statistically significant due to the low number of affected teeth (1) and measurements in this patient (Table 1; n = 6).
FIG 3.
qPCR analysis of S. mutans and V. parvula/dispar in plaque-isolated DNA from seven root caries patients (n = 6 to 8). Given previous studies suggesting Veillonella species can be associated with increased caries risk or lower caries risk (20, 21), we sought to assess Veillonella levels in our root caries samples. In 8 out of 9 cases where one tooth was more highly colonized by Veillonella (shown with an asterisk), that tooth was carious. Furthermore, in 5 out of 8 cases, that higher level of Veillonella was associated with an increased level of S. mutans (shown with an asterisk). Data were analyzed by paired-tooth analysis (A) or as aggregates of carious or healthy teeth in each patient (B). *, P ≤ 0.05 by Student’s t test (A) and Mann-Whitney test for multiple teeth (B). Student’s t test was used in panel B if only one carious and one healthy tooth was present in the patient (patients 1 and 4).
TABLE 1.
S. mutans and Veillonella parvula/dispar correlation coefficients for carious teetha
| Group | r | P value | No. of bacteria |
|---|---|---|---|
| Aggregate | 0.389 | <0.001 | 84 |
| Patient 1 | 0.271 | 0.603 | 6 |
| Patient 2 | 0.607 | 0.002 | 24 |
| Patient 3 | 0.180 | 0.575 | 12 |
| Patient 4 | −0.392 | 0.442 | 6 |
| Patient 5 | 0.585 | 0.046 | 12 |
| Patient 6 | 0.909 | <0.001 | 12 |
| Patient 7 | −0.047 | 0.884 | 12 |
Colonization levels for S. mutans and V. parvula/dispar on all diseased teeth from each person or the aggregate of all 7 patients together were compared using the Pearson’s correlation coefficient (r). If r is equal to 0.3 to 0.39, there is a moderate positive correlation between the species. When r equals 0.4 to 0.69, there is a strong positive correlation between the species, and if r is ≥0.7, there is a very strong correlation. All values ≥0.3 are in bold. In 3 out of 7 patients, there was a strong or very strong correlation between the levels of S. mutans and V. parvula/dispar on carious teeth. Furthermore, when all patients were combined (aggregate), there was still a correlation between the levels of S. mutans and V. parvula/dispar. For the one case (patient 4) where a negative correlation was found, the difference was not significant (P = 0.442).
Assessment of other species more recently found associated with root caries confirms the role of some, but not all, in disease and health.
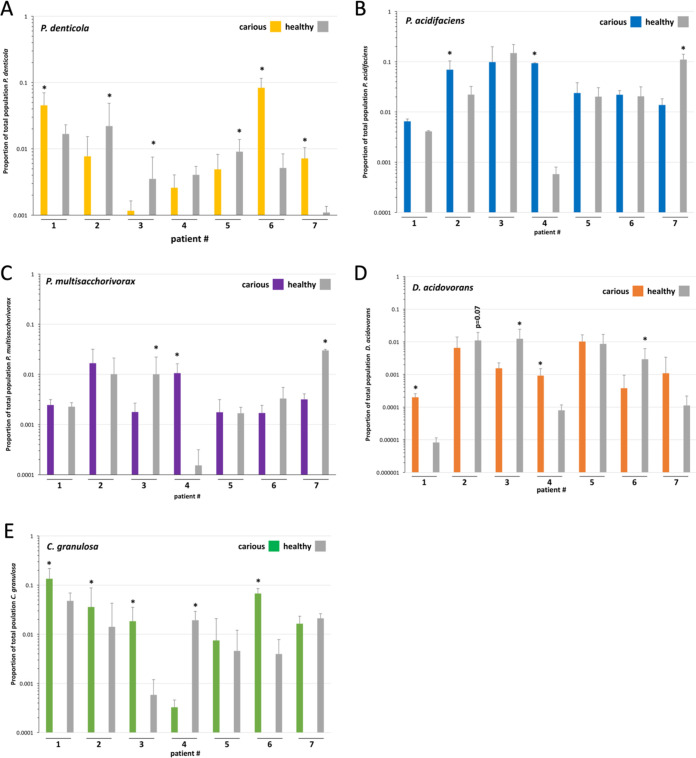
Chen et al. studied 21 patients with root caries and found several species associated with root caries or health (12). While their findings supported the role of S. mutans and Actinomyces species identified in previous studies, they also identified three new species associated with root caries, Prevotella multisaccharivorax, Prevotella denticola, and Propionibacterium acidifaciens, and two species associated with health, Delftia acidovorans, and Capnocytophaga granulosa. We designed PCR primers to detect these species as well and assessed the levels of these species in our patient population. Compared at the patient level, we found P. denticola was elevated on carious teeth in 3 out of 7 patients, yet it was also elevated on healthy teeth in 3 out of 7 patients (Fig. 4A). Furthermore, when aggregated across all patients, there was no statistical difference in P. denticola levels on carious or noncarious teeth (Fig. 2). This finding illustrates the importance of looking at colonization levels at various levels of grouping, as trends in multiple patients can be lost when all patients are aggregated together. Correlations between P. acidifaciens and P. multisaccharivorax and caries were less dramatic, with elevated levels on carious teeth in 2 out of 7 and 1 out of 7 patients, respectively (Fig. 4B and C). As with P. denticola, neither P. multisaccharivorax nor P. acidifaciens showed a higher association with caries when data from all patients were aggregated (Fig. 2). For D. acidovorans, a species previously associated with health (12), at the patient level, 2 out of 7 times, higher levels of this organism were associated with a patient’s healthy teeth, yet 2 out of 7 times, it was associated with diseased teeth (Fig. 4D). However, if data are combined from all patients’ carious teeth and healthy teeth, we find a trend toward D. acidovorans and health (Fig. 2), but the association does not reach statistical significance (P = 0.08, using logistic regression to account for the variability between patients). Contrary to previous findings (12), we found C. granulosa to be associated with caries rather than health when teeth from all patients were compared (Fig. 2). This was also true at the patient level, with 4 out of 7 patients showing higher levels of C. granulosa on carious teeth and only 1 out of 7 having increased levels of C. granulosa on healthy teeth (Fig. 4E).
FIG 4.
qPCR analysis of plaque-isolated DNA from seven root caries patients for Prevotella denticola (A), Propionibacterium acidifaciens (B), Prevotella multisaccharivorax (C), Delftia acidovorans (D), and Capnocytophaga granulosa (E) (n = 6). All data are presented as aggregates of carious or healthy tooth colonization levels per patient. Tooth pair analysis is shown in Fig. S5 in the supplemental material. *, P ≤ 0.05 by the Mann-Whitney test for multiple teeth and Student’s t test if only one carious and one healthy tooth was present in the patient (patients 1 and 4).
Microbiome analyses confirm findings from qPCR.
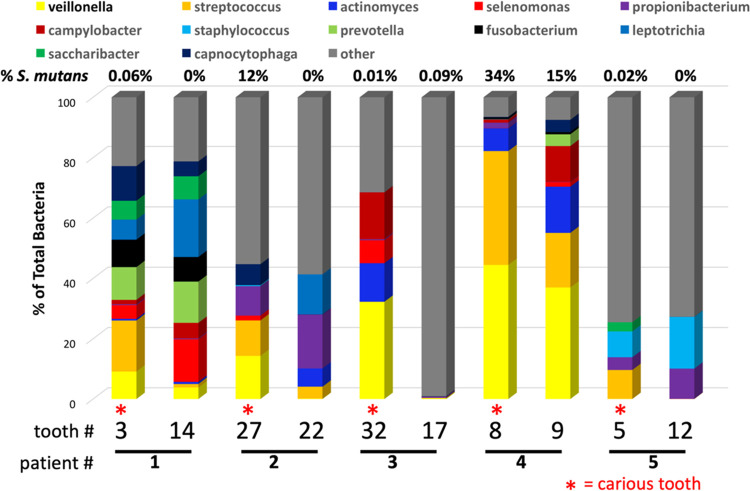
Since quantitative PCR (qPCR) data are limited to species targeted for PCR based on previous studies, we also performed whole-microbiome analyses on 10 plaque samples (patients 1 to 5, paired carious and noncarious samples) using the entire 16S rRNA sequence (Fig. 5; Table S3). There were no clearly predominant genera from the microbiome analysis that we missed in our qPCR analysis (using a cutoff of 5% of the total microbiome). Furthermore, at the species level, the abundance of S. mutans found in the microbiome analysis generally reflected the qPCR findings (Fig. 1 and 5; Table S3). Furthermore, while our species-specific PCR primers would not be expected to distinguish between S. mutans and Streptococcus sobrinus (both members of the mutans streptococci group), microbiota analysis indicated that the majority of the mutans streptococci present were S. mutans (Table S3). Similarly, when V. dispar was identified by microbiome analysis, it was never >1% of the total microbiome, suggesting the species indicated by Veillonella spp. in the microbiota analysis may be primarily V. parvula rather than V. dispar (Table S3). Unfortunately, while Actinomyces species were well represented in the microbiome analysis, the most prominent species could not be clearly identified at the species level (Actinomyces sp.) (Table S3), although we suspect it is A. naeslundii or A. viscosus based on our species-specific PCR results.
FIG 5.
Microbiome analysis of 10 samples from patients 1 to 5. Following PCR amplification of the plaque DNA samples by using universal 16S rRNA primers, the entire 16S rRNA locus was sequenced and analyzed by Molecular Research DNA Analysis, Inc. Any genus that represented >5% of the total population in either the carious or health toothy was illustrated. A complete table of microbiome results at the genus and species levels of identification can be found in Table S3 in the supplemental material.
Since our Selenomonas CS002-specific primer only recognizes certain strains of Selenomonas, microbiome analysis was key in identifying S. noxia in a number of plaque samples, although in some cases, the highest level was found on the healthy tooth in the pair.
Finally, as indicated by our species-specific PCR, Lactobacillus species were notably absent from the microbiota analysis, indicating their relatively low abundance in the patients tested.
Co-culturing of S. mutans and V. parvula leads to enhanced biofilm formation.
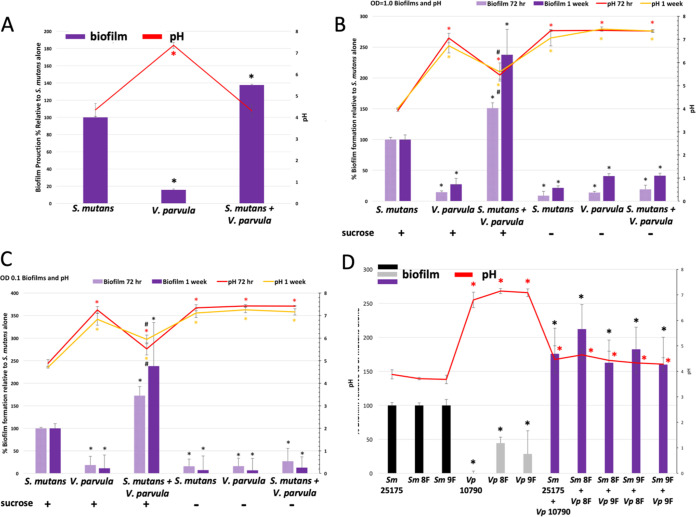
Numerous studies have shown important metabolic interactions between S. mutans and Veillonella species (20, 21, 23–28). Since we found a correlation between high levels of S. mutans and high levels of V. parvula/dispar (Table 1), we used an in vitro biofilm system to assess biofilm formation by S. mutans and V. parvula alone and in combination as well as assessed the impact of V. parvula on culture acidification by S. mutans. S. mutans strain ATCC 25175, when combined with V. parvula (ATCC 10790), led to a 50% increase in biofilm formation relative to S. mutans alone in rich medium (Brucella broth with vitamin K and hemin) (Fig. 6A). V. parvula incorporation in the biofilm did not reduce acidification by S. mutans in rich medium with 0.5% sucrose (Fig. 6A). V. parvula alone was not able to establish a robust biofilm in our system (Fig. 6A). However, when grown as part of a mixed biofilm, V. parvula made up ~50% of bacteria present in the biofilm (Table S4). The robust growth of V. parvula in mixed biofilms was due to metabolism of lactate produced by S. mutans to acetate and propionate by V. parvula as determined by quantitative nuclear magnetic resonance (NMR) analysis of culture supernatants (Table S5).
FIG 6.
Co-culturing of S. mutans with V. parvula leads to enhanced biofilm formation. Cultures were grown for 24 h, 72 h, or 1 week in an anaerobic chamber prior to processing the biofilms and measuring the pH of the supernatants. Biofilms were formed in BB+ broth with 0.5% sucrose (A) or artificial saliva with 0.5% sucrose (B to D) (29). Bacteria were inoculated at a starting OD600 of 1.0 (A, B, and D) or 0.1 (C) by using S. mutans ATCC 25175 and V. parvula ATCC 10790. (D) S. mutans and V. parvula clinical isolates from patient 4 (teeth 8 and 9) were also assessed for biofilm formation and acidification. *, P ≤ 0.05 by Student’s t test.
When artificial saliva (29) was used rather than rich medium, we saw a similar enhancement in biofilm formation due to the addition of V. parvula (Fig. 6B). Since the buffering capacity of artificial saliva is more similar to natural saliva than rich medium and bacterial growth was less robust, S. mutans-mediated strong acidification (pH ~4) took as long as 72 h, and V. parvula co-culturing reduced the acidity by ~1.5 pH units, even in week-old biofilms (Fig. 6B). If biofilms were initiated with a reduced inoculum (optical density at 600 nm [OD600] of 0.1 rather than 1.0), almost no acidification occurred by 24 h, even with S. mutans alone (data not shown), but by 72 h, biofilm levels were similar to those inoculated at an OD of 1.0, and V. parvula still led to decreased acidity when combined with S. mutans (Fig. 6C).
To determine if paired clinical isolates of S. mutans and V. parvula from one of our patients with high Veillonella levels (patient 4) (Fig. 5) also form interactive biofilms, we isolated pure clinical isolates of S. mutans and V. parvula from teeth 8 and 9 of patient 4 (Materials and Methods). Culturing paired clinical isolates from the carious lesion in patient 4 (tooth 8) also led to enhanced biofilm formation when S. mutans and V. parvula were combined compared to biofilms by S. mutans from that patient alone (Fig. 6D). S. mutans and V. parvula from the healthy contralateral (neighboring) control tooth of patient 4 (tooth 9) were also able to form enhanced biofilms (Fig. 6D). As with ATCC type strains (S. mutans 25175 and V. parvula 10790), the presence of a V. parvula clinical isolate led to a statistically significant reduction in the acidity of S. mutans biofilms from S. mutans isolated from the same plaque sample and even V. parvula from the neighboring tooth (Fig. 6D). It should be noted that pairing of S. mutans from tooth 8 with V. parvula from tooth 8 gave the most robust biofilm (>100% increase over S. mutans alone), and this biofilm enhancement was statistically higher than that found when S. mutans from tooth 8 (diseased) and V. parvula tooth 9 (healthy) were combined (P = 0.03) (Fig. 6D).
The presence of V. parvula reduces S. mutans membrane permeability.
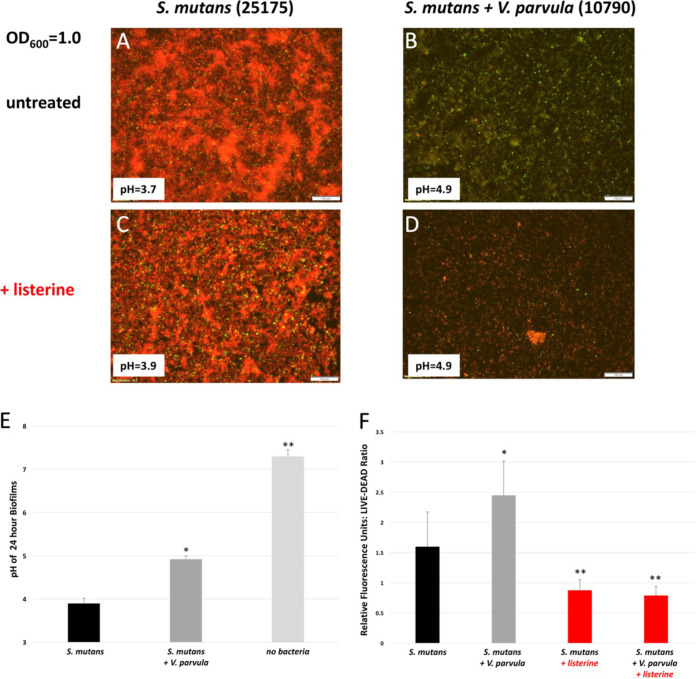
Since, in the presence of V. parvula, S. mutans biofilms were less acidic and potentially presented a more hospitable environment for both species, we assessed membrane permeability of biofilm-grown cells using a traditional LIVE/DEAD stain of SYTO 9 (live, green) and propidium iodide (PI) (red, dead). Twenty-four-hour biofilms initiated with S. mutans with or without V. parvula at an OD600 of 1.0 in artificial saliva with 0.5% sucrose were grown in 4-well microscope chamber slides. In the presence of V. parvula, mixed biofilms were less permeable to the membrane-impermeant dye PI and thus were greener than S. mutans grown alone (Fig. 7A and B). This increase in green staining in the presence of V. parvula was also reflected in the calculation of a LIVE/DEAD (green/red) ratio in a parallel 24-well plate treated identically to the 4-well chamber slides (Fig. 7F). The presence of V. parvula also resulted in an increase in pH from pH 3.7 to 4.9 (less acidic), as expected (Fig. 7A, B, and E). Control biofilms treated with Listerine Naturals for 10 min were completely red, as expected for dead cells. CFU analysis confirmed Listerine-treated biofilms lost >106-fold viability (data not shown).
FIG 7.
V. parvula enhances the health of S. mutans biofilms. S. mutans biofilms with or without V. parvula were initiated with cultures at an OD600 of 1.0 and grown for 24 h in artificial saliva with 0.5% sucrose. Biofilms were stained with SYTO 9 (green, live) and propidium iodide (red, dead) for LIVE/DEAD staining followed by fluorescence microscopy (A to D) or LIVE/DEAD ratio measurements in a plate reader (F). (E) pH levels from both microscope slide wells and 24-well plates were measured. As a control for dead cells, Listerine Naturals was used to kill biofilms for 10 min prior to imaging. The pH for individual wells used for imaging are shown as inserts to the image. (A to D) Shown are images from the same day, representative of results from 3 different days. pH measurements constitute measurements averaged from 16 wells over 8 experiments. (E) *, P < 0.01 relative to S. mutans 25175; **, P < 0.01 relative to S. mutans 25175 with V. parvula 10790 by Student’s t test. (F) *, P < 0.01 relative to S. mutans 25175; **, P < 0.01 relative to S. mutans 25175 or S. mutans 25175 with V. parvula 10790 without Listerine by Student’s t test.
Co-culturing of S. mutans with V. parvula does not result in upregulation of glucosyltransferases in artificial saliva.
One possible cause of the enhanced biofilm mass observed when S. mutans and V. parvula are cocultured is the upregulation of glucosyltransferase genes of S. mutans (gtfB, gtfC, and gtfD) leading to enhanced extracellular polysaccharide (glucan) production and a supplemented biofilm architecture. S. mutans gtfB and gtfC have been proposed to be upregulated in rich media when grown with V. parvula previously (28). We assessed the levels of gtfB, gtfC, and gtfD in S. mutans relative to the RNA polymerase β-subunit-encoding gene rpoB in the presence or absence of V. parvula strains ATCC 10790 and ATCC 17745. We found no increase in the expression of gtfB, gtfC, and gtfD relative to rpoB in the presence of either V. parvula strain (Fig. S6). The only statistically significant change we observed was that gtfC was significantly reduced when S. mutans 25175 was grown in the presence of V. parvula 17745 (Fig. S6).
DISCUSSION
Our data reveal that high levels of S. mutans and V. parvula/dispar are predictive of root caries and that these two species cooperate to form a more robust biofilm than S. mutans alone (Fig. 1, 2, 3, and 6; see Fig. S2 in the supplemental material). While Actinomyces naeslundii/viscosus showed a slight increase in colonization levels on carious teeth when all patients were aggregated (Fig. 2), no single patient had a statistically significant increase in Actinomyces naeslundii/viscosus. Previous studies have provided conflicting evidence regarding the association of Actinomyces species and root caries (6, 8–10).
While previous studies had suggested a protective role for C. granulosa in root caries (12), we found a higher level of C. granulosa on carious teeth when all patients were aggregated (Fig. 2), and 3 out of 7 patients had higher levels of C. granulosa on carious teeth (Fig. 4). Future studies will investigate this potential contribution of C. granulosa to root caries. We note our V6F C. granulosa primer can also efficiently amplify the 16S rRNA gene from Capnocytophaga gingivalis, but not Capnocytophaga sputigena (Fig. S1E). We did find a trend toward increased levels of D. acidovorans, and health was as previously suggested (12), but the trend did not reach statistical significance (P = 0.08) (Fig. 2). Two out of seven patients had higher levels of D. acidovorans on healthy teeth than on carious teeth (Fig. 4).
Lactobacillus species, while implicated in root caries in some previous studies (6, 8, 10, 12), were present at very low levels on all teeth examined in our study and are thus unlikely to be contributing to root caries (Fig. 1 and Fig. S2). This was confirmed by microbiome analysis as well (Table S3). It should be noted our Lactobacillus casei and general Lactobacillus species’ species-specific primers reacted modestly with control L. casei DNA (Fig. S1 and data not shown). However, even if the levels of Lactobacillus were adjusted by increasing them 25- to 50-fold to account for this, all Lactobacillus levels would remain <0.1% of the total species with the exception of a carious tooth in patient 2 (Fig. 1 and Fig. S2). Olsenella and Selenomonas species that had also been found to be previously associated with root caries (10) were not associated with root caries in our study, with the possible exception of elevated levels of Olsenella uli on a carious tooth of patient 5 (Fig. S2 and S3). Microbiome analysis indicated an additional Selenomonas species that would not be detected by our Selenomonas CS002 primers, S. noxia, was present in some patients. This species could potentially contribute to root caries, although it was found at high levels on noncarious teeth in some patients (Fig. 5 and Table S3).
Species from the Bifidobacteriaceae that were found at high levels associated with root caries previously that were not specifically tested in this study (Bifidobacterium dentium, Parascardovia denticolens, and Scardovia genomo species C1 [30]) were not identified in our microbiome analysis (Fig. 5 and Table S3).
Another major observation is that since people vary dramatically in their microbiome composition, the way one analyzes such microbiome data can dramatically affect whether differences are statistically different. By comparing diseased and healthy contralateral teeth for bacterial colonization in a single patient, one can control for the host microbiome and even different environmental conditions in the oral cavity. However, results from one person may vary dramatically from another person since many microorganisms can generate acid and lead to tooth demineralization and caries. By looking at multiple teeth within a single individual, one can determine whether, with multiple lesions in a single host, one can observe an association between certain microbes and increased caries. From our perspective, this patient level of analysis (Fig. 1B and C, Fig. 3B, and Fig. 4) may be the most informative, as it controls for the available host microbiome. This is highlighted by the fact that if a single patient has a high level of a particular organism on a carious tooth, this can give the appearance of generally high levels of that organism in the whole population when all patients are aggregated (Fig. 2 and Fig. S3; see Olsenella uli), even if it was only elevated in one patient. This is especially important to consider in small sample sizes. One should also consider that one patient may lack a key microbe that can contribute to root caries, but they may have an alternative organism that can contribute to disease. Finally, aggregation of data from all patients at various sites in the oral cavity gives an overall picture of major contributors to root caries, but with such analyses, differences that were clear with some patients can lack significance in the fully aggregated data. This does not mean those organisms did not contribute to disease in those patients, but, rather, since some people have low levels of certain organisms, caries, in general, is not consistently associated with those organisms. Again, this suggests that looking at patient-level data can be especially informative.
One other method of analysis to consider is the threshold hypothesis. One can assume that an organism must be present in the plaque biofilm at some critical level to contribute to demineralization and caries (31). Thus, even though a certain bacterium is higher on a carious tooth than on a healthy tooth, it does not mean there are enough bacteria present to contribute to disease. Where this threshold lies with organisms that are significant contributors to caries disease is unclear, but one might imagine a ≥1% colonization level may start to provide the critical mass of a particular cariogenic organism to initiate disease. This threshold hypothesis also relates to the fact that perhaps a consortium of acid-generating organisms (like S. mutans and Actinomyces naeslundii/viscosus) can each be present at reduced levels and still cause disease if the total acidogenic biomass is of sufficient density. One possible example of this phenomenon in our study was the carious lesion on tooth 3 in patient 1. This tooth had modest levels of both S. mutans and C. granulosa (both statistically higher than on the healthy tooth) (Fig. 1, Fig. 4E, and Fig. 5). Since both species are capable of producing acids from carbohydrates (32), together, they may have combined to produce root caries. As a correlate to this idea, bacteria that degrade acids, like D. acidovorans, could reduce acidity and lead to health. We observed a trend toward health with D. acidovorans (P = 0.08) (Fig. 2), and others have seen an association between higher D. acidovorans and health (12).
One unanticipated finding was that S. sanguinis is not protective against root caries. This suggests a different mode of competition for tooth colonization on the root surface from the coronal surface (17, 19). This may reflect the more proteinaceous nature of tooth root cementum and the underlying dentin.
In agreement with other studies (12, 21, 33, 34), we also found an association between Veillonella parvula/dispar and caries. Given that Veillonella species can metabolize lactic acid produced by S. mutans to propionic and acetic acids, we hypothesize this metabolic interaction allows S. mutans to grow to higher levels before reaching a pH that inhibits further bacterial growth and impacts S. mutans viability (Fig. 7). While acetic and propionic acids are weaker acids than lactic acid, they can still result in acidification below pH ~5.5 (Fig. 6) and thus initiate hydroxyapatite demineralization and dental caries (35). Our results and the work of others support the idea that co-culturing of S. mutans and Veillonella parvula leads to more robust biofilms (Fig. 6 and 7) (28). DNA analysis of these mixed biofilms demonstrated that V. parvula makes up about 50% of the biofilm (Table S4), even though, alone, V. parvula does not form robust biofilms in our system (Fig. 6). Thus, generation of lactic acid by S. mutans (via sucrose metabolism) allows V. parvula to thrive in the mixed culture, and V. parvula may even adhere to S. mutans in the biofilm, perhaps via the induction of V. parvula autotransporter adhesins (36). Acidification of S. mutans and V. parvula co-species biofilms depended on the growth conditions used. When the rich medium Brucella broth supplemented with vitamin K, hemin, and 0.5% sucrose was used, the biofilm acidified rapidly, and the presence of V. parvula had a modest impact on acidification measured at 24 h (Fig. 6A), similar to previous reports using rich medium (28). However, when we switched to a more biologically relevant growth medium of artificial saliva containing 2 mg/mL mucin supplemented with 0.5% sucrose, the presence of V. parvula had a dramatic effect on the acidity of the biofilm at 24 h, 72 h, and 1 week (Fig. 7A, B, and E and Fig. 6B and C). For biofilms initiated at an OD600 of 1.0 in artificial saliva, the difference in pH went from pH ~4 for S. mutans alone to pH ~5 to 5.5 for S. mutans with V. parvula (Fig. 6B and Fig. 7E). This less acidic environment leads to less membrane damage to S. mutans biofilms containing V. parvula as measured by LIVE/DEAD staining of biofilms (Fig. 7). Biofilm studies using S. mutans and V. parvula clinical samples from a patient in this study also showed enhanced biofilms and reduced acidification when the two species were mixed, although the level of biofilm formation by V. parvula clinical isolates on their own was higher than with the laboratory type strain ATCC 10790 (Fig. 6D).
It should be noted that while our V6F detection primer for mutans streptococci (S. mutans and S. sobrinus) is quite species specific (Fig. S1C), our V6F primer designed to detect V. parvula and V. dispar, two common colonizers or teeth, can also detect Veillonella atypica and Veillonella denticariosi, but not Veillonella montpellierensis. While clinical isolates from patient 4 confirmed the presence of V. parvula based on combined V3 and V6 sequencing, some patients may harbor other Veillonella species. These other PCR-reactive Veillonella species also metabolize lactate produced by S. mutans (37).
Finally, the enhanced level of biofilm we see in S. mutans with V. parvula biofilms, relative to S. mutans alone, does not appear to be due to enhanced expression of glycan-producing glucosyltransferases, as gene expression analysis of the S. mutans gtfB, gtfC, and gtfD in biofilms grown in artificial saliva with sucrose did not demonstrate increased levels of any of these alleles in mixed biofilms relative to S. mutans alone (Fig. S6). This again suggests a difference in physiological responses in rich medium from artificial saliva (28). A recent report of transcriptional responses of V. parvula in association with Streptococcus gordonii indicated that oxidoreductase activity and other components in the oxidation-reduction process were upregulated in V. parvula in mixed cultures compared to V. parvula monocultures (38), but such studies have not been performed on long-term co-cultures of S. mutans and V. parvula. Such genome-wide analysis would enhance our understanding of the paired physiological responses when S. mutans and V. parvula are grown together to form mature biofilms.
Our study supports the idea that although V. parvula/dispar can metabolize lactic acid generated by S. mutans from carbohydrates to weaker acids, the presence of S. mutans and V. parvula together in root caries plaque is associated with a higher likelihood of root caries. Whether this is due to development of a thicker, healthier biofilm and thus more continuous acid generation or some other physiological process remains to be determined. These findings highlight the importance of interspecies metabolic interactions in the shift from healthy to diseased states in dental plaque.
MATERIALS AND METHODS
Patient population.
Seven adult patients with root caries (6 male and 1 female), ranging from 65 to 83 years of age, were enlisted as research subjects. Dental plaque was obtained using a sterile curette and removing supragingival plaque on root surfaces. Within each patient, we obtained a plaque sample from the diseased tooth and the contralateral healthy tooth to compare the microbiota of diseased versus healthy roots within the same patient. In cases where the contralateral tooth was missing, we obtained plaque from the closest available tooth. Exclusion criteria for this study were any systemic disease (diabetes, HIV, autoimmune diseases, and infectious diseases), smoking, alcohol or recreational drug abuse, plaque index >50%, and antibiotic use within the previous 3 months. This study was approved by the University of Detroit Mercy institutional review board (IRB) committee, UDM IRB no. 1415-06. All study participants signed informed consent forms according to UDM IRB no.1415-06.
Isolation and processing of dental plaque samples.
Plaque samples were placed in a sterile tube containing 2 mL phosphate-buffered saline (PBS) buffer and 10% glycerol as a cryoprotective agent, and the sample was stored at −80°C until processing. Upon thawing, 0.5 mL of the sample was subjected to lysis and chromosomal DNA purification using a Qiagen blood and tissue DNA purification kit with the modification that after resuspending the plaque sample in buffer ATL, digestion with proteinase K and addition of buffer AL, 500 μL of sterile zirconia beads (Fisher Scientific) were added, and the samples were sonicated for 2.75 min prior to proceeding with column purification of the extracted DNA (Mini Bead Beater; BioSpec Products). Purified DNA was quantified and used as a template for PCR analysis with 16S rRNA gene primers designed to detect all bacteria (universal primers flanking the V6 region) or detect each organism of interest (species-specific primers within the V6 region) including Streptococcus mutans/sobrinus, Actinomyces naeslundii/viscosus, Lactobacillus casei, or more diverse Lactobacillus species, Selenomonas species, Olsenella uli, Streptococcus sanguinis, Veillonella parvula/dispar, Prevotella multisaccharivorax, Prevotella denticola, Propionibacterium acidifaciens, Capnocytophaga granulosa, and Delftia acidovorans. Primers used for quantitative PCR are found in Table S1 in the supplemental material. Quantification of the level of each organism in plaque samples was determined using real-time PCR (Bio-Rad; CFX96) and comparison to a positive-control DNA standard for each organism of interest (Table S1). Reaction mixtures contained 10 μL SYBR Green supermix (Bio-Rad; catalog no. 1708882), 0.05 μL of each species primer (from 50 μM stock), 1 μL template DNA (≤1 ng/μL), and 9 μL H2O. Fifty cycles of amplification were performed with 10 s at 95°C (melting), 30 s at 52°C (annealing), and 30 s at 72°C (elongation). PCRs were performed in duplicate wells on at least 3 separate days.
Biofilm assays.
S. mutans was grown overnight in 10 mL brain heart infusion at 37°C and 5% CO2. V. parvula was grown overnight for 2 to 3 days in Brucella broth with 1 μg/mL vitamin K and 5 μg/mL hemin (BB+) in an anaerobic chamber (Coy Laboratories; 85% N2, 5% CO2, and 10% H2 gas mix). S. mutans and V. parvula were then pelleted and resuspended in BB+ or artificial saliva (29) at an OD600 of 1.0 or 0.1 for biofilm studies. Biofilms were prepared in 24-well tissue culture-treated plates (CytoOne). We placed 740 μL of BB+ or artificial saliva into each well containing a single strain along with 250 μL of culture with an OD600 of 1.0 or 0.1. We placed 490 μL of BB+ or artificial saliva into each well containing mixed biofilms along with 250 μL of each culture with an OD600 of 1.0 or 0.1. We also added 10 μL of H2O or 50% sucrose to each well (1 mL total) to achieve a final sucrose concentration of 0 or 0.5%, respectively, in BB+ or artificial saliva. Biofilms were grown for approximately 24 h, 72 h, or 1 week at 37°C in an anaerobic chamber.
After biofilm growth, the supernatant was removed from each well, filtered, and saved for pH measurements (Mettler Toledo; FiveEasy). Wells were washed carefully twice with 1 mL PBS and heat fixed at 80°C for 30 min. Five hundred microliters of 0.5% crystal violet (Sigma Chemical) was added to each well and placed on an orbital shaker for 30 min. Crystal violet was removed, wells were washed carefully with five consecutive submersions in water, and any remaining liquid was removed from the wells. We added 1 mL of 33% acetic acid (Fisher Scientific) to each well to dissolve the biofilm, and the plate was placed on an orbital shaker for 5 to 10 min to ensure that the entire biofilm was dissolved. Samples were then diluted in a 96-well plate at 1:2 dilutions. The absorbance of the plate reader (Molecular Devices; SpectraMax M3) was set to 570 nm, the density of the purple color from the lysed biofilm was measured, and data were corrected for the dilution.
Fluorescence microscopy.
Overnight cultures of S. mutans 25175 grown in BHI at 37°C, 5% CO2, and V. parvula 10790 grown for 1 to 2 days in BB+ at 37°C anaerobically were spun for 10 min at 4,500 rpm at 25°C, washed once with PBS, and resuspended in artificial saliva at an OD600 of 1.0. Four-well chamber slides (Nunc Lab-Tek II; catalog no. 154526) were inoculated with 250 μL bacteria (S. mutans 25175 alone or S. mutans 25175 with V. parvula 10790), 740 μL or 490 μL artificial saliva, respectively, and 10 μL 50% sucrose (1 mL total) and grown 24 h at 37°C anaerobically. Wells were washed gently twice with 1 mL PBS, and 250 μL of PBS containing 2 μM SYTO 9 dye (Thermo Fisher; catalog no. S34854) and 20 μM propidium iodide (Sigma; catalog no. P4170) was added to each well for 15 min in the dark prior to imaging on an Olympus BX63 fluorescence microscope. Control wells were treated with 1 mL Listerine Naturals for 10 min prior to processing for LIVE/DEAD staining.
LIVE/DEAD 24-well plate staining.
Twenty-four-well tissue culture-treated plates set up identically to the 4-well chamber slides described above were also grown to assess the ratio of live (SYTO 9) to dead (propidium iodine) staining of S. mutans and S. mutans plus V. parvula biofilms. Green (excitation, 485 nm; emission, 530 nm) and red (excitation, 485 nm; emission, 630 nm) signals were read on a Tecan Spark plate reader, and relative fluorescence units were used to calculate the LIVE/DEAD (green/red) ratio.
DNA analysis of biofilms.
To determine the proportion of S. mutans and V. parvula in mixed biofilms, DNA was extracted as described for plaque samples from bacteria growing in 24-well plates like the biofilm-processed wells. PCR analysis was then performed with S. mutans- and V. parvula-specific primers within the V6 region and compared to 16S rRNA gene universal primers flanking the V6 region (Table S1), and the percentage of each species in the biofilm was determined.
Microbiome analysis of clinical plaque samples.
The 16S rRNA gene PCR primers 27F (AGRGTTTGATCMTGGCTCAG) and 1492R (GGGTTACCTTGTTACGACTT) with a barcode on the forward primer were used in a 33-cycle PCR on root caries, and healthy plaque DNA samples were prepared as described above using the HotStarTaq Plus master mix kit (Qiagen, USA) under the following conditions: 94°C for 3 min, followed by 33 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, after which a final elongation step at 72°C for 5 min was performed. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. 16S rRNA gene PCR products from all 10 root caries samples were pooled in equal proportions based on their molecular weight and DNA concentrations and purified using Ampure PB beads (Pacific Biosciences). SMRTbell libraries (Pacific Biosciences) were prepared following the manufacturer’s user guide, and sequencing was performed at MR DNA (Shallowater, TX, USA; https://www.mrdnalab.com) on a PacBio Sequel following the manufacturer’s guidelines. After completion of initial DNA sequencing, each library underwent a secondary analysis, circular consensus sequencing (CCS), using PacBio’s CCS2 algorithm, to correct stochastic errors generated in the initial analysis. Sequence data were then processed using the MR DNA analysis pipeline. In summary, the CCS sequencing data were depleted of barcodes, sequences <150 bp were removed, and sequences with ambiguous base calls were removed. Sequences were denoised, operational taxonomic units (OTUs) generated, and chimeras removed. Operational taxonomic units were defined by clustering at 1% divergence (99% similarity). Final OTUs were taxonomically classified using BLASTn against a curated database derived from RDPII and NCBI (https://www.ncbi.nlm.nih.gov; http://rdp.cme.msu.edu).
Isolation of S. mutans and Veillonella parvula clinical strains from dental plaque.
To isolate strains of S. mutans from our clinical plaque samples, about 200 μL of the glycerol stocks containing the plaque samples were plated onto mitis-salivarius-bacitracin (MSB) agar to enrich for S. mutans isolates. Dark blue-staining colonies were then restruck for purification, and chromosomal DNA was prepared using the Qiagen blood and tissue DNA purification kit with the addition of a zirconia bead sonication step as described above. The 16S rRNA gene of each S. mutans candidate was then amplified with primers 16S full-length 5′ and 16S full-length 3′, and products were sent to Genewiz for DNA sequencing using primers designed to sequence the V3 and V6 regions. Those isolates with a perfect match to S. mutans strains in the NCBI BLAST database were stored as glycerol stocks and used for mixed biofilms from clinical isolates.
To isolate Veillonella strains from dental plaque, plaque samples that indicated high levels of Veillonella colonization based on microbiome analysis (Fig. 5) were struck onto Brucella blood agar plates with 1 μg/mL vitamin K and 5 μg/mL hemin and grown anaerobically for 3 to 5 days. Based on similar colony morphologies to laboratory Veillonella strains and the fact that in some cases, 30 to 40% of the total microbiome was from the genus Veillonella (Fig. 5, patient 4, teeth 8 and 9), numerous colonies were screened by real-time PCR using diagnostic Veillonella parvula/V. dispar V6 primers (Table S1). Those Veillonella candidates that reacted strongly (nearly 1:1 with the 16S universal PCR primer pair) were then subjected to chromosomal DNA isolation (Qiagen blood and tissue DNA purification kit), and the 16S sequence was determined as described for S. mutans above. Those strains verified to be Veillonella parvula by NCBI BLAST analysis were stored as glycerol stocks and used for mixed biofilms from clinical isolates.
Analysis of gtf expression in S. mutans.
To assess expression of the three glucosyltransferase genes in S. mutans (gtfB, gtfC, and gtfD), S. mutans and S. mutans with V. parvula biofilms were grown in artificial saliva with 0.5% sucrose in 24-well plates in duplicate as described for biofilms above. After 24 h, unbound bacteria were removed by gentle rinsing with 1 mL PBS, and plate-bound bacteria were lysed with 400 μL TRI reagent (Sigma; catalog no. T3934) per well. We added 80 μL chloroform to the cell extract in an Eppendorf tube, and the tube was inverted and then vortexed for 15 s. The tube was left to rest for 15 min followed by 15 min centrifugation at 13,000 rpm and 4°C. The upper aqueous layer was transferred to a fresh tube, and 200 μL isopropanol was added, vortexed, left to rest for 5 min, and then centrifuged at 13,000 rpm. The pellet was rinsed with 75% ethanol and centrifuged for 5 min at 13,000 rpm. The supernatant was removed, the pellet was dried by SpeedVac, and the RNA was resuspended in 50 μL DNase/RNase-free water (Invitrogen/Life Technologies; catalog no. 10977-015). We used 1 μL of the RNA as the template for cDNA generation with an iScript cDNA synthesis kit (Bio-Rad; catalog no. 1708891) in the following reaction mixture: 4 μL iScript reaction mixture, 1 μL iScript reverse transcriptase, 14 μL H2O, and 1 μL RNA. After generation of cDNA, 1 μL of cDNA was used in a real-time PCR with 0.25 μL S. mutans-specific rpoB primers (Table S1, + control) and gtfB, gtfC, or gtfD primers (Table S1) to determine the levels of expression of each gtf gene relative to rpoB for biofilms containing S. mutans (ATCC 25175) alone or in combination with V. parvula (ATCC 10790 or ATCC 17745). gtf expression levels relative to rpoB were calculated with rpoB levels set to 1. The following protocol for real-time PCR of the cDNA was the same as that used on plaque samples: 50 cycles of amplification with 10 s at 95°C (melting), 30 s at 52°C (annealing), and 30 s at 72°C (elongation). cDNA PCRs were performed in duplicate wells on 2 separate days.
Quantitative NMR profiling of bacterial fermentation.
Bacterial fermented media were centrifuged, and the obtained supernatants were prepared with phosphate buffer (pH 7.4) containing 10% D2O (Cambridge Isotope Laboratories, Inc., Andover, MA, USA), 0.5 mM sodium salt of 3-trimethylsilylpropionic acid (TSP; Sigma-Aldrich, St. Louis, MO, USA), and 1.5 mM NaN3 (Sigma-Aldrich) (39). The proton nuclear magnetic resonance (NMR) spectra were acquired on a Bruker 600 MHz NMR instrument (Billerica, MA, USA), with δ in ppm related to TSP and J (the coupling constant) in Hz. A water suppression pulse sequence was used to suppress water peak, and the number of scans was 64. Metabolites were quantified through integration areas referring to those of 0.5 mM TSP.
Statistical analysis of qPCR results.
qPCR data were analyzed by individual tooth, by patient (aggregate of teeth within that patient), or by combining data from the entire population in the study, comparing microbial colonization results from carious teeth to noncarious teeth. Student’s t test was used when comparing single carious teeth versus noncarious teeth to each other, as the data are expected to conform to a normal distribution from duplicate PCR measurements on at least 3 different days. When multiple carious and diseased teeth from a single patient were aggregated and compared, we used the nonparametric Mann-Whitney test, as one would not expect different teeth to have a normal distribution of bacterial colonization levels relative to each other. Finally, when analyzing data from all 7 patients aggregated together to compare colonization levels between all 14 carious and 13 noncarious teeth, we used multivariate logistic regression to account for the high variability in colonization levels between the people in the study.
ACKNOWLEDGMENTS
We would like to thank Joshua Thomson and Sarah Plecha for helpful discussions regarding these studies and David Sherman for helpful discussions and for facilitating the collaboration between E.S.K. and A.T. In addition, we thank Maribel Okiye for help with delivering samples for processing by NMR.
Work reported in this publication was supported by the National Institutes of Health Common Fund and Office of Scientific Workforce Diversity under three linked awards, RL5GM118981, TL4GM118983, and 1UL1GM118982, administered by the National Institute of General Medical Sciences. This work was also supported by the University of Detroit Mercy School of Dentistry FRG grant numbers 2015-01 and 2017-01 to E.S.K. and J.G.P.
Footnotes
Supplemental material is available online only.
Contributor Information
Eric S. Krukonis, Email: krukones@udmercy.edu.
Marvin Whiteley, Georgia Institute of Technology School of Biological Sciences.
REFERENCES
- 1.Hoppenbrouwers PM, Driessens FC, Borggreven JM. 1987. The mineral solubility of human tooth roots. Arch Oral Biol 32:319–322. 10.1016/0003-9969(87)90085-9. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi N, Nyvad B. 2016. Ecological hypothesis of dentin and root caries. Caries Res 50:422–431. 10.1159/000447309. [DOI] [PubMed] [Google Scholar]
- 3.de Soet JJ, Nyvad B, Kilian M. 2000. Strain-related acid production by oral streptococci. Caries Res 34:486–490. 10.1159/000016628. [DOI] [PubMed] [Google Scholar]
- 4.Zambon JJ, Kasprzak SA. 1995. The microbiology and histopathology of human root caries. Am J Dent 8:323–328. [PubMed] [Google Scholar]
- 5.Fure S, Romaniec M, Emilson CG, Krasse B. 1987. Proportions of Streptococcus mutans, lactobacilli and Actinomyces spp in root surface plaque. Scand J Dent Res 95:119–123. 10.1111/j.1600-0722.1987.tb01817.x. [DOI] [PubMed] [Google Scholar]
- 6.Bowden GH, Ekstrand J, McNaughton B, Challacombe SJ. 1990. Association of selected bacteria with the lesions of root surface caries. Oral Microbiol Immunol 5:346–351. 10.1111/j.1399-302x.1990.tb00439.x. [DOI] [PubMed] [Google Scholar]
- 7.Ellen RP, Banting DW, Fillery ED. 1985. Streptococcus mutans and Lactobacillus detection in the assessment of dental root surface caries risk. J Dent Res 64:1245–1249. 10.1177/00220345850640101301. [DOI] [PubMed] [Google Scholar]
- 8.Nyvad B, Kilian M. 1990. Microflora associated with experimental root surface caries in humans. Infect Immun 58:1628–1633. 10.1128/iai.58.6.1628-1633.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Houte J, Lopman J, Kent R. 1994. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res 73:1727–1734. 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 10.Preza D, Olsen I, Aas JA, Willumsen T, Grinde B, Paster BJ. 2008. Bacterial profiles of root caries in elderly patients. J Clin Microbiol 46:2015–2021. 10.1128/JCM.02411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preza D, Olsen I, Willumsen T, Boches SK, Cotton SL, Grinde B, Paster BJ. 2009. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis 28:509–517. 10.1007/s10096-008-0662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Qin B, Du M, Zhong H, Xu Q, Li Y, Zhang P, Fan M. 2015. Extensive description and comparison of human supra-gingival microbiome in root caries and health. PLoS One 10:e0117064. 10.1371/journal.pone.0117064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schupbach P, Osterwalder V, Guggenheim B. 1995. Human root caries: microbiota in plaque covering sound, carious and arrested carious root surfaces. Caries Res 29:382–395. 10.1159/000262097. [DOI] [PubMed] [Google Scholar]
- 14.Schupbach P, Osterwalder V, Guggenheim B. 1996. Human root caries: microbiota of a limited number of root caries lesions. Caries Res 30:52–64. 10.1159/000262137. [DOI] [PubMed] [Google Scholar]
- 15.Brailsford SR, Shah B, Simons D, Gilbert S, Clark D, Ines I, Adams SE, Allison C, Beighton D. 2001. The predominant aciduric microflora of root-caries lesions. J Dent Res 80:1828–1833. 10.1177/00220345010800091101. [DOI] [PubMed] [Google Scholar]
- 16.Mikx FH, van der Hoeven JS, Konig KG, Plasschaert AJ, Guggenheim B. 1972. Establishment of defined microbial ecosystems in germ-free rats. I. The effect of the interactions of Streptococcus mutans or Streptococcus sanguis with Veillonella alcalescens on plaque formation and caries activity. Caries Res 6:211–223. 10.1159/000259801. [DOI] [PubMed] [Google Scholar]
- 17.Caufield PW, Dasanayake AP, Li Y, Pan Y, Hsu J, Hardin JM. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun 68:4018–4023. 10.1128/IAI.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, Griffen AL. 2002. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40:1001–1009. 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Hoeven JS, Toorop AI, Mikx RH. 1978. Symbiotic relationship of Veillonella alcalescens and Streptococcus mutans in dental plaque in gnotobiotic rats. Caries Res 12:142–147. 10.1159/000260324. [DOI] [PubMed] [Google Scholar]
- 21.Noorda WD, Purdell-Lewis DJ, van Montfort AM, Weerkamp AH. 1988. Monobacterial and mixed bacterial plaques of Streptococcus mutans and Veillonella alcalescens in an artificial mouth: development, metabolism, and effect on human dental enamel. Caries Res 22:342–347. 10.1159/000261134. [DOI] [PubMed] [Google Scholar]
- 22.Qudeimat MA, Alyahya A, Karched M, Behbehani J, Salako NO. 2021. Dental plaque microbiota profiles of children with caries-free and caries-active dentition. J Dent 104:103539. 10.1016/j.jdent.2020.103539. [DOI] [PubMed] [Google Scholar]
- 23.Mikx FH, Van der Hoeven JS. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch Oral Biol 20:407–410. 10.1016/0003-9969(75)90224-1. [DOI] [PubMed] [Google Scholar]
- 24.Palmer RJ, Jr., Diaz PI, Kolenbrander PE. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J Bacteriol 188:4117–4124. 10.1128/JB.01958-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kara D, Luppens SB, Cate JM. 2006. Differences between single- and dual-species biofilms of Streptococcus mutans and Veillonella parvula in growth, acidogenicity and susceptibility to chlorhexidine. Eur J Oral Sci 114:58–63. 10.1111/j.1600-0722.2006.00262.x. [DOI] [PubMed] [Google Scholar]
- 26.Luppens SB, Kara D, Bandounas L, Jonker MJ, Wittink FR, Bruning O, Breit TM, Ten Cate JM, Crielaard W. 2008. Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm. Oral Microbiol Immunol 23:183–189. 10.1111/j.1399-302X.2007.00409.x. [DOI] [PubMed] [Google Scholar]
- 27.Mashima I, Nakazawa F. 2014. The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe 28:54–61. 10.1016/j.anaerobe.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Chen M, Wang Y, Zhou X, Peng X, Ren B, Li M, Cheng L. 2020. Effect of Veillonella parvula on the physiological activity of Streptococcus mutans. Arch Oral Biol 109:104578. 10.1016/j.archoralbio.2019.104578. [DOI] [PubMed] [Google Scholar]
- 29.Roger P, Delettre J, Bouix M, Beal C. 2011. Characterization of Streptococcus salivarius growth and maintenance in artificial saliva. J Appl Microbiol 111:631–641. 10.1111/j.1365-2672.2011.05077.x. [DOI] [PubMed] [Google Scholar]
- 30.Mantzourani M, Fenlon M, Beighton D. 2009. Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol Immunol 24:32–37. 10.1111/j.1399-302X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- 31.Zickert I, Emilson CG, Krasse B. 1983. Correlation of level and duration of Streptococcus mutans infection with incidence of dental caries. Infect Immun 39:982–985. 10.1128/iai.39.2.982-985.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto T, Kajiura S, Hirai Y, Watanabe T. 1994. Capnocytophaga haemolytica sp. nov. and Capnocytophaga granulosa sp. nov., from human dental plaque. Int J Syst Bacteriol 44:324–329. 10.1099/00207713-44-2-324. [DOI] [PubMed] [Google Scholar]
- 33.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417. 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. 2012. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7:e47722. 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawes C. 2003. What is the critical pH and why does a tooth dissolve in acid? J Can Dent Assoc 69:722–724. [PubMed] [Google Scholar]
- 36.Bechon N, Jimenez-Fernandez A, Witwinowski J, Bierque E, Taib N, Cokelaer T, Ma L, Ghigo JM, Gribaldo S, Beloin C. 2020. Autotransporters drive biofilm formation and autoaggregation in the diderm firmicute Veillonella parvula. J Bacteriol 202:e00461-20. 10.1128/JB.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Manoil D, Belibasakis GN, Kotsakis GA. 2021. Veillonellae: beyond bridging species in oral biofilm ecology. Front Oral Health 2:774115. 10.3389/froh.2021.774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mutha NVR, Mohammed WK, Krasnogor N, Tan GYA, Wee WY, Li Y, Choo SW, Jakubovics NS. 2019. Transcriptional profiling of coaggregation interactions between Streptococcus gordonii and Veillonella parvula by Dual RNA-Seq. Sci Rep 9:7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. 2007. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2:2692–2703. 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6 and Tables S1 to S5. Download iai.00355-22-s0001.pdf, PDF file, 3.7 MB (3.8MB, pdf)