ABSTRACT
A complete 30,616-nucleotide Cervid atadenovirus A genome was determined from the tissues of black-tailed deer that had died in 2020 in British Columbia, Canada. Unique, nonsynonymous single-nucleotide polymorphisms in the E1B, Iva2, and E4.3 coding regions and deletions totaling 74 nucleotides that were not observed in moose and red deer isolates were present.
ANNOUNCEMENT
Adenovirus hemorrhagic disease (AHD) is an acute, infectious, and usually fatal viral disease of several cervid species. AHD was first described in black-tailed deer in California in 1993, although retrospective analysis of tissues suggested that AHD was present in California in 1981 (1). The disease was previously reported in Canada (2), although no genome sequences are available. Deer deaths were reported on Galiano Island, British Columbia, Canada, in September 2020, with subsequent analyses (histopathology, adenovirus consensus PCR, and amplicon sequencing) confirming AHD (3). Since then, hundreds of black-tailed deer on many Gulf Islands, including Vancouver Island, have died of confirmed AHD (4). The disease was also observed in neighboring Washington state (in the United States) in 2021 (5). The 10 currently available complete genomes for Deer atadenovirus A (Odocoileus adenovirus 1 [OdAdV-1]), the causative agent of AHD, were all obtained from cervids in the United States (Fig. 1c).
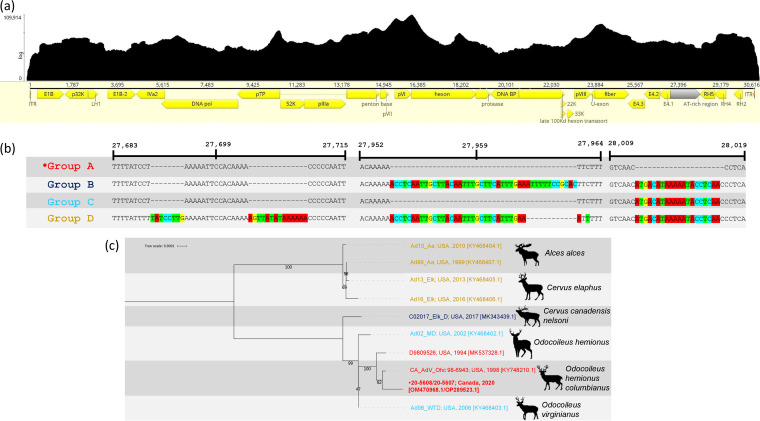
FIG 1.
(a) Genome organization of Cervid atadenovirus A, showing coverage depth based on mapping of reads from one sample to the assembled complete genome. (b) Nucleotide alignment of the variable region of the AT-rich noncoding region of 10 Deer atadenovirus A genomes. Genome position numbering, insertions, and SNPs are relative to the Cervid atadenovirus A from this study (20-5608), with insertions and SNPs highlighted. Genomes with identical sequences are grouped together. Group A contains accession numbers OM470968, OP289523, MK537328, and KY748210, group B contains accession number MK343439, group C contains accession numbers KY468402 and KY468403, and group D contains accession numbers KY468407, KY468404, KY468405, and KY468406. (c) Midpoint-rooted maximum likelihood phylogenetic tree of Cervid atadenovirus A (Deer atadenovirus A) whole genomes. The genomes for the tree are the same as the accessions listed in panel B. Alignment was performed using MAFFT with default parameters (9), and the tree was generated using IQ-TREE (12) with the find best model setting with ModelFinder (13) (model HKY-F was chosen) and 1,000 ultrafast bootstraps (14). Isolate names of sequences, as well as the location and year of collection and the GenBank accession numbers, are shown, and the species of the host is indicated on the right. The genome presented here is indicated with a bullet.
Total DNA that had been extracted using the QIAamp DNA minikit (Qiagen) from frozen lung tissue samples from three black-tailed deer that had died of AHD on Galiano Island (two initial samples collected on 11 September 2020 and one subsequent sample collected on 16 September 2020) was submitted for high-throughput sequencing (HTS). Preparation of cDNA and subsequent HTS were performed, followed by viral sequence enrichment with a custom capture-probe set (6), following a previously published method (7). Enriched libraries were quantified, pooled, and sequenced on an Illumina MiSeq sequencer with a v2 flow cell and a 500-cycle kit (Illumina).
Default parameters were used for all bioinformatic analyses. The initial exploratory metagenomic analysis was carried out using a custom Nextflow workflow, nf-villumina v2.0.0 (https://github.com/CFIA-NCFAD/nf-villumina), which automates low-quality read removal, read classification, de novo assembly, and BLAST homology searches of assembled contigs, as detailed in a previous publication (8). From these results, de novo assembled contigs with homology to cervid atadenoviruses were found in all three samples. The presence of complete 40-bp inverted terminal repeats (ITRs) in two of the assembled contigs (one sample from each submission date) indicated that they represented complete genomes. All three contigs were aligned using MAFFT (9) and were identical in overlapping regions. Reference mapping total reads (6,536,978 reads, 7,576,032 reads, and 9,124,356 reads) to the assembled adenovirus genome resulted in mean coverage depths of 109.4× and 4,038.8× for the earlier samples and 4,590.5× for the later sample.
The 30,616-bp assembled adenovirus genomes had a GC content of 33.2%. Comparison with the most closely related OdAdV-1 reference (GenBank accession number KY748210.1) from BLAST and phylogenetic analyses (Fig. 1) showed 99.96% pairwise nucleotide identity, including 100% identity in the ITR. Geneious v9.1.8 (Biomatters) (10) was used for reference mapping, determination of coverage depth, and comparison to a reference sequence. A single-nucleotide polymorphism (SNP) at position 1,409, causing a potential 12-amino-acid truncation, was observed in the viral replication-critical E1B protein (11). Nonsynonymous substitutions were also found in Iva2 (T5486C) and E4.3 (C25817T) coding regions (Table 1). The variable region of the noncoding AT-rich region contained four deletions totaling 74 nucleotides, compared to isolates from moose and red deer, and appears to cluster with black-tailed deer and mule deer isolates (Fig. 1).
TABLE 1.
SNPs observed between the Cervid atadenovirus A genome and the most closely related publicly available match, OdAdV-1 (GenBank accession number KY748210.1)
| Genome positiona | Gene | Nucleotide(s) in OdAdV-1 (GenBank accession no. KY748210.1) | Nucleotide(s) in Cervid atadenovirus A 20-5608 | Effect on coding sequence |
|---|---|---|---|---|
| 267 | Noncoding region | A | C | No effect |
| 1,409 | E1B | G | T | 12-amino-acid truncation |
| 5,486 | IVa2 | T | C | Nonsynonymous (Lys to Glu) |
| 12,796 | pIIIa | G | A | Synonymous |
| 14,961 | pVII | Y (C or T) | C | No effect if reference is C, but if reference is T then nonsynonymous (Leu to Phe) |
| 15,134 | pVII | T | A | Synonymous |
| 16,238 | Hexon | C | T | Synonymous |
| 25,817 | E4.3 | C | T | Nonsynonymous (Met to Ile) |
| 27,054–27,056 | AT-rich region | TAT | – – –b | No effect |
| 27,968 | AT-rich region | C | – | No effect |
| 28,364 | RH5 | C | T | Synonymous |
| 29,295 | RH4 | K (G or T) | G | No effect if G in reference, but if reference is T then nonsynonymous (His to Asn) |
Genome positions are relative to OdAdV-1 (GenBank accession number KY748210.1).
Dashes indicate a deletion in the sequence relative to the reference.
Data availability.
Nucleotide sequences, including annotations and raw sequencing reads for the two complete genome sequences, were deposited in GenBank and the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA803320 with the following GenBank, SRA, and BioSample accession numbers: OM470968, SRX14039924, and SAMN25644053 and OP289523, SRX17188812, and SAMN30469495, respectively. The versions described in this paper are the first versions.
ACKNOWLEDGMENTS
M.N. was supported in part by the LabsCanada Equipment Sharing and Scientific Platforms pilot project and a Canadian Safety and Security Program Project CSSP-2018-CP-2339.
We acknowledge Dr. Oksana Vernygora and Josip Rudar for feedback on the manuscript and Dr. Leslie Woods (University of California, Davis) for advice regarding the samples collected from Galiano Island. We also thank staff members from the necropsy, virology, and molecular diagnostics sections of the British Columbia Animal Health Centre for their technical assistance and the staff members of the British Columbia Wildlife Health Program, volunteers, and the public for reporting dead and sick deer and collecting samples.
We declare no conflicts of interest.
O.L. and T.J. conceived the study. M.F. prepared the next-generation sequencing library. O.L., M.F., and M.N. analyzed the data. H.S. coordinated sample collection and submission, and G.M. and T.J. performed laboratory analyses. All coauthors contributed to the manuscript.
Contributor Information
Oliver Lung, Email: oliver.lung@inspection.gc.ca.
Kenneth M. Stedman, Portland State University
REFERENCES
- 1.Woods LW, Schumaker BA, Pesavento PA, Crossley BM, Swift PK. 2018. Adenoviral hemorrhagic disease in California mule deer, 1990–2014. J Vet Diagn Invest 30:530–537. doi: 10.1177/1040638718766036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canadian Cooperative Wildlife Health Centre. 2006. Wildlife health alert: adenoviral hemorrhagic disease of deer. Wildlife Health Centre Newsletter, Volume 12-1, Spring & Summer 2006 12(1):3. https://digitalcommons.unl.edu/icwdmccwhcnews/25. [Google Scholar]
- 3.Ministry of Forests, Lands, Natural Resource Operations, and Rural Development. 2020. New deer disease suspected in B.C. https://news.gov.bc.ca/releases/2020FLNR0061-001863.
- 4.Laird K. 2021. Fast-spreading disease baffles wildlife experts:“hundreds” of deer dropping dead on Vancouver Island. Vancouver Island Free Daily https://www.vancouverislandfreedaily.com/news/fast-spreading-disease-baffles-wildlife-experts. [Google Scholar]
- 5.Washington Department of Fish and Wildlife. 2021. WDFW confirms AHD in San Juan Islands deer. https://wdfw.wa.gov/news/wdfw-confirms-ahd-san-juan-islands-deer.
- 6.Wylie TN, Wylie KM, Herter BN, Storch GA. 2015. Enhanced virome sequencing using targeted sequence capture. Genome Res 25:1910–1920. doi: 10.1101/gr.191049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papineau A, Berhane Y, Wylie TN, Wylie KM, Sharpe S, Lung O. 2019. Genome organization of Canada goose coronavirus, a novel species identified in a mass die-off of Canada geese. Sci Rep 9:5954. doi: 10.1038/s41598-019-42355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher M, Harrison TMR, Nebroski M, Kruczkiewicz P, Rothenburger JL, Ambagala A, Macbeth B, Lung O. 2020. Discovery and comparative genomic analysis of elk circovirus (ElkCV), a novel circovirus species and the first reported from a cervid host. Sci Rep 10:19548. doi: 10.1038/s41598-020-75577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidalgo P, Ip WH, Dobner T, Gonzalez RA. 2019. The biology of the adenovirus E1B 55K protein. FEBS Lett 593:3504–3517. doi: 10.1002/1873-3468.13694. [DOI] [PubMed] [Google Scholar]
- 12.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Nucleotide sequences, including annotations and raw sequencing reads for the two complete genome sequences, were deposited in GenBank and the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA803320 with the following GenBank, SRA, and BioSample accession numbers: OM470968, SRX14039924, and SAMN25644053 and OP289523, SRX17188812, and SAMN30469495, respectively. The versions described in this paper are the first versions.