ABSTRACT
Placental immunity is critical for fetal health during pregnancy, as invading pathogens spread from the parental blood to the fetus through this organ. However, inflammatory responses in the placenta can adversely affect both the fetus and the pregnant person, and the balance between protective placental immune response and detrimental inflammation is poorly understood. Extracellular vesicles (EVs) are membrane-enclosed vesicles that play a critical role in placental immunity. EVs produced by placental trophoblasts mediate immune tolerance to the fetus and to the placenta itself, but these EVs can also activate detrimental inflammatory responses. The regulation of these effects is not well characterized, and the role of trophoblast EVs (tEVs) in the response to infection has yet to be defined. The Gram-positive bacterial pathogen Listeria monocytogenes infects the placenta, serving as a model to study tEV function in this context. We investigated the effect of L. monocytogenes infection on the production and function of tEVs, using a trophoblast stem cell (TSC) model. We found that tEVs from infected TSCs can induce the production of the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) in recipient cells. Surprisingly, this tEV treatment could confer increased susceptibility to subsequent L. monocytogenes infection, which has not been reported previously as an effect of EVs. Proteomic analysis and RNA sequencing revealed that tEVs from infected TSCs had altered cargo compared with those from uninfected TSCs. However, no L. monocytogenes proteins were detected in tEVs from infected TSCs. Together, these results suggest an immunomodulatory role for tEVs during prenatal infection.
KEYWORDS: extracellular vesicles, Listeria monocytogenes, trophoblast, placental immunology
INTRODUCTION
The placenta is a remarkable organ in which the immune system plays a precarious role, balancing protective responses and potentially deleterious inflammation. The maternal decidua is an altered immune environment that permits the development of the semiallogeneic placental tissues and fetus, and some pathogens exploit this immunosuppressed site, invading and replicating inside the placenta. Placental pathogens include viruses, parasites, and bacteria (1). One such organism is the Gram-positive bacterium Listeria monocytogenes (2). This facultative intracellular parasite is the causative agent of listeriosis, an illness that affects approximately 1,600 people annually in the United States, resulting in around 300 deaths (3). Listeriosis typically afflicts the immunocompromised, with pregnant people being especially at risk (4). Prenatal listeriosis can lead to spontaneous abortions, stillbirths, and birth defects, while the pregnant person may show only mild symptoms (5). People initially ingest L. monocytogenes with contaminated food, such as deli meats, soft cheeses, and other dairy products. It escapes from the gastrointestinal tract and into the bloodstream, where it disseminates throughout the body and invades the liver, spleen, and the placenta (6). This pathogen has a well-characterized intracellular life cycle which allows it to spread throughout host tissues, within monocytes and other cells. L. monocytogenes enters the cell either by phagocytosis or by means of internalins, which are virulence factors that bind host surface proteins and induce uptake (7). Once inside the cell, L. monocytogenes is first contained in a phagosomal vacuole, which it lyses by means of the cholesterol-dependent cytolysin listeriolysin O (LLO), gaining access to the cytosol (8). In the cytoplasm, L. monocytogenes scavenges the host for nutrients and replicates. Eventually, the bacterium hijacks and polymerizes host actin to create actin rockets, which facilitate intracellular motility and entry into neighboring cells, where it restarts this process (9, 10). Importantly, the ability of L. monocytogenes to replicate intracellularly allows it to undergo cell-to-cell spread in trophoblasts, breaching the placental barrier while minimizing exposure to the extracellular environment (6).
A coordinated cell-mediated immune response is critical to resolve L. monocytogenes infection (11). A recently discovered mechanism that could play a role in mediating cellular immunity is intercellular communication mediated by extracellular vesicles (EVs). EVs are small (50 to 1,000 nm) membrane-enclosed vesicles that are secreted by nearly every type of cell in the human body and across all domains of life (12). Two of the major types of EVs in eukaryotes have been historically designated exosomes and microvesicles, which are differentiated based on how they are formed. Exosomes are smaller (50 to 150 nm) and form by the inward folding of the plasma membrane. Multiple vesicles form in multivesicular endosomes (MVEs), which translocate to the cell membrane, leading to membrane fusion and the release of the exosomes to the extracellular environment. Microvesicles, which have a wider range in size (100 to 1,000 nm), are formed by the outward budding of the plasma membrane directly into the extracellular space (13).
EVs play a critical role in placental development and immune regulation during pregnancy. The number of EVs per volume of blood in a healthy mother greatly increases during pregnancy, and the majority of these EVs originate from fetal trophoblasts (14). Trophoblast EVs (tEVs) have been found to carry immunoregulatory molecules, presumably suppressing the immune response to allow for the successful development of the fetus (15). However, tEVs also play a detrimental role during placental disease, such as preeclampsia (16, 17).
The role that EV-mediated communication plays during intracellular bacterial infections has been explored only recently, with a select number of pathogens, and the role of tEVs during prenatal infection is unknown. Mycobacterium-infected macrophages produce EVs that carry bacterial components, such as RNA, proteins, and glycopeptidolipids (18–20). EVs from Salmonella enterica serovar Typhimurium-infected macrophages carry Salmonella proteins and induce the production of proinflammatory cytokines through Toll-like receptor 2 (TLR2)- and TLR4-dependent mechanisms (21). Additionally, mice treated with EVs from S. enterica-infected cells generated antibodies against proteins found on the EVs, specifically Salmonella outer membrane proteins (22). EVs from L. monocytogenes-infected macrophages carry bacterial DNA, and this response is dependent on the DNA-sensing cGAS-STING system (23). These findings show that EVs play a potential role in immune responses to intracellular bacterial infection. The purpose of the work presented here is to begin to decipher the immunological role of tEVs produced in response to L. monocytogenes placental infection. Toward that end, we determined the effects of L. monocytogenes infection on tEV production, on the ability of tEVs to alter target immune cells, and on tEV RNA and protein content.
We used an L. monocytogenes-infected trophoblast stem cell (TSC) system to model placental infections and tEV production and function. tEVs isolated from infected TSCs stimulated the production of the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) in recipient macrophage-like cells. Unexpectedly, we observed that cells became more susceptible to L. monocytogenes infection after the tEV treatment. Using an untargeted proteomics approach, we found that tEVs from infected and uninfected TSCs had distinct protein profiles, with the infected tEVs containing more unique protein signatures than tEVs from uninfected TSCs. Ribosomal and other RNA binding proteins were increased in the tEVs by infection. However, in contrast to previous studies using macrophages, no bacterial proteins were found. RNA sequencing on the EVs revealed many mRNAs that were overrepresented in the tEVs from infected cells, including genes involved vasculogenesis and morphogenesis, which are processes involved in placental development. These data suggest that infection changes the tEVs produced in the placenta, which may lead to an altered immune response.
RESULTS
Characterization of tEVs from infected TSCs.
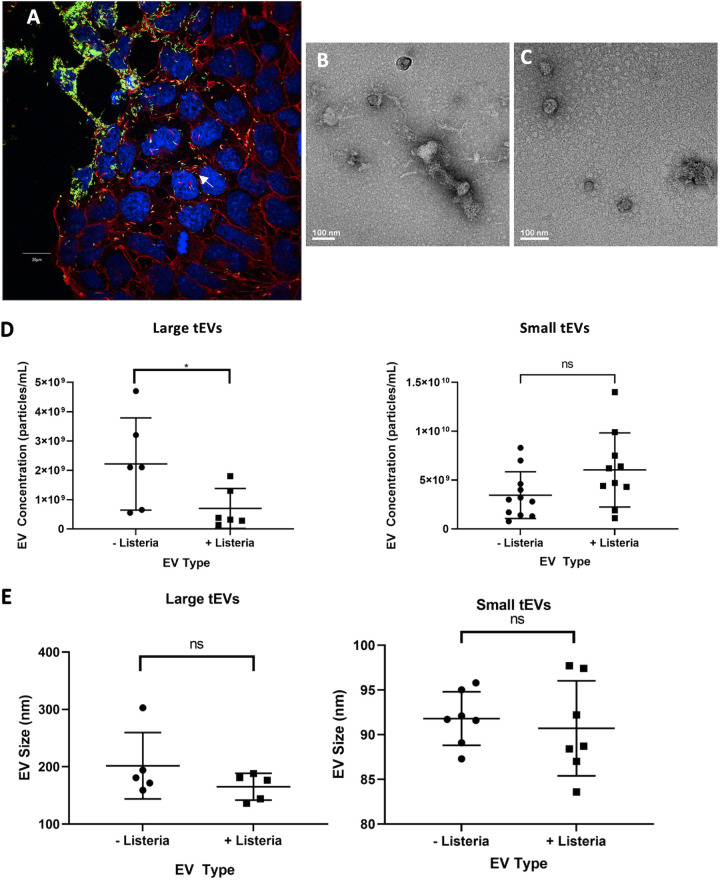
Trophoblast stem cells (TSCs) from C57BL/6 mice were used to model placental infections. TSCs were infected with L. monocytogenes at a multiplicity of infection (MOI) of 100, and fluorescence microscopy was used to visualize the infection at 24 h postinfection (hpi), confirming that L. monocytogenes can indeed infect these cells, as well as replicate and polymerize actin in this time frame (Fig. 1A). Additional infection of TSCs with a bioluminescent L. monocytogenes strain confirmed that the bacteria are replicating at 24 hpi and continue to replicate beyond this time point (see Fig. S1A and B in the supplemental material). These results show that TSCs are infected readily with L. monocytogenes, albeit in a longer time frame than J774 macrophages or other professional phagocytes (24).
FIG 1.
Extracellular vesicles from L. monocytogenes-infected TSCs. (A) Trophoblast stem cells (TSCs) from C57BL/6 mice were infected with GFP-expressing L. monocytogenes at a multiplicity of infection (MOI) of 100:1. At 24 h postinfection, the cells were fixed and stained with DAPI (blue) and rhodamine phalloidin (Red) which bind to DNA and polymerized actin, respectively. The cells were later imaged with an Olympus FluoView scanning confocal light microscope. The scale bar is 20 μm. (B, C) Transmission electron microscopy images of L-tEVs (B) and S-tEVs (C) from L. monocytogenes-infected TSCs. (D, E) TSC-derived EVs were analyzed by nanoparticle tracking analysis that gives the concentration and size distribution of the nanoparticles. The concentration (D) and mean size (E) of the tEVs with and without infection are given. Comparisons were conducted using Student’s t test; *, P < 0.05; ns, not significant.
Differentiated trophoblasts.
In the mouse placenta, TSCs differentiate into different trophoblast types, such as syncytiotrophoblasts (SynTs) and trophoblast giant cells (TGCs), to serve various functions. Previous studies found that the removal of the growth factors FGF-4 and activin, which are used to continue the proliferation of TSCs, pushes the cells toward the TGC phenotype. Additionally, the addition of retinoic acid (RA), the active derivative of vitamin A, leads cells toward TGC differentiation (25). Similarly, the activation of the Wnt pathway differentiates TSCs to SynTs (26). Here, we tested if the differentiation of TSCs altered their susceptibility to L. monocytogenes infection. We treated TSCs with either growth factors (GFs), base media (BM) alone, 5 μM RA, or 3 μM CHIR99021 (CHIR). The cells that received RA or CHIR did not receive any GFs. CHIR is an activator of the Wnt pathway, a cascade that is involved in many functions throughout the body, is required for proper placental development, and leads to the development of SynTs, where BM and RA treatments produced TGCs (25, 26). After 96 h, we infected the differentiated trophoblasts with bioluminescent L. monocytogenes to determine if there was any difference in susceptibility. We found that the differentiated trophoblasts were much more resistant to L. monocytogenes than TSCs grown with GFs, whether the TSCs were treated with BM alone, RA, or CHIR (Fig. S1C). Because L. monocytogenes replicates in trophoblasts in vivo (27), we wished to determine the effects of L. monocytogenes replication on tEV production, so we used undifferentiated TSCs for the characterization here. However, the effect of L. monocytogenes treatment on differentiated TSCs is of interest and is the subject of ongoing studies.
TSCs produce tEVs.
At 24 hpi, the medium from uninfected and L. monocytogenes-infected TSCs was collected, and large tEVs (L-tEVs) were isolated by collecting the vesicles from the top of the 0.22-μm filter, while small tEVs (S-tEVs) were purified by ultracentrifugation at 100,000 × g. EV preparations have often been referred to as microvesicles and exosomes, but as these entities are formed by distinct processes and are not just differentiated based on their size, we will refer to our separated samples as L-tEVs and S-tEVs throughout this report (28). Transmission electron microscopy (TEM) on the tEV preparations show the distinctive round shape in both vesicle preparations (Fig. 1B and C). The TEM images do show S-tEVs and L-tEVs that appear to be a similar size to each other despite the different isolation methods. A possible explanation for this finding is that the L-tEV preparations have a more diverse population in the size of EVs isolated. To determine if the infection altered tEV production, we performed nanoparticle tracking analysis on them using a Zetaview instrument (see Fig. S2A to D in the supplemental material). We found that infection decreased the number of L-tEVs produced by TSCs but did not affect the number of S-tEVs (Fig. 1D). In addition, infection did not alter the size of either L-tEVs or S-tEVs (Fig. 1E). Thus, L. monocytogenes differentially affected tEV production, decreasing the number of L-tEVs isolated.
tEV-mediated stimulation of macrophages.
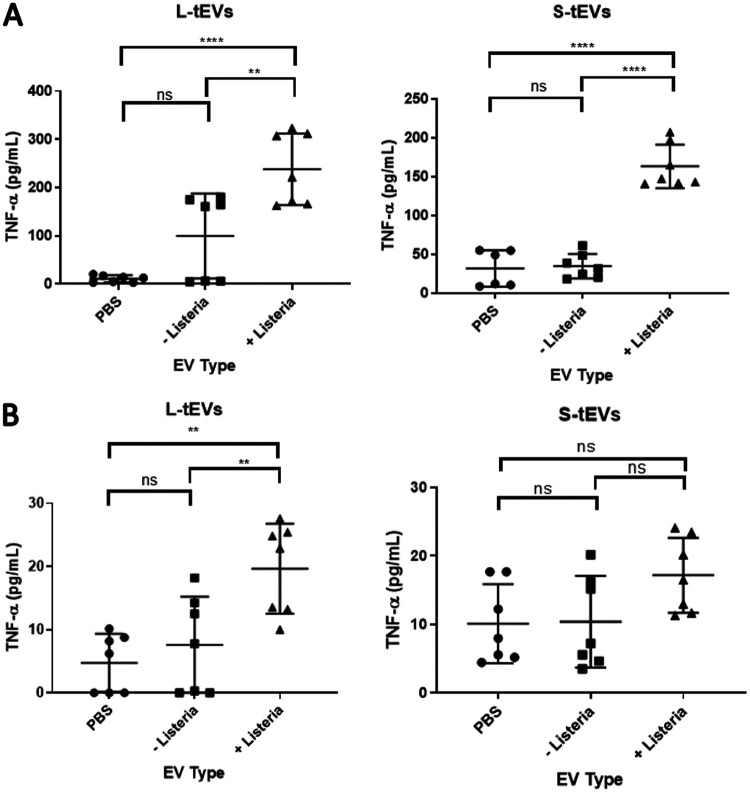
L. monocytogenes infections lead to a proinflammatory response that is necessary to control the infection, with infected cells producing cytokines, such as TNF-α (11). We hypothesized that tEVs can induce a similar proinflammatory response to L. monocytogenes infection. To test this hypothesis, we treated J774 and RAW 264.7 macrophage-like cells with 5 × 106 tEVs derived from uninfected and infected TSCs, and after 24 h, the TNF-α levels were measured using the enzyme-linked immunosorbent assay (ELISA). We found that RAW 264.7 cells treated with L-tEVs or S-tEVs derived from L. monocytogenes-infected cells resulted in the induction of TNF-α, while the treatment with phosphate-buffered saline (PBS) or tEVs from uninfected cells showed little TNF-α production (Fig. 2A). J774 cells, on the other hand, showed a significant increase of TNF-α only when treated with L-tEVs derived from infected cells, but not with S-tEVs from infected cells at the same concentration (Fig. 2B). These results demonstrate that tEV function is altered by L. monocytogenes infection of TSCs and that this alteration is tEV subtype specific and cell specific.
FIG 2.
tEVs from L. monocytogenes-infected TSCs induce TNF-α production. A total of 5 × 105 RAW 264.7 (A) and J774 (B) macrophage-like cells were treated with 5 × 106 tEVs from uninfected and L. monocytogenes-infected TSCs, and the production of tumor necrosis factor α (TNF-α) was measured using enzyme-linked immunosorbent assay (ELISA). Comparisons were done using the Student’s t test; **, P < 0.01; ****, P < 0.0001.
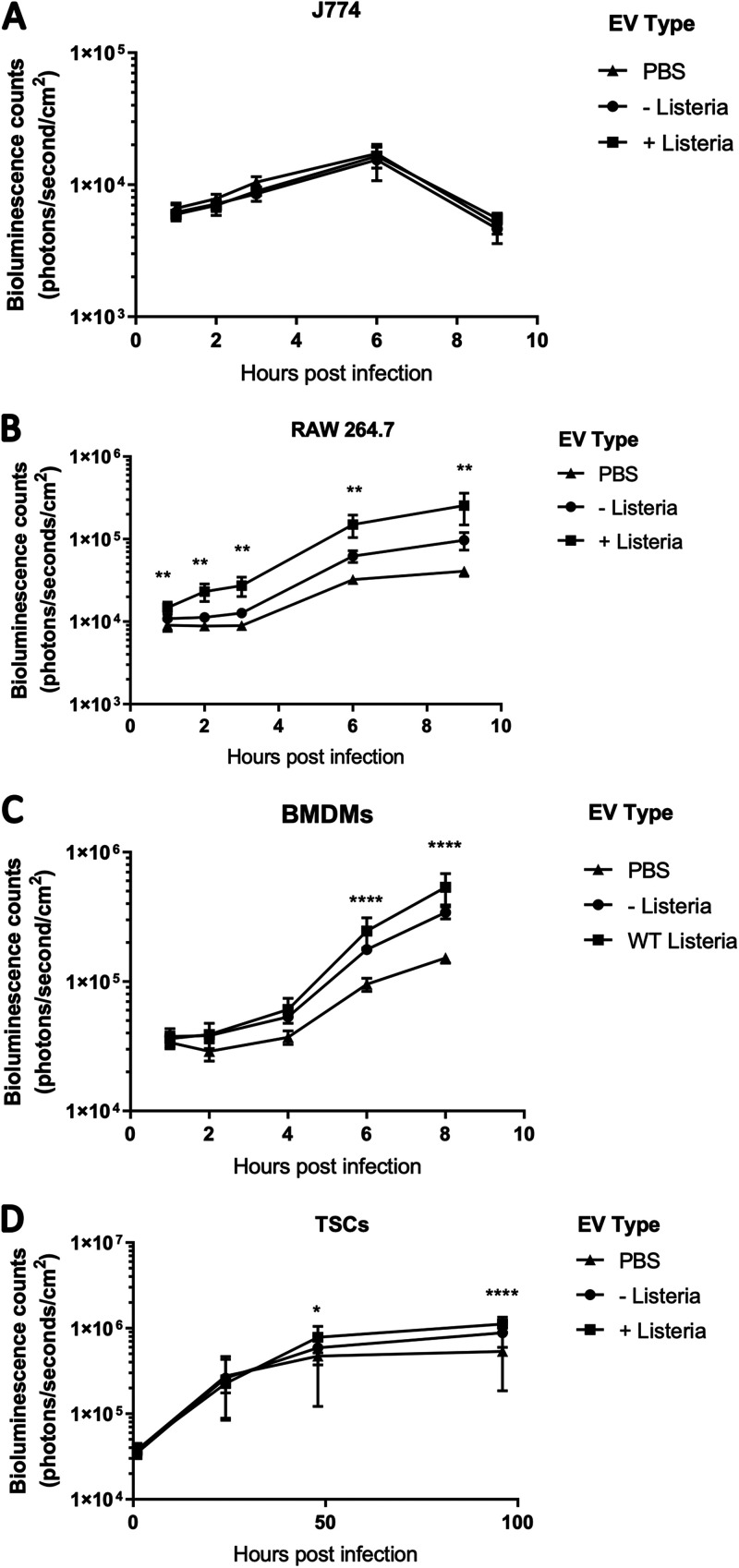
EVs have been proposed as vaccines because of their ability to stimulate macrophages and induce antigen-specific memory (29–31). Additionally, macrophage activation causes increased resistance to L. monocytogenes infection (32). We hypothesized that macrophages activated by the tEVs would become more resistant to L. monocytogenes infection. We again treated RAW 264.7 and J774 cells with 5 × 106 S-tEVs, and after 24 h, they were infected with bioluminescent L. monocytogenes. In J774 cells, there was no difference in L. monocytogenes growth (Fig. 3A). This result was expected since these tEVs failed to induce TNF-α production in J774 cells. However, we found that treatment with S-tEVs from uninfected TSCs increased the susceptibility of RAW 264.7 cells to infection. Surprisingly, treatment with tEVs from infected cells made the RAW 264.7 cells even more susceptible to infection (Fig. 3B). This result was unexpected, and as far as we are aware, this report is the first to demonstrate that EVs can induce macrophages to become more susceptible to infection. This effect was not limited to cell lines. Bone marrow-derived macrophages (BMDMs) also became more susceptible to infection with tEV treatment, especially with those from L. monocytogenes-infected cells; albeit the differences were not seen until later time points (Fig. 3C). Additionally, tEV treatment also increased TSC susceptibility to L. monocytogenes infection (Fig. 3D), although this effect was less prominent than that of macrophage-like cells. Overall, we found that S-tEVs from infected TSCs can increase susceptibility to infection, which was an unexpected effect.
FIG 3.
tEVs from L. monocytogenes-infected TSCs make cells susceptible to L. monocytogenes infection. (A to D) A total of 5 × 105 J774 cells (A), RAW 264.7 cells (B), BMDMs (C), and TSCs (D) were treated with 5 × 106 S-tEVs. After 24 h, the cells were infected at an MOI of 10 (A, B, C) or an MOI of 100 (D) with mid-log phase bioluminescent L. monocytogenes. The cells were imaged using the PerkinElmer in vivo imaging system (IVIS). Each group consisted of six replicates. At each time point, the EV groups were compared using Tukey’s post hoc multiple-comparison test. The stars indicate the any statistical difference between the + Listeria and PBS groups; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Treatment of mice with tEVs.
Our in vitro results suggest that tEVs from L. monocytogenes-infected TSCs make macrophages more susceptible to L. monocytogenes infection. However, tEV treatment also induced TNF-α. We therefore sought to determine the effect of tEV pretreatment on in vivo infection and whether tEVs would exacerbate the infection as indicated by the macrophage result or lessen the infection due to the induction of cytokines. We treated BALB/c mice intravenously (i.v.) with 108 S-tEVs from uninfected or infected TSCs or an equivalent amount of PBS. At 24 h later, 104 bioluminescent L. monocytogenes was inoculated into the mice through i.v. injection. At 72 h postinfection, the animals were imaged using in vivo imaging system (IVIS) imaging to quantify bacterial growth (see Fig. S3A in the supplemental material). The spleens from the animals were also collected and serially plated to determine CFU. There was no difference in the bioluminescence of the infection with any of the tEV conditions (Fig. S3B), and although there was a slight decrease in spleen CFU recovered with tEVs from infected TSCs, this finding was not statistically significant (Fig. S3C). Taken together, tEV pretreatment does not significantly affect the susceptibility of nonpregnant mice to L. monocytogenes infection. Whether tEVs affect infection in the context of pregnancy is the subject of ongoing experiments.
Proteomic analysis of the effect on infection on tEVs.
EVs carry a wide range of signaling molecules to deliver to recipient cells (33). We performed a proteomic analysis to determine if L. monocytogenes infection alters the protein profile of S-tEVs. Using shotgun tandem mass spectrometry, we found that there were many more unique proteins identified in S-tEVs from infected TSCs (331 proteins) than those in S-tEVs from uninfected cells (13 proteins). Additionally, in proteins that were shared between the 2 tEV groups (187 proteins), there were often more peptides identified in the infection condition, indicating larger amounts of that protein being transported in the tEVs from infected TSCs. Ribosomes, histones, and tubulin proteins are some of the categories where there were increased amounts in the infected tEVs. The full list of proteins that had at least a 2-fold increase in peptide signature in the tEVs from the infected TSCs are listed in Table S1 in the supplemental material. However, only four proteins saw a 2-fold higher content in peptide signatures in S-tEVs from uninfected cells that that in infected cells (see Table S2 in the supplemental material).
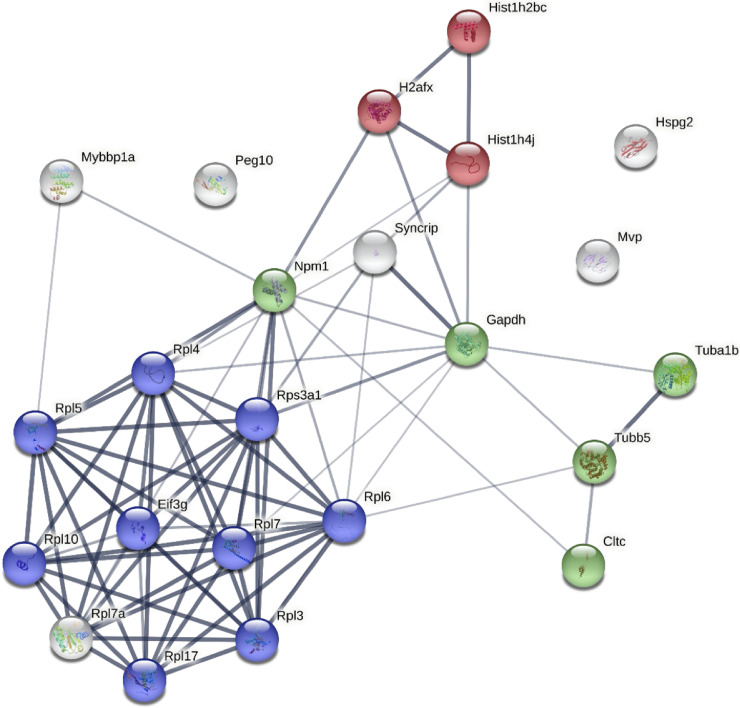
Gene ontology (GO) enrichment analysis was performed to determine biological functions that were represented by the proteins that were increased in S-tEVs from L. monocytogenes-infected TSCs. The main processes that were seen were related to RNA processing and translation, which is not an unexpected result given the many ribosomal and other RNA-binding proteins in these samples (Table S2). A protein interaction map was created using the proteins that had at least a 2-fold increase in peptides in the S-tEVs from infected cells (Fig. 4). We found that these proteins have high levels of interaction with each other, and we can see clusters of ribosomal, cytoskeleton, and histone proteins. There were not enough proteins that were increased in the tEVs from uninfected TSCs to perform a GO analysis. Altogether, our results show that L. monocytogenes infection does lead to different proteins loaded in tEVs, with more unique proteins seen in tEVs from infected cells. In contrast to other reports of EVs isolated from infected cells, no L. monocytogenes proteins were detected in tEVs from any sample.
FIG 4.
Protein interaction networks of proteins found in S-tEVs from L. monocytogenes-infected TSCs. The proteins of the S-tEVs from uninfected and L. monocytogenes-infected TSCs were determined by mass spectrometry. Protein interaction networks of the proteins identified in the S-tEVs from L. monocytogenes-infected TSCs were generated using the STRING program. The proteins involved in translation, microtubule cytoskeleton organization, and nucleosome core pathways are highlighted in blue, green, and red, respectively. Proteins that had twice the number of peptides identified in the L. monocytogenes-infected tEV samples versus the uninfected tEV samples were used for the analysis. The thickness of the line represents the confidence of the interaction between the proteins.
RNA sequencing on S-tEVs from L. monocytogenes-infected TSCs.
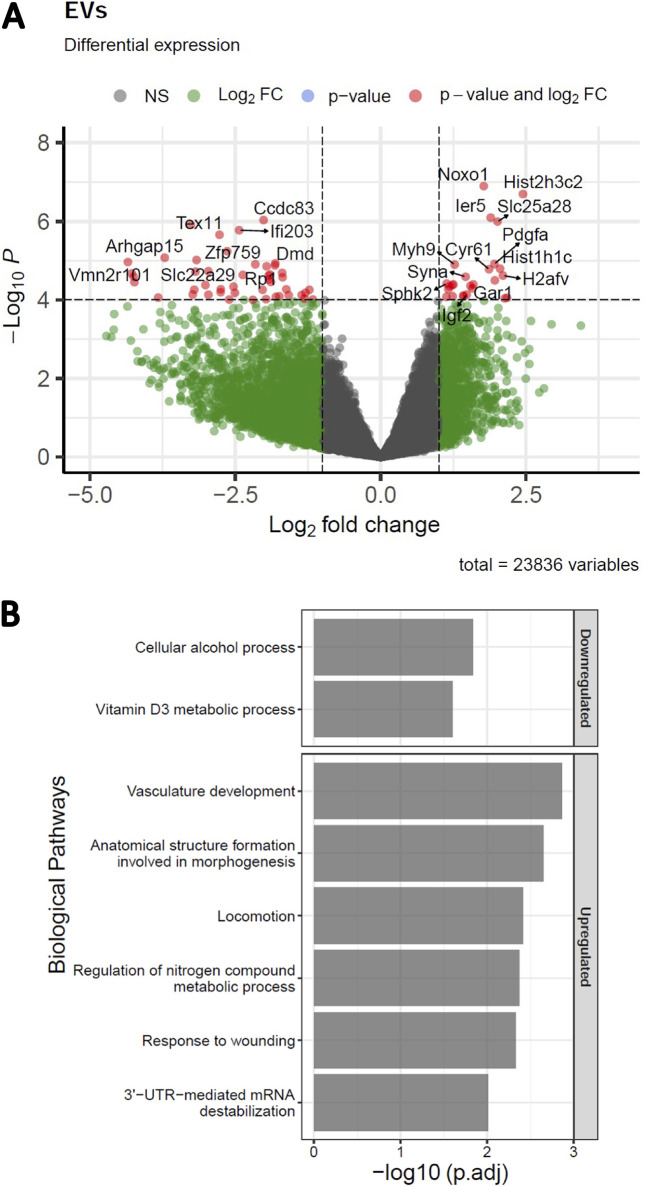
Our finding that S-tEVs from infected TSCs have increased amounts of RNA-binding proteins led us to suspect that these EVs could also carry different RNAs. We performed RNA sequencing on S-tEVs from uninfected and L. monocytogenes-infected TSCs (Fig. 5). We identified 22,836 genes in the mRNAs from the S-tEVs, with 68 genes being overrepresented in the S-tEVs from infected cells and 116 genes underrepresented in the S-tEV mRNAs from those cells (Fig. 5A). These differentially represented genes were used for the GO analysis (Fig. 5B). Interestingly, two of the pathways upregulated in the genes of L. monocytogenes-infected S-tEVs involved vascular development and morphogenesis, which are functions involved in placenta implantation and development. The pathways downregulated in the S-tEVs from infected TSCs involved metabolic processes. Overall, we found that L. monocytogenes infection of TSCs altered the host mRNAs found in the S-tEVs.
FIG 5.
L. monocytogenes-infected TSCs produce S-tEVs with altered RNA profiles. (A) Volcano plot of differentially expressed genes in S-tEVs from L. monocytogenes-infected and uninfected TSCs. Red dots represent statistical significance (P < 10−5) and log2 fold change (FC) greater or less than 1. Total variables represent the number of genes that were used to generate a volcano plot. (B) Gene ontology analysis of differentially expressed genes (p-adj of <0.05) was used to investigate the biological process pathways that were downregulated and upregulated in L. monocytogenes-infected cells using gProfileR.
DISCUSSION
EVs are the subject of exciting new research that offers the potential for novel approaches for the diagnosis and treatment of many diseases. A primary function of EVs appears to be the stimulation of cell signaling pathways in recipient cells, including immune cells, activating them in certain instances and dampening responses in others. This delicate balance can be altered during disease and infection. For example, certain cancers secrete EVs that suppress immune cell activity, allowing the malignancy to proliferate (34). Conversely, cells infected with intracellular pathogens secrete proinflammatory EVs that help to control infection, although EVs from infected cells may sometimes have the opposite effect (35).
During normal, healthy pregnancy, the number of EVs in the maternal bloodstream increases greatly (14). These EVs, produced mainly by fetal trophoblasts of the placenta, modulate immune responses but also can cause inflammation, leading to diseases, such as the life-threatening preeclampsia. However, the function of placental EVs in prenatal bacterial infection remains unknown. Considering their large number and the abovementioned effects on immune cells, placental EVs may play an important role in either ameliorating or exacerbating prenatal infection. We sought to study the effect of L. monocytogenes infection on EV production and function by trophoblasts because L. monocytogenes replicates in cytotrophoblasts in vivo (27). Whereas L. monocytogenes does not infect syncytiotrophoblasts, the infection of cytotrophoblasts is likely to have effects on pregnancy (6), perhaps mediated by tEVs.
Modeling placental function in vitro is a challenge. Replicating trophoblast cell lines, such as BeWo and HTR8/SVneo, are convenient, but their genetic alterations conferring proliferation may compromise their response to infection (36). Primary human or mouse cells or explants are a superior representation of in vivo trophoblasts, but they are expensive and laborious to isolate and cannot be maintained in culture, as they require repeated isolation (37). TSCs offer an alternative between the two extremes. They are isolated from mouse blastocysts and are naturally replicating with the addition of growth factors, facilitating repeated controlled experiments while also maintaining many biological properties (38, 39).
As EVs are much smaller than eukaryotic cells or even bacteria, their isolation is complex and has been the subject of controversy. Currently, differential ultracentrifugation is the most used method for isolating S-EVs. Low-speed centrifugation removes cells and larger cell debris, and subsequent high-speed ultracentrifugation (100,000 × g minimum) pools the tiny vesicles separate from the cells. Recent literature suggests that different isolation methods can affect the profile of EVs. Precipitation, density gradients, and filtration have all been used to isolate EVs, although each of these methods have their own positives and drawbacks (40). We chose to use differential centrifugation paired with filtration to ensure preparations free of soluble protein and bacteria because it is considered the gold standard method. It is important to note, though, that the EV isolation method can potentially influence the findings (28).
L. monocytogenes is an intracellular pathogen that normally grows rapidly in macrophages in vitro unless the macrophage has been activated toward a proinflammatory M1 phenotype. For example, RAW 264.7 macrophage-like cells treated with IFN-γ are resistant to infection and kill intracellular L. monocytogenes (32). Unexpectedly, we found that treatment with tEVs isolated from L. monocytogenes-infected TSCs did not lead to increased L. monocytogenes death; conversely, this treatment made the cells more permissible for growth of the bacteria. This result occurred despite the induction of TNF-α, which normally indicates the stimulation of macrophages resulting in greater resistance to infection. As far as the authors are aware, this is the first account of an EV treatment that causes cells to become more susceptible to L. monocytogenes infection. Interestingly, this phenotype was not observed in J774 cells. Both RAW 264.7 and J774 macrophage-like cell lines originate from BALB/c mice and have long been used in infection studies, although RAW 264.7 cells are from a male and J774 cells are from a female. J774 cells are more permissive for the growth of L. monocytogenes, and we expected activation by tEVs to reduce bacterial replication in these cells. The observation that these cells were equally permissive for L. monocytogenes growth under all conditions was unexpected. The mechanisms of the increased susceptibility in RAW 264.7 cells (and other cell types) but not in J774 cells remain of interest for future work. In addition, how the tEVs from infected TSCs stimulate recipient cells in the absence of bacterial products is under investigation and will require the directed manipulation of the altered contents. One hypothesis is that the tEVs are transmitting damage-associated molecular pattern (DAMP) molecules. The presence of histones, which elicit inflammation through DAMP recognition (41) in the tEVs from infected TSCs, supports this hypothesis. This process could result in the alteration of macrophage function, including possible enhanced phagocytic uptake, resulting in the increased susceptibility to infection. However, the mechanisms will require more extensive analysis than we provide here. Should the hypothesis be true, the implications for placental immunity could be very interesting.
EVs represent a potential strategy by L. monocytogenes to spread throughout the host by rendering recipient cells more susceptible to the bacteria. L. monocytogenes invades humans through epithelial cells that line the intestines, and from there, the pathogen relies on cell-to-cell spread to access the rest of the body. A previous report indicated that inhibiting EV formation attenuated L. monocytogenes growth in the spleens of mice, further suggesting that L. monocytogenes could be hijacking host EV function for its own benefit (23). The investigators reported that EVs from L. monocytogenes-infected cells carried Listeria DNA and activated the cGAS-STING pathway. This result is notable because this pathway leads to the release of type I interferons, which enhance L. monocytogenes growth in vivo (42–44). Further studies will be needed to identify these pathways and define the role tEVs play during placental infection.
In mice, tEVs from infected TSCs detectably reduced infection in the spleen, but the result was not statistically significant to a P value of 0.05. The reduction was modest, which could have been due to many factors. The number of tEVs administered (108 tEVs), single versus multiple tEV injections, whether the mice are pregnant, the timing and infectious dose of L. monocytogenes, and the strain of mouse used may all be important, and these parameters need further exploration.
There were several interesting proteins detected in the tEVs. We were particularly interested to see an enrichment of ribosomal proteins in tEVs derived from infected cells. Other groups have also found ribosomal proteins when performing a proteomic analysis of EVs (45–47). These studies usually focus on the RNA-binding aspects of these proteins, as RNA has been associated heavily with EVs (13). However, the appearance of ribosomes in EVs could be independent of RNA. A potential explanation is that translation levels are increased during cell stress (such as an infection), which could lead to higher ribosome numbers and eventually more ribosomal proteins in EVs. Another RNA binding protein identified in our infection tEVs is PEG10. PEG10 is a Gag-like protein that is required for trophoblast differentiation and placental development (48). This protein can also selectively bind and load mRNA into exosomes, and these EVs alter the gene expression of the recipient cell (49). The ability of EV-associated PEG10 to alter gene expression could be an explanation for the altered behavior of cells treated with tEVs from L. monocytogenes-infected cells. Surprisingly, tEVs from infected TSCs lacked any L. monocytogenes proteins in our analysis, contrasting previous EV studies of macrophage infection (19, 22, 50, 51). Most previous reports used macrophages as the initially infected cells, and the lack of L. monocytogenes proteins may reflect reduced bactericidal mechanisms of trophoblasts compared with macrophages. This difference in protein processing and EV protein content could have important implications for the function of tEVs during pregnancy. The lack of any L. monocytogenes proteins also indicates that bacteria or significant amounts of bacterial components were not likely to be present in the tEV preparations, either inside or as contaminants, at least at the level of detection of mass spectrometry.
In addition to altered proteins found in the tEVs, we also saw a change in mRNAs during infection. One notable type of mRNAs observed in the S-tEVs from infected cells corresponds to histone protein products. As noted above, we also found histone proteins themselves in the tEVs from L. monocytogenes-infected TSCs. Interestingly, histones have been identified in EVs in response to treatment with the Gram-negative bacterial component lipopolysaccharide (LPS) (45, 52). Additionally, histones have been identified in the bloodstreams of animal models of sepsis and patients with sepsis (53–55). These results together with our findings suggest that packaging of histones, and potentially host cell DNA along with them, may be a general mechanism to communicate infections via DAMP recognition. Further work is needed to determine if histones are responsible for the tEV activation of macrophages.
Some of the other genes that we saw overrepresented in the mRNAs from the infected S-tEVs involved vasculature development and morphogenesis. Vasculogenesis of fetal and maternal vessels are required steps for placenta implantation and development (56). Previous work with tEVs found that they recruit vascular smooth muscle cells and promote the invasion of extravillious trophoblasts, which are key steps that are essential for remodeling the decidua surrounding the placenta (57, 58). EVs have also been found to play a direct role in implantation of the embryo (59), and inflammation is a key part of implantation (60). Additionally, morphogenesis of the placenta is required for the proper development of the organ (61, 62). One intriguing gene that had increased mRNA in the tEVs from infected TSCs is syncytin-A, which is responsible for the cellular fusion necessary for the development of the multinucleated synciotrophoblasts (63, 64). It is possible that L. monocytogenes invasion of the placenta activates the release of tEVs required to carry out these processes, with the mRNAs housed in the vesicles acting on the recipient cells. Otherwise, the RNA profiles of the tEVs could represent the mRNAs being transcribed in the TSCs during infection, although the mRNAs we identified in the tEVs differ compared with those seen in human trophoblasts infected with L. monocytogenes (65). Additionally, EVs have been found previously to contain mRNA profiles that differ significantly from the mRNAs in the cell of origin, and EV mRNAs can be translated in recipient cells (66, 67). Much more work will be required to determine the exact function of tEV RNAs during prenatal infection.
Overall, we show that infection of TSCs with L. monocytogenes alters tEV production and function in unexpected ways. tEVs from L. monocytogenes-infected TSCs elicit TNF-α production in RAW 264.7 macrophage-like cells. We also found that tEVs can make certain cell types less resistant to subsequent infection, which was unexpected. A multiomics approach showed that L. monocytogenes treatment greatly altered the components loaded into the tEVs, resulting in increased RNA and nucleic acid binding proteins and unique mRNAs in the tEVs from infected cells. The observation that there were no L. monocytogenes proteins present in tEVs from infected TSCs suggests a host factor or factors altered in the tEVs may be mediating the stimulation of target macrophages. The mechanism of this interaction is of great interest and is the subject of current investigation.
MATERIALS AND METHODS
Bacterial cultures.
Listeria monocytogenes 10403S bioluminescent strain 2C (Xen32) was used throughout the study (68). This strain has a lux-kan insertion in the flaA locus and has a 4-fold increase in intravenous 50% lethal dose compared with wild-type 10403S. Constitutively green fluorescent protein (GFP)-expressing L. monocytogenes (10403S wild-type strain transformed with pMB2044) was provided kindly by Daniel A. Portnoy (University of California, Berkeley, CA). All strains were grown in brain heart infusion medium (BHI) to mid-logarithmic phase for infection.
Cell culture.
Trophoblast stem cells (TSCs) were isolated originally from C57BL/6 mice and were graciously provided by Julie Baker (Standford University, Palo Alto, CA) (38). They were grown in RPMI 1640 medium with GlutaMAX, 20% fetal bovine serum (FBS), and 1 μM sodium pyruvate, as well as 35 μg/mL fibroblast growth factor 4 (FGF-4), 10 ng/mL activin, and 1 μg/mL heparin to maintain TSC replication (39). RAW 264.7 and J774 cells were obtained from ATCC and were grown in RPMI medium with GlutaMAX, 10% fetal bovine serum (FBS), and 1 μM sodium pyruvate.
Isolation of extracellular vesicles.
A total of 107 TSCs in a 150-cm2 flask were infected with L. monocytogenes at a multiplicity of infection (MOI) of 100 or treated with an equivalent volume of BHI. After 1 h, the cells were washed three times with PBS and fresh medium that was depleted of EVs by centrifugation, and 5 μg/mL gentamicin was added to ensure that there were no extracellular bacteria (24). At 24 h of infection, the conditioned medium from the infected and uninfected TSCs was collected and centrifuged at 4,000 × g for 20 min in 50-mL conical tubes. The supernatants were transferred to fresh conical tubes and centrifuged again at 4,000 × g for 30 min. The supernatants were then filtered with a 0.22-μm filter using the Steriflip system. To collect large vesicles (L-tEVs), the filter was washed once with phosphate-buffered saline (PBS), and then 1 mL of PBS was added repeatedly to the top of the filter, which resuspended the tEVs from the filter. This preparation was then stored at −80°C. To collect small tEVs (S-tEVs), the flow through from the filter was ultracentrifuged at 100,000 × g for 2 h. The supernatant was removed carefully so that there was about 0.5 mL left at the bottom of the tube, then 25 mL of PBS was added, and the preparation was ultracentrifuged again at 100,000 × g for 2 h. Once again, the supernatant was removed carefully, and the pellet was resuspended in an additional 1 mL PBS. The preparation was stored at −80°C.
Transmission electron microscopy.
A total of 108 tEVs were fixed in 2% paraformaldehyde for 5 min. A total of 5 µL of the sample solution was placed on carbon-coated EM grids, and tEVs were immobilized for 1 min. The grids were washed by transferring to five 100-µL drops of distilled water and letting them dry for 2 min on each drop. The samples were stained with 1% uranyl acetate. Excess uranyl acetate was removed gently with filter paper, and the grids were air dried. The grids were imaged with a JEOL 100CXII transmission electron microscope operating at 100 kV. Images were captured on a Gatan Orius Digital Camera.
Nanoparticle tracking analysis.
tEV preparations were diluted 1:100 in PBS and were injected into a Zetaview machine (Particle Metrix). The Zetaview was set to a sensitivity of 89, a shutter speed of 300, and a frame rate of 30 frames per second. Cutoffs of a 10-nm minimum and 1,200-nm maximum were used.
TNF-α quantification.
J774 and RAW 264.7 macrophages were plated into a 24-well plate at 5 × 104 cells/well. After 24 h, the cells were treated with 5 × 106 of tEVs from either uninfected or infected TSCs or an equal volume of PBS. After 24 h, the conditioned medium was collected and TNF-α was quantified by enzyme-linked immunosorbent (ELISA) assay from R&D Systems according to the instructions of the manufacturer.
Listeria intracellular growth assay.
Cells were plated into a 24-well plate at 5 × 10(4) cells/well. After approximately enough time to allow the cells to replicate and reach confluence, they were washed three times with PBS. Medium with L. monocytogenes was added at the given multiplicity of infection (MOI) for CFU of L. monocytogenes per cell. After 1 h, the wells were washed three times with PBS and medium with 5 μg/mL gentamicin added. Bioluminescence images were taken at the given time points using an in vivo imaging system (IVIS) Lumina System (Perkin Elmer, Inc.), with 5 min of exposure and large binning, starting upon infection. The signal was quantified using Living Image software (Perkin Elmer). Images were again taken at the given time points. To determine the CFU of the infected cells, TSCs were infected with L. monocytogenes in the same manner as above. At the indicated time points, the cells were lysed in Millipore water with 10 mM Tris (Invitrogen) and 0.1% Triton X-100 (Sigma). The cells were rocked in the lysis buffer for 5 min. The lysates were then diluted in PBS and plated onto BHI plates with 50 µg/mL kanamycin.
Fluorescence microscopy.
Flame-sterilized glass coverslips were placed into a 6-well dish. A total of 105 cells were seeded into each well. The cells were infected 24 h later with mid-log phase green fluorescence protein-expressing L. monocytogenes at an MOI of 100 as reported previously (69). After 1 h, the media were replaced with medium containing 5 μg/mL gentamicin. At the listed periods postinfection, the cells were fixed with 4% paraformaldehyde and solubilized with 0.1% Triton X-100. The coverslips were treated with rhodamine phalloidin (Invitrogen) for 30 min. The coverslips were then mounted to slides with DAPI Fluoromount-G (SouthernBiotech). The slides were imaged with an Olympus Filter FV1000 confocal microscope, and images were taken at ×60 magnification.
Proteomics.
The protein profile of the purified tEVs was determined using untargeted mass spectrometry performed at the Michigan State University (MSU) Genomics Core Facility. Briefly, three independent EV preparations each of 109 S-tEVs from uninfected and infected TSCs were lysed, and the proteins were precipitated using acetone and digested with trypsin. Nanospray liquid chromatography with tandem mass spectrometry (LC-MS/MS) was used to determine the peptide profiles. The peptide data were analyzed using the Scaffold proteome software, which mapped the identified peptides back to the mouse and L. monocytogenes references to determine the originating proteins.
Gene ontology (GO) enrichment analysis for proteomics.
GO analysis was performed using the Gene Ontology Resource (http://geneontology.org/) and protein Analysis Through Evolutionary Relationships (PANTHER) program to identify the biological processes of the proteins seen in the S-tEVs from infected TSCs (70). Additionally, protein interaction networks were generated using the STRING program (71). Proteins that had twice the number of peptides identified in the + Listeria EV samples versus the – Listeria EV samples were used for the analysis.
RNA sequencing.
RNA extraction, RNA library preparations, sequencing reactions, and initial bioinformatics analysis were conducted at Genewiz, LLC (South Plainfield, NJ). Three independent EV preparations each of 109 S-tEVs from uninfected and infected TSCs were used. Total RNA was extracted following the TRIzol reagent user guide (Thermo Fisher Scientific). RNA was quantified using a Qubit fluorometer (Life Technologies, Carlsbad, CA), and RNA integrity was checked with the TapeStation system (Agilent Technologies, Palo Alto, CA). The SMART-Seq v4 ultra low input kit for sequencing was used for full-length cDNA synthesis and amplification (Clontech, Mountain View, CA), and the Illumina Nextera XT library was used for sequencing library preparation. Briefly, cDNA was fragmented, an adaptor was added using transposase, and limited-cycle PCR was conducted to enrich and add indexes to the cDNA fragments. The final library was assessed with the Agilent TapeStation system. The sequencing libraries were multiplexed and clustered on one lane of a flow cell. After the clustering step, the flow cell was loaded on the Illumina HiSeq instrument according to the manufacturer’s instructions. The samples were sequenced using a 2-bp × 150-bp paired-end (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from the Illumina HiSeq system were converted into fastq files and demultiplexed using the Illumina bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
The raw PE read sequencing data were uploaded to the Galaxy Web platform, and we used the public server at http://www.usegalaxy.org to process the transcriptome sequencing (RNA-seq) data (72). Briefly, PE reads were processed to trim sequencing adapters and low-quality bases using Trimmomatic (73). The clean PE RNA-seq reads were mapped to the mouse reference genome (Mus musculus 10) using HISAT2 (74). The gene expression of mapped reads was then measured with featureCounts (75).
Differential gene expression analysis was performed using DESEq2 v1.32.0 in R v4.1.1 (76). Genes with a minimum of 5 reads in at least 4 samples were filtered out, resulting in a total of 23,836 genes. Differentially expressed genes with p-adj of <0.05 were used to perform gene ontology analysis using the g:Profiler system (https://biit.cs.ut.ee/gprofiler/gost) (77). Biological processes with a p-adj of <0.05 were considered significant. The volcano plot was generated using the EnhancedVolcano package in R using fold change of >1 and P value of <10−5 parameters (78) (Fig. 5).
Mouse infections.
All animal experiments were performed under IACUC-approved animal protocol 201800030 in accordance with biosafety level 2 (BSL-2) guidelines established by Michigan State University Campus Animal Resources. Michigan State is an AAALAC International-accredited institution. The 5- to 8-week-old BALB/c mice were obtained from Charles River Laboratories. They were housed in the Clinical Center Animal Wing at Michigan State University for 2 weeks to acclimate them. The mice were treated with either 200 μL of PBS, 108 tEVs from uninfected TSCs, or 108 tEVs from L. monocytogenes-infected TSC through tail vein injection. After 24 h, the mice were infected with 104 L. monocytogenes through tail vein infection. At 96 h postinfection, the mice were imaged using the IVIS imaging system as described previously (68, 79) and sacrificed humanely using cervical dislocation in accordance with approved procedures while the animals were anesthetized. The spleens were harvested, mashed, serial diluted, and plated onto BHI plates with 50 µg/mL kanamycin. Each spleen was diluted and plated in duplicate.
Data availability.
Gene expression data, including raw sequencing files and read counts, can be accessed on the public NCBI Gene Expression Omnibus platform (GEO accession no. GSE212928).
ACKNOWLEDGMENTS
We thank the Institute for Quantitative Health Science and Engineering (IQ) at MSU for providing the facility and resources for executing this work. We gratefully acknowledge Melinda Frame and Alicia Withrow at the Center for Advanced Microscopy at MSU. Additionally, we also gratefully acknowledge Douglas Whitten at the Research Technology Support Facility Proteomics Core at MSU. Finally, we are most grateful for the team at Azenta/Genewiz, including Michael Bullard, Sabbir Siddiqui, and Aleksandar Janjic, for their expert consultation and sequencing.
Footnotes
Supplemental material is available online only.
Contributor Information
Jonathan W. Hardy, Email: hardyjon@msu.edu.
Nancy E. Freitag, University of Illinois at Chicago
REFERENCES
- 1.Arora N, Sadovsky Y, Dermody TS, Coyne CB. 2017. Microbial vertical transmission during human pregnancy. Cell Host Microbe 21:561–567. 10.1016/j.chom.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, González-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pohl AM, Pouillot R, Bazaco MC, Wolpert BJ, Healy JM, Bruce BB, Laughlin ME, Hunter JC, Dunn JR, Hurd S, Rowlands JV, Saupe A, Vugia DJ, Van Doren JM. 2019. Differences among incidence rates of invasive listeriosis in the U.S. FoodNet population by age, sex, race/ethnicity, and pregnancy status, 2008–2016. Foodborne Pathog Dis 16:290–297. 10.1089/fpd.2018.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vázquez-Boland JA, Krypotou E, Scortti M. 2017. Listeria placental infection. mBio 8:e00949-17. 10.1128/mBio.00949-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakardjiev AI, Theriot JA, Portnoy DA. 2006. Listeria monocytogenes traffics from maternal organs to the placenta and back. PLoS Pathog 2:e66. 10.1371/journal.ppat.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722–1725. 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 8.Camilli A, Paynton CR, Portnoy DA. 1989. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc Natl Acad Sci U S A 86:5522–5526. 10.1073/pnas.86.14.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilney LG, Portnoy DA. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109:1597–1608. 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portnoy DA, Auerbuch V, Glomski IJ. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol 158:409–414. 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pamer EG. 2004. Immune responses to Listeria monocytogenes. Nat Rev Immunol 4:812–823. 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 12.Ñahui Palomino RA, Vanpouille C, Costantini PE, Margolis L. 2021. Microbiota-host communications: bacterial extracellular vesicles as a common language. PLoS Pathog 17:e1009508. 10.1371/journal.ppat.1009508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raposo G, Stoorvogel W. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell MD, Peiris HN, Kobayashi M, Koh YQ, Duncombe G, Illanes SE, Rice GE, Salomon C. 2015. Placental exosomes in normal and complicated pregnancy. Am J Obstet Gynecol 213:S173–S181. 10.1016/j.ajog.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Sabapatha A, Gercel-Taylor C, Taylor DD. 2006. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am J Reprod Immunol 56:345–355. 10.1111/j.1600-0897.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 16.Han C, Wang C, Chen Y, Wang J, Xu X, Hilton T, Cai W, Zhao Z, Wu Y, Li K, Houck K, Liu L, Sood AK, Wu X, Xue F, Li M, Dong J-F, Zhang J. 2020. Placenta-derived extracellular vesicles induce preeclampsia in mouse models. Haematologica 105:1686–1694. 10.3324/haematol.2019.226209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill M, Motta-Mejia C, Kandzija N, Cooke W, Zhang W, Cerdeira AS, Bastie C, Redman C, Vatish M. 2019. Placental syncytiotrophoblast-derived extracellular vesicles carry active NEP (neprilysin) and are increased in preeclampsia. Hypertension 73:1112–1119. 10.1161/HYPERTENSIONAHA.119.12707. [DOI] [PubMed] [Google Scholar]
- 18.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. 2007. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 110:3234–3244. 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giri PK, Kruh NA, Dobos KM, Schorey JS. 2010. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics 10:3190–3202. 10.1002/pmic.200900840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Schorey JS. 2019. Extracellular vesicles deliver Mycobacterium RNA to promote host immunity and bacterial killing. EMBO Rep 20:e46613. 10.15252/embr.201846613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui WW, Hercik K, Belsare S, Alugubelly N, Clapp B, Rinaldi C, Edelmann MJ. 2018. Salmonella enterica serovar Typhimurium alters the extracellular proteome of macrophages and leads to the production of proinflammatory exosomes. Infect Immun 86:e00386-17. 10.1128/IAI.00386-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hui WW, Emerson LE, Clapp B, Sheppe AE, Sharma J, del Castillo J, Ou M, Maegawa GHB, Hoffman C, Larkin J, III, Pascual DW, Edelmann MJ. 2021. Antigen encapsulating host extracellular vesicles derived from Salmonella-infected cells stimulate pathogen-specific Th1-type responses in vivo. PLoS Pathog 17:e1009465. 10.1371/journal.ppat.1009465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nandakumar R, Tschismarov R, Meissner F, Prabakaran T, Krissanaprasit A, Farahani E, Zhang B-C, Assil S, Martin A, Bertrams W, Holm CK, Ablasser A, Klause T, Thomsen MK, Schmeck B, Howard KA, Henry T, Gothelf KV, Decker T, Paludan SR. 2019. Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signalling. Nat Microbiol 4:701–713. 10.1038/s41564-019-0367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J, Tanaka S, Oda M, Makino T, Ohgane J, Shiota K. 2001. Retinoic acid promotes differentiation of trophoblast stem cells to a giant cell fate. Dev Biol 235:422–432. 10.1006/dbio.2001.0300. [DOI] [PubMed] [Google Scholar]
- 26.Zhu D, Gong X, Miao L, Fang J, Zhang J. 2017. Efficient induction of syncytiotrophoblast layer II cells from trophoblast stem cells by canonical Wnt signaling activation. Stem Cell Rep 9:2034–2049. 10.1016/j.stemcr.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakardjiev AI, Stacy BA, Fisher SJ, Portnoy DA. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect Immun 72:489–497. 10.1128/IAI.72.1.489-497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach J-M, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, et al. 2018. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7:1535750. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammadzadeh R, Ghazvini K, Farsiani H, Soleimanpour S. 2020. Mycobacterium tuberculosis extracellular vesicles: exploitation for vaccine technology and diagnostic methods. Crit Rev Microbiol 47:13–33. 10.1080/1040841X.2020.1830749. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Hammer LA, Liu W, Hobbs MM, Zielke RA, Sikora AE, Jerse AE, Egilmez NK, Russell MW. 2017. Experimental vaccine induces Th1-driven immune responses and resistance to Neisseria gonorrhoeae infection in a murine model. Mucosal Immunol 10:1594–1608. 10.1038/mi.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Y, Schorey JS. 2013. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur J Immunol 43:3279–3290. 10.1002/eji.201343727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portnoy DA, Schreiber RD, Connelly P, Tilney LG. 1989. Gamma interferon limits access of Listeria monocytogenes to the macrophage cytoplasm. J Exp Med 170:2141–2146. 10.1084/jem.170.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kugeratski FG, Hodge K, Lilla S, McAndrews KM, Zhou X, Hwang RF, Zanivan S, Kalluri R. 2021. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat Cell Biol 23:631–641. 10.1038/s41556-021-00693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. 2018. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol 15:617–638. 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 35.White JR, Dauros-Singorenko P, Hong J, Vanholsbeeck F, Phillips A, Swift S. 2021. The complex, bidirectional role of extracellular vesicles in infection. Biochem Soc Trans 49:881–891. 10.1042/BST20200788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothbauer M, Patel N, Gondola H, Siwetz M, Huppertz B, Ertl P. 2017. A comparative study of five physiological key parameters between four different human trophoblast-derived cell lines. Sci Rep 7:5892. 10.1038/s41598-017-06364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrish DW, Whitley G, Cartwright JE, Graham CH, Caniggia I. 2002. In vitro models to study trophoblast function and dysfunction—a workshop report. Placenta 23:S114–S118. 10.1053/plac.2002.0798. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075. 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 39.Erlebacher A, Price KA, Glimcher LH. 2004. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol 275:158–169. 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 40.Coughlan C, Bruce KD, Burgy O, Boyd TD, Michel CR, Garcia-Perez JE, Adame V, Anton P, Bettcher BM, Chial HJ, Königshoff M, Hsieh EWY, Graner M, Potter H. 2020. Exosome isolation by ultracentrifugation and precipitation and techniques for downstream analyses. Curr Protoc Cell Biol 88:e110. 10.1002/cpcb.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsman G, Zeerleder S, Luken BM. 2016. Extracellular histones, cell-free DNA, or nucleosomes: differences in immunostimulation. Cell Death Dis 7:e2518. 10.1038/cddis.2016.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaabani N, Vartabedian VF, Nguyen N, Honke N, Huang Z, Teijaro JR. 2021. IFN-β, but not IFN-α, is responsible for the pro-bacterial effect of type I interferon. Cell Physiol Biochem 55:256–264. 10.33594/000000370. [DOI] [PubMed] [Google Scholar]
- 43.O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med 200:437–445. 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med 200:527–533. 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Boza-Serrano A, Dunning CJR, Clausen BH, Lambertsen KL, Deierborg T. 2018. Inflammation leads to distinct populations of extracellular vesicles from microglia. J Neuroinflammation 15:168. 10.1186/s12974-018-1204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sork H, Corso G, Krjutskov K, Johansson HJ, Nordin JZ, Wiklander OPB, Lee YXF, Westholm JO, Lehtiö J, Wood MJA, Mäger I, El Andaloussi S. 2018. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci Rep 8:10813. 10.1038/s41598-018-28485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzbekova S, Almiñana C, Labas V, Teixeira-Gomes A-P, Combes-Soia L, Tsikis G, Carvalho AV, Uzbekov R, Singina G. 2020. Protein cargo of extracellular vesicles from bovine follicular fluid and analysis of their origin from different ovarian cells. Front Vet Sci 7:584948. 10.3389/fvets.2020.584948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abed M, Verschueren E, Budayeva H, Liu P, Kirkpatrick DS, Reja R, Kummerfeld SK, Webster JD, Gierke S, Reichelt M, Anderson KR, Newman RJ, Roose-Girma M, Modrusan Z, Pektas H, Maltepe E, Newton K, Dixit VM. 2019. The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLoS One 14:e0214110. 10.1371/journal.pone.0214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segel M, Lash B, Song J, Ladha A, Liu CC, Jin X, Mekhedov SL, Macrae RK, Koonin EV, Zhang F. 2021. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 373:882–889. 10.1126/science.abg6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrami L, Brandi L, Moayeri M, Brown MJ, Krantz BA, Leppla SH, van der Goot FG. 2013. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Rep 5:986–996. 10.1016/j.celrep.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García-Martínez M, Vázquez-Flores L, Álvarez-Jiménez VD, Castañeda-Casimiro J, Ibáñez-Hernández M, Sánchez-Torres LE, Barrios-Payán J, Mata-Espinosa D, Estrada-Parra S, Chacón-Salinas R, Serafín-López J, Wong-Baeza I, Hernández-Pando R, Estrada-García I. 2019. Extracellular vesicles released by J774A.1 macrophages reduce the bacterial load in macrophages and in an experimental mouse model of tuberculosis. Int J Nanomedicine 14:6707–6719. 10.2147/IJN.S203507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nair RR, Mazza D, Brambilla F, Gorzanelli A, Agresti A, Bianchi ME. 2018. LPS-challenged macrophages release microvesicles coated with histones. Front Immunol 9:1463. 10.3389/fimmu.2018.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalbitz M, Grailer JJ, Fattahi F, Jajou L, Herron TJ, Campbell KF, Zetoune FS, Bosmann M, Sarma JV, Huber-Lang M, Gebhard F, Loaiza R, Valdivia HH, Jalife J, Russell MW, Ward PA. 2015. Role of extracellular histones in the cardiomyopathy of sepsis. FASEB J 29:2185–2193. 10.1096/fj.14-268730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ekaney ML, Otto GP, Sossdorf M, Sponholz C, Boehringer M, Loesche W, Rittirsch D, Wilharm A, Kurzai O, Bauer M, Claus RA. 2014. Impact of plasma histones in human sepsis and their contribution to cellular injury and inflammation. Crit Care 18:543. 10.1186/s13054-014-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, Taylor FB, Esmon NL, Lupu F, Esmon CT. 2009. Extracellular histones are major mediators of death in sepsis. Nat Med 15:1318–1321. 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geva E, Ginzinger DG, Zaloudek CJ, Moore DH, Byrne A, Jaffe RB. 2002. Human placental vascular development: vasculogenic and angiogenic (branching and nonbranching) transformation is regulated by vascular endothelial growth factor-A, angiopoietin-1, and angiopoietin-2. J Clin Endocrinol Metab 87:4213–4224. 10.1210/jc.2002-020195. [DOI] [PubMed] [Google Scholar]
- 57.Salomon C, Yee S, Scholz-Romero K, Kobayashi M, Vaswani K, Kvaskoff D, Illanes SE, Mitchell MD, Rice GE. 2014. Extravillous trophoblast cells-derived exosomes promote vascular smooth muscle cell migration. Front Pharmacol 5:175. 10.3389/fphar.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salomon C, Kobayashi M, Ashman K, Sobrevia L, Mitchell MD, Rice GE. 2013. Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PLoS One 8:e79636. 10.1371/journal.pone.0079636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen K, Liang J, Qin T, Zhang Y, Chen X, Wang Z. 2022. The role of extracellular vesicles in embryo implantation. Front Endocrinol (Lausanne) 13:809596. 10.3389/fendo.2022.809596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mor G, Cardenas I, Abrahams V, Guller S. 2011. Inflammation and pregnancy: the role of the immune system at the implantation site. Ann N Y Acad Sci 1221:80–87. 10.1111/j.1749-6632.2010.05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cross JC, Nakano H, Natale DRC, Simmons DG, Watson ED. 2006. Branching morphogenesis during development of placental villi. Differentiation 74:393–401. 10.1111/j.1432-0436.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 62.Mihu CM, Suşman S, Rus Ciucă D, Mihu D, Costin N. 2009. Aspects of placental morphogenesis and angiogenesis. Rom J Morphol Embryol 50:549–557. [PubMed] [Google Scholar]
- 63.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 64.Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V, Heidmann T. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A 102:725–730. 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson LJ, Azari S, Webb A, Zhang X, Gavrilin MA, Marshall JM, Rood K, Seveau S. 2021. Human placental trophoblasts infected by Listeria monocytogenes undergo a pro-inflammatory switch associated with poor pregnancy outcomes. Front Immunol 12:709466. 10.3389/fimmu.2021.709466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10:1470–1476. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 68.Hardy J, Francis KP, DeBoer M, Chu P, Gibbs K, Contag CH. 2004. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303:851–853. 10.1126/science.1092712. [DOI] [PubMed] [Google Scholar]
- 69.Rocha CE, Mol JPS, Garcia LNN, Costa LF, Santos RL, Paixão TA. 2017. Comparative experimental infection of Listeria monocytogenes and Listeria ivanovii in bovine trophoblasts. PLoS One 12:e0176911. 10.1371/journal.pone.0176911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426. 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 76.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a Web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:W191–W198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blighe K, Rana S, Lewis M. 2018. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. [Google Scholar]
- 79.Hardy J, Kirkendoll B, Zhao H, Pisani L, Luong R, Switzer A, McConnell MV, Contag CH. 2012. Infection of pregnant mice with Listeria monocytogenes induces fetal bradycardia. Pediatr Res 71:539–545. 10.1038/pr.2012.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3 and Tables S1 to S3. Download iai.00347-22-s0001.pdf, PDF file, 0.5 MB (484.8KB, pdf)
Data Availability Statement
Gene expression data, including raw sequencing files and read counts, can be accessed on the public NCBI Gene Expression Omnibus platform (GEO accession no. GSE212928).