ABSTRACT
A 1.488-Gb draft genome sequence was assembled for the fungus Massospora cicadina, an obligate parasite of periodical cicadas. The M. cicadina genome has experienced massive expansion via transposable elements (TEs), which account for 92% of the genome.
ANNOUNCEMENT
Massospora and other Entomophthorales (Zoopagomycota) are grossly understudied due to their ephemeral and fastidious lifestyles, as well as the complicated disease and host life cycles (1–4). The recent discovery of cathinone and psilocybin in Massospora-infected cicadas has raised questions about their biosynthesis, which have proven difficult to answer due to unwieldy metagenomes derived from field-collected cicadas (5). The generation of high-quality genomic resources is fundamental to answering these and other questions regarding Massospora’s unique biology and evolution.
Conidia and azygospores of Massospora cicadina strain MCPNR19 (ARSEF14555) were collected from M. cicadina-infected Magicicada septendecim in Pennsylvania in June 2019 (Fig. 1). The spores were liberated from harvested posterior fungal plugs (conidia) or by scraping abdominal walls (azygospores) of frozen infected cicada cadavers stored at −80°C. Azygospores were further isolated using 40- and 25-μm soil sieves to remove host tissue and provide sufficient fungal biomass. Genomic DNA (gDNA) was extracted from the spore pools using a fungal cell lysis and cetyltrimethylammonium bromide (CTAB) gDNA purification protocol (6). Oxford Nanopore (ONT) DNA libraries generated using the SQK-LSK109 ligation kit were sequenced on a MinION instrument with five R9.4.1 flow cells (2 for conidia, 3 for azygospores) and base called using Guppy v6.0.1-GPU (Table 1) to produce 29.7 Gb (coverage, 20×). Illumina sequencing of 1 azygospore library on a NovaSeq instrument (2× 150 bp) using a Covaris-sheared DNA library produced 26.2 Gb (coverage, ~18×). Table 1 details the library preparation, sequencing, and assembly details obtained using NanoStat v1.4.0, wtdbg2 v2.5, BBMap v38.86, and AAFTF v0.2.6 (7–11). Bacterial contamination was removed by inspection of the Blobtools2 results (12, 13), iterative taxonomic searches using MMseqs2 v13-45111 (13) with UniRef50 (14), and analysis of the fungal transposable element (TE) content (15, 16). Metagenome-assembled bacterial genomes were analyzed separately (17). A 1.488 Gbp assembly in 19,694 scaffolds was constructed from a combined read coverage of 38× (L50, 139 kb; N50, 3,261; mean GC content, 41.13%). A BUSCO v5.2.2 (18) completeness assessment identified 182 complete markers (71%) out of 255 markers in the Eukaryota Odb10 data set and 491 (65%) of 758 markers in the Fungi Odb10 data set (Table 1).
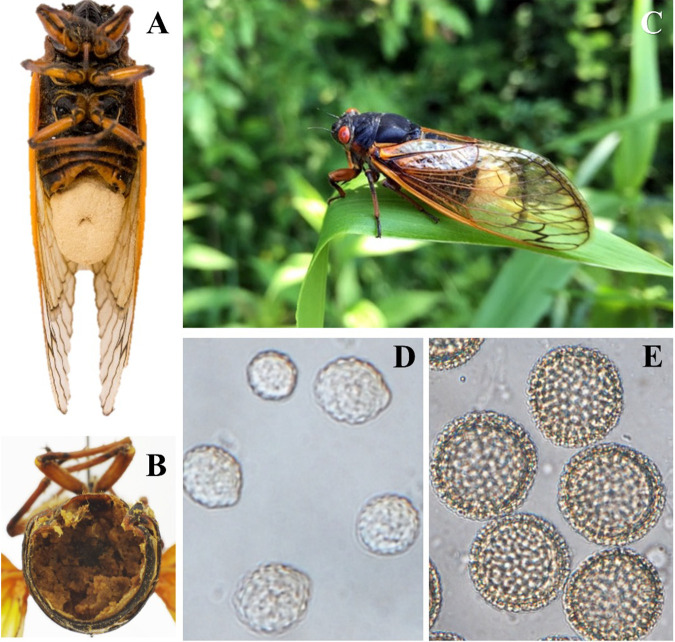
FIG 1.
Photographs of Massospora cicadina-infected Pharaoh cicadas (Magicicada septendecim) and associated spore stages. (A) Adult female with conspicuous conidia “plug” protruding from the posterior end of the abdomen. (B) Adult male with inconspicuous azygospore (resting spore) infection. (C) Active male with conidial plug. (D) Close-up of M. cicadina verrucose (“warty”) conidia. (E) Close-up of M. cicadina thick-walled reticulated (“net-like”) azygospores. The photos in panels A, B, D, and E are from brood V, Morgantown, WV (2016). The photo in panel C is of a live infected cicada included in the sampling for strain MCPNR19 (ARSEF14555). Photos in panels A and B are by Cameron Stauder. Photos in panels C to E are by Matt Kasson.
TABLE 1.
Genome strain information, statistics, and methods for Massospora cicadina
| Characteristic | Data for strain: |
|
|---|---|---|
| MICH 231384 | MCPNR19 (ARSEF 14555) | |
| Spore type | Conidia | Conidia, azygospores |
| Yr/location/cicada brood | 2016/OH, USA/brood V | 2019/PA, USA/brood VIII |
| Sampling location | I-77S rest area (Summit County) | Carnegie Museum of Natural History Powdermill Nature Reserve (Westmoreland County) |
| Sampling coordinates (lat, long) | 41.194492, −81.624714 | 40.164837, −79.265278 |
| Sequencing technology | Illumina HiSeq | Illumina NovaSeq 6000 + Oxford Nanopore MinION R9 LSK109 |
| Assembly method | SPAdes v. 3.10.0 | wtdbg2 v. 2.5, target assembly size of 1.1 Gb |
| Assembly polishing | wtpoa-cns using Illumina reads and Racon | |
| Assembly contig extension | BBMap extend.sh using Illumina reads | |
| Assembly adaptor and contamination screening | AAFTF v. 0.2.6 | |
| Genome size (Mbp) | 766.56 | 1,488.88 |
| GC content (%) | 39.3 | 41.13 |
| Scaffold N50 | 3,457 | 3,261 |
| Scaffold L50 (bp) | 67,854 | 139,493 |
| No. of scaffolds | 272,193 | 19,694 |
| Longest scaffold (kbp) | 225 | 1,107 |
| No. of contigs | 373,021 | 19,694 |
| Avg coverage (×) | 6.5 | 38.0 |
| Total Illumina sequence data (Gbp) | 16.6 | 26.22 |
| Total Nanopore sequence data (Gbp) | 29.73 | |
| Avg Nanopore coverage (×) | 20 | |
| Nanopore read N50 (bp) | 5,209 | |
| Longest Nanopore reads (kbp) | 265, 179, 159 | |
| Nanopore read quality scorea | 9.9 million (85.1%) of the 11 million reads had a mean quality score of 10 (>Q10) | |
| No. of BUSCOs (Eukaryota/Fungi Odb10 data sets [n {%}]) | ||
| Complete | 107 (42)/301 (40) | 182 (71)/491 (65) |
| Complete and single-copy | 107 (42)/301 (40) | 177 (69)/481 (63) |
| Complete and duplicated | 0 (0)/4 (1) | 5 (2)/10 (1) |
| Fragmented | 56 (22)/110 (15) | 29 (11)/74 (10) |
| Total no. of genes (protein-coding genes/tRNAs) | 9,889 (9,510/379) | 7,532 (5,435/2,079) |
| GenBank accession no. | QMCF00000000 | JAKSWZP000000000 |
| GenBank assembly accession no. | GCA_006912075.1 | GCA_022478985.1 |
| SRA accession no | SRR7045068 (Illumina) | SRR17553520–SRR17553524 (ONT); SRR17553525, SRR17553526 (Illumina) |
| BioSample accession no. | SAMN08956764 | SAMN24722893 |
| BioProject accession no. | PRJNA451007 | PRJNA795459 |
| Supporting data | ||
| Sanger sequencing data (GenBank accession no.) | See reference 3 | Representative sequences deposited as M. cicadina strains 8PA01 and 8PA02: 28S (MN706567, MN706568); 18S (MN706543, MN706544); EFL (MT044284, MT044285). |
| Specimen collection and storage | Collected alive individually in 15-mL Falcon tubes, transported on ice to lab, and stored at −80°C until sequencing. Shipped on dry ice from WVU to UCR. | |
| Spore cleanup methods | Both: spores were aseptically removed from cicada host and placed in sterile secondary tubes for transport. Azygospores were passed through sterile 40- and 25-μm soil sieves to reduce host tissue and specifically enrich for spores. | |
| Library prep and parameters | Illumina: NEB Ultra II FS (New England Biolabs, Ipswich, MA) (fragmentation enzyme) libraries prepared from DNA sheared with a Covaris S220 (Woburn, MA). Oxford Nanopore: constructed for R9.4.1 flow cells and the LSK109 kit. The NEBNext end-repair/dA-tailing module (E7546), NEBNext FFPE DNA repair mix (M6630), and NEBNext quick ligation module (E6056) kits (NEB) were used with the ONT SQK-LSK109 ligation sequencing kit. Sequence reads were base called using Guppy v6.0.1-GPU with the parameters “--min_qscore 7 --detect_adapter --detect_primer --trim_strategy dna.” |
|
| Contamination assessment and removal | Blobtools2 combined with manual assessment of predicted bacterial contigs using analysis of the repetitive content, which was diagnostic for fungal versus bacteria, depth of coverage, and bacterium taxonomy. | |
| Bioinformatics methods for assembly, bacterium removal, repeat identification, and annotation | https://github.com/zygolife/Massospora_cicadina_ONT_asm (7), https://github.com/zygolife/Massospora_cicadina_MCPNR19 (8) | |
Computed using NanoStat.
The genome was masked using RepeatMasker v4-1-1 (19) with Repbase (20) fungi repeats and a species-specific library generated using RepeatModeler v2.0.1 (21, 22). The repeats were screened manually to remove protein-coding genes using a DIAMOND v2.0.13 (23, 24) search of the Swiss-Prot v2021_04 database (DB) (25). The best (373 total) BUSCO-derived models were used to train the ab initio predictors SNAP v2013_11_29 (26) and AUGUSTUS v3.3.3 (27), with additional predictions from the self-trained programs GeneMark-ES v4.68 (28) and GlimmerHMM v3.0.4 (29). Exon evidence was generated to improve gene predictions using DIAMOND BLASTX and Exonerate v2.4.0 to align Swiss-Prot DB proteins (30). EVidenceModeler v1.1.1 (31) was used via Funannotate to generate consensus gene models with default evidence weights. tRNA genes were predicted using tRNAscan-SE v2.0.9 (32). Putative protein functions were assigned based on sequence similarity to the InterProScan v5.51-85.0 (33, 34), Pfam v35.0 (35), eggNOG v2.1.6-d35afda (36), dbCAN2 v9.0 (37), and MEROPS v12.0 (38) databases, relying on NCBI BLAST v2.9.0+ (39) and HMMER v3.3.2 (40). Secretion signals and transmembrane domains were annotated using Phobius (41) and SignalP v5.0b (42). A total of 7,532 gene models (5,453 protein-coding genes and 2,079 tRNAs) were predicted.
The genome of M. cicadina strain MCPNR19 is a significant improvement over the previously sequenced strain MICH 231384 (5). Similarly to the 1.018-Gbp myrtle rust genome (43) and the 1.25-Gbp soybean rust genome (44), 92% (1.369 Gbp) of the MCPNR19 genome consists of TEs, 73% of which are LTR Ty3 retrotransposons. The low predicted protein-coding gene count likely reflects gene undercalling in the absence of transcriptome sequencing (RNA-seq) data and efforts to avoid overpredicting TEs as genes (Table 1). Future work incorporating transcriptomic data is needed to validate these findings.
Data availability.
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAKSZP000000000. The version described in this paper is version JAKSZP010000000. The sequence reads have been deposited under SRA project accession numbers SRR17553520 to SRR17553526 and BioProject accession number PRJNA795459.
ACKNOWLEDGMENTS
J.E.S. is a CIFAR fellow in the program “Fungal Kingdom: Threats and Opportunities” and was supported by the U.S. Department of Agriculture National Institute of Food and Agriculture Hatch project CA-R-PPA-211-5062-H and National Science Foundation (NSF) award DEB-1441715. Illumina library preparation was completed at the University of California—Riverside in the Institute of Integrative Genome Biology (IIGB), and sequencing was conducted at the UC Berkeley Vincent J. Coates facility. Genome assembly was performed on the IIGB High-Performance Computing Cluster supported by NSF grant DBI-1429826 and NIH grant S10-OD016290.
We thank John Wenzel (Carnegie Museum of Natural History [CMNH]) for permission to sample the cicadas, Amy Metheny and Angie Macias for assistance with collection, and Matthew Collin and Holly Clark (UCR, IIGB) for library preparation. We also thank Kathryn Bushley, ARSEF curator, for assistance depositing the M. cicadina-infected cicadas at ARSEF.
Funding Statement
J.E.S. is a CIFAR fellow in the program "Fungal Kingdom: Threats and Opportunities" and was supported by the U.S. Department of Agriculture, National Institute of Food and Agriculture Hatch project CA-R-PPA-211-5062-H and National Science Foundation (NSF) award DEB-1441715. Genome assembly was performed on the IIGB High-Performance Computing Cluster supported by NSF DBI-1429826 and NIH S10-OD016290 grants.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/MRA.00413-22.
Contributor Information
Jason E. Stajich, Email: jason.stajich@ucr.edu.
Antonis Rokas, Vanderbilt University.
REFERENCES
- 1.Cooley JR, Marshall DC, Hill KBR. 2018. A specialized fungal parasite (Massospora cicadina) hijacks the sexual signals of periodical cicadas (Hemiptera: Cicadidae: Magicicada). Sci Rep 8:1432. doi: 10.1038/s41598-018-19813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovett B, Macias A, Stajich JE, Cooley J, Eilenberg J, de Fine Licht HH, Kasson MT. 2020. Behavioral betrayal: how select fungal parasites enlist living insects to do their bidding. PLoS Pathog 16:e1008598. doi: 10.1371/journal.ppat.1008598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macias AM, Geiser DM, Stajich JE, Łukasik P, Veloso C, Bublitz DC, Berger MC, Boyce GR, Hodge K, Kasson MT. 2020. Evolutionary relationships among Massospora spp. (Entomophthorales), obligate pathogens of cicadas. Mycologia 112:1060–1074. doi: 10.1080/00275514.2020.1742033. [DOI] [PubMed] [Google Scholar]
- 4.Elya C, De Fine Licht HH. 2021. The genus Entomophthora: bringing the insect destroyers into the twenty-first century. IMA Fungus 12:34. doi: 10.1186/s43008-021-00084-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce GR, Gluck-Thaler E, Slot JC, Stajich JE, Davis WJ, James TY, Cooley JR, Panaccione DG, Eilenberg J, De Fine Licht HH, Macias AM, Berger MC, Wickert KL, Stauder CM, Spahr EJ, Maust MD, Metheny AM, Simon C, Kritsky G, Hodge KT, Humber RA, Gullion T, Short DPG, Kijimoto T, Mozgai D, Arguedas N, Kasson MT. 2019. Psychoactive plant- and mushroom-associated alkaloids from two behavior modifying cicada pathogens. Fungal Ecol 41:147–164. doi: 10.1016/j.funeco.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter-House D, Stajich JE, Unruh S, Kurbessoian T. 26 October 2020. Fungal CTAB DNA extraction. protocols.io. doi: 10.17504/protocols.io.bhx8j7rw. [DOI]
- 7.De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods 17:155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stajich JE, Palmer J. 2019. Automatic assembly for the fungi (AAFTF): genome assembly pipeline. Zenodo. doi: 10.5281/zenodo.1620526. [DOI]
- 11.Bushnell B. 2020. BBMap. http://sourceforge.net/projects/bbmap/.
- 12.Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. 2020. BlobToolKit—interactive quality assessment of genome assemblies. G3 (Bethesda) 10:1361–1374. doi: 10.1534/g3.119.400908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirdita M, Steinegger M, Breitwieser F, Söding J, Levy Karin E. 2021. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics 37:3029–3031. doi: 10.1093/bioinformatics/btab184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 15.Stajich JE. 2022. zygolife/Massospora_cicadina_ONT_asm: v1.0—first release associated with MRA. Zenodo. doi: 10.5281/zenodo.6964872. [DOI]
- 16.Stajich JE. 2022. zygolife/Massospora_cicadina_MCPNR19: v1.0—first release associated with MRA. Zenodo. doi: 10.5281/zenodo.6964878. [DOI]
- 17.Ettinger L, Kasson S. 2022. Metagenome-assembled genomes of bacteria associated with Massospora cicadina fungal plugs from infected brood VIII periodical cicadas. Microbiol Resour Announc. doi: 10.1128/mra.00413-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol 1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 19.Smit AFA, Hubley R, Green P. 2013. RepeatMasker. http://www.repeatmasker.org.
- 20.Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA 117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storer JM, Hubley R, Rosen J, Smit AFA. 2021. Curation guidelines for de novo generated transposable element families. Curr Protoc 1:e154. doi: 10.1002/cpz1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 24.Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.UniProt Consortium. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 28.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 30.Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49:D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L, Zhang H, Wu P, Entwistle S, Li X, Yohe T, Yi H, Yang Z, Yin Y. 2018. dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res 46:D516–D521. doi: 10.1093/nar/gkx894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. 2018. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 43.Tobias PA, Schwessinger B, Deng CH, Wu C, Dong C, Sperschneider J, Jones A, Lou Z, Zhang P, Sandhu K, Smith GR, Tibbits J, Chagné D, Park RF. 2021. Austropuccinia psidii, causing myrtle rust, has a gigabase-sized genome shaped by transposable elements. G3 (Bethesda) 11:jkaa015. doi: 10.1093/g3journal/jkaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta YK, Marcelino-Guimarães FC, Lorrain C, Farmer AD, Haridas S, Ferreira EGC, Lopes-Caitar VS, Oliveira LS, Morin E, Widdison S, Cameron C, Innoue Y, Thor K, Robinson K, Drula E, Henrissat B, LaButti K, Bini AMR, Paget E, Singan V, Daum C, Dorme C, van Hoek M, Janssen A, Chandat L, Tarriotte Y, Richardson J, do Vale Araújo Melo B, Wittenberg A, Schneiders H, Peyrard S, Zanardo LG, Holtman VC, Chauvel FC, Link TI, Balmer D, Müller AN, Kind S, Bohnert S, Wirtz L, Chen C, Yan M, Ng V, Gautier P, Meyer MC, Vögele RT, Liu Q, Grigoriev IV, Conrath U, Brommonschenkel SH, et al. 2022. The soybean rust pathogen Phakopsora pachyrhizi displays transposable element proliferation that correlates with broad host-range adaptation on legumes. bioRxiv. doi: 10.1101/2022.06.13.495685. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAKSZP000000000. The version described in this paper is version JAKSZP010000000. The sequence reads have been deposited under SRA project accession numbers SRR17553520 to SRR17553526 and BioProject accession number PRJNA795459.