Abstract
Genome-wide association studies (GWASs) are a research approach used to identify genetic variants associated with common diseases, like COVID-19. The lead genetic variants (n = 41) reported by the eleven largest COVID-19 GWASs are mapped to 22 different chromosomal regions. The loci 3q21.31 (LZTFL1 and chemokine receptor genes) and 9q34.2 (ABO), associated with disease severity and susceptibility to infection, respectively, were the most replicated findings across studies. Genes involved with mucociliary clearance (CEP97, FOXP4), viral-entry (ACE2, SLC6A20) and mucosal immunity (MIR6891) are associated with the risk of SARS-CoV-2 infection while genes of antiviral immune response (IFNAR2, OAS1), leukocyte trafficking (CCR9, CXCR6) and lung injury (DPP9, NOTCH4) are associated with severe disease. The biological processes underlying the risk of infection occur prominently, but not exclusively, in the upper airways whereas the severe COVID-19-associated processes in alveolar-capillary interface. The COVID-19 GWASs has unraveled key genetic mechanisms of SARS-CoV-2 pathogenesis, although the genetic basis of other COVID-19 related phenotypes (long COVID and neurological impairment) remains to be elucidated.
Keywords: GWAS, COVID-19, SARS-CoV-2, Host, Genetic variants
1. Introduction
In 2020, before the introduction of vaccines against SARS-CoV-2, almost 90 million people were infected, resulting in approximately 2 million deaths worldwide, a global death toll similar to that caused by lower respiratory infections in 2019 (2.6 million) (WHO, 2022). The pathogenesis of SARS-CoV-2 seems to be dependent on structural features of the respiratory tract and on qualitative, quantitative and temporal aspects of the host immune response (Lamers and Haagmans, 2022). All of those features are determined, at least partially, by the host genetics.
It has been observed, throughout this ongoing COVID-19 pandemic, that apparently low-risk individuals (young without co-morbidities) may evolve with very severe disease, whereas supposedly high-risk individuals may have only mild SARS-CoV-2 infection outcomes without the need of hospitalization or additional medical care (Gao et al., 2021; Oran and Topol, 2020). One of the main causes underlying these two opposite outcomes relies in part on specific sequence variations in the genome of individuals (van der Made et al., 2020). Indeed, the variability in COVID-19 outcomes is mostly determined by a complex interplay between host genetic variants and non-genetic factors (e.g. age, sex, body mass index, socioeconomic features) (Zhang et al., 2022).
The continuum of clinical and molecular phenotypes in between extreme COVID-19 outcomes (asymptomatic vs death), is virtually influenced by the whole set of genetic variants in a given genome. The fact that every human genome (size: 3.4 × 109 base pairs) has millions of genetic variants, many of them with extremely low minor allele frequency (Karczewski et al., 2020; Lek et al., 2016), poses a challenge to unravel the genetic architecture of complex diseases like COVID-19. In addition, allele frequencies as well as social, cultural and demographic factors, vary across populations what might influence the differential pandemic burden in a global context (Prohaska et al., 2019). Rare genetic variants (allele frequencies <0.001) in type I IFN immunity-related genes (e.g. IFNRA, TLR3, IRF7) have been identified in individuals with life-threatening COVID-19 pneumonia (Zhang et al., 2020). Conversely, Kosmicki et al did not find any variant associated with COVID-19 outcomes by analyzing whole exome sequences from ∼20,000 COVID-19 cases and half a million controls across five continental ancestries (Kosmicki et al., 2021). This study analyzed seven different COVID outcomes, of which five related to infection susceptibility and two related to disease severity. Recent studies have demonstrated rare genetic variants as an important component in the etiology of complex diseases (Wang et al., 2021), although its identification is still challenging. Fallerini et al., has recently developed a machine-learning model to describe the contribution of both common and rare genetic variants on COVID-19 severity (Fallerini et al., 2022).
Genome Wide Association Studies (GWASs) are a Genetic Epidemiology tool suitable to leverage environmental/lifestyle and genetic information to estimate disease risk. In a typical GWAS, thousands of diseased and non-diseased individuals are genotyped for over a million genetic variants across the genome in order to identify genomic regions (i.e. loci) associated with disease (Box 1 ). Association does not necessarily imply causation, since most GWAS signal comes from genotyped variants in linkage disequilibrium (LD) with unobserved causal locus (i.e. indirect association) (Balding, 2006). While the analysis and results interpretation of COVID-19 GWAS might be tricky for specialties outside the Genetics field, its meaning is relevant to the scientific community broadly. Here we reviewed eleven COVID-19 GWASs, aiming to summarize the genomic loci associated with COVID-19 risk and to integrate the genetic variants into disease mechanisms underlying the SARS-CoV-2 infection outcomes.
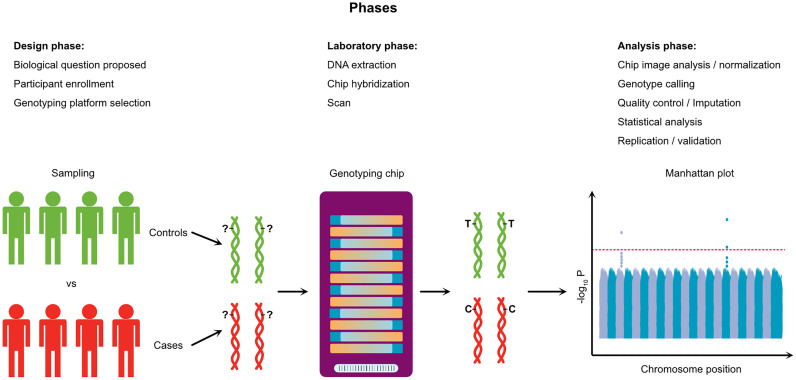
Box 1. Typical workflow of a genome-wide association study.
Genome Wide Association Studies (GWASs) rely on the general hypothesis that the disease/trait variability in a given population is influenced by common genetic variants. In the design phase, biological samples of thousands of research participants with different disease status (Cases vs Controls) are obtained. In the laboratory, the DNA is extracted and hybridized against oligonucleotide probes targeting specific single nucleotide variants (SNPs) across the whole genome. Millions of probes are attached to a solid support (Chip/Array) where a single base extension reaction adds labeled nucleotides in each specific locus. When excited by a laser, those nucleotides emit signals that are scanned and read by imaging software. Thus, this technology is able to genotype millions of genetic variants in a single individual at once. Each variant is identified by a unique arbitrary “rs” number, and correspond to a chromosomal position, according to the genome reference assembly (e.g. GRCh38). This massive amount of genetic data (millions of genotypes for thousands of individuals) is subject to statistical modeling in order to estimate the effect of a given allele on the disease risk, taking into account the influence of potential confounders such as age and sex. A typical statistical model (logistic model) for genetic association studies regresses the disease status D on observed covariates:
,
where the genetic information (rs number) is usually coded as 0, 1 or 2 according to the number of effect alleles a given person carriers. The effect size and direction of association between the effect allele and disease risk is reflected in the odds ratio (OR), calculated from the coefficient β 1. If OR is <1.0 or >1.0 the effect allele is called protective or risk allele, respectively. The precision and significance for the OR estimate is characterized by two parameters, respectively, the confidence interval (CI) and p-value. A typical GWAS analyzes over a million genetic variants, generating an equal number of hypothesis tests. Assuming a significance level α of 0.05 for each test, GWASs need to adopt multiple test adjustment procedures (e.g. Bonferroni correction) leading to local significance level of 5 × 10−8 (= 0.05/106) (Prof. Dr. Andreas and König, 2010). GWAS results are classically shown in a Manhattan Plot, with the chromosomal position of genetic variants at x axis and the p-values for each variant test (in -log10 scale) at y axis.
Box 1.
Alt-text: Box 1
2. Genome-wide association studies of COVID-19
For this Review, we included the eleven largest peer-reviewed GWAS (case sample size >1500) published until 25 September 2022. Given the pandemic-emergency context, these studies were conducted in a record time for GWAS standards, thanks to collaborative work from hundreds of scientists around the world. In order to accomplish that, genetic information about control samples was mostly obtained from preexisting genetic databases, such as the population-base cohort of UK biobank and the direct-to-consumer genetic testing companies 23andMe and AncestryDNA. The pinnacle of scientific collaborative spirit is represented by the Host Genetics Initiative (HGI), a project that currently has brought together over 100 studies from dozens of countries (The COVID-19 Host Genetics Initiative, 2020). The majority of the GWASs performed trans-ancestry meta-analysis, which consists in: 1) classifying individuals in ancestry categories (European-EUR, African-AFR, South Asian-SAS, East Asian-EAS, Ameridian/Latin American/Hispanic-AME) based on their genetic profile; 2) performing a GWAS for each ancestry group separately; and 3) meta-analyzing the results of all ancestry GWASs. The eleven GWASs are briefly described below and summarized in Table 1 .
Table 1.
Summary of the COVID-19 GWASs.
| Study | Publication date | Population | Sample sizea, n |
Phenotype description (Case vs Control) | |
|---|---|---|---|---|---|
| Case | Control | ||||
| GWAS 1 (PMID: 32558485) | 17 − 06-2020 | Italy, Spain | 1980 | 2381 | COVID-19 positive test and supplementary oxygen or mechanical ventilation vs Population-based blood donors with unknown COVID-19 status |
| GWAS 2 (PMID: 33307546) | 11-12-2020 | UK | 2244 | 64,784 | Critically ill vs Population-based controls with unknown COVID-19 status |
| GWAS 3 (PMID: 33888907) | 22-04-2021 | US | 12,972 | 101,268 | COVID-19 positive test vs COVID-19 negative test |
| 802 | 797,153 | COVID-19 positive test and hospitalization vs Unknown COVID-19 status | |||
| 1286 | 797,084 | COVID-19 positive test and pneumonia vs Unknown COVID-19 status | |||
| 636 | 797,180 | COVID-19 positive test and supplementary oxygen or ventilation vs Unknown COVID-19 status | |||
| 1447 | 796,151 | COVID-19 positive test and respiratory support or pneumonia vs Unknown COVID-19 status | |||
| GWAS 4 (PMID: 34237774) | 08-07-2021 | 19 countries | 6179 | 1,483,780 | Critically ill vs Population-based controls with unknown COVID-19 status |
| 13,641 | 2,070,709 | COVID-19 positive test and hospitalization vs Population-based controls with unknown COVID-19 status | |||
| 49,562 | 1,770,206 | Reported SARS-CoV-2 infection vs Population-based controls with unknown COVID-19 status | |||
| GWAS 5 (PMID: 35241825) | 03-03-2022 | US, UK | 52,630 | 704,016 | COVID-19 positive vs COVID-19 negative or unknown |
| 52,630 | 109,605 | COVID-19 positive vs COVID-19 negative | |||
| 45,641 | 704,016 | COVID-19 positive and not hospitalized vs COVID-19 negative or unknown | |||
| 6911 | 689,620 | COVID-19 positive and hospitalized vs COVID-19 negative or unknown |
|||
| 2184 | 689,620 | COVID-19 positive and severe vs COVID-19 negative or unknown | |||
| 6911 | 45,185 | COVID-19 positive and hospitalized vs COVID-19 positive and not hospitalized |
|||
| 2184 | 45,185 | COVID-19 positive and severe vs COVID-19 positive and not hospitalized | |||
| GWAS 6 (PMID: 35410379) | 11-04-2022 | US | 5373 | 35,901 | COVID-19 positive test vs COVID-19 negative test |
| 5373 | 95,027 | COVID-19 positive test vs Population-based controls with unknown COVID-19 status | |||
| 474 | 4159 | COVID-19 positive and hospitalized vs COVID-19 positive and not hospitalized |
|||
| 474 | 99,198 | COVID-19 positive and hospitalized vs Population-based controls with unknown COVID-19 status |
|||
| 2022 | 1060 | COVID-19 positive and had a cohabitant with a confirmed COVID-19 diagnosis vs COVID-19 negative and had a cohabitant with a confirmed COVID-19 diagnosis | |||
| 1060 | 98,507 | COVID-19 negative and had a cohabitant with a confirmed COVID-19 diagnosis vs Population-based controls with unknown COVID-19 status | |||
| 4353 | 391 | COVID-19 positive and symptomatic vs COVID-19 positive and asymptomatic or paucisymptomatic | |||
| 4952 | – | COVID-19 positive score that combines nine different measures of COVID-19 symptom severity | |||
| GWAS 7 (PMID: 35368071) | 15-09-2022 | Brazil | 3533 | 1700 | COVID-19 positive and hospitalized vs COVID-19 positive and not hospitalized |
| GWAS 8 (PMID: 35708486) | 16-06-2022 | Spain | 5966 | 11,916 | COVID-19 positive and hospitalized vs COVID-19 positive and not hospitalized + Population-based controls with unknown COVID-19 status |
| 2379 | 14,375 | COVID-19 positive and hospitalized with severity signal b vs COVID-19 positive without severity signal + Population-based controls with unknown COVID-19 status | |||
| 1128 | 16,754 | Critically ill vs COVID-19 positive but not critically ill + Population-based controls with unknown COVID-19 status | |||
| 11,939 | 5943 | COVID-19 positive vs Population-based controls with unknown COVID-19 status | |||
| GWAS 9 (PMID: 35848942) | 15-07-2022 | Italy, Spain, Norway and Germany/Austria | 3255 | 12,488 | COVID-19 positive and hospitalized with respiratory support vs Population-based controls with unknown COVID-19 status |
| 1911 | 12,226 | Critically ill vs Population-based controls with unknown COVID-19 status | |||
| GWAS 10 (PMID: 34465887) | 31-08-2021 | China | 598 d | 2260 d | Critically ill or other severe conditions c vs COVID-19 positive without severe symptoms + Population-based controls with unknown COVID-19 status |
| 474 e | 1615 e | Critically ill or other severe conditions c vs COVID-19 positive without severe symptoms + Population-based controls with unknown COVID-19 status | |||
| GWAS 11 (PMID: 35940203) | 08-08-2022 | Japan | 2393 | 3289 | COVID-19 positive and hospitalized vs Population-based controls with unknown COVID-19 status |
| 990 | 3289 | COVID-19 positive and hospitalized with respiratory support and/or intensive care vs Population-based controls with unknown COVID-19 status | |||
Numbers represent the total sample size of the discovery cohorts. The Host Genetics Initiative (HGI) samples used in meta-analyses were not considered, except for the HGI study itself (GWAS 4).
Severity was defined based on the following criteria: PaO2 < 65 mmHg or SaO2 < 90% OR PaO2/FiO2 < 30 OR SaO2/FiO2 < 440 OR Dyspnea OR Respiratory frequency ≥ 22 bpm OR Infiltrates affecting > 50% of the lungs.
At least one of the following: respiratory rate ≥ 30 times/min OR oxygen saturation ≤ 93% at resting state OR arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤300 mmHg OR pulmonary imaging examination showed that the lesions significantly progressed by >50%.
Samples used in the GWAS.
Samples used in the Whole Genome Sequencing study.
2.1. GWAS 1 (Severe Covid-19 GWAS Group et al., 2020)
This was the first GWAS of COVID-19 to be published. It enrolled, from February to May 2020, 1980 severe cases with respiratory failure, from the pandemic epicenters-cities in Italy and Spain. The controls were blood donors from Italy and Spain, most of which with unknown SARS-CoV-2 infection status. It is worth to point out that European Medicines Agency (EMA) endorsed the life-saving treatment with Dexamethasone only in September 2020 (Gozzo et al., 2020), therefore, this GWAS1 cohort tends to better represent the natural history of the disease. The meta-analysis involving Italian (n cases = 835; n controls = 1255) and Spanish cohorts (n cases = 775; n controls = 950) identified two disease-associated loci: 3p21.31 (lead variant: rs11385942) and 9q34.2 (lead variant: rs657152), the locus harboring the ABO gene. The authors also detected a protective effect of blood group O against severe disease (odds ratio = 0.65; p = 0.00001).
2.2. GWAS 2 GenOMICC (Pairo-Castineira et al., 2021)
This study also involved critically ill COVID-19 cases (n = 2244) mostly recruited from 208 intensive care units in the United Kingdom (UK) and controls selected from the UK biobank (n = 11,220). Eight independent association loci were detected, including the locus 3p21.31, and validated through analysis with two additional cohorts of controls (100,000 Genomes Project, n = 45,875; and Generation Scotland, n = 7689). Despite of being more powered (larger sample size) than GWAS 1, GWAS 2 did not detect statistical association with 9q34.2 at genome-wide significance level, suggesting that ABO locus is not associated with severe disease. These apparently discordant findings raised the hypothesis that the limited knowledge about the clinical management of COVID-19 patients, given the incipient pandemic time, contributed to the worsening in disease outcomes for cases from GWAS 1 rather than a direct effect of genetic variants at ABO on COVID-19 severity.
2.3. GWAS 3 (Shelton et al., 2021)
This study was conducted by the company 23andMe, by recruiting over one million research participants from their preexisting genetic cohort. They identified via online surveys 101,268 self-reported COVID-19 test-negative individuals, and 15,434 COVID-19 test-positive, of which 1131 needed hospitalization. Eighty percent of the study participants were of European ancestry, followed by 11.3% of Latino, 2.7% of African American, and 5.7% of other non-European ancestry. This study identified 9q34.2 (ABO) as infection susceptibility locus (P = 5.3 × 10−20) and 3p21.31 as severe disease locus (P = 1.6 × 10−18).
2.4. GWAS 4 (COVID-19 Host Genetics Initiative, 2021)
This study meta-analyzed GWAS results of 46 distinct studies, spanning 19 countries, and encompassing approximately 50,000 cases and 2 million controls, defined as individuals without known SARS-CoV-2 infection. For analysis purposes, COVID-19 cases were classified in three phenotypes: 1) Critically ill (hospitalization + respiratory support); 2) Moderate COVID-19 (hospitalization); and 3) Positive COVID-19 test (regardless of symptoms). The three cases phenotypes were compared to genetically ancestry-matched controls. Even in this large trans-ancestry GWAS, only 22–29% of samples were of non-European origin. Thirteen independent association loci were identified, of which nine had stronger association (i.e. larger odds ratios) with severe COVID-19. Of note, GWAS 4 replicated five associated loci from GWAS 2, in addition to the ABO locus detected in GWAS 1 and GWAS 3. In addition, this study contains cohorts from the first three GWASs, what is expected given the global collaborative nature of HGI. Lastly, this study used Release 5 data, the summary statistics for the most up to date Release 7 data is available at https://www.covid19hg.org/results/r7/.
2.5. GWAS 5 (Horowitz et al., 2022)
This study, published on March 3rd 2022, analyzed genetic data from 52,630 COVID-19 cases and 704,016 individuals with no record of SARS-CoV-2 infection (Controls) from four different cohorts: Geisinger Health System (GHS), Penn Medicine BioBank (PMBB), UK Biobank (UKB) and AncestryDNA. This GWAS used four COVID-19 positive phenotypes (“positive”, “positive and not hospitalized”, “positive and hospitalized”, and “positive and severe”) and two COVID-19 negative phenotypes (“negative”, and “negative or unknown”), combined in seven different comparison schemes, of which five informed about susceptibility to infection and two comparisons informed about risk of severe disease. With regard to the type of variants, they analyzed 13 millions of common variants and 76 millions of rare variants. As results, in addition to validate six previously reported associations (two loci associated with susceptibility to infection and four loci associated with severe disease risks), they also reported a new association between SARS-CoV-2 infection susceptibility and rs190509934, a rare variant located 60 bp upstream of the ACE2 gene and associated with its differential expression levels.
2.6. GWAS 6 (Roberts et al., 2022)
Published along with GWAS 5 at the same issue of Nature Genetics (Volume 54, Issue 4, April 2022), this study explored eight comparison schemes involving 736,723 AncestryDNA customers that have consented to research and responded the web-survey from 22 April to 3 August 2020. Interestingly, the authors used comparisons such as household exposed individuals who tested positive vs negative for COVID-19 to deeply investigate susceptibility to infection. With this study design, they confirmed that ABO variants influence the risk of infection rather than COVID-19 severity. In addition, 12 independent genetic markers from GWAS 4 and GWAS 5 were replicated, with a caveat that 12% and 46% of GWAS 6 samples overlapped samples from GWAS 4 and GWAS 5, respectively.
2.7. GWAS 7 (Pereira et al., 2022)
This study used a Brazilian cohort (BRACOVID) of 3533 hospitalized COVID-19 cases and 1700 non-hospitalized COVID-19 controls, all recruited from São Paulo, Brazil. In addition to replicate the previously reported findings for loci 3p21.31 and 21q22.11, the authors reported a novel association between variant rs11240388 (1q32.1) and disease severity. The ABO locus was not replicated in this study.
2.8. GWAS 8 (Cruz et al., 2022)
This GWAS analyzed 11,939 cases and 5943 general population controls, most of which obtained from the Spanish DNA biobank (https://www.bancoadn.org/). Of note, there were no overlapping samples between this Spanish cohort and the other GWASs reviewed here. This study investigated both the risk of infection and severity by transforming cases phenotype into a five-level severity scale: asymptomatic, mild, moderate, severe, and critical. In addition to replicate the 3p21.31 and 21q22.11 associated loci, Cruz et al. identified novel variants at chromosomes 9 and 17. They also performed sex-disaggregated analysis to identify an unreported variant rs1826292621 (9q21.32) associated with hospitalization risk in females only. While acknowledging the relevance of this analysis strategy, we will not discuss this finding throughout the manuscript in order to keep consistency regarding the analytical procedures across studies.
2.9. GWAS 9 (Degenhardt et al., 2022)
This study expanded the case and control cohorts from GWAS 1 by recruiting additional participants from Italy (n = 6270), Spain (n = 5798), Norway (n = 324), and Germany/Austria (n = 3351) to achieve a total of 3255 severe COVID-19 cases and 12,488 population controls. All cases were hospitalized and further divided in two sub-phenotypes (1-respiratory support and 2-mechanical ventilation). Variants at loci 3p21.31, 9q34.2, and 19p13.2 passed the threshold of P < 5 × 10−8 and were once again replicated.
2.10. GWAS 10 (Wu et al., 2021)
This was the first published genetic association study in Asian population. A total of 4947 Chinese individuals were evaluated by GWAS (controls = 1401; Mild COVID-19 = 859; Severe COVID-19 = 598) and by Whole Genome Sequencing (controls = 948; Mild COVID-19 = 667; Severe COVID-19 = 474). The rare Chinese-specific variant rs74490654 (19p13.11), within MEF2B gene, was associated with the risk of severe disease (P = 1.2 × 10−8).
2.11. GWAS 11 (Namkoong et al., 2022, p. 2)
From April 2020 to January 2021, 2393 COVID-19 cases that required hospitalization were enrolled as part of The Japan COVID-19 Task Force. By performing a GWAS with population controls (n = 3289), the authors found the variant rs60200309 (5q35) associated severe disease, especially in young individuals (P = 1.2 × 10−8). This finding was replicated in an additional cohort of 1243 patients with severe COVID-19 and 3769 controls. By using in vitro and in vivo experiments, the authors functionally validated the effect of this population-specific variant, unraveling the role of DOCK2 gene on the host immune response to SARS-CoV-2.
3. Susceptibility genes to COVID-19
In order to identify the main genes that confer susceptibility to COVID-19, we selected the lead variants reported by the eleven GWASs and systematically applied the steps described in Table 2 .
Table 2.
Methods and resources used in this Review to investigate variants and genes identified by the COVID-19 GWASs.
| Step | Resource | Features |
|---|---|---|
| 1. Variant genomic location and functional mapping | Ensembl https://www.ensembl.org/index.html |
Genome build GRCh38 |
| 2. Variant effect on transcript abundance across human tissues | Genotype-Tissue Expression (GTEx) https://gtexportal.org/home/ |
|
| 3. Gene function and tissue/cell specificity | Human Protein Atlas (HPA) https://www.proteinatlas.org/ |
None |
| 4. Assessment of linkage disequilibrium (LD) and allele frequencies patterns in global populations | LDlink https://ldlink.nci.nih.gov/ |
|
| 5. Gene role on COVID-19 pathogenesis | PubMed https://pubmed.ncbi.nlm.nih.gov/ |
Literature review. Search terms: “Gene name/symbol” AND “COVID-19” AND “SARS-CoV-2”. |
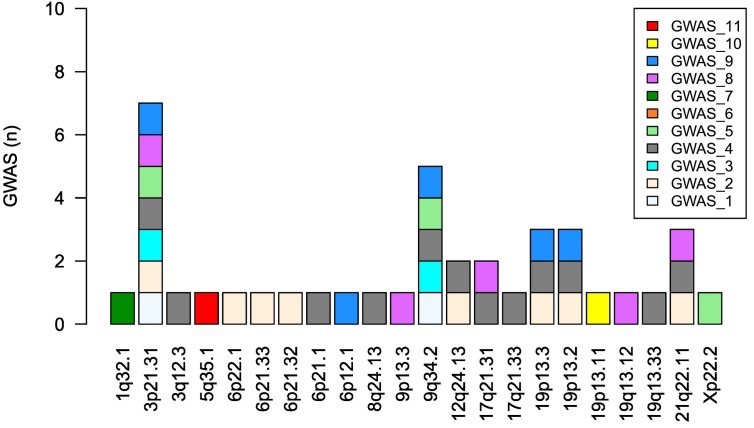
The lead genetic variants reported by the reviewed GWASs are listed in Table 3 . We included only the variants that surpassed the genome-wide significance P-value threshold as defined by each study. With the exception of GWAS 1, the remaining studies performed meta-analysis with HGI data, in addition to the GWAS with the discovery cohort. Thus, Table 3 includes only the variants detected in the discovery cohort analysis with the populations listed in Table 1. Fig. 1 summarizes the chromosome regions associated with COVID-19 across studies.
Table 3.
Lead genetic variants associated with COVID-19 in eleven published GWASs.
| GWAS | Lead variant | Chr | Position | Pheno | Effect Allele | Alternate Allele | OR | p-value |
|---|---|---|---|---|---|---|---|---|
| 7 | rs11240388 | 1q32.1 | 205,208,489 | DS | G | A | 1.35 | 3.9 × 10−08 |
| 8 | rs17213127 | 3p21.31 | 45,756,734 | DS | T | C | 1.52 | 3.8 × 10−09 |
| 4 | rs2271616 | 3p21.31 | 45,796,521 | IS | T | G | 1.15 | 1.8 × 10−34 |
| 5 | rs2531743 | 3p21.31 | 45,796,808 | IS | G | A | 0.94 | 2.9 × 10−12 |
| 3 | rs13078854 | 3p21.31 | 45,820,440 | DS | G | A | 0.59 | 1.6 × 10−18 |
| 4 | rs10490770 | 3p21.31 | 45,823,240 | DS | C | T | 1.89 | 2.2 × 10−61 |
| 1 | rs11385942 | 3p21.31 | 45,834,968 | DS | AAA | AA | 1.77 | 1.1 × 10−10 |
| 2,5,9 | rs73064425 | 3p21.31 | 45,859,597 | DSa | T | C | 2.10 | 3.6 × 10−32 |
| 8 | rs1994492 | 3p21.31 | 45,919,154 | DS | C | T | 1.46 | 8.6 × 10−15 |
| 8 | rs1994490 | 3p21.31 | 45,966,210 | DS | T | C | 0.85 | 3.9 × 10−08 |
| 4 | rs11919389 | 3q12.3 | 101,705,614 | IS | C | T | 0.94 | 3.5 × 10−15 |
| 11 | rs60200309 | 5q35.1 | 170,092,608 | DS | A | G | 2.01 | 1.2 × 10−08 |
| 2 | rs9380142 | 6p22.1 | 29,831,017 | DS | A | G | 1.30 | 1.8 × 10−08 |
| 2 | rs143334143 | 6p21.33 | 31,153,649 | DSa | A | G | 1.80 | 2.6 × 10−24 |
| 2 | rs3131294 | 6p21.32 | 32,212,369 | DS | G | A | 1.50 | 1.3 × 10−10 |
| 4 | rs1886814 | 6p21.1 | 41,534,945 | DS | C | A | 1.26 | 1.1 × 10−09 |
| 9 | rs78531133 | 6p12.1 | 56,842,705 | DS | A | G | 1.67 | 2.7 × 10−09 |
| 4 | rs72711165 | 8q24.13 | 124,324,323 | DS | C | T | 1.37 | 2.1 × 10−09 |
| 8 | rs10813976 | 9p13.3 | 33,426,577 | DS | A | G | 0.18b | 2.7 × 10−08 |
| 9 | rs687289 | 9q34.2 | 133,261,703 | DS | A | G | 1.24 | 4.5 × 10−10 |
| 1 | rs657152 | 9q34.2 | 133,263,862 | DS | A | C | 1.32 | 4.9 × 10−08 |
| 3 | rs9411378 | 9q34.2 | 133,270,015 | IS | C | A | 0.86 | 5.3 × 10−20 |
| 5 | rs879055593 | 9q34.2 | 133,271,182 | IS | T | C | 1.10 | 7.1 × 10−34 |
| 4 | rs912805253 | 9q34.2 | 133,274,084 | IS | T | C | 0.91 | 1.4 × 10−39 |
| 4 | rs10774671 | 12q24.13 | 112,919,388 | DS | A | G | 1.20 | 4.1 × 10−13 |
| 2 | rs10735079 | 12q24.13 | 112,942,203 | DS | A | G | 1.30 | 2.8 × 10−09 |
| 8 | rs1230082 | 17q21.31 | 45,422,978 | DS | C | G | 0.16b | 2.1 × 10−08 |
| 4 | rs1819040 | 17q21.31 | 46,142,465 | DS | A | T | 0.88 | 1.8 × 10−10 |
| 4 | rs77534576 | 17q21.33 | 49,863,303 | DS | T | C | 1.45 | 4.4 × 10−09 |
| 9 | rs12610495 | 19p13.3 | 4,717,660 | DS | G | A | 1.29 | 2.9 × 10−08 |
| 2,4 | rs2109069 | 19p13.3 | 4,719,431 | DSa | A | G | 1.26 | 9.7 × 10−22 |
| 2,4 | rs74956615 | 19p13.2 | 10,317,045 | DS | A | T | 1.43 | 9.7 × 10−12 |
| 9 | rs11085725 | 19p13.2 | 10,351,837 | DS | T | C | 1.31 | 3.2 × 10−09 |
| 2 | rs11085727 | 19p13.2 | 10,355,447 | DS | T | C | 1.30 | 1.3 × 10−07 |
| 10 | rs74490654 | 19p13.11 | 19,163,581 | DS | G | C | 8.73 | 1.2 × 10−08 |
| 8 | rs77127536 | 19q13.12 | 35,687,796 | DS | G | A | -0.22b | 1.3 × 10−08 |
| 4 | rs4801778 | 19q13.33 | 48,867,352 | IS | T | G | 0.95 | 1.2 × 10−08 |
| 8 | rs9636867 | 21q22.11 | 33,237,639 | IS | G | A | 1.21 | 3.5 × 10−10 |
| 2,4 | rs13050728 | 21q22.11 | 33,242,905 | DS | C | T | 0.82 | 1.1 × 10−16 |
| 2 | rs2236757 | 21q22.11 | 33,252,612 | DSa | A | G | 1.30 | 5.0 × 10−08 |
| 5 | rs190509934 | Xp22.2 | 15,602,217 | IS | C | T | 0.60 | 4.5 × 10−13 |
Chr: chromosome. Pheno: associated phenotype. IS: infection susceptibility. DS: disease severity. Position: chromosome coordinate based on GRCh38 genome build. OR: odds ratio.
For variants being identified by more than one study, we used OR and P-value for the most significant finding. There is no genetic variant for GWAS 6 since none of them achieved genome-wide significance.
These variants were also associated with infection susceptibility.
Beta coefficient from multinomial logistic regression, using a three-level phenotype scheme (mild, intermediate and severe).
Fig. 1.
Chromosome locations associated with COVID-19 according to the eleven published GWASs. Genetic association with loci 3p21.31 and 9q34.2 were the most replicated findings. GWAS 6 did not find any variant-phenotype association at genome-wide significance (P < 1.25 × 10−9).
The section bellow provides a brief description of the genes under the influence of variants within each chromosome location, by taking into account the tissue-specific effect of genetic variants on transcript abundance and the linkage disequilibrium pattern among variants, according to the steps described in Table 2. Throughout the manuscript preparation, we have been continuously searching for those genes across the most up-to-date literature concerning COVID-19 pathogenesis as an attempt to leverage genetic data interpretation.
3.1. 1q32.1
The variant rs11240388, detected in the Brazilian GWAS, locates in a promoter flank region of DSTYK, a gene encoding a kinase involved with cell death. According to GTEx, this variant influences the expression of several transcripts (CNTN2, DSTYK, TMCC2, TMEM81, RBBP5) in different tissues. The Brazilian group suggests DSTYK as the most important gene based on Transcription-Wide Association Analysis (Pereira et al., 2022), however there is no experimental study linking the gene to SARS-CoV-2 pathogenesis. Interestingly, rs11240388 is in LD with variants rs6664706 and rs1172149, associated with distribution width of red blood cells (Chen et al., 2020) and platelets (Astle et al., 2016), respectively.
3.2. 3p21.31
Seven out of the 11 GWASs have identified nine lead variants at 3p21.31 with genome-wide significance (Table 3), spanning a region of 204 Kb. Some of these variants are in LD and is associated with severe disease, while others with infection susceptibility. The 3p21.31 locus harbors several genes potentially implicated with COVID-19 mechanism, so it is likely that these variants are tagging different causal signals.
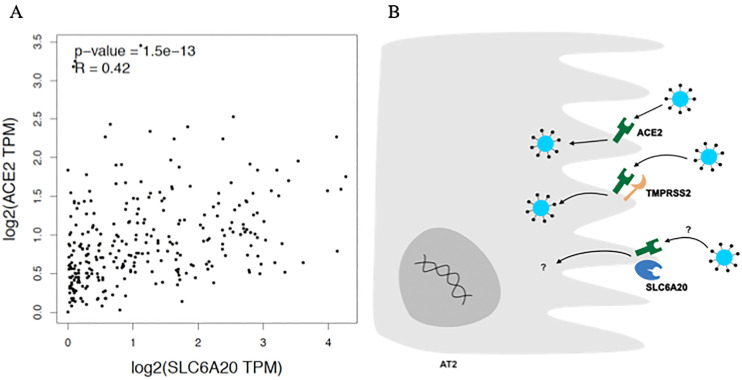
Variants rs17213127 and rs2271616 locate at 3’UTR and promoter of SLC6A20 gene, also known as SIT1. This gene encodes a protein involved with transport of imino acids (e.g. proline and hydroxyproline) across cell membranes. The risk allele rs2271616(T) is associated with increased transcript levels of SLC6A20, according to GTEx data. Of importance, SLC6A20 expression is enhanced in several cell types, including the epithelial alveolar type 2 cells (AT2), according to Human Protein Atlas (https://www.proteinatlas.org/ENSG00000163817-SLC6A20). The main link between SLC6A20 and COVID-19 comes from the functional interaction between SLC6A20 and ACE2 in the brush border membrane of small intestine enterocytes (Camargo et al., 2020). Interestingly, SLC6A20 and ACE2 expression levels are positively correlated (R = 0.42, p = 1.5 × 10−13) in lungs (Fig. 2 ), which is consistent with a recent finding showing that both genes are predominantly expressed in AT2 (Wang et al., 2020). Yan et al. (Yan et al., 2020) elucidated the ternary structure of a complex involving the Spike receptor binding domain (RBD), ACE2, and SLC6A19, another amino acid transporter with 46% identity with SLC6A20. Thus, despite the lack of experimental data demonstrating the role of SLC6A20 on SARS-CoV-2 entry mechanism, it is reasonable to assume a role for this locus on infection susceptibility rather than disease severity, which is consistent with GWASs findings (Table 3).
Fig. 2.
Potential role of SLC6A20 on susceptibility to SARS-CoV-2 infection.
A. Gene expression levels between SLC6A20 and ACE2 are positively correlated in lungs (Spearman correlation test. Data from GTEx via GEPIA (Tang et al., 2017)). TPM: transcript per million. B. Proposed functional interaction of proteins in alveolar type 2 cell (AT2). Unlike ACE2 and TMPRSS2, the role of SLC6A20 on SARS-CoV-2 infection remains to be elucidated.
With regard to the variant rs2531743, despite of being only 287 bp distant from rs2271616, it is located out of SLC6A20, but within an SLC6A20-overlaping long non-coding RNA (lncRNA) gene (ENSG00000288720). Surprisingly, the two variants are in linkage equilibrium in all 1KG populations (r2 < 0.07) but in EAS (r2 = 0.30), where the risk allele of rs2271616(T) is correlated with the protective allele of rs2531743(G) (LDpair Tool), possibly softening the effect of these variants on SARS-CoV-2 infection risk in EAS populations. According to GTEx, rs2531743 alters the expression of FYCO1 with no effect on SLC6A20 transcript levels, thus suggesting the two variants might be related to different causal pathways. FYCO1 encodes a protein involved with autophagy and endoplasmic reticulum-derived double membrane vesicle (DMV), a primary replication site for SARS-CoV-2 (Parkinson et al., 2020, p.).
The remaining variants (rs13078854, rs10490770, rs11385942, rs73064425, rs1994492, and rs1994490) partially overlap the chromosomal segment that includes the haplotype inherited from Neanderthals (chr3:45825948–45,867,532), that has been associated with severe disease (Zeberg and Pääbo, 2020). Most of these variants are in LD with each other and distributed in a region containing CC chemokine receptor genes (CCRs), FYCO1, LZTFL1 and two LZTFL1-overlapping lncRNAs (ENSG00000288720 and ENSG00000285788). A Transcriptome-Wide Association Study (TWAS) showed that risk alleles are associated with increased expression of CCR2 (pro-inflammatory) and decreased expression of CCR3 and CXCR6 (Pairo-Castineira et al., 2021). Recently, a Phenome-Wide Association Study with over 300,000 European individuals, suggested that these genetic variants influence COVID-19 risk by altering blood cell traits with CCRs as mediators (CCR6 for eosinophil and neutrophil counts and CCR2/CCR1 for monocyte count) (Zhou et al., 2021). Some of these variants are also eQTL for LZTFL1, SLC6A20 and FYCO1. LZTFL1 regulates protein trafficking to the ciliary membrane (Seo et al., 2011) and is highly expressed in nasopharynx and bronchi (https://www.proteinatlas.org/ENSG00000163818-LZTFL1). Therefore, this locus seems to harbor genes that confer susceptibility to infection (LZTFL1 and SLC6A20) and severe COVID-19 risk (chemokine receptors genes). Recently, Yao et al confirmed CCR9 and SLC6A20 as the causal genes from 3q21.21 locus by applying CRISPR/Cas genome editing tool in bronchial epithelium and immune cell types (Yao et al., 2021), while Downes et al., using a similar approach, found LZTFL1 as the effector gene and rs17713054 as the causative variant (Downes et al., 2021). Of note, this variant is in LD (r2 > 0.8, LDlink) with the other four variants (rs13078854, rs10490770, rs11385942, rs73064425).
3.3. 3q12.3
There was only one genetic variant (rs11919389) found in this locus. This variant is in LD with rs11720745 (r2 > 0.9), a regulatory variant located in the promoter of CEP97. This gene encodes the centrosomal protein 97 involved with ciliogenesis (Dobbelaere et al., 2020, p. 97)(Pearson, 2011) and highly expressed in nasopharynx and bronchus tissues (https://www.proteinatlas.org/ENSG00000182504-CEP97). Thus, CEP97 along with LZTFL1 seem to play role in the interaction between BBsome and basal body proteins during cilia formation, a process relevant to SARS-CoV-2 pathogenesis. The viral protein ORF10 causes cilia loss by downregulating host proteins of BBsome complex (e.g. BBS4) involved with ciliogenesis (Wang et al., 2022, p. 10). Interestingly, GWAS 4 showed that the variant rs11919389 was not associated with severe disease (P > 10−07) but with susceptibility to infection (P = 3.5 × 10−15).
3.4. 5q35.1
This locus was associated with COVID-19 in the Japanese GWAS, through the identification of rs60200309, an intergenic variant near DOCK2 gene. There is no eQTL for this variant in GTEx, but Namkoong et al. found the risk allele rs60200309(A) associated with decreased expression of DOCK2 in the blood of patients with age < 65 years (n = 270; β = −2.15, P = 0.0030) (Namkoong et al., 2022, p. 2). The Japanese group extensively demonstrated that downregulation of DOCK2 impairs macrophage migration, decreases expression of type I IFN genes (IFNA and IFNAB) and increases expression of IFNG, IL-6 and CCL5, in both humans and hamsters infected with SARS-CoV-2 (Namkoong et al., 2022, p. 2).
3.5. 6p22.1
The variant rs9380142 locates at the 3’-UTR of HLA-G, a gene involved with immunetolerance in pregnancy but also in antiviral response by modulating NK, CD8 T cells and B cells (LeMaoult and Yan, 2021). According to GTEx, rs9380142 is associated with HLA-G alternative splicing transcripts (sQTL) in both whole blood (P = 10−57) and lung (P = 10−36). Interestingly, patients with severe COVID-19 present higher serum levels of soluble HLA-G, a molecule produced via alternative splicing (Al-Bayatee and Adhiah, 2021). In addition, the variant is also eQTL for HLA-A, with the risk allele rs9380142(A) being associated with lower levels of HLA-A in whole blood (P = 1.6 × 10−22, GTEx). Viral-derived peptides are presented by HLA-A-restricted CD8 T cells via IFNG-induced immunoproteasome pathway. Recently, Shkurnikov et al reported HLA-A alleles as associated with early death from COVID-19 (before 60 years old) in Russian and Spanish cohorts (Shkurnikov et al., 2021).
3.6. 6p21.33
Despite being physically located within CCHCR1, the variant rs143334143 does not alter CCHCR1 expression in whole blood and lung, according to GTEx. Instead, the risk allele A is associated with increased expression of MIR6891 in whole blood (NES = 0.92; P = 1.2 × 10−36) and lung (NES = 0.89; P = 7.4 × 10−26). This gene encodes a micro-RNA that targets IgA-related mRNAs (IGHA1 and IGHA2) for degradation (Chitnis et al., 2017). Thus, it is likely that individuals carrying rs143334143(A) allele produce low levels of IgA, the immunoglobulin type that dominates the neutralizing antibody response against SARS-CoV-2 (Sterlin et al., 2021).
3.7. 6p21.32
Rs3131294 is an intronic variant that is in LD (r2 = 0.99) with rs3132946, a variant associated with fibrotic idiopathic interstitial pneumonias (Fingerlin et al., 2013). Notch4 was recently reported to contribute to lung inflammation in severe COVID-19 patients by impairing the action of the tissue repair cytokine amphiregulin (Areg) (Harb et al., 2021, p. 4). In addition, rs3131294 is eQTL for several COVID-19 candidate genes, including AGER (inflammation, highly expressed in lungs' alveolar cells type 1), complement system genes (C4A, C4B, CFB), antigen-processing genes (MHC class II genes, TAP1), MIR6891, and NOTCH4. Once again, another link between locus 6p21 and IgA appears (in addition to MIR6891), since rs3131294 is in LD with rs9271366 (r2 = 0.66; Population: EUR), a variant associated with serum IgA levels (Ferreira et al., 2010). Of note, the risk allele rs3131294(G) for severe COVID-19 (Table 2) is correlated with the risk allele rs9271366(A) for IgA deficiency (Chi-sq = 667, P < 0.0001, LDpair Tool).
3.8. 6p21.1
The variant rs1886814 is located within FOXP4-AS, a lncRNA gene that upregulates FOXP4 (Wu et al., 2019), a transcriptional repressor that modulates the expression of lung-specific genes involved with epithelial injury response and mucus production (Chokas et al., 2010)(Li et al., 2012). In line with this, rs1886814 is eQTL for a single gene in a single tissue: FOXP4 in lung (P = 0.000003, GTEx). Another variant in FOXP4-AS1 (rs1853837) was also reported as associated with severe COVID-19 in the Chinese study (GWAS 10) (Wu et al., 2021). Interestingly, these two variants were more strongly correlated in EAS and AMR (r2 ∼ 0.65) populations compared to others 1KG populations (r2 < 0.2 for AFR, EUR and SAS).
3.9. 6p12.1
The variant rs78531133 is located in the protein coding gene DST, but within the intronic promoter for its antisense lncRNA (DST-AS). DST encodes for dystonin, a cytoskeleton linker protein with role in herpes simplex virus 1 intracellular transport (McElwee et al., 2013). Rs78531133 is not in LD with any variant present in the GWAS Catalog and there was no eQTL for this varint in GTEx.
3.10. 8q24.13
Rs72711165 is an intron variant in TMEM65, a gene encoding the transmembrane protein 65. The depletion of TMEM65 in human cultured cells induced oxidative stress, apoptosis and activation of mitochondrial unfolded protein response pathway (Urushima et al., 2020, p. 65). There is no eQTL reported in GTEx database nor any disease/trait associated with this variant in GWAS Catalog. Thus, we consider premature to speculate any mechanism linking the locus 8q24.13 to COVID-19.
3.11. 9p13.3
Rs10813976 is an intergenic variant located between the aquaporin coding genes AQP7 and AQP3. The top eQTL for this variant is AQP3 in lung, with the risk allele A being associated with the lowest transcript levels (GTEx, P = 3.3 × 10−20). Zhu et al. experimentally demonstrate that the self-healing capacity of lung airway was impaired in AQP3-knockout mice (Zhu et al., 2016). Interestingly, Bayraktar et al. found increased levels of aquaporin-1 in the serum of critically ill COVID-19 patients (Bayraktar et al., 2022).
3.12. 9q34.2
This is the ABO locus, one of the most replicated association findings, representing the genetic basis for the relationship between ABO blood groups and COVID-19 risk (Liu et al., 2021). Rs657152, rs9411378, and rs879055593 are intronic variants, but only the former is located within transcription factors binding site, whereas rs912805253 is located in the promoter of ABO gene, and is eQTL for ABO, according to GTEx. Interestingly, rs657152 and rs912805253 are in LD (r2 > 0.8) with two relevant variants: 1) rs643434, a variant associated with inflammatory biomarkers including IL-6 and C-reactive protein (Naitza et al., 2012); and 2) rs687289, a variant associated with coagulation factor levels (Factor VIII, von Willebrand factor) (Sabater-Lleal et al., 2019) and venous thromboembolism risk (Lindström et al., 2019). This variant is also associated with monocyte and lymphocyte counts (data from GWAS catalog through LDtrait tool).
3.13. 12q24.13
OAS genes encode an interferon-induced enzyme that synthetizes dimers of 2′-5′-oligoadenylates which is involved in the activation of ribonuclease L (RNase L), which in turn degrades viral RNA (Schoggins, 2021). OAS1 is highly expressed across the lung tissue, especially in respiratory epithelial cells and lung macrophages (https://www.proteinatlas.org/ENSG00000089127-OAS1/tissue). The variant rs10735079 is located within OAS3 while rs10774671, a splice acceptor variant, within OAS1. In line with this, rs10774671 is sQTL for OAS1, with the risk allele rs10774671(A) being associated with a dramatically reduction in OAS1 expression in whole blood (P = 6.6 × 10−227) and lung (P = 4.0 × 10−168), according to GTEx. Therefore, it is reasonable to assume rs10774671 as the causal variant of 12q24.13 locus since it has been experimentally validated in patients with severe COVID-19 (Wickenhagen et al., 2021).
3.14. 17q21.31
Two variants (rs1230082 and rs1819040), 719 Kb apart from each other, were found in this locus. Rs1230082 localizes within ARHGAP27, a gene enconding a Rho GTPase activating protein involved with clathrin-mediated endocytosis. Rs1819040 is an intron variant of KANSL1 that is located in a high-density gene region. This gene may control the transcription of its neighbor genes through acetylation of nucleosomal histone H4. Despite no clear relationship between the genes at 17q21.31 and COVID-19, rs1819040 is in LD with rs56108300, a variant associated with neutrophil counts (Chen et al., 2020). The high number of blood neutrophils is associated with severe COVID-19 likely through the role of neutrophils in immunothrombosis (Reusch et al., 2021). Lastly, the two variants are in linkage equilibrium (r2 varying from 0.06 to 0.19 in all 1KG populations), thus suggesting they are tagging distinct causal variants.
3.15. 17q21.33
The variant rs77534576 is located in a regulatory region and influences the expression of DLX3 in lung (GTEx). DLX3 codes for a homeobox transcription regulator known to upregulate proinflammatory cytokines and favoring the accumulation of macrophages through STAT3 signaling in skin (Bhattacharya et al., 2018, p. 3). We speculate that this gene may play similar function in the lung tissue. There was no trait associated with this variant in the GWAS catalog.
3.16. 19p13.3
Variants rs12610495 and rs2109069 are in LD and both located within DPP9, a gene enconding the cytosolic dipeptidyl peptidase 9, known to inhibit the caspase-1-dependent pyroptosis by interacting with the inflammasome proteins NLRP1 (Hollingsworth et al., 2021) and CARD8 (Sharif et al., 2021, p. 8). Of note, the rs2109069(A) risk allele is associated with reduced expression of DPP9 (P = 1.7 × 10−9, tissue: lung, GTEx), therefore, favoring lung injury due to the exacerbation of inflammatory response. In addition, rs2109069 is in LD with rs12610495 (r2 = 0.88), a missense variant reported as associated to idiopathic pulmonary fibrosis risk in several independent studies (Allen et al., 2020; Fingerlin et al., 2013).
3.17. 19p13.2
The rs74956615 is located in the 3’UTR region of RAVER1, a gene encoding a ribonucleoprotein involved with alternative splicing regulation of tropomyosin 1 (TPM1) pre-mRNA (Rideau et al., 2006, p. 1). Bradbury et al., recently demonstrated that tropomyosin isoform 2.1 (Tpm2.1) is a key element during the TGF-β1-driven extracellular matrix (ECM) remodeling that leads to pulmonary fibrosis (Bradbury et al., 2021). In addition, according to GTEx database, rs74956615 is eQTL for TYK2 (P = 9.1 × 10−5, tissue: whole blood) and SHFL/C19orf66 (P = 9.4 × 10−5, tissue: whole blood), two relevant genes for COVID-19 pathogenesis. The former gene codes for the tyrosine kinase that phosphorylates the interferon-alpha/beta receptor while SHFL is an RNA binding protein known to inhibit the translation of viral RNA and has antiviral activity against SARS-CoV-2 (Lee et al., 2021). Therefore, it is not clear if this locus influence COVID-19 risk by either Raver1-Tpm1-ECM remodeling or by other unknown mechanism leading to TYK2 and SHFL impaired expression. Lastly, rs74956615 is associated with platelet indices (plateletcrit) such as mean platelet volume and platelet distribution width (Astle et al., 2016) and chronic inflammatory diseases (Ellinghaus et al., 2016) in European-ancestry individuals, what increases the likelihood of 19p13.2 being a true causal locus.
The other two variants found within 19p13.2 (rs11085725 and rs11085727) are distant by 3.6 Kb, and in LD with each other, but not correlated with rs74956615 (r2 < 0.1 in all 1KG population). Rs11085727 is located within TYK2 and both variants are eQTL for TYK2 (P = 8.4 × 10−16, tissue: whole blood). In addition, these variants are also eQTL for intercellular adhesion molecules (ICAM3, p = 9.8 × 10−12, tissue: whole blood; ICAM5, p = 1.9 × 10−07, tissue: Heart-Left ventricle), key molecules involved in both T-cell and neutrophils trans endothelial migration and immune cells- and antibody-mediated cytotoxicity (Ostermann et al., 2002). In line with this, rs11085727 is associated with neutrophil percentage and lymphocyte counts (Astle et al., 2016).
3.18. 19p13.11
Rs74490654 is an intron variant of MEF2B, a gene encoding an immune-metabolism transcriptional regulator, with role on lymphocyte development and activation (Clark et al., 2013). Recently, Wu et al. identified MEF2C as one of the mediators of ORAI1-mediated Ca2+ signaling effect on the expression of antiviral genes involved with resistance to SARS-CoV-2 infection in vitro ( Wu et al., 2022 , p. 1).
3.19. 19p13.12
The variant rs77127536, detected in GWAS 8, is eQTL for several genes in whole blood (UPK1A, ARHGAP33, ETV2, KMT2B, U2AF1L4), although it is localized in an intergenic region. This variant is not associated with any trait in GWAS Catalog according to the LDtrait search criteria. Cruz et al. identified ARHGAP33 as one of the most biologically plausible gene (Cruz et al., 2022), and just as rs1230082 (17q21.31), the variant rs77127536 is also related to a Rho GTPase activating protein (ARHGAP27). However, the mechanism by which these kind of proteins contributes to SARS-CoV-2 pathogenesis remains to be elucidated.
3.20. 19q13.33
In lung, the variant rs4801778 is eQTL for TULP2, HSD17B14, and PLEKHA4, according to GTEx. Although with unknown function, TULP2 belongs to the tubby-like protein (TULP) family that is involved with ciliary protein trafficking (Hong et al., 2021), thus a potential component of the respiratory primary barrier. HSD17B14 is an enzyme that catalyzes the conversion of estradiol to estrone, in addition to promote inflammation through NFkB (Qureshi et al., 2020). PLEKHA4 codes for a protein that binds specifically to phosphatidylinositol 3-phosphate (PI3P), the precursor of phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2). The inhibition of PI-3P-5-kinase, the enzyme that converts PI3P in PI(3,5)P2, prevents the SARS-CoV-2 infection (Kang et al., 2020). Therefore, we speculate that Plekha4 binding would render PI3P unavailable to PI-3P-5-kinase, thus protecting the cell from SARS-CoV-2 infection likely via endosomal trafficking disruption.
3.21. 21q22.11
The three variants found in this locus (rs9636867, rs13050728, rs2236757) are located within IFNAR2 introns, and correlated with each other (r2 > 0.8). Curiously, these variants are associated with differential IFNAR2 transcript processing according to the tissue type (sQTL), with opposite direction effects in blood vs lung. The risk alleles rs13050728(T) and rs2236757(A) are associated with reduced expression of IFNAR2 in whole blood and increased IFNAR2 expression in lung, based on GTEx data. In addition, the three variants are also eQTL for the antiviral gene IL10RB in several tissues, including lung.
3.22. Xp22.2
The genetic variant rs190509934 is located within a transcription factor-binding site of ACE2 regulatory region. This gene encodes the membrane-bound receptor used by SARS-CoV-2 to infect host cells. The protective allele rs190509934(C) of this genetic variant, is associated with lower levels of ACE2 expression and with 40% reduction in the risk of infection for those carrying one copy of the protective allele (Horowitz et al., 2022). Of note, the protective allele was not found in East Asian populations, and its frequency varies, respectively, from 0.14% to 0.65% in European and African populations, according to gnomAD database (Karczewski et al., 2020).
4. Key mechanisms linking genetic variants to COVID-19
In this section, we functionally integrated the susceptibility genes described in the previous section, taking into account its cellular location, function, and pathways of relevance to COVID-19 pathogenesis. We also leveraged the COVID-19 phenotype associated with the variants from Table 3 (infection susceptibility or disease severity) aiming to have better insights about disease mechanism.
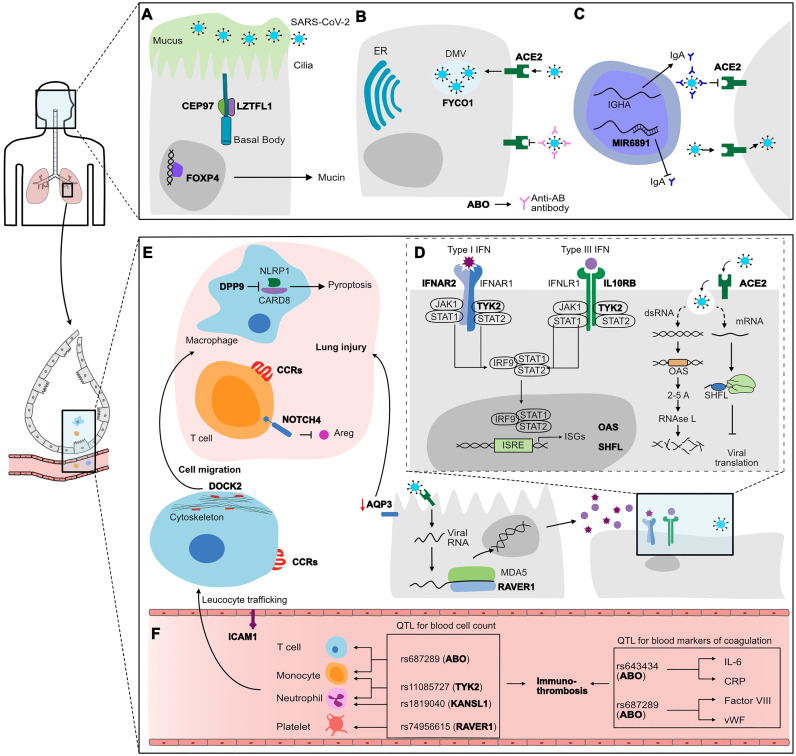
The genetic component underlying the risk of infection and/or mild disease seems to be related to biological processes occurring in the upper airways, such as mucociliary clearance, mucosal immunity and the initial phases of viral infection (Robinot et al., 2021) (Fig. 3 ). Some of the variants associated with genes influencing infection susceptibility (Table 3) are involved with ciliogenesis and mucus production (LZTFL1, CEP97, FOXP4, TULP2) or genes with function in viral entry and viral replication processes (SLC6A20, FYCO1, ACE2). In addition, two independent variants at 6p21 (rs143334143 and rs3131294) seems to influence antigen presentation and IgA production. According to the results from GWAS 4, the variants rs2271616 (SLC6A20), rs11919389 (CEP97) and rs4801778 (PLEKHA4, TULP2) were associated with susceptibility to infection rather then disease severity (p < 0.004 for the test of effect size difference between phenotypes Hospitalized vs Reported infection). The ABO gene at 9q34.2 was also consistently associated with infection susceptibility in GWASs 3–5. Interestingly, ABO locus was associated with critical COVID-19 cases in GWAS 1, however, those patients did not had the opportunity to be treated with corticosteroids, a game changer COVID-19 therapy. ABO encodes the glycosyltransferase responsible for the ABO blood group system. There is evidence that Anti-A and anti-B antibodies in individuals of blood group O might target glycoproteins in the virus surface, impairing the viral-entry process (Pendu et al., 2021). In addition, the variants at the ABO locus influence the concentration of blood markers of coagulation and thus may be involved with the risk of thromboembolitic events in COVID-19 patients.
Fig. 3.
Mechanisms underlying COVID-19 based on the genetic variants identified by GWASs. The genes associated with COVID-19 phenotypes are highlighted in bold. In the top panel, biological processes predominantly occurring in the upper respiratory tract, such as mucociliary clearance (A), viral-entry (B) and mucosal immunity (C). The genetic variants related to those genes are associated with susceptibility to infection (IS phenotype, Table 3). Impairment in these processes caused by the combined effect of risk alleles may favor the passage of higher viral loads to the lower airways until the alveolar space, contributing to the disease worsening. Immune modulatory processes, including antiviral immune response (D), inflammasome/piroptosis, leukocyte trafficking and migration (E), drive the mechanism underlying severe disease. The genetic variants controlling those processes are associated with disease severity (DS phenotype, Table 3). Variants rs643434 and rs687289 are in LD with the variants in ABO locus (9q34.2), and associated with the concentration of blood markers of coagulation (F). The other variants are associated with the amount of circulating leukocytes and platelets. Altogether, this quantitative effect on blood cells and proteins potentially links these genetic loci to immunothrombosis (F), an event frequently found in COVID-19 patients.
Conversely, several genes playing roles in antiviral immune response (RAVER1, IFNAR2, OAS, SHFL, HLA-A, HLA-G) were under the control of some of the association loci summarized in this Review. Of note, several COVID-19-associated variants influence the expression of genes within IFN pathway while other loci targeted genes involved with inflammation and/or lung injury response (CCR1, CCR9, CXCR6, NOTCH4, FOXP4, DPP9) (Fig. 3). Interestingly, locus 3q21.31 harbors variants associated with both infection susceptibility (rs2271616, rs2531743), as well as severe disease (rs13078854, rs10490770, rs11385942, rs1994492). In line with this, Nakanishi et al showed that individuals younger than 60 years old carrying the risk allele rs10490770(C) are almost two times more likely to develop severe COVID-19 (death or severe respiratory failure) when compared to those carriers aged above 60 (Nakanishi et al., 2021).
QTL: quantitative trait locus. ER: endoplasmic reticulum. DMV: double membrane vesicle. CCRs: chemokine receptors. Areg: amphiregulin. ISRE: interferon-sensitive response element. ISG: Interferon-stimulated genes.
5. Limitations
The findings summarized in this Review should be interpreted with caution since most of the associated variants came from studies biased towards European-ancestry samples and thus might not be generalizable to other non-European populations. The identification of population-specific variants (e.g. Chinese - rs74490654/MEF2B and Japanese - rs60200309/DOCK2) underscores the importance to conduct large genomic studies in diverse populations. In addition, most of the genetic studies reviewed here have used GWAS approach, which focuses on variants with minor allele frequencies >0.01, not accounting for the effect of rare genetic variants on disease risk. Therefore, future studies using whole genome sequencing approach will be crucial to fully dissect the genetic architecture of COVID-19.
6. Conclusions
The unprecedented effort from the scientific community to unravel the genetic basis of COVID-19 has consistently identified genes with high biological plausibility. The genes involved with infection susceptibility participate in biological processes more relevant to the upper airways, while the genes associated with severe outcomes are related to antiviral and inflammatory responses in the lower airways, especially in alveolar type 2 cells and leukocytes. This should be considered when selecting target genes for development of new therapeutic strategies. Indeed, some of the genes listed here (ICAM1, CCR9), especially those from IFN pathway (IFNAR2, IL10RB, TYK2), encode actionable proteins targeted by drug candidates currently in clinical trials or already approved (Gaziano et al., 2021). Lastly, further researches should also expand the studied phenotypes, in order to understand the genetic component underlying COVID-related complications such as thrombotic events, neurological impairment, and long COVID.
Author contribution
L.C.F. contributed to conceptualizing the Review, analyzing the COVID-19 GWAS results, reviewing the literature, and writing the manuscript. C.E.M.G. contributed to creating the Figures, critically revising the manuscript, and providing expertise in Genetics. J.F.R.N. contributed to critically revising the manuscript, providing key references and expertise in Immunology. S.M.B.J. contributed to critically revising the manuscript, and coordinating COVID-19 research projects as director of the Institute of Tropical Medicine at Federal University of Rio Grande do Norte.
Declaration of Competing Interest
The authors declare no competing interests.
Data availability
All data used in this Review was obtained from published articles and public databases.
References
- COVID-19 Host Genetics Initiative Mapping the human genetic architecture of COVID-19. Nature. 2021;1–8 doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bayatee N.T., Adhiah A.H. Soluble HLA-G is upregulated in serum of patients with severe COVID-19. Hum. Immunol. 2021 doi: 10.1016/j.humimm.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R.J., Guillen-Guio B., Oldham J.M., Ma S.-F., Dressen A., Paynton M.L., Kraven L.M., Obeidat M., Li X., Ng M., Braybrooke R., Molina-Molina M., Hobbs B.D., Putman R.K., Sakornsakolpat P., Booth H.L., Fahy W.A., Hart S.P., Hill M.R., Hirani N., Hubbard R.B., McAnulty R.J., Millar A.B., Navaratnam V., Oballa E., Parfrey H., Saini G., Whyte M.K.B., Zhang Y., Kaminski N., Adegunsoye A., Strek M.E., Neighbors M., Sheng X.R., Gudmundsson G., Gudnason V., Hatabu H., Lederer D.J., Manichaikul A., Newell J.D., O’Connor G.T., Ortega V.E., Xu H., Fingerlin T.E., Bossé Y., Hao K., Joubert P., Nickle D.C., Sin D.D., Timens W., Furniss D., Morris A.P., Zondervan K.T., Hall I.P., Sayers I., Tobin M.D., Maher T.M., Cho M.H., Hunninghake G.M., Schwartz D.A., Yaspan B.L., Molyneaux P.L., Flores C., Noth I., Jenkins R.G., Wain L.V. Genome-wide association study of susceptibility to idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2020;201:564–574. doi: 10.1164/rccm.201905-1017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas Ziegler, König Inke R. 2010. A Statistical Approach to Genetic Epidemiology. [Google Scholar]
- Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A., Lambourne J.J., Sivapalaratnam S., Downes K., Kundu K., Bomba L., Berentsen K., Bradley J.R., Daugherty L.C., Delaneau O., Freson K., Garner S.F., Grassi L., Guerrero J., Haimel M., Janssen-Megens E.M., Kaan A., Kamat M., Kim B., Mandoli A., Marchini J., Martens J.H.A., Meacham S., Megy K., O’Connell J., Petersen R., Sharifi N., Sheard S.M., Staley J.R., Tuna S., van der Ent M., Walter K., Wang S.-Y., Wheeler E., Wilder S.P., Iotchkova V., Moore C., Sambrook J., Stunnenberg H.G., Di Angelantonio E., Kaptoge S., Kuijpers T.W., Carrillo-de-Santa-Pau E., Juan D., Rico D., Valencia A., Chen L., Ge B., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yang Y., Guigo R., Beck S., Paul D.S., Pastinen T., Bujold D., Bourque G., Frontini M., Danesh J., Roberts D.J., Ouwehand W.H., Butterworth A.S., Soranzo N. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding D.J. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- Bayraktar N., Bayraktar M., Ozturk A., Ibrahim B. Evaluation of the relationship between Aquaporin-1, Hepcidin, zinc, copper, and İron levels and oxidative stress in the serum of critically ill patients with COVID-19. Biol. Trace Elem. Res. 2022 doi: 10.1007/s12011-022-03400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Kim J.-C., Ogawa Y., Nakato G., Nagle V., Brooks S.R., Udey M.C., Morasso M.I. DLX3-dependent STAT3 signaling in keratinocytes regulates skin immune homeostasis. J Invest Dermatol. 2018;138:1052–1061. doi: 10.1016/j.jid.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P., Nader C.P., Cidem A., Rutting S., Sylvester D., He P., Rezcallah M.C., O’Neill G.M., Ammit A.J. Tropomyosin 2.1 collaborates with fibronectin to promote TGF-β1-induced contraction of human lung fibroblasts. Respir. Res. 2021;22:129. doi: 10.1186/s12931-021-01730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo S.M.R., Vuille-dit-Bille R.N., Meier C.F., Verrey F. ACE2 and gut amino acid transport. Clin. Sci. 2020;134:2823–2833. doi: 10.1042/CS20200477. [DOI] [PubMed] [Google Scholar]
- Chen M.-H., Raffield L.M., Mousas A., Sakaue S., Huffman J.E., Moscati A., Trivedi B., Jiang T., Akbari P., Vuckovic D., Bao E.L., Zhong X., Manansala R., Laplante V., Chen M., Lo K.S., Qian H., Lareau C.A., Beaudoin M., Hunt K.A., Akiyama M., Bartz T.M., Ben-Shlomo Y., Beswick A., Bork-Jensen J., Bottinger E.P., Brody J.A., van Rooij F.J.A., Chitrala K., Cho K., Choquet H., Correa A., Danesh J., Di Angelantonio E., Dimou N., Ding J., Elliott P., Esko T., Evans M.K., Floyd J.S., Broer L., Grarup N., Guo M.H., Greinacher A., Haessler J., Hansen T., Howson J.M.M., Huang Q.Q., Huang W., Jorgenson E., Kacprowski T., Kähönen M., Kamatani Y., Kanai M., Karthikeyan S., Koskeridis F., Lange L.A., Lehtimäki T., Lerch M.M., Linneberg A., Liu Y., Lyytikäinen L.-P., Manichaikul A., Martin H.C., Matsuda K., Mohlke K.L., Mononen N., Murakami Y., Nadkarni G.N., Nauck M., Nikus K., Ouwehand W.H., Pankratz N., Pedersen O., Preuss M., Psaty B.M., Raitakari O.T., Roberts D.J., Rich S.S., Rodriguez B.A.T., Rosen J.D., Rotter J.I., Schubert P., Spracklen C.N., Surendran P., Tang H., Tardif J.-C., Trembath R.C., Ghanbari M., Völker U., Völzke H., Watkins N.A., Zonderman A.B., Million Veteran Program V.A., Wilson P.W.F., Li Y., Butterworth A.S., Gauchat J.-F., Chiang C.W.K., Li B., Loos R.J.F., Astle W.J., Evangelou E., van Heel D.A., Sankaran V.G., Okada Y., Soranzo N., Johnson A.D., Reiner A.P., Auer P.L., Lettre G. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182:1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis N., Clark P.M., Kamoun M., Stolle C., Brad Johnson F., Monos D.S. An expanded role for HLA genes: HLA-B encodes a microRNA that regulates IgA and other immune response transcripts. Front. Immunol. 2017;8:583. doi: 10.3389/fimmu.2017.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokas A.L., Trivedi C.M., Lu M.M., Tucker P.W., Li S., Epstein J.A., Morrisey E.E. Foxp1/2/4-NuRD interactions regulate gene expression and epithelial injury response in the lung via regulation of Interleukin-6 *. J. Biol. Chem. 2010;285:13304–13313. doi: 10.1074/jbc.M109.088468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.I., Tan S.W.S., Péan C.B., Roostalu U., Vivancos V., Bronda K., Pilátová M., Fu J., Walker D.W., Berdeaux R., Geissmann F., Dionne M.S. MEF2 is an in vivo immune-metabolic switch. Cell. 2013;155:435–447. doi: 10.1016/j.cell.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz R., Diz-de Almeida S., López de Heredia M., Quintela I., Ceballos F.C., Pita G., Lorenzo-Salazar J.M., González-Montelongo R., Gago-Domínguez M., Sevilla Porras M., Tenorio Castaño J.A., Nevado J., Aguado J.M., Aguilar C., Aguilera-Albesa S., Almadana V., Almoguera B., Alvarez N., Andreu-Bernabeu Á., Arana-Arri E., Arango C., Arranz M.J., Artiga M.-J., Baptista-Rosas R.C., Barreda-Sánchez M., Belhassen-Garcia M., Bezerra J.F., Bezerra M.A.C., Boix-Palop L., Brion M., Brugada R., Bustos M., Calderón E.J., Carbonell C., Castano L., Castelao J.E., Conde-Vicente R., Cordero-Lorenzana M.L., Cortes-Sanchez J.L., Corton M., Darnaude M.T., De Martino-Rodríguez A., del Campo-Pérez V., Diaz de Bustamante A., Domínguez-Garrido E., Luchessi A.D., Eiros R., Estigarribia Sanabria G.M., Carmen Fariñas M., Fernández-Robelo U., Fernández-Rodríguez A., Fernández-Villa T., Gil-Fournier B., Gómez-Arrue J., González Álvarez B., Gonzalez Bernaldo de Quirós F., González-Peñas J., Gutiérrez-Bautista J.F., Herrero M.J., Herrero-Gonzalez A., Jimenez-Sousa M.A., Lattig M.C., Liger Borja A., Lopez-Rodriguez R., Mancebo E., Martín-López C., Martín V., Martinez-Nieto O., Martinez-Lopez I., Martinez-Resendez M.F., Martinez-Perez A., Mazzeu J.F., Merayo Macías E., Minguez P., Moreno Cuerda V., Silbiger V.N., Oliveira S.F., Ortega-Paino E., Parellada M., Paz-Artal E., Santos N.P.C., Pérez-Matute P., Perez P., Pérez-Tomás M.E., Perucho T., Pinsach-Abuin M.L., Pompa-Mera E.N., Porras-Hurtado G.L., Pujol A., Ramiro León S., Resino S., Fernandes M.R., Rodríguez-Ruiz E., Rodriguez-Artalejo F., Rodriguez-Garcia J.A., Ruiz Cabello F., Ruiz-Hornillos J., Ryan P., Soria J.M., Souto J.C., Tamayo E., Tamayo-Velasco A., Taracido-Fernandez J.C., Teper A., Torres-Tobar L., Urioste M., Valencia-Ramos J., Yáñez Z., Zarate R., Nakanishi T., Pigazzini S., Degenhardt F., Butler-Laporte G., Maya-Miles D., Bujanda L., Bouysran Y., Palom A., Ellinghaus D., Martínez-Bueno M., Rolker S., Amitrano S., Roade L., Fava F., Spinner C.D., Prati D., Bernardo D., Garcia F., Darcis G., Fernández-Cadenas I., Holter J.C., Banales J.M., Frithiof R., Duga S., Asselta R., Pereira A.C., Romero-Gómez M., Nafría-Jiménez B., Hov J.R., Migeotte I., Renieri A., Planas A.M., Ludwig K.U., Buti M., Rahmouni S., Alarcón-Riquelme M.E., Schulte E.C., Franke A., Karlsen T.H., Valenti L., Zeberg H., Richards B., Ganna A., Boada M., de Rojas I., Ruiz A., Sánchez-Juan P., Real L.M., SCOURGE Cohort Group, HOSTAGE Cohort Group, GRA@CE Cohort Group, Guillen-Navarro E., Ayuso C., González-Neira A., Riancho J.A., Rojas-Martinez A., Flores C., Lapunzina P., Carracedo A. Novel genes and sex differences in COVID-19 severity. Human Mol. Gen. 2022:ddac132. doi: 10.1093/hmg/ddac132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt F., Ellinghaus D., Juzenas S., Lerga-Jaso J., Wendorff M., Maya-Miles D., Uellendahl-Werth F., ElAbd H., Rühlemann M.C., Arora J., Özer O., Lenning O.B., Myhre R., Vadla M.S., Wacker E.M., Wienbrandt L., Blandino Ortiz A., de Salazar A., Garrido Chercoles A., Palom A., Ruiz A., Garcia-Fernandez A.-E., Blanco-Grau A., Mantovani A., Zanella A., Holten A.R., Mayer A., Bandera A., Cherubini A., Protti A., Aghemo A., Gerussi A., Ramirez A., Braun A., Nebel A., Barreira A., Lleo A., Teles A., Kildal A.B., Biondi A., Caballero-Garralda A., Ganna A., Gori A., Glück A., Lind A., Tanck A., Hinney A., Carreras Nolla A., Fracanzani A.L., Peschuck A., Cavallero A., Dyrhol-Riise A.M., Ruello A., Julià A., Muscatello A., Pesenti A., Voza A., Rando-Segura A., Solier A., Schmidt A., Cortes B., Mateos B., Nafria-Jimenez B., Schaefer B., Jensen B., Bellinghausen C., Maj C., Ferrando C., de la Horra C., Quereda C., Skurk C., Thibeault C., Scollo C., Herr C., Spinner C.D., Gassner C., Lange C., Hu C., Paccapelo C., Lehmann C., Angelini C., Cappadona C., Azuure C., COVICAT study group, A.S. (COVAS), Bianco C., Cea C., Sancho C., Hoff D.A.L., Galimberti D., Prati D., Haschka D., Jiménez D., Pestaña D., Toapanta D., Muñiz-Diaz E., Azzolini E., Sandoval E., Binatti E., Scarpini E., Helbig E.T., Casalone E., Urrechaga E., Paraboschi E.M., Pontali E., Reverter E., Calderón E.J., Navas E., Solligård E., Contro E., Arana-Arri E., Aziz F., Garcia F., García Sánchez F., Ceriotti F., Martinelli-Boneschi F., Peyvandi F., Kurth F., Blasi F., Malvestiti F., Medrano F.J., Mesonero F., Rodriguez-Frias F., Hanses F., Müller F., Hemmrich-Stanisak G., Bellani G., Grasselli G., Pezzoli G., Costantino G., Albano G., Cardamone G., Bellelli G., Citerio G., Foti G., Lamorte G., Matullo G., Baselli G., Kurihara H., Neb H., My I., Kurth I., Hernández I., Pink I., de Rojas I., Galván-Femenia I., Holter J.C., Afset J.E., Heyckendorf J., Kässens J., Damås J.K., Rybniker J., Altmüller J., Ampuero J., Martín J., Erdmann J., Banales J.M., Badia J.R., Dopazo J., Schneider J., Bergan J., Barretina J., Walter J., Hernández Quero J., Goikoetxea J., Delgado J., Guerrero J.M., Fazaal J., Kraft J., Schröder J., Risnes K., Banasik K., Müller K.E., Gaede K.I., Garcia-Etxebarria K., Tonby K., Heggelund L., Izquierdo-Sanchez L., Bettini L.R., Sumoy L., Sander L.E., Lippert L.J., Terranova L., Nkambule L., Knopp L., Gustad L.T., Garbarino L., Santoro L., Téllez L., Roade L., Ostadreza M., Intxausti M., Kogevinas M., Riveiro-Barciela M., Berger M.M., Schaefer M., Niemi M.E.K., Gutiérrez-Stampa M.A., Carrabba M., Figuera Basso M.E., Valsecchi M.G., Hernandez-Tejero M., Vehreschild M.J.G.T., Manunta M., Acosta-Herrera M., D’Angiò M., Baldini M., Cazzaniga M., Grimsrud M.M., Cornberg M., Nöthen M.M., Marquié M., Castoldi M., Cordioli M., Cecconi M., D’Amato M., Augustin M., Tomasi M., Boada M., Dreher M., Seilmaier M.J., Joannidis M., Wittig M., Mazzocco M., Ciccarelli M., Rodríguez-Gandía M., Bocciolone M., Miozzo M., Imaz Ayo N., Blay N., Chueca N., Montano N., Braun N., Ludwig N., Marx N., Martínez N., Norwegian SARS-CoV-2 Study group, Cornely O.A., Witzke O., Palmieri O., Pa Study Group, Faverio P., Preatoni P., Bonfanti P., Omodei P., Tentorio P., Castro P., Rodrigues P.M., España P.P., Hoffmann P., Rosenstiel P., Schommers P., Suwalski P., de Pablo R., Ferrer R., Bals R., Gualtierotti R., Gallego-Durán R., Nieto R., Carpani R., Morilla R., Badalamenti S., Haider S., Ciesek S., May S., Bombace S., Marsal S., Pigazzini S., Klein S., Pelusi S., Wilfling S., Bosari S., Volland S., Brunak S., Raychaudhuri S., Schreiber S., Heilmann-Heimbach S., Aliberti S., Ripke S., Dudman S., Wesse T., Zheng T., The STORM Study group, T.H.T.F., The Humanitas Gavazzeni Task Force, Bahmer T., Eggermann T., Illig T., Brenner T., Pumarola T., Feldt T., Folseraas T., Gonzalez Cejudo T., Landmesser U., Protzer U., Hehr U., Rimoldi V., Monzani V., Skogen V., Keitel V., Kopfnagel V., Friaza V., Andrade V., Moreno V., Albrecht W., Peter W., Poller W., Farre X., Yi X., Wang X., Khodamoradi Y., Karadeniz Z., Latiano A., Goerg S., Bacher P., Koehler P., Tran F., Zoller H., Schulte E.C., Heidecker B., Ludwig K.U., Fernández J., Romero-Gómez M., Albillos A., Invernizzi P., Buti M., Duga S., Bujanda L., Hov J.R., Lenz T.L., Asselta R., de Cid R., Valenti L., Karlsen T.H., Cáceres M., Franke A. Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Human Molecular Genetics. 2022:ddac158. doi: 10.1093/hmg/ddac158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Schmidt Cernohorska M., Huranova M., Slade D., Dammermann A. Cep97 is required for centriole structural integrity and cilia formation in Drosophila. Curr. Biol. 2020;30:3045–3056.e7. doi: 10.1016/j.cub.2020.05.078. [DOI] [PubMed] [Google Scholar]
- Downes D.J., Cross A.R., Hua P., Roberts N., Schwessinger R., Cutler A.J., Munis A.M., Brown J., Mielczarek O., de Andrea C.E., Melero I., Gill D.R., Hyde S.C., Knight J.C., Todd J.A., Sansom S.N., Issa F., Davies J.O.J., Hughes J.R. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat. Genet. 2021;53:1606–1615. doi: 10.1038/s41588-021-00955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Jostins L., Spain S.L., Cortes A., Bethune J., Han B., Park Y.R., Raychaudhuri S., Pouget J.G., Hübenthal M., Folseraas T., Wang Y., Esko T., Metspalu A., Westra H.-J., Franke L., Pers T.H., Weersma R.K., Collij V., D’Amato M., Halfvarson J., Jensen A.B., Lieb W., Degenhardt F., Forstner A.J., Hofmann A., Schreiber S., Mrowietz U., Juran B.D., Lazaridis K.N., Brunak S., Dale A.M., Trembath R.C., Weidinger S., Weichenthal M., Ellinghaus E., Elder J.T., Barker J.N.W.N., Andreassen O.A., McGovern D.P., Karlsen T.H., Barrett J.C., Parkes M., Brown M.A., Franke A. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallerini C., Picchiotti N., Baldassarri M., Zguro K., Daga S., Fava F., Benetti E., Amitrano S., Bruttini M., Palmieri M., Croci S., Lista M., Beligni G., Valentino F., Meloni I., Tanfoni M., Minnai F., Colombo F., Cabri E., Fratelli M., Gabbi C., Mantovani S., Frullanti E., Gori M., Crawley F.P., Butler-Laporte G., Richards B., Zeberg H., Lipcsey M., Hultström M., Ludwig K.U., Schulte E.C., Pairo-Castineira E., Baillie J.K., Schmidt A., Frithiof R., WES/WGS Working Group Within the HGI, GenOMICC Consortium, GEN-COVID Multicenter Study, Mari F., Renieri A., Furini S. Common, low-frequency, rare, and ultra-rare coding variants contribute to COVID-19 severity. Hum. Genet. 2022;141:147–173. doi: 10.1007/s00439-021-02397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R.C., Pan-Hammarström Q., Graham R.R., Gateva V., Fontán G., Lee A.T., Ortmann W., Urcelay E., Fernández-Arquero M., Núñez C., Jorgensen G., Ludviksson B.R., Koskinen S., Haimila K., Clark H.F., Klareskog L., Gregersen P.K., Behrens T.W., Hammarström L. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat. Genet. 2010;42:777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- Fingerlin T.E., Murphy E., Zhang W., Peljto A.L., Brown K.K., Steele M.P., Loyd J.E., Cosgrove G.P., Lynch D., Groshong S., Collard H.R., Wolters P.J., Bradford W.Z., Kossen K., Seiwert S.D., du Bois R.M., Garcia C.K., Devine M.S., Gudmundsson G., Isaksson H.J., Kaminski N., Zhang Y., Gibson K.F., Lancaster L.H., Cogan J.D., Mason W.R., Maher T.M., Molyneaux P.L., Wells A.U., Moffatt M.F., Selman M., Pardo A., Kim D.S., Crapo J.D., Make B.J., Regan E.A., Walek D.S., Daniel J.J., Kamatani Y., Zelenika D., Smith K., McKean D., Pedersen B.S., Talbert J., Kidd R.N., Markin C.R., Beckman K.B., Lathrop M., Schwarz M.I., Schwartz D.A. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat. Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Piernas C., Astbury N.M., Hippisley-Cox J., O’Rahilly S., Aveyard P., Jebb S.A. Associations between body-mass index and COVID-19 severity in 6·9 million people in England: a prospective, community-based, cohort study. The Lancet Diabetes & Endocrinology. 2021;9:350–359. doi: 10.1016/S2213-8587(21)00089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziano L., Giambartolomei C., Pereira A.C., Gaulton A., Posner D.C., Swanson S.A., Ho Y.-L., Iyengar S.K., Kosik N.M., Vujkovic M., Gagnon D.R., Bento A.P., Barrio-Hernandez I., Rönnblom L., Hagberg N., Lundtoft C., Langenberg C., Pietzner M., Valentine D., Gustincich S., Tartaglia G.G., Allara E., Surendran P., Burgess S., Zhao J.H., Peters J.E., Prins B.P., Angelantonio E.D., Devineni P., Shi Y., Lynch K.E., DuVall S.L., Garcon H., Thomann L.O., Zhou J.J., Gorman B.R., Huffman J.E., O’Donnell C.J., Tsao P.S., Beckham J.C., Pyarajan S., Muralidhar S., Huang G.D., Ramoni R., Beltrao P., Danesh J., Hung A.M., Chang K.-M., Sun Y.V., Joseph J., Leach A.R., Edwards T.L., Cho K., Gaziano J.M., Butterworth A.S., Casas J.P. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat. Med. 2021;27:668–676. doi: 10.1038/s41591-021-01310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozzo L., Longo L., Vitale D.C., Drago F. Dexamethasone treatment for Covid-19, a curious precedent highlighting a regulatory gap. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.621934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb H., Benamar M., Lai P.S., Contini P., Griffith J.W., Crestani E., Schmitz-Abe K., Chen Q., Fong J., Marri L., Filaci G., Del Zotto G., Pishesha N., Kolifrath S., Broggi A., Ghosh S., Gelmez M.Y., Oktelik F.B., Cetin E.A., Kiykim A., Kose M., Wang Z., Cui Y., Yu X.G., Li J.Z., Berra L., Stephen-Victor E., Charbonnier L.-M., Zanoni I., Ploegh H., Deniz G., De Palma R., Chatila T.A. Notch4 signaling limits regulatory T-cell-mediated tissue repair and promotes severe lung inflammation in viral infections. Immunity. 2021;54:1186–1199.e7. doi: 10.1016/j.immuni.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth L.R., Sharif H., Griswold A.R., Fontana P., Mintseris J., Dagbay K.B., Paulo J.A., Gygi S.P., Bachovchin D.A., Wu H. DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature. 2021;592:778–783. doi: 10.1038/s41586-021-03350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.J., Kim K.E., Park S.Y., Bok J., Seo J.T., Moon S.J. Differential roles of tubby family proteins in ciliary formation and trafficking. Mol Cells. 2021;44:591–601. doi: 10.14348/molcells.2021.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz J.E., Kosmicki J.A., Damask A., Sharma D., Roberts G.H.L., Justice A.E., Banerjee N., Coignet M.V., Yadav A., Leader J.B., Marcketta A., Park D.S., Lanche R., Maxwell E., Knight S.C., Bai X., Guturu H., Sun D., Baltzell A., Kury F.S.P., Backman J.D., Girshick A.R., O’Dushlaine C., McCurdy S.R., Partha R., Mansfield A.J., Turissini D.A., Li A.H., Zhang M., Mbatchou J., Watanabe K., Gurski L., McCarthy S.E., Kang H.M., Dobbyn L., Stahl E., Verma A., Sirugo G., Ritchie M.D., Jones M., Balasubramanian S., Siminovitch K., Salerno W.J., Shuldiner A.R., Rader D.J., Mirshahi T., Locke A.E., Marchini J., Overton J.D., Carey D.J., Habegger L., Cantor M.N., Rand K.A., Hong E.L., Reid J.G., Ball C.A., Baras A., Abecasis G.R., Ferreira M.A.R. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat. Genet. 2022;1–11 doi: 10.1038/s41588-021-01006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.-L., Chou Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. PNAS. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Gauthier L.D., Brand H., Solomonson M., Watts N.A., Rhodes D., Singer-Berk M., England E.M., Seaby E.G., Kosmicki J.A., Walters R.K., Tashman K., Farjoun Y., Banks E., Poterba T., Wang A., Seed C., Whiffin N., Chong J.X., Samocha K.E., Pierce-Hoffman E., Zappala Z., O’Donnell-Luria A.H., Minikel E.V., Weisburd B., Lek M., Ware J.S., Vittal C., Armean I.M., Bergelson L., Cibulskis K., Connolly K.M., Covarrubias M., Donnelly S., Ferriera S., Gabriel S., Gentry J., Gupta N., Jeandet T., Kaplan D., Llanwarne C., Munshi R., Novod S., Petrillo N., Roazen D., Ruano-Rubio V., Saltzman A., Schleicher M., Soto J., Tibbetts K., Tolonen C., Wade G., Talkowski M.E., Neale B.M., Daly M.J., MacArthur D.G. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmicki J.A., Horowitz J.E., Banerjee N., Lanche R., Marcketta A., Maxwell E., Bai X., Sun D., Backman J.D., Sharma D., Kury F.S.P., Kang H.M., O’Dushlaine C., Yadav A., Mansfield A.J., Li A.H., Watanabe K., Gurski L., McCarthy S.E., Locke A.E., Khalid S., O’Keeffe S., Mbatchou J., Chazara O., Huang Y., Kvikstad E., O’Neill A., Nioi P., Parker M.M., Petrovski S., Runz H., Szustakowski J.D., Wang Q., Wong E., Cordova-Palomera A., Smith E.N., Szalma S., Zheng X., Esmaeeli S., Davis J.W., Lai Y.-P., Chen X., Justice A.E., Leader J.B., Mirshahi T., Carey D.J., Verma A., Sirugo G., Ritchie M.D., Rader D.J., Povysil G., Goldstein D.B., Kiryluk K., Pairo-Castineira E., Rawlik K., Pasko D., Walker S., Meynert A., Kousathanas A., Moutsianas L., Tenesa A., Caulfield M., Scott R., Wilson J.F., Baillie J.K., Butler-Laporte G., Nakanishi T., Lathrop M., Richards J.B., Jones M., Balasubramanian S., Salerno W., Shuldiner A.R., Marchini J., Overton J.D., Habegger L., Cantor M.N., Reid J.G., Baras A., Abecasis G.R., Ferreira M.A.R. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am. J. Hum. Genet. 2021;108:1350–1355. doi: 10.1016/j.ajhg.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Haagmans B.L. SARS-CoV-2 pathogenesis. Nat Rev Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee Y., Choi Y., Son A., Park Y., Lee K.-M., Kim J., Kim J.-S., Kim V.N. The SARS-CoV-2 RNA interactome. Mol. Cell. 2021;81:2838–2850.e6. doi: 10.1016/j.molcel.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Tukiainen T., Birnbaum D.P., Kosmicki J.A., Duncan L.E., Estrada K., Zhao F., Zou J., Pierce-Hoffman E., Berghout J., Cooper D.N., Deflaux N., DePristo M., Do R., Flannick J., Fromer M., Gauthier L., Goldstein J., Gupta N., Howrigan D., Kiezun A., Kurki M.I., Moonshine A.L., Natarajan P., Orozco L., Peloso G.M., Poplin R., Rivas M.A., Ruano-Rubio V., Rose S.A., Ruderfer D.M., Shakir K., Stenson P.D., Stevens C., Thomas B.P., Tiao G., Tusie-Luna M.T., Weisburd B., Won H.-H., Yu D., Altshuler D.M., Ardissino D., Boehnke M., Danesh J., Donnelly S., Elosua R., Florez J.C., Gabriel S.B., Getz G., Glatt S.J., Hultman C.M., Kathiresan S., Laakso M., McCarroll S., McCarthy M.I., McGovern D., McPherson R., Neale B.M., Palotie A., Purcell S.M., Saleheen D., Scharf J.M., Sklar P., Sullivan P.F., Tuomilehto J., Tsuang M.T., Watkins H.C., Wilson J.G., Daly M.J., MacArthur D.G. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaoult J., Yan W.-H. Editorial: the biological and clinical aspects of HLA-G. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.649344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang Y., Zhang Y., Lu M.M., DeMayo F.J., Dekker J.D., Tucker P.W., Morrisey E.E. Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development. 2012;139:2500–2509. doi: 10.1242/dev.079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström S., Wang L., Smith E.N., Gordon W., van Hylckama Vlieg A., de Andrade M., Brody J.A., Pattee J.W., Haessler J., Brumpton B.M., Chasman D.I., Suchon P., Chen M.-H., Turman C., Germain M., Wiggins K.L., MacDonald J., Braekkan S.K., Armasu S.M., Pankratz N., Jackson R.D., Nielsen J.B., Giulianini F., Puurunen M.K., Ibrahim M., Heckbert S.R., Damrauer S.M., Natarajan P., Klarin D., Program Million Veteran, de Vries P.S., Sabater-Lleal M., Huffman J.E., CHARGE Hemostasis Working Group, Bammler T.K., Frazer K.A., McCauley B.M., Taylor K., Pankow J.S., Reiner A.P., Gabrielsen M.E., Deleuze J.-F., O’Donnell C.J., Kim J., McKnight B., Kraft P., Hansen J.-B., Rosendaal F.R., Heit J.A., Psaty B.M., Tang W., Kooperberg C., Hveem K., Ridker P.M., Morange P.-E., Johnson A.D., Kabrhel C., Trégouët D.-A., Smith N.L. Genomic and transcriptomic association studies identify 16 novel susceptibility loci for venous thromboembolism. Blood. 2019;134:1645–1657. doi: 10.1182/blood.2019000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhang T., Ma L., Zhang H., Wang H., Wei W., Pei H., Li H. The impact of ABO blood group on COVID-19 infection risk and mortality: a systematic review and meta-analysis. Blood Rev. 2021;48 doi: 10.1016/j.blre.2020.100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee M., Beilstein F., Labetoulle M., Rixon F.J., Pasdeloup D. Dystonin/BPAG1 promotes plus-end-directed transport of herpes simplex virus 1 capsids on microtubules during entry. J. Virol. 2013;87:11008–11018. doi: 10.1128/JVI.01633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naitza S., Porcu E., Steri M., Taub D.D., Mulas A., Xiao X., Strait J., Dei M., Lai S., Busonero F., Maschio A., Usala G., Zoledziewska M., Sidore C., Zara I., Pitzalis M., Loi A., Virdis F., Piras R., Deidda F., Whalen M.B., Crisponi L., Concas A., Podda C., Uzzau S., Scheet P., Longo D.L., Lakatta E., Abecasis G.R., Cao A., Schlessinger D., Uda M., Sanna S., Cucca F. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T., Pigazzini S., Degenhardt F., Cordioli M., Butler-Laporte G., Maya-Miles D., Bujanda L., Bouysran Y., Niemi M.E., Palom A., Ellinghaus D., Khan A., Martínez-Bueno M., Rolker S., Amitrano S., Roade Tato L., Fava F., FinnGen, COVID-19 Host Genetics Initiative (HGI), Spinner C.D., Prati D., Bernardo D., Garcia F., Darcis G., Fernández-Cadenas I., Holter J.C., Banales J.M., Frithiof R., Kiryluk K., Duga S., Asselta R., Pereira A.C., Romero-Gómez M., Nafría-Jiménez B., Hov J.R., Migeotte I., Renieri A., Planas A.M., Ludwig K.U., Buti M., Rahmouni S., Alarcón-Riquelme M.E., Schulte E.C., Franke A., Karlsen T.H., Valenti L., Zeberg H., Richards J.B., Ganna A. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. J. Clin. Invest. 2021;131 doi: 10.1172/JCI152386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkoong H., Edahiro R., Takano T., Nishihara H., Shirai Y., Sonehara K., Tanaka H., Azekawa S., Mikami Y., Lee H., Hasegawa T., Okudela K., Okuzaki D., Motooka D., Kanai Masahiro, Naito Tatsuhiko, Yamamoto K., Wang Q.S., Saiki R., Ishihara R., Matsubara Y., Hamamoto J., Hayashi H., Yoshimura Y., Tachikawa N., Yanagita E., Hyugaji T., Shimizu E., Katayama Kotoe, Kato Y., Morita T., Takahashi Kazuhisa, Harada N., Naito Toshio, Hiki M., Matsushita Y., Takagi H., Aoki R., Nakamura A., Harada S., Sasano Hitoshi, Kabata H., Masaki K., Kamata H., Ikemura S., Chubachi S., Okamori S., Terai H., Morita A., Asakura T., Sasaki J., Morisaki H., Uwamino Y., Nanki K., Uchida S., Uno S., Nishimura T., Ishiguro T., Isono T., Shibata S., Matsui Y., Hosoda C., Takano K., Nishida T., Kobayashi Y., Takaku Y., Takayanagi N., Ueda Soichiro, Tada A., Miyawaki M., Yamamoto Masaomi, Yoshida E., Hayashi R., Nagasaka T., Arai S., Kaneko Y., Sasaki K., Tagaya E., Kawana M., Arimura K., Takahashi Kunihiko, Anzai T., Ito S., Endo A., Uchimura Y., Miyazaki Yasunari, Honda T., Tateishi T., Tohda S., Ichimura N., Sonobe K., Sassa C.T., Nakajima J., Nakano Y., Nakajima Y., Anan R., Arai R., Kurihara Y., Harada Y., Nishio K., Ueda T., Azuma M., Saito R., Sado T., Miyazaki Yoshimune, Sato R., Haruta Y., Nagasaki T., Yasui Y., Hasegawa Y., Mutoh Y., Kimura Tomoki, Sato Tomonori, Takei R., Hagimoto S., Noguchi Y., Yamano Y., Sasano Hajime, Ota S., Nakamori Y., Yoshiya K., Saito Fukuki, Yoshihara T., Wada D., Iwamura H., Kanayama S., Maruyama S., Yoshiyama T., Ohta Ken, Kokuto H., Ogata H., Tanaka Y., Arakawa K., Shimoda M., Osawa T., Tateno H., Hase I., Yoshida Shuichi, Suzuki S., Kawada M., Horinouchi H., Saito Fumitake, Mitamura K., Hagihara M., Ochi J., Uchida T., Baba R., Arai D., Ogura Takayuki, Takahashi Hidenori, Hagiwara S., Nagao G., Konishi S., Nakachi I., Murakami K., Yamada M., Sugiura H., Sano H., Matsumoto S., Kimura N., Ono Y., Baba H., Suzuki Y., Nakayama S., Masuzawa K., Namba S., Suzuki K., Naito Y., Liu Y.-C., Takuwa A., Sugihara F., Wing J.B., Sakakibara S., Hizawa N., Shiroyama T., Miyawaki S., Kawamura Y., Nakayama A., Matsuo H., Maeda Y., Nii T., Noda Y., Niitsu T., Adachi Y., Enomoto T., Amiya S., Hara R., Yamaguchi Y., Murakami T., Kuge T., Matsumoto Kinnosuke, Yamamoto Y., Yamamoto Makoto, Yoneda M., Kishikawa T., Yamada S., Kawabata S., Kijima N., Takagaki M., Sasa N., Ueno Y., Suzuki M., Takemoto N., Eguchi H., Fukusumi T., Imai T., Fukushima M., Kishima H., Inohara H., Tomono K., Kato K., Takahashi Meiko, Matsuda F., Hirata H., Takeda Y., Koh H., Manabe T., Funatsu Y., Ito F., Fukui T., Shinozuka K., Kohashi S., Miyazaki M., Shoko T., Kojima M., Adachi T., Ishikawa M., Takahashi Kenichiro, Inoue Takashi, Hirano T., Kobayashi K., Takaoka H., Watanabe K., Miyazawa N., Kimura Y., Sado R., Sugimoto H., Kamiya A., Kuwahara N., Fujiwara A., Matsunaga T., Sato Yoko, Okada T., Hirai Y., Kawashima H., Narita A., Niwa K., Sekikawa Y., Nishi K., Nishitsuji M., Tani M., Suzuki J., Nakatsumi H., Ogura Takashi, Kitamura H., Hagiwara E., Murohashi K., Okabayashi H., Mochimaru T., Nukaga S., Satomi R., Oyamada Y., Mori N., Baba T., Fukui Yasutaka, Odate M., Mashimo S., Makino Y., Yagi K., Hashiguchi M., Kagyo J., Shiomi T., Fuke S., Saito H., Tsuchida T., Fujitani S., Takita M., Morikawa D., Yoshida T., Izumo T., Inomata M., Kuse N., Awano N., Tone M., Ito A., Nakamura Y., Hoshino K., Maruyama J., Ishikura H., Takata T., Odani T., Amishima M., Hattori T., Shichinohe Y., Kagaya T., Kita T., Ohta Kazuhide, Sakagami S., Koshida K., Hayashi K., Shimizu T., Kozu Y., Hiranuma H., Gon Y., Izumi N., Nagata K., Ueda K., Taki R., Hanada S., Kawamura K., Ichikado K., Nishiyama Kenta, Muranaka H., Nakamura K., Hashimoto N., Wakahara K., Sakamoto K., Omote N., Ando A., Kodama N., Kaneyama Y., Maeda S., Kuraki T., Matsumoto T., Yokote K., Nakada T.-A., Abe R., Oshima T., Shimada T., Harada M., Takahashi T., Ono H., Sakurai T., Shibusawa T., Kimizuka Y., Kawana A., Sano T., Watanabe C., Suematsu R., Sageshima H., Yoshifuji A., Ito K., Takahashi S., Ishioka K., Nakamura M., Masuda M., Wakabayashi A., Watanabe Hiroki, Ueda Suguru, Nishikawa M., Chihara Y., Takeuchi M., Onoi K., Shinozuka J., Sueyoshi A., Nagasaki Y., Okamoto M., Ishihara Sayoko, Shimo M., Tokunaga Y., Kusaka Y., Ohba T., Isogai S., Ogawa A., Inoue Takuya, Fukuyama S., Eriguchi Y., Yonekawa A., Kan-o K., Matsumoto Koichiro, Kanaoka K., Ihara S., Komuta K., Inoue Y., Chiba S., Yamagata K., Hiramatsu Y., Kai H., Asano K., Oguma T., Ito Y., Hashimoto S., Yamasaki M., Kasamatsu Y., Komase Y., Hida N., Tsuburai T., Oyama B., Takada M., Kanda H., Kitagawa Yuichiro, Fukuta T., Miyake T., Yoshida Shozo, Ogura S., Abe S., Kono Y., Togashi Y., Takoi H., Kikuchi R., Ogawa Shinichi, Ogata T., Ishihara Shoichiro, Kanehiro A., Ozaki S., Fuchimoto Y., Wada S., Fujimoto N., Nishiyama Kei, Terashima M., Beppu S., Yoshida K., Narumoto O., Nagai H., Ooshima N., Motegi M., Umeda A., Miyagawa K., Shimada H., Endo M., Ohira Y., Watanabe M., Inoue S., Igarashi A., Sato M., Sagara H., Tanaka A., Ohta S., Kimura Tomoyuki, Shibata Y., Tanino Y., Nikaido T., Minemura H., Sato Yuki, Yamada Y., Hashino T., Shinoki M., Iwagoe H., Takahashi Hiroshi, Fujii K., Kishi H., Kanai Masayuki, Imamura T., Yamashita T., Yatomi M., Maeno T., Hayashi S., Takahashi Mai, Kuramochi M., Kamimaki I., Tominaga Y., Ishii T., Utsugi M., Ono A., Tanaka T., Kashiwada T., Fujita K., Saito Y., Seike M., Watanabe Hiroko, Matsuse H., Kodaka N., Nakano C., Oshio T., Hirouchi T., Makino S., Egi M., Omae Y., Nannya Y., Ueno T., Katayama Kazuhiko, Ai M., Fukui Yoshinori, Kumanogoh A., Sato Toshiro, Hasegawa N., Tokunaga K., Ishii M., Koike R., Kitagawa Yuko, Kimura A., Imoto S., Miyano S., Ogawa Seishi, Kanai T., Fukunaga K., Okada Y. DOCK2 is involved in the host genetics and biology of severe COVID-19. Nature. 2022:1–11. doi: 10.1038/s41586-022-05163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann G., Weber K.S.C., Zernecke A., Schröder A., Weber C. JAM-1 is a ligand of the β2 integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 2002;3:151–158. doi: 10.1038/ni755. [DOI] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., Furniss J., Richmond A., Gountouna E., Wrobel N., Harrison D., Wang B., Wu Y., Meynert A., Griffiths F., Oosthuyzen W., Kousathanas A., Moutsianas L., Yang Z., Zhai R., Zheng C., Grimes G., Beale R., Millar J., Shih B., Keating S., Zechner M., Haley C., Porteous D.J., Hayward C., Yang J., Knight J., Summers C., Shankar-Hari M., Klenerman P., Turtle L., Ho A., Moore S.C., Hinds C., Horby P., Nichol A., Maslove D., Ling L., McAuley D., Montgomery H., Walsh T., Pereira A.C., Renieri A., Shen X., Ponting C.P., Fawkes A., Tenesa A., Caulfield M., Scott R., Rowan K., Murphy L., Openshaw P.J.M., Semple M.G., Law A., Vitart V., Wilson J.F., Baillie J.K. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Parkinson N., Rodgers N., Head Fourman M., Wang B., Zechner M., Swets M.C., Millar J.E., Law A., Russell C.D., Baillie J.K., Clohisey S. Dynamic data-driven meta-analysis for prioritisation of host genes implicated in COVID-19. Sci. Rep. 2020;10:22303. doi: 10.1038/s41598-020-79033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G. A kinesin in command of primary Ciliogenesis. Cell. 2011;145:817–819. doi: 10.1016/j.cell.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Pendu J.L., Breiman A., Rocher J., Dion M., Ruvoën-Clouet N. ABO blood types and COVID-19: spurious, anecdotal, or truly important relationships? A reasoned review of available data. Viruses. 2021;13:160. doi: 10.3390/v13020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A.C., Bes T.M., Velho M., Marques E., Jannes C.E., Valino K.R., Dinardo C.L., Costa S.F., Duarte A.J.S., Santos A.R., Mitne-Neto M., Medina-Pestana J., Krieger J.E. Genetic risk factors and COVID-19 severity in Brazil: results from BRACOVID study. Hum. Mol. Genet. 2022;31:3021–3031. doi: 10.1093/hmg/ddac045. [DOI] [PubMed] [Google Scholar]
- Prohaska A., Racimo F., Schork A.J., Sikora M., Stern A.J., Ilardo M., Allentoft M.E., Folkersen L., Buil A., Moreno-Mayar J.V., Korneliussen T., Geschwind D., Ingason A., Werge T., Nielsen R., Willerslev E. Human disease variation in the light of population genomics. Cell. 2019;177:115–131. doi: 10.1016/j.cell.2019.01.052. [DOI] [PubMed] [Google Scholar]
- Qureshi R., Picon-Ruiz M., Aurrekoetxea-Rodriguez I., Nunes de Paiva V., D’Amico M., Yoon H., Radhakrishnan R., Morata-Tarifa C., Ince T., Lippman M.E., Thaller S.R., Rodgers S.E., Kesmodel S., Vivanco M.D.M., Slingerland J.M. The major pre- and postmenopausal estrogens play opposing roles in obesity-driven mammary inflammation and breast Cancer development. Cell Metab. 2020;31:1154–1172.e9. doi: 10.1016/j.cmet.2020.05.008. [DOI] [PubMed] [Google Scholar]
- Reusch N., De Domenico E., Bonaguro L., Schulte-Schrepping J., Baßler K., Schultze J.L., Aschenbrenner A.C. Neutrophils in COVID-19. Front. Immunol. 2021;12:952. doi: 10.3389/fimmu.2021.652470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideau A.P., Gooding C., Simpson P.J., Monie T.P., Lorenz M., Hüttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W.J. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Roberts G.H.L., Partha R., Rhead B., Knight S.C., Park D.S., Coignet M.V., Zhang M., Berkowitz N., Turrisini D.A., Gaddis M., McCurdy S.R., Pavlovic M., Ruiz L., Sass C., Haug Baltzell A.K., Guturu H., Girshick A.R., Ball C.A., Hong E.L., Rand K.A. Expanded COVID-19 phenotype definitions reveal distinct patterns of genetic association and protective effects. Nat. Genet. 2022;54:374–381. doi: 10.1038/s41588-022-01042-x. [DOI] [PubMed] [Google Scholar]
- Robinot R., Hubert M., de Melo G.D., Lazarini F., Bruel T., Smith N., Levallois S., Larrous F., Fernandes J., Gellenoncourt S., Rigaud S., Gorgette O., Thouvenot C., Trébeau C., Mallet A., Duménil G., Gobaa S., Etournay R., Lledo P.-M., Lecuit M., Bourhy H., Duffy D., Michel V., Schwartz O., Chakrabarti L.A. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 2021;12:4354. doi: 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabater-Lleal M., Huffman J.E., de Vries P.S., Marten J., Mastrangelo M.A., Song C., Pankratz N., Ward-Caviness C.K., Yanek L.R., Trompet S., Delgado G.E., Guo X., Bartz T.M., Martinez-Perez A., Germain M., de Haan H.G., Ozel A.B., Polasek O., Smith A.V., Eicher J.D., Reiner A.P., Tang W., Davies N.M., Stott D.J., Rotter J.I., Tofler G.H., Boerwinkle E., de Maat M.P.M., Kleber M.E., Welsh P., Brody J.A., Chen M.-H., Vaidya D., Soria J.M., Suchon P., van Hylckama Vlieg A., Desch K.C., Kolcic I., Joshi P.K., Launer L.J., Harris T.B., Campbell H., Rudan I., Becker D.M., Li J.Z., Rivadeneira F., Uitterlinden A.G., Hofman A., Franco O.H., Cushman M., Psaty B.M., Morange P.-E., McKnight B., Chong M.R., Fernandez-Cadenas I., Rosand J., Lindgren A., INVENT Consortium; MEGASTROKE Consortium of the International Stroke Genetics Consortium (ISGC), Gudnason V., Wilson J.F., Hayward C., Ginsburg D., Fornage M., Rosendaal F.R., Souto J.C., Becker L.C., Jenny N.S., März W., Jukema J.W., Dehghan A., Trégouët D.-A., Morrison A.C., Johnson A.D., O’Donnell C.J., Strachan D.P., Lowenstein C.J., Smith N.L. Genome-wide association Transethnic Meta-analyses identifies novel associations regulating coagulation factor VIII and von Willebrand factor plasma levels. Circulation. 2019;139:620–635. doi: 10.1161/CIRCULATIONAHA.118.034532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J. Defective viral RNA sensing linked to severe COVID-19. Science. 2021;374:535–536. doi: 10.1126/science.abm3921. [DOI] [PubMed] [Google Scholar]
- Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., Sheffield V.C. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and smoothened. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe Covid-19 GWAS Group, Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., Asselta R., Grimsrud M.M., Milani C., Aziz F., Kässens J., May S., Wendorff M., Wienbrandt L., Uellendahl-Werth F., Zheng T., Yi X., de Pablo R., Chercoles A.G., Palom A., Garcia-Fernandez A.-E., Rodriguez-Frias F., Zanella A., Bandera A., Protti A., Aghemo A., Lleo A., Biondi A., Caballero-Garralda A., Gori A., Tanck A., Carreras Nolla A., Latiano A., Fracanzani A.L., Peschuck A., Julià A., Pesenti A., Voza A., Jiménez D., Mateos B., Nafria Jimenez B., Quereda C., Paccapelo C., Gassner C., Angelini C., Cea C., Solier A., Pestaña D., Muñiz-Diaz E., Sandoval E., Paraboschi E.M., Navas E., García Sánchez F., Ceriotti F., Martinelli-Boneschi F., Peyvandi F., Blasi F., Téllez L., Blanco-Grau A., Hemmrich-Stanisak G., Grasselli G., Costantino G., Cardamone G., Foti G., Aneli S., Kurihara H., ElAbd H., My I., Galván-Femenia I., Martín J., Erdmann J., Ferrusquía-Acosta J., Garcia-Etxebarria K., Izquierdo-Sanchez L., Bettini L.R., Sumoy L., Terranova L., Moreira L., Santoro L., Scudeller L., Mesonero F., Roade L., Rühlemann M.C., Schaefer M., Carrabba M., Riveiro-Barciela M., Figuera Basso M.E., Valsecchi M.G., Hernandez-Tejero M., Acosta-Herrera M., D’Angiò M., Baldini M., Cazzaniga M., Schulzky M., Cecconi M., Wittig M., Ciccarelli M., Rodríguez-Gandía M., Bocciolone M., Miozzo M., Montano N., Braun N., Sacchi N., Martínez N., Özer O., Palmieri O., Faverio P., Preatoni P., Bonfanti P., Omodei P., Tentorio P., Castro P., Rodrigues P.M., Blandino Ortiz A., de Cid R., Ferrer R., Gualtierotti R., Nieto R., Goerg S., Badalamenti S., Marsal S., Matullo G., Pelusi S., Juzenas S., Aliberti S., Monzani V., Moreno V., Wesse T., Lenz T.L., Pumarola T., Rimoldi V., Bosari S., Albrecht W., Peter W., Romero-Gómez M., D’Amato M., Duga S., Banales J.M., Hov J.R., Folseraas T., Valenti L., Franke A., Karlsen T.H. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H., Hollingsworth L.R., Griswold A.R., Hsiao J.C., Wang Q., Bachovchin D.A., Wu H. Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Immunity. 2021;54:1392–1404.e10. doi: 10.1016/j.immuni.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Sonmez T., Coker D., Symons A., Esparza-Gordillo J., The 23andMe COVID-19 Team, Chubb A., Fitch A., Kung A., Altman A., Kill A., Tan J., Pollard J., McCreight J., Bielenberg J., Matthews J., Lee J., Tran L., Agee M., Royce M., Tang N., Gandhi P., d’Amore R., Tennen R., Dvorak S., Hadly S., Park S., Morrow T., Le T., Zheng Y., Aslibekyan S., Auton A. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- Shkurnikov M., Nersisyan S., Jankevic T., Galatenko A., Gordeev I., Vechorko V., Tonevitsky A. Association of HLA class I genotypes with severity of coronavirus Disease-19. Front. Immunol. 2021;12:423. doi: 10.3389/fimmu.2021.641900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., Gunn C., Hockett R., Mudumba S., Guihot A., Luyt C.-E., Mayaux J., Beurton A., Fourati S., Bruel T., Schwartz O., Lacorte J.-M., Yssel H., Parizot C., Dorgham K., Charneau P., Amoura Z., Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Li Chenwei, Kang B., Gao G., Li Cheng, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The COVID-19 Host Genetics Initiative The COVID-19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushima Y., Haraguchi M., Yano M. Depletion of TMEM65 leads to oxidative stress, apoptosis, induction of mitochondrial unfolded protein response, and upregulation of mitochondrial protein import receptor TOMM22. Biochem Biophys Rep. 2020;24 doi: 10.1016/j.bbrep.2020.100870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M., Hofste T., Astuti G., Corominas Galbany J., van der Schoot V., van der Hoeven H., Hagmolen Of Ten Have, W, Klijn E., van den Meer C., Fiddelaers J., de Mast Q., Bleeker-Rovers C.P., Joosten L.A.B., Yntema H.G., Gilissen C., Nelen M., van der Meer J.W.M., Brunner H.G., Netea M.G., van de Veerdonk F.L., Hoischen A. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Chiou J., Poirion O.B., Buchanan J., Valdez M.J., Verheyden J.M., Hou X., Kudtarkar P., Narendra S., Newsome J.M., Guo M., Faddah D.A., Zhang K., Young R.E., Barr J., Sajti E., Misra R., Huyck H., Rogers L., Poole C., Whitsett J.A., Pryhuber G., Xu Y., Gaulton K.J., Preissl S., Sun X., NHLBI LungMap Consortium Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife. 2020;9 doi: 10.7554/eLife.62522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Dhindsa R.S., Carss K., Harper A.R., Nag A., Tachmazidou I., Vitsios D., Deevi S.V.V., Mackay A., Muthas D., Hühn M., Monkley S., Olsson H., Wasilewski S., Smith K.R., March R., Platt A., Haefliger C., Petrovski S. Rare variant contribution to human disease in 281,104 UK biobank exomes. Nature. 2021;597:527–532. doi: 10.1038/s41586-021-03855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu C., Yang B., Zhang H., Jiao J., Zhang R., Liu S., Xiao S., Chen Y., Liu B., Ma Y., Duan X., Guo Y., Guo M., Wu B., Wang X., Huang X., Yang H., Gui Y., Fang M., Zhang L., Duo S., Guo X., Li W. SARS-CoV-2 ORF10 impairs cilia by enhancing CUL2ZYG11B activity. J. Cell Biol. 2022;221 doi: 10.1083/jcb.202108015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO The Top 10 Causes of Death. 2022. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-deathhttps://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death [WWW Document]. URL. (accessed 5.11.22)
- Wickenhagen A., Sugrue E., Lytras S., Kuchi S., Noerenberg M., Turnbull M.L., Loney C., Herder V., Allan J., Jarmson I., Cameron-Ruiz N., Varjak M., Pinto R.M., Lee J.Y., Iselin L., Palmalux N., Stewart D.G., Swingler S., Greenwood E.J.D., Crozier T.W.M., Gu Q., Davies E.L., Clohisey S., Wang B., Trindade Maranhão Costa F., Freire Santana M., de Lima Ferreira L.C., Murphy L., Fawkes A., Meynert A., Grimes G., ISARIC4C Investigators, Da Silva Filho J.L., Marti M., Hughes J., Stanton R.J., Wang E.C.Y., Ho A., Davis I., Jarrett R.F., Castello A., Robertson D.L., Semple M.G., Openshaw P.J.M., Palmarini M., Lehner P.J., Baillie J.K., Rihn S.J., Wilson S.J. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021;374:eabj3624. doi: 10.1126/science.abj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Xiao Y., Zhou Y., Zhou Z., Yan W. LncRNA FOXP4-AS1 is activated by PAX5 and promotes the growth of prostate cancer by sequestering miR-3184-5p to upregulate FOXP4. Cell Death Dis. 2019;10:1–14. doi: 10.1038/s41419-019-1699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Peng, Ding L., Li X., Liu S., Cheng F., He Q., Xiao M., Wu Ping, Hou H., Jiang M., Long P., Wang H., Liu Linlin, Qu M., Shi X., Jiang Q., Mo T., Ding W., Fu Y., Han S., Huo X., Zeng Y., Zhou Y., Zhang Q., Ke J., Xu Xi, Ni W., Shao Z., Wang J., Liu P., Li Z., Jin Y., Zheng F., Wang Fang, Liu Lei, Li W., Liu K., Peng R., Xu Xuedan, Lin Y., Gao H., Shi L., Geng Z., Mu X., Yan Y., Wang K., Wu D., Hao X., Cheng S., Qiu G., Guo H., Li K., Chen G., Sun Z., Lin X., Jin X., Wang Feng, Sun C., Wang C. Trans-ethnic genome-wide association study of severe COVID-19. Commun Biol. 2021;4:1–10. doi: 10.1038/s42003-021-02549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Ramaiah A., Garcia G., Hasiakos S., Arumugaswami V., Srikanth S. ORAI1 limits SARS-CoV-2 infection by regulating tonic type I IFN signaling. J. Immunol. 2022;208:74–84. doi: 10.4049/jimmunol.2100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Ye F., Li K., Xu P., Tan W., Feng Q., Rao S. Genome and epigenome editing identify CCR9 and SLC6A20 as target genes at the 3p21.31 locus associated with severe COVID-19. Sig Transduct Target Ther. 2021;6:85. doi: 10.1038/s41392-021-00519-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Pen J.L., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Voyer T.L., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Turki S.A., Hasanato R., Beek D., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Clinicians, C.-S., Clinicians, C., Group, I.C., Group, F.C.C.S., Cohort, C.-C., Biobank, A.U.C.-19, Effort, C.H.G., Group, N.-U.C.I, Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Cobat A., Casanova J.-L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603:587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Sun Y., Huang W., Ye K. Altered blood cell traits underlie a major genetic locus of severe COVID-19. J. Gerontol. A Biol. Sci. Med. Sci. 2021;76:e147–e154. doi: 10.1093/gerona/glab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.-X., Zhou J.-B., Zhu X.-D., Zhou J., Li J., Song Y.-L., Bai C.-X. Impaired self-healing capacity in airway epithelia lacking aquaporin-3. Respir. Physiol. Neurobiol. 2016;233:66–72. doi: 10.1016/j.resp.2016.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this Review was obtained from published articles and public databases.