Abstract
Backgound:
New-onset diabetes (NOD) has been suggested as an early indicator of pancreatic cancer. However, the definition of NOD by American Diabetes Association requires two simultaneous or consecutive elevated glycemic measures. We aimed to apply a machine-learning approach using electronic health records to predict the risk in patients with recent onset hyperglycemia.
Methods:
In this retrospective cohort study, health plan enrollees 50–84 years of age who had an elevated (6.5%+) glycated hemoglobin (HbA1c) tested in January 2010–September 2018 with recent onset hyperglycemia were identified. 102 potential predictors were extracted. Ten imputation datasets were generated to handle missing data. Random survival forests approach was used to develop and validate risk models. Performance was evaluated by c-index, calibration plot, sensitivity, specificity, and positive predictive value (PPV).
Results:
The cohort consisted of 109,266 patients (mean age 63.6 years). The three-year incidence rate was 1.4 (95% CI 1.3–1.6)/1,000 person-years of follow-up. The three models containing age, weight change in one year, HbA1c, and one of the three variables (HbA1c change in one year, HbA1c in the prior 6 months, or HbA1c in the prior 18 months) appeared most often out of the 50 training samples. The c-indexes were in the range of 0.81–0.82. The sensitivity, specificity, PPV in patients who had the top 20% of the predicted risks were 56–60%, 80%, and 2.5–2.6%, respectively.
Conclusions:
Targeting evaluation at the point of recent hyperglycemia based on elevated HbA1c could offer an opportunity to identify pancreatic cancer early and possibly impact survival in cancer patients.
Keywords: risk prediction, pancreatic cancer, machine learning, hyperglycemia
INTRODUCTION
Pancreatic cancer is the third leading cause of cancer-related death in the United States with a 5-year survival of 10%.[1] A factor contributing to the lethal nature of this disease is often the advanced stage at diagnosis. Widespread population-based screening is unlikely to be effective based on the relatively low incidence of pancreatic cancer (13.1 per 100,000 person-years),[1] and is not currently recommended by the United States Preventative Services Task Force.[2] Therefore, targeted screening of high-risk populations represents an important opportunity to impact the natural history of pancreatic cancer.
New-onset diabetes after age 50 has been suggested as an early indicator of pancreatic cancer with estimated 3-year risk from 0.25–1.0%.[3–5] While this represents a much higher risk compared to that of the general population, it is widely recognized that further risk stratification is still needed to identify a suitable high-risk subgroup of patients for targeted screening. Several clinical prediction models have been proposed to further identify a subgroup of patients with new onset diabetes at increased risk for pancreatic cancer,[3,5,6] and a risk-based screening strategy “is likely to be considered an acceptable value”.[7] Recent data indicate that hyperglycemia can precede the diagnosis of diabetes as well as pancreatic cancer by up to 24 and 36 months, respectively.[8] In addition, glycated hemoglobin has recently emerged as the predominant form of glycemic testing for diabetic screening and monitoring across many health systems in the United States.[9] Targeting evaluation at the point of initial hyperglycemia based on elevation in glycated hemoglobin offers an opportunity to identify patients with sufficient lead-time to impact survival for patients with pancreatic cancer.
The aim of the present study was to develop a clinical prediction model for risk of pancreatic cancer in patients with recent onset hyperglycemia. Specifically, we sought to apply a machine-learning approach to risk prediction at the point of elevated glycated hemoglobin that could be broadly applicable across racial/ethnic groups.
MATERIALS AND METHODS
Study design and setting
Utilizing multi-ethnic health plan enrollees of Kaiser Permanente Southern California (KPSC), a retrospective cohort study was conducted. Study setting can be found in the Methods Section of a previous study.[10] The study protocol was approved by the KPSC’s institutional review board.
Cohort Identification and follow-up
First, all elevated (6.5% or higher) HbA1c tested in a KPSC medical facility between 1/1/2010 and 09/30/2018 for patients 50–84 years of age were identified. The elevated HbA1c tests that met the following criteria were excluded.
Evidence of diabetes (Supplemental Digital Content 1) beyond six-months prior to the index date.
History of pancreatic cancer
Not actively enrolled in the KPSC health plan on the test date or in the past 12 months (gaps 45 days or less were allowed)
The selection of index HbA1c threshold was motivated by an earlier study.[10] For patients with multiple qualifying elevated HbA1c tests during the time period, a random one was selected (index lab test), and the date of the index test was referred to as the index date (t0).
For each patient in the cohort, follow-up started on t0 and ended with the earliest of the following events: disenrollment from the health plan, end of the study (December 31, 2018), reached the maximum length of follow-up (3 years), non-PDAC related death, or PDAC diagnosis or death (outcome).
Outcome identification
The primary outcome was defined as the diagnosis of pancreatic ductal adenocarcinoma (PDAC) or death with pancreatic cancer in the 3 years after the index date. PDAC was captured from the KPSC Cancer Registry by using the Tenth Revision of International Classification of Diseases, Clinical Modification (ICD-10-CM) code C25.x and histology codes listed in Supplemental Digital Content 1. The KPSC Cancer Registry is part of the Surveillance, Epidemiology, and End Results (SEER) program. The pancreatic cancer deaths were derived from the linkage with the California State Death Master files and captured using ICD-10-CM codes C25.x. The utilization of the State files allowed the identification of pancreatic cancer cases that were not otherwise captured in the registry.[11]
Patient demographic and clinical features at baseline
A complete list of features included in the analyses is shown in Table 1. The definitions of derived variables can be found in Supplemental Digital Content 2. A flexible R package called ‘missRanger’ was applied to impute the missing values if the frequency of missing for a feature was less than 60%.[12] We used predictive mean matching method[13] with k=5. Laboratory and weight-related measures with 60% or more missingness or change/change rate measures with 80% or more missingness were not included in the model development process. Ten imputed datasets were generated.
Table 1.
Characteristics of study subjects at baseline (n=109,266).
| Patient characteristics | n (%) unless otherwise stated |
|---|---|
| Demographics and life-style characteristics | |
|
| |
| Age, mean (SD) | 63.6 (8.9) |
|
| |
| Sex, female | 56328 (51.6) |
|
| |
| Race/ethnicity | |
| Non-Hispanic white | 36730 (33.6) |
| African American | 14837 (13.6) |
| Hispanic | 37537 (34.3) |
| Asian and Pacific Islanders | 17157 (15.7) |
| Multiple/Other/Unknown | 3005 (2.8) |
|
| |
| Tobacco use ever | |
| Yes | 43481 (39.8) |
| No | 65546 (60.0) |
| Unknown | 239 (0.2) |
|
| |
| Diagnosis of alcohol abuse in the past year | 2217 (2.0) |
|
| |
| Diagnosis of alcohol abuse any time in the past | 5543 (5.1) |
|
| |
| Medical insurance (one or more) | |
| Commercial | 64966 (59.5) |
| Medicare | 38601 (35.3) |
| Medi-CAL and other State programs | 3955 (3.6) |
| Private pay | 28964 (26.5) |
|
| |
| Years since first enrollment, mean (SD) | 21.1 (13.7) |
|
| |
| Family history of pancreatic cancer | 1681 (1.5) |
|
| |
| Weight, median (IQR) | 189.4 (160.9, 223.3) |
| Weight group defined by body mass index (kg/m2) | |
| Underweight (<18.5) | 455 (0.4) |
| Normal weight (18.5–24.9) | 13617 (12.5) |
| Overweight (25–29.9) | 34314 (31.4) |
| Obese (30+) | 60343 (55.2) |
| Unknown | 537 (0.5) |
| Weight change in one year (kg), median (IQR) | 0.9 (−4.0, 5.5) |
| ≤ −6 kg | 6152 (5.6) |
| > −6 & ≤ −4 kg | 5137 (4.7) |
| > −4 & ≤ −2 kg | 10605 (9.7) |
| > −2 & < 2 kg | 43894 (40.2) |
| ≥ 2 & < 4 kg | 14093 (12.9) |
| ≥ 4 kg | 13974 (12.8) |
| Unknown | 15411 (14.1) |
|
| |
| Clinical characteristics | |
|
| |
| Gallstone disorders | 9954 (9.1) |
|
| |
| Acute pancreatitis (ever) | 1,765 (1.6) |
|
| |
| Chronic pancreatitis | 310 (0.3) |
|
| |
| Benign pancreatic disease | 458 (0.4) |
|
| |
| Biliary tract disease | 10991 (10.1) |
|
| |
| Depression | 23297 (21.3) |
|
| |
| Deep vein thrombosis | 1972 (1.8) |
| HbA1c on index date, median (IQR) | 6.6 (6.5, 7.0) |
| HbA1c change in 1y, median (IQR), n=75,674 | 0.3 (0.1, 0.5) |
| HbA1c 6.5% or higher in the past 6m | |
| Yes | 21691 (19.9) |
| No | 9129 (8.4) |
| Unknown | 78446 (71.8) |
| HbA1c 6.5% or higher in the past 18m | |
| Yes | 32309 (29.6) |
| No | 36745 (33.6) |
| Unknown | 40212 (36.8) |
| Alanine transaminase (ALT) in prior 6m, median (IQR), n=87,567 | 26.0 (20.0, 37.0) |
| Alanine transaminase (ALT) change in 1y, median (IQR) | 0.0 (−4.0, 5.0) |
| < 0 IU/L | 14296 (13.1) |
| ≥ 0 | 17522 (16.0) |
| Unknown | 77448 (70.9) |
|
| |
| Alkaline phosphatase (ALP) in prior 6m, median (IQR) | 73.0 (59.0, 90.0) |
| High (> 125 IU/L) | 1696 (1.6) |
| Low/Medium (≤ 125) | 25812 (23.6) |
| Unknown | 81758 (74.8) |
|
| |
| Bilirubin (total) in prior 6m, median (IQR) | 0.7 (0.5, 0.9) |
| Low (≤ 0.1 mg/dL) | 94 (0.1) |
| Medium (≤ 1.0 & > 0.1) | 22993 (21.0) |
| High (> 1.0) | 3966 (3.6) |
| Unknown | 82213 (75.2) |
|
| |
| Hemoglobin (HGB) in prior 6m, median (IQR), g/dl, n=85,589 | 14.0 (13.1, 15.0) |
| Hemoglobin, male (HGB) in prior 6m, median (IQR), n=40,711 | 14.8 (13.9, 15.6) |
| Hemoglobin, female (HGB) in prior 6m, median (IQR), n=44,877 | 14.8 (13.9, 15.6) |
|
| |
| Red blood cell (RBC) counts in prior 6m, median (IQR), million/mm3, n=84,338 | 4.7 (4.4, 5.0) |
|
| |
| Sodium in prior 6m, median (IQR), mEq/L, n=90,914 | 139.0 (137.0, 141.0) |
|
| |
| Symptoms prior to the index date | |
|
| |
| Abdominal pain | |
| Within 6m | 8257 (7.6) |
| 7–12m | 5704 (5.2) |
| 13–23m | 10106 (9.2) |
| More than 24m | 35198 (32.2) |
|
| |
| Chest pain | |
| Within 6m | 5646 (5.2) |
| 7–12m | 3843 (3.5) |
| 13–23m | 7110 (6.5) |
| More than 24m | 30624 (28.0) |
|
| |
| Constipation | |
| Within 6m | 3104 (2.8) |
| 7–12m | 2557 (2.1) |
| 13–23m | 4168 (3.8) |
| More than 24m | 15093 (13.8) |
|
| |
| Diarrhea | |
| Within 6m | 2447 (2.2) |
| 7–12m | 1841 (1.7) |
| 13–23m | 3443 (3.2) |
| More than 24m | 15676 (14.3) |
|
| |
| Itching | |
| Within 6m | 3661 (3.4) |
| 7–12m | 2618 (2.4) |
| 13–23m | 4883 (4.5) |
| More than 24m | 18147 (16.6) |
|
| |
| Jaundice | |
| Within 6m | 53 (0.0) |
| 7–12m | 6 (0.0) |
| 13–23m | 40 (0.0) |
| More than 24m | 164 (0.2) |
|
| |
| Malaise fatigue | |
| Within 6m | 10347 (9.5) |
| 7–12m | 6282 (5.7) |
| 13–23m | 10737 (9.8) |
| More than 24m | 32715 (29.9) |
|
| |
| Melena | |
| Within 6m | 797 (0.7) |
| 7–12m | 609 (0.6) |
| 13–23m | 1210 (1.1) |
| More than 24m | 6673 (6.1) |
|
| |
| Nausea or vomiting | |
| Within 6m | 3842 (3.5) |
| 7–12m | 2636 (2.4) |
| 13–23m | 4943 (4.5) |
| More than 24m | 18553 (17.0) |
Note: Medians were estimated without including unknown values.
Model training and validation
To overcome the limitations of regression-based models that are traditionally used for analysis of time-to-event data, we applied ‘random survival forests’ (RSF), a nonparametric machine learning method [14–16], to pre-select features and train/validate models. To avoid model over-fitting, we applied a cross-validation process by using 50 training and 50 validation datasets. The process of model training and validation can be found in Supplemental Digital Content 3. To increase clinical utility of the risk prediction models, we limited the model training and validation process to patients whose follow-up was at least 90 days.
Performance measures
The discriminative power for each of the winning models was evaluated by c-index, averaged across all the relevant validation datasets. Calibration was assessed by calibration plots.[17]
To estimate sensitivity, specificity, positive predictive value (PPV), and fold increase in risk using the incidence rate in the entire cohort as the reference were examined for patients whose predicted risks were at the top 20, 15, 10, 5 and 2.5%, we restricted the analyses to a subset of patients who had a complete follow up or developed PDAC in 36 months. The results were averaged across the validation datasets for each winning model. The mean area under the curve (AUC) was reported and the AUC curve was plotted for each winning model.
Exploratory analyses (Supplemental Digital Content 4)
Statistical analysis
All the analyses were performed using SAS (Version 9.4 for Unix; SAS Institute, Cary, NC) except for the R packages mentioned previously. All computations and analyses carried out in R were based on R Version 3.6.0 (R Foundation, Vienna, Austria).
RESULTS
Characteristics of the study cohort
Out of a total of 504,801 patients with at least one elevated (6.5%+) glycosylated hemoglobin measure during the study period, 109,266 were eligible (Figure 1). Patient characteristics at baseline are shown in Table 1. In the eligible patients, 33.6% were white, 34.3% were Hispanic and 51.6% were females. Majority (59.5%) were on commercial insurance, and slightly over one-third (35.3%) were on Medicare (Table 1). On the average, the patients were 63.6 years of age and had been with KPSC for 21 years. Tobacco use was common with 39.8% ever smoked. Alcohol abuse was diagnosed in 2.0% of the patients in the past year and 5.1% any time in the past. Only 1.5% had a family history of pancreatic cancer, while 55.2% of the patients were obese and additional 31.4% were overweight. About one-fifth of the patients lost at least 2 kilograms in the year before t0 (index date). The median glycosylated hemoglobin was 6.6% on t0 (index date) and the median increase in the past year was 0.3%.
Fig 1.
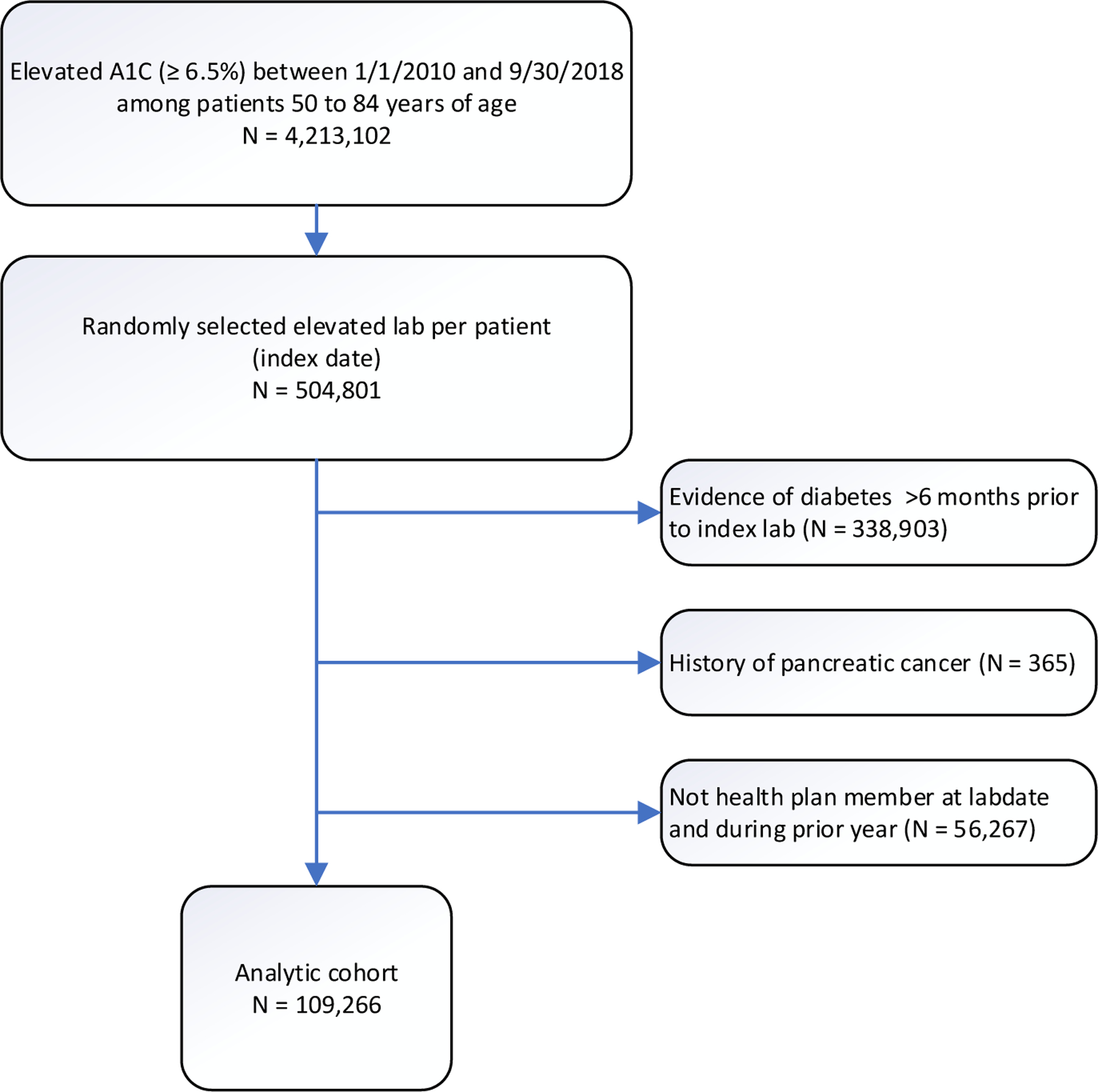
Consort diagram.
Incidence of PDAC
Of the 319 PDAC cases identified in 109,266 eligible patients, 234 (73%) were identified from the Cancer Registry and the rest (85 or 27%) died of pancreatic cancer based on the State death files. 4479 (4.1%) patients died of causes other than pancreatic cancer. The three-year incidence rate was 1.4 (95% CI 1.3–1.6)/1,000 person-years of follow-up time (Table 2). Incidence rates appeared higher in older 2.4 (2.1–2.7) and non-Hispanic white 2.2 (1.8–2.5) patients. A glycated hemoglobin measure at index of 7.5% at or higher increased the incidence rate to 3.2 (2.6–3.9), compared to 1.0 (0.9–1.2)/1,000 person-years in patients whose index glycosylated hemoglobin measure were within 6.5% and 6.9%. In addition, an increase of glycosylated hemoglobin measure of 0.3% or higher in 1 year, or an elevated glycosylated hemoglobin measure in the prior 6 months or 18 months also appeared to increase the incidence rates.
Table 2.
Total follow-up (f/u) time, number and incidence rate of PDAC per 1,000 person-years (PY) and 95% CI (n=109,266).
| Total f/u time (years) | No. of PDAC | Incidence rate of PDAC/1000-PY (95% CI) |
Time to PDAC (days) (median, IQR) |
|
|---|---|---|---|---|
| Total | 224,573 | 319 | 1.4 (1.3, 1.6) | 141 (39, 406) |
| Age group | ||||
| 50–64 | 129,369 | 91 | 0.7 (0.6, 0.9) | 147 (32, 395) |
| 65–84 | 95,204 | 228 | 2.4 (2.1, 2.7) | 137.5 (40, 409) |
| Sex | ||||
| Female | 118,485 | 154 | 1.3 (1.1, 1.5) | 123.5 (35, 385) |
| Male | 106,088 | 165 | 1.6 (1.3, 1.8) | 157 (44, 415) |
| Race/ ethnicity | ||||
| Non-Hispanic white | 76,150 | 164 | 2.2 (1.8, 2.5) | 137.5 (40, 397) |
| African American | 32,105 | 40 | 1.2 (0.9, 1.7) | 114.5 (42, 292) |
| Asian/Pacific Islanders | 36,048 | 33 | 0.9 (0.6, 1.3) | 249 (42, 435) |
| Hispanic | 75,045 | 79 | 1.1 (0.8, 1.3) | 135 (35, 417) |
| HbA1c values on index date | ||||
| 6.5–6.9% | 171,833 | 173 | 1.0 (0.9, 1.2) | 157 (40, 444) |
| 7.0–7.4% | 23,087 | 51 | 2.2 (1.7, 2.9) | 134 (48, 359) |
| ≥7.5% | 29,653 | 95 | 3.2 (2.6, 3.9) | 105 (26, 307) |
| HbA1c change in 1y | ||||
| < 0.3 | 92,307 | 94 | 1.0 (0.8, 1.2) | 250.5 (48, 494) |
| ≥ 0.3 | 62,816 | 137 | 2.2 (1.8, 2.6) | 112 (32, 295) |
| None | 69,450 | 88 | 1.3 (1, 1.6) | 152 (44.5, 415.5) |
| HbA1c values in prior 6m | ||||
| ≥6.5% | 43,899 | 101 | 2.3 (1.9, 2.8) | 135 (46, 404) |
| <6.5% | 19,572 | 29 | 1.5 (1.0, 2.1) | 249 (68, 444) |
| None | 161,102 | 189 | 1.2 (1.0, 1.3) | 135 (34, 399) |
| HbA1c values in prior 18m | ||||
| ≥6.5% | 65,920 | 122 | 1.9 (1.5, 2.2) | 137.5 (38, 404) |
| <6.5% | 76,099 | 91 | 1.2 (1.0, 1.5) | 141 (31, 400) |
| None | 82,553 | 106 | 1.3 (1.1, 1.5) | 152 (48, 414) |
Winning models
The preselection process identified 24–29 potential predictors from the 10 imputed datasets. Age, weight change in 1 year and HbA1c change in 1 year formed the initial predictor set. Out of the 50 training datasets, the models with HbA1c change rate, HbA1c in prior 6 months and HbA1c in prior 18 months as the fourth predictor appeared 20, 8 and 6 times, respectively (Table 3).
Table 3.
Predictors selected to form prediction models by at least one of the 50 training datasets on top of the initial model*, frequency of each prediction model, the final winning models** and the c-index of each winning model based on the holdout validation datasets.
|
Model number |
Training | Validation | ||
|---|---|---|---|---|
| Predictors | Frequency (%), n=50 | Winning model** | Mean c-index (SD) | |
| 1 | Age, weight change, HbA1c change, HgA1c change rate | 20 (40) | x | 0.8124 (0.01774) |
| 2 | Age, weight change, HbA1c change, HgA1c in prior 6 mos | 8 (16) | x | 0.8079 (0.01687) |
| 3 | Age, weight change, HbA1c change, HgA1c in prior 18 mos | 6 (12) | x | 0.8220 (0.01035) |
| 4 | Age, weight change, HbA1c change, HgA1c at index | 3 (6) | ||
| 5 | Age, weight change, HbA1c change, abdominal pain | 3 (6) | ||
| 6 | Age, weight change, HbA1c change, BMI | 2 (4) | ||
| 7 | Age, weight change, HbA1c change, alcohol consumption | 2 (4) | ||
| 8 | Age, weight change, HbA1c change, sodium value | 1 (2) | ||
| 9 | Age, weight change, HbA1c change, chest pain | 1 (2) | ||
| 10 | Age, weight change, HbA1c change, ALT change | 1 (2) | ||
| 11 | Age, weight change, HbA1c change, Hgb value | 1 (2) | ||
| 12 | Age, weight change, HbA1c change, race/ ethnicity | 1 (2) | ||
| 13 | Age, weight change, HbA1c change, years of health plan enrollment | 1 (2) | ||
The initial model included age, weight change in 1 year and HbA1c change in 1 year.
The three most frequent models.
Model performance
The c-indexes were comparable among the three winning models (Table 3). The calibration plot based on Model 3 was displayed in Supplemental Digital Content 5. The differences between the average predicted and averaged observed differences were small for the three lowest risk groups. Although the differences appeared to be somewhat large in the two highest risk groups, the ranges of the absolute difference between the predicted and the observed were only 0.33% and 1.10%, respectively (data not shown). The calibration plots for Model 1 and Model 2 were similar to that of Model 3 (data not shown).
Sensitivity, specificity, PPV and fold increase in risk were also comparable among the three winning models (Table 4). Take Model 1 as an example, the sensitivity, specificity, PPV and the fold increase in patients who had the top 20% of the predicted risks were 60%, 80%, 2.5% and 3.0 times, respectively. The corresponding figures in patients with top 5% of the predicted risk were 27.3%, 95.2%, 4.5% and 5.5 times (Table 4). The area under the curves (AUC) of the three models were comparable (0.81 for Models 1 and 2, and 0.82 for Model 3, Figure 2).
Table 4.
Percent of patients whose risk was among the top 20%, 15%, 10%, 5% and 2.5%, sensitivity, specificity, positive predicted Value (PPV), and risk fold increase for each of the three models.
| Model 1 | Model 2 | Model 3 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High risk patients | High risk patients | High risk patients | |||||||||||||
| Top 20% | Top 15% | Top 10% | Top 5% | Top 2.5% | Top 20% | Top 15% | Top 10% | Top 5% | Top 2.5% | Top 20% | Top 15% | Top 10% | Top 5% | Top 2.5% | |
| No. of eligible patients whose risk was above each risk threshold | 2278 | 1708 | 1139 | 569 | 285 | 2267 | 1701 | 1134 | 567 | 284 | 2278 | 1709 | 1139 | 570 | 285 |
| Sensitivity (%) | 60.0 | 52.0 | 42.0 | 27.3 | 17.3 | 56.2 | 48.7 | 37.1 | 25.3 | 16.0 | 60.0 | 52.7 | 39.0 | 24.2 | 14.7 |
| Specificity (%) | 80.3 | 85.3 | 90.3 | 95.2 | 97.6 | 80.3 | 85.3 | 90.2 | 95.2 | 97.6 | 80.3 | 85.3 | 90.3 | 95.2 | 97.6 |
| PPV (%) | 2.5 | 2.9 | 3.5 | 4.5 | 5.7 | 2.3 | 2.6 | 3.0 | 4.1 | 5.2 | 2.6 | 3.0 | 3.4 | 4.2 | 5.1 |
| Fold increase in risk* | 3.0 | 3.5 | 4.2 | 5.5 | 6.9 | 2.8 | 3.2 | 3.7 | 5.1 | 6.4 | 3.0 | 3.5 | 3.9 | 4.8 | 5.9 |
Compared the incidence rate in the entire cohort.
Model 1: Age, weight change, HbA1c change, HgA1c change rate
Model 2: Age, weight change, HbA1c change, HgA1c in prior 6 months
Model 3: Age, weight change, HbA1c change, HgA1c in prior 18 months
All reported numbers were average across 20, 8 and 6 validation datasets for Models 1, 2 and 3, respectively, estimated in patients with complete 36 months follow up or those who developed PDAC in 36 months (n=11,379).
Fig 2.
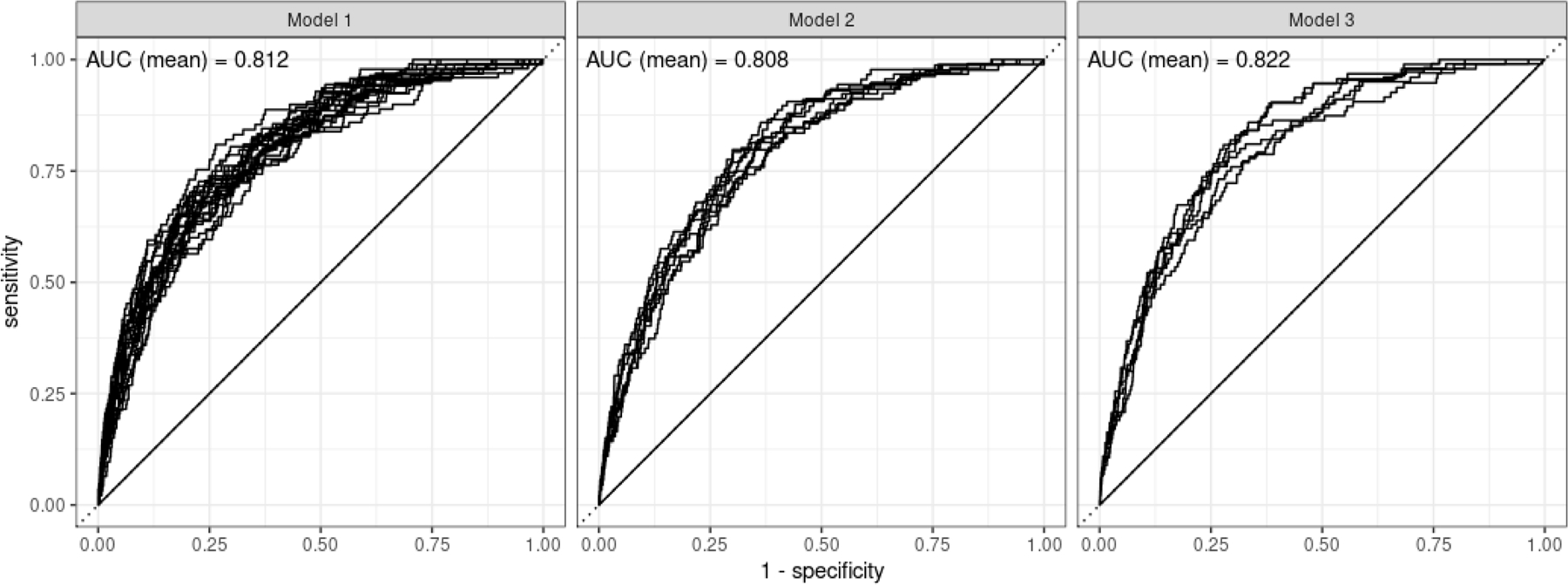
ROC curves in patients with complete 36 months follow up or those who developed PDAC or died.
Exploratory analysis (Supplemental Digital Content 4)
Implementation
The predicted three-year risks of PDAC based on Model 1 can be estimated using the R codes shown in Supplemental Digital Content 6. For a 70 (75) year old patient with an increase of hemoglobin A1c value 0.5% (1.0%) in 200 days between the two measurements (and thus a change rate per day of 0.0025 (0.005)), no prior history of diabetes, and reduction in weight of 4 lbs. (8 lbs.) in the past year, the predicted three-year risks of PDAC was 0.4% (0.9%) (Supplemental Digital Content 6).
As a demonstration, the decision rules based on one of the trees built for Model 1 is displayed in Supplemental Digital Content 7.
DISCUSSION
In this retrospective cohort study, we developed and validated three risk prediction models through a cross-validation process in patients with an elevated (6.5% or higher) glycosylated hemoglobin (defined as recent onset hyperglycemia), but without long standing diabetes. Age, weight loss, HbA1c change and HbA1c change rate (or HbA1c in prior 6 or 18 months) were important predictors of PDAC in this population. This means incorporating the prior glycemic status in addition to rate of change enhanced the utility of the algorithm. The model performance for all three models appeared reasonable. Take Model 1 as an example, targeting patients with the top 20% of risk of developing PDAC could capture 60% of PDAC cases in the study population, with a PPV of 2.5%. The risk of PDAC was three times as that of the entire cohort. The findings suggested that artificial intelligence and electronic health records offer opportunities to provide risk stratification and thus allow physicians to engage with patients for further cancer surveillance. A PPV of 2.5% means that a physician needs to screen 40 patients to identify 1 PDAC case. Users can adjust the risk threshold to screen more or less patients depending on clinical as well as cost-benefit considerations (e.g. the aggressiveness and cost of a particular screening procedure in mind).
Models to predict risk of PDAC in NOD have been attempted in the past.[3,5] A model consisting of three predictors, Enriching New-Onset Diabetes for Pancreatic Cancer (ENDPAC),[5] reached an AUC of 0.75 when it was validated against the electronic health records of KPSC.[18] Compared to the more restricted eligibility criteria of ENDPAC (new onset of diabetes: two elevated glucose tests within 90 days and a previous non-elevated HbA1c), the current study identified a group of patients with only one single elevated HbA1c. The relaxation of the eligibility criteria translates into a much broader population and a more practical application of the model and thus, much higher level of identification of all PDAC cases. As shown in Table 5, an application of the ENDPAC criteria based on the same study period (1/1/2010–09/30/2018) resulted in a sample size of 18k (with 95 PDAC cases), which is less than 17% of the size of the current sample (n=109k with 319 cases) (Table 5). In our previous validation, we showed that a risk score of 3+ (corresponding to 22% of the study population) estimated by the ENDPAC model achieved a sensitivity of 62%, a specificity of 78%, a PPV of 2.0% and a 2.9-fold increase in risk,[18] (Table 2) which are comparable to those estimated in the current study.
Table 5.
Comparisons between current and ENDPAC models.
| Current Model | ENDPAC Model | |
|---|---|---|
| Predictors | Age, weight change, HbA1c change, HbA1c change rates (or HbA1c in prior 6 or 18 months) | Age, weight change, glycemic change |
| Number of predictors | 4 | 3 with information from 4 dimensions* |
| Population | • Single elevated HbA1c • No long-standing diabetes • No history of pancreatic cancer • 12 months continuous health plan enrollment |
New onset of diabetes:
• Two consecutive elevated glycemic tests within 90 days or one elevated glycemic lab followed by diabetes treatment • No history of diabetes treatment • No history of pancreatic cancer • Normal glycemic test* in prior 18 months • 12 months continuous health plan enrollment Implementation in the current study for the purpose of comparison with the PAC-Glycemia Model: • Two consecutives elevated HgA1c within 90 days • No history of pancreatic cancer • Normal HgA1c in prior 18 months • 12 months continuous health plan enrollment |
| Time period | 1/1/2010 and 09/30/2018 | 1/1/2010 and 09/30/2018 |
| Number of eligible patients | 109k | 18k |
| Number of PDAC events | 319 | 95 |
| Incidence rate | 1.4/1,000 person-years of follow-up | 2.0/1,000 person-years of follow-up |
See more details in DISCUSSION Section.
Although the ENDPAC model only contains three predictors (age, weight change and glycemic change), the glycemic change predictor was defined not only based on the change of the categorized values but also the actual baseline measure (also categorized), assigning a higher score for patients with lower baseline value.[5] (Table 1). Therefore, the ENDPAC model essentially contains the information derived from four dimensions. As a handy tool without needing a calculator, ENDPAC model categorizes all the three predictors, allowing users to calculate a final score which then can be converted into a risk category (high, medium, low). In contrast, our model keeps all the original values of the predictors to avoid loss of information. To avoid the tedious manual calculation, a web-based tool will be made available.
Boursi et al, recently published a risk model based on individuals with impaired fasting glucose (IFG), defined as having fasting glucose level between 100 and 125 mg/dL (equivalent to HbA1c 5.1–6.0%).[6] Compared to the PDAC model the same authors reported in 2017 for NOD patients,[3] the new model no longer carries BMI change and glycemia parameters, but includes alanine transaminase (ALT), a liver function measure. An elevated ALT may indicate a liver problem such as jaundice, a late symptom of PDAC. The difference in selected features between the two models clearly indicated the difference between the NOD and the IFG populations. The current study is more similar to the ENDPAC model in terms of the types of patients being identified and the features being selected.
In the current study, targeting patients who had the top 20% of risk of developing PDAC could capture more the 50% of all the PDAC cases associated with hyperglycemia in all the racial/ethnic groups (Supplemental Digital Content 4). Non-Hispanic white had the highest incidence rate (4) in all the ethnic groups. The PPVs were 3.4% and 2.2% in non-Hispanic whites and Hispanics, respectively, with the highest 20% risks. These estimates appeared to be comparable or even slightly higher compared to what were previously reported in the ENDPAC model validation.[18] (Table 4) In the ENDPAC model validation study, the top 23% (20%) of the non-Hispanic white (Hispanic) patients had a PPV of 3.0% (1.4%).
Several previous studies of PDAC risk prediction in NOD populations used the first qualifying NOD date as the index date [5,18]. Therefore, we explored a design in which the first measure of elevated HbA1c (6.5%+) was defined as the index date. Under this alternative design, the three-year incidence rate was 1.1 (95% CI 1.0–1.2)/1,000 person-years of follow-up time and the 18-month incidence rate is 1.5 (1.3–1.6), about 21% lower compared to the corresponding incidence rates reported in Table 2. The findings revealed that the selection of a random qualifying HbA1c measure date instead of the first one avoided the underestimation of risk because the risk of developing the outcome increases overtime for a specific individual as the patient becomes older. In addition, the selection of a random qualifying measure instead of relying on the first one reflects the reality. When patients are assessed for the risk of PDAC, an elevated HbA1C could occur any time during the course of their enrollment with KPSC, and not necessarily the first elevated one during the study period.
The algorithms of our models are stored in a public website for easy implementation. A web application will be available to estimate the risk of PDAC for individual patients. For organizations that have ample resources (e.g. data, computing power), it is recommended that the RSF models be updated using local data before implementation.[19–21] The re-produced decision trees are likely to yield a better performance, especially when there are differences in the incidence rate, patient case mix, and clinical practice between the KPSC population and the population of interest.
Readers need to aware of the following limitations. First, pancreatic cancer cases identified from the California State Death Master files only (n=85) were all considered as PDAC. An evaluation based on the Cancer Registry data included in the current study showed that about 90% of pancreatic cancer cases were PDAC. Second, to estimate sensitivity, specificity, PPV and fold of risk increase, we relied on a subset of patients (~70% of the total patients) who had a complete follow up unless they died of pancreatic cancer. This restriction over-estimated the risk of PDAC, because the patients who were excluded from this analysis were at-risk for some period of time. Third, our model training and validation process included patients with index dates at least three months prior to cancer diagnosis. This restriction may less than optimal from the perspective of early detection. Finally, the implementation of the algorithms requires imputation for missing data. This could be a computational burden for organizations that implement the algorithms within their electronic health systems.
In conclusion, we developed and validated a tool to stratify patients with one single elevated glycemic measure. It is essential that the predictive performance be externally validated. The implementation of the relaxed of glycemic criteria in comparison with the standard NOD definition will provide a more practical environment for real-time recruitment studies involving various screening solutions such as imaging or biomarkers.
Supplementary Material
Supplementary digital content 1:
ICD-O-3 Histology Codes Used to Identify Pancreatic Ductal Adenocarcinoma (PDAC)
Definition of diabetes
Supplementary digital content 2:
Definitions of Weight/Laboratory Value Changes and Symptom Related Features
Supplementary digital content 3:
Model Training and Validation
Supplementary digital content 4:
Methods and Results of the Exploratory Analyses
Supplemental Digital Content 6:
Case Example
Supplemental Digital Content 7:
Decision rules based on one of the trees built for Model 1
Supplemental Digital Content 5:
Calibration Plot for Model 3
eFig 1 in Supplemental Digital Content 5. Calibration plot for Model 3. The five clusters represent the five risk groups defined by the ranges of predicted risks: <50th, 50–74th, 75–90th, 90–94th, 95–100th percentiles. Within each cluster, there are 6 dots representing six pairs of predicted and observed risks, calculated from 6 validation datasets.
Acknowledgements
The authors thank Sole Cardoso for the assistance with formatting.
Source of funding
Research reported in this publication was supported by a grant from the National Cancer Institute (5U01CA200468–04). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding.
Abbreviations
- ALT
Alanine transaminase
- ALP
Alkaline phosphatase
- AUC
area under the curve
- CI
confidence interval
- END-PAC
Enriching New-Onset Diabetes for Pancreatic Cancer
- NOD
new onset of diabetes
- KPSC
Kaiser Permanente Southern California
- SEER
Surveillance, Epidemiology, and End Results
- ICD-9-CM
Ninth Revision of International Classification of Diseases, Clinical Modification
- ICD-10-CM
Tenth Revision of International Classification of Diseases, Clinical Modification
- PDAC
pancreatic ductal adenocarcinoma
- PPV
positive predictive value
- RSF
Random Survival Forest
Footnotes
Conflicts of interest
None declared conflict of interest.
References
- 1.SEER.cancer.gov/statfacts. Cancer Stat Facts: Common Cancer Sites https://seer.cancer.gov/statfacts/html/common.htm. Accessed March 18, 2021.
- 2.Owens DK, Davidson KW, Krist AH, et al. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA 2019;322(5):438–444. [DOI] [PubMed] [Google Scholar]
- 3.Boursi B, Finkelman B, Giantonio BJ, et al. A Clinical Prediction Model to Assess Risk for Pancreatic Cancer Among Patients With New-Onset Diabetes. Gastroenterology 2017;152(4):840–850.e843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chari ST, Leibson CL, Rabe KG, et al. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology 2008;134(1):95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma A, Kandlakunta H, Nagpal SJS, et al. Model to Determine Risk of Pancreatic Cancer in Patients With New-Onset Diabetes. Gastroenterology 2018;155(3):730–739.e733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boursi B, Finkelman B, Giantonio BJ, et al. A clinical prediction model to assess risk for pancreatic cancer among patients with prediabetes. Eur J Gastroenterol Hepatol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz NRM, Matrisian LM, Shrader EE, Feng Z, Chari S, Roth JA. Potential Cost-Effectiveness of Risk-Based Pancreatic Cancer Screening in Patients With New-Onset Diabetes. J Natl Compr Canc Netw 2021:1–9. [DOI] [PubMed] [Google Scholar]
- 8.Sharma A, Smyrk TC, Levy MJ, Topazian MA, Chari ST. Fasting Blood Glucose Levels Provide Estimate of Duration and Progression of Pancreatic Cancer Before Diagnosis. Gastroenterology 2018;155(2):490–500.e492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nichols GA, Schroeder EB, Karter AJ, et al. Trends in diabetes incidence among 7 million insured adults, 2006–2011: the SUPREME-DM project. Am J Epidemiol 2015;181(1):32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu BU, Butler RK, Lustigova E, Lawrence JM, Chen W. Association of Glycated Hemoglobin Levels With Risk of Pancreatic Cancer. JAMA Netw Open 2020;3(6):e204945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Yao J, Liang Z, et al. Temporal Trends in Mortality Rates among Kaiser Permanente Southern California Health Plan Enrollees, 2001–2016. Perm J 2019;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright MN, Ziegler A. A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J Stat Softw 2017;77(1):1–17. [Google Scholar]
- 13.Little RJA. Missing-data adjustments in large surveys. J Bus Econ Stat 1988;6(3):287–296. [Google Scholar]
- 14.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat 2008;2(3):841–860. [Google Scholar]
- 15.Dietrich S, Floegel A, Troll M, et al. Random Survival Forest in practice: a method for modelling complex metabolomics data in time to event analysis. Int J Epidemiol 2016;45(5):1406–1420. [DOI] [PubMed] [Google Scholar]
- 16.Ishwaran H, Kogalur UB. randomForestSRC: Fast Unified Random Forests for Survival, Regression, and Classification (RF-SRC) http://web.ccs.miami.edu/~hishwaran/https://github.com/kogalur/randomForestSRC/. Published 2021. Accessed 10/20/2020.
- 17.Demler OV, Paynter NP, Cook NR. Tests of calibration and goodness-of-fit in the survival setting. Stat Med 2015;34(10):1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Butler RK, Lustigova E, Chari ST, Wu BU. Validation of the Enriching New-Onset Diabetes for Pancreatic Cancer Model in a Diverse and Integrated Healthcare Setting. Dig Dis Sci 2021;66(1):78–87. [DOI] [PubMed] [Google Scholar]
- 19.Su TL, Jaki T, Hickey GL, Buchan I, Sperrin M. A review of statistical updating methods for clinical prediction models. Stat Methods Med Res 2018;27(1):185–197. [DOI] [PubMed] [Google Scholar]
- 20.Davis SE, Greevy RA, Lasko TA, Walsh CG, Matheny ME. Comparison of Prediction Model Performance Updating Protocols: Using a Data-Driven Testing Procedure to Guide Updating. AMIA Annual Symposium proceedings AMIA Symposium 2019;2019:1002–1010. [PMC free article] [PubMed] [Google Scholar]
- 21.Davis SE, Greevy RA, Fonnesbeck C, Lasko TA, Walsh CG, Matheny ME. A nonparametric updating method to correct clinical prediction model drift. J Am Med Inform Assoc 2019;26(12):1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary digital content 1:
ICD-O-3 Histology Codes Used to Identify Pancreatic Ductal Adenocarcinoma (PDAC)
Definition of diabetes
Supplementary digital content 2:
Definitions of Weight/Laboratory Value Changes and Symptom Related Features
Supplementary digital content 3:
Model Training and Validation
Supplementary digital content 4:
Methods and Results of the Exploratory Analyses
Supplemental Digital Content 6:
Case Example
Supplemental Digital Content 7:
Decision rules based on one of the trees built for Model 1
Supplemental Digital Content 5:
Calibration Plot for Model 3
eFig 1 in Supplemental Digital Content 5. Calibration plot for Model 3. The five clusters represent the five risk groups defined by the ranges of predicted risks: <50th, 50–74th, 75–90th, 90–94th, 95–100th percentiles. Within each cluster, there are 6 dots representing six pairs of predicted and observed risks, calculated from 6 validation datasets.


