Abstract
Our understanding of the genetic architecture of phenotypic traits has experienced drastic growth over the last years. Nevertheless, the majority of studies associating genotypes and phenotypes have been conducted at the ontogenetic level. Thus, we still have an elusive knowledge of how these genetic-developmental architectures evolve themselves and how their evolution is mirrored in the phenotypic change across evolutionary time. We tackle this gap by reconstructing the evolution of male genital size, one of the most complex traits in insects, together with its underlying genetic architecture. Using the order Hemiptera as a model, spanning over 350 million years of evolution, we estimate the correlation between genitalia and three features: development rate, body size, and rates of DNA substitution in 68 genes associated with genital development. We demonstrate that genital size macro-evolution has been largely dependent on body size and weakly influenced by development rate and phylogenetic history. We further revealed significant correlations between mutation rates and genital size for 19 genes. Interestingly, these genes have diverse functions and participate in distinct signaling pathways, suggesting that genital size is a complex trait whose fast evolution has been enabled by molecular changes associated with diverse morphogenetic processes. Our data further demonstrate that the majority of DNA evolution correlated with the genitalia has been shaped by negative selection or neutral evolution. Thus, in terms of sequence evolution, changes in genital size are predominantly facilitated by relaxation of constraints rather than positive selection, possibly due to the high pleiotropic nature of the morphogenetic genes.
Keywords: development, DNA, genitals, morphogenesis, sequence evolution
Introduction
How phenotypic changes are facilitated or constrained in the long term is a pillar of the evolutionary biology research. Studies on the evo-devo era have shown that determining how genes coordinate morphogenesis during development is a fundamental step to understand phenotypic evolution (Carroll 2008; Mallarino and Abzhanov 2012). A symbolic example is the segmental origins of insect wings, which has been debated for centuries with no consensus (Averof and Cohen 1995; Medved et al. 2015). The elegant evo-devo study by Linz and Tomoyasu (2018) with Tribolium beetles revealed a possible “dual origin” for these structures, suggesting wings are in part a modified leg and simultaneously a tergal expansion (Linz and Tomoyasu 2018). Undeniably, individual-level studies as such provide invaluable data to study phenotypic evolution. Nevertheless, single-species approaches are insufficient to explore the evolutionary processes that act after the emergence of novel traits. Our understanding of these processes is elusive, and we have poor knowledge of how the genetic-developmental architecture of morphological traits evolve themselves and how their evolution is mirrored in the phenotypic change across large time scales (Baker et al. 2021). There is an urgent need for more studies associating morphological, developmental, and genetic data in comparative frameworks (Sanger and Rajakumar 2019).
An illustrative example of this gap is the study of genital evolutionary development in insects. There has been growing research scrutinizing the genes and pathways underlying genital development (e.g., Vincent et al. 2019; Xu et al. 2019; Smith et al. 2020). Notably, recent studies with Drosophila and Carabus beetles revealed sequence divergence of distinct genomic regions associated with the divergence of genital traits in recently separated species (Fujisawa et al. 2019; Hagen et al. 2021). These data may have started to illuminate the role of genital morphology and their underlying genetics in the speciation process. A direct prediction from these observations is that the outstanding rates of genital evolution in insects may have left genomic signatures over time. For example, we might expect that accelerated rates of genital diversification correlate with changes in the rates of molecular evolution or with the strength of molecular adaptation. One avenue to test this hypothesis is to estimate the degree of association between genitalia and molecular substitution rates across a phylogeny. The advantage of this approach in our understanding of the evolutionary development of genitalia would be 2-fold. First, studying the genotype–phenotype association at the macro-evolutionary scale allows for investigating the predominant mechanisms that have led to diversification (Lartillot and Poujol 2011; Jones et al. 2020). For instance, has rapid genital evolution been facilitated by positive selection acting in key developmental genes? Second, the comparative approach provides a way to evaluate the developmental and genetic bases of morphological evolution across an entire clade instead of a single species (Organ et al. 2015). In this study, we tackle this topic by reconstructing the evolution of male genital size across the order Hemiptera, spanning over 350 million years of evolution (Johnson et al. 2018). Following, we link the phenotype with the genotype by estimating the association between genital size and the rates of sequence evolution of several developmental genes.
Size is one of the most important attributes of genitalia in insects. A proper genital size must be guaranteed during development since it allows the perfect fit between males and females and avoids mechanical mismatches in copula (Usami et al. 2006; Tanaka et al. 2018). As a result, genital size tends to develop distinctly from other traits, usually showing a negative allometric scaling—that is, a relatively constant size despite variations in overall organism size (Lupše et al. 2016). There is solid evidence that genital size develops more independently through an insensitivity to systemic regulators of body size development, for example, the insulin signaling cascade (Tang et al. 2011; Emlen et al. 2012; Dreyer and Shingleton 2019). While insulin insensitivity may partially explain patterns of genital growth during ontogeny, which genes and pathways underlie this special mode of genital development are yet poorly understood (Terada et al. 2021). By extension, we know even less about the genetic-developmental mechanisms that may allow, facilitate or constrain genital size diversification on the evolutionary scale. For example, a handful of studies have shown that knocking down central developmental genes may result in smaller genitals, like genes involved in sex determination, segmental organization, and appendage patterning (Aspiras et al. 2011; Macagno and Moczek 2015). However, if those master regulatory genes or, alternatively, more specific downstream genes, are the ones whose structures are shaped by selection in the evolution of genital size, is largely unknown.
Here, we employ a comparative approach to investigate the evolution of male genital size across 92 hemipteran species (fig. 1), aiming at the evolution of its underlying genetic-developmental machinery. We chose to study genital size, more specifically width, due to its demonstrated role in mating for the Hemiptera. Studies with different families have documented a correspondence between the width of the external genitalia in males and females (Moreno-García and Cordero 2008; Wang et al. 2009; Genevcius et al. 2017; Ruschel et al. 2019). The male capsule attaches horizontally to the female plates providing the mechanical stability in copula (Genevcius and Schwertner 2017). Furthermore, the overall organization of the external genitalia is conserved within Hemiptera (Singh-Pruthi 1925), comprising a rigid capsule surrounding additional structures like claspers and the proctiger. The facts that external genitalia show constant bauplan and functions indicate that genital width has an important and conserved functional/evolutionary role order-wise. We use phylogenetic methods and publicly available genome-scale data to estimate the rates of evolution of 68 genital-developmental genes and correlate those rates with changes in genital size and development rate. We further evaluated the most likely selective regimes associated with these genes by estimating site-wise non-synonymous and synonymous mutation ratios. Our analyses revealed that male genital size in hemipterans has evolved in dissociation with development rate but, in contrast with former observations, in close association with body size. We also found 19 genes whose substitution rates were significantly correlated with genital size. Lastly, we show that only a few sites in these genes have evolved under positive selective pressures, while the majority of sequence change has been driven by negative selection and neutral evolution. We discuss how our data fit in or disagree with the current theory of genital evolution, and how our analyses illuminate the mechanisms underlying the macro-evolution of genital size and its genetic architecture.
Fig. 1.
Phylogenetic diversity of genital size (inner circle), body size (middle circle), and development rate (outer circle) across the Hemiptera, distributed in the ultrametric super-tree constructed here. There is a clear trend of larger species exhibiting larger genitals (e.g., Pentatomoidea and Sternorrhyncha), while development rate seems less correlated with both morphological traits.
Results
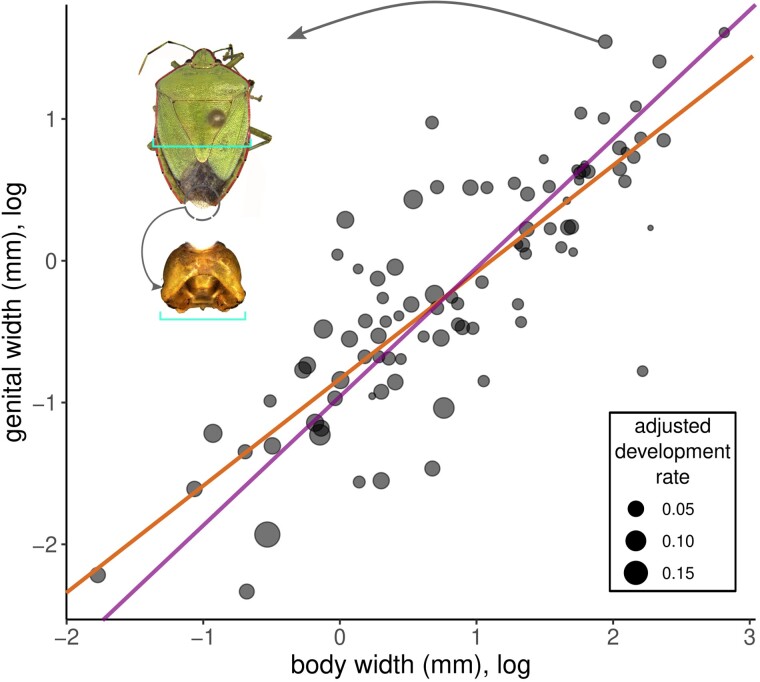
We conducted an extensive literature search to gather data on development, male genital size, and body size in Hemiptera. Our final dataset revealed great variation for the three traits across the order (fig. 1). We also observed high heterogeneity across the phylogeny. For example, genital size exhibited the largest values and were relatively constant in pentatomoids, while extremely variable in nepomorphans and lygaeoids + allies. Results of the Standardize Major Axis analysis revealed that genital size follows an isometric pattern, with an estimated slope of 0.906 (lower limit: 0.806, upper limit: 1.020) and not statistically different from 1 (P = 0.101). There is a clear visual trend of larger species exhibiting larger genitals (fig. 1), which is consistent with our models. The simplest generalized least squares (GLS) model, where genital size is explained exclusively by body size (“pure allometric model”), revealed a significant correlation (R2 = 0.68, P < 0.001, fig. 2) and had the lowest Akaike information criterion (AIC) value (table 1). Nevertheless, we also observed that adding development rate to this model resulted in a slightly higher AIC value. In addition, the model combining body size with development rate had much better fit than the same model with the inclusion of the phylogeny (table 1). These results, combined, indicate that the great majority of genital size variation is explained by body size, while development plays a secondary role and the phylogeny is the least relevant.
Fig. 2.
Relationship between body and genital size (given in mm, log-transformed), where each point is a hemipteran species and point size represents the development rate. Best fit line (orange) is from the GLS analyses of the pure allometric model (see table 1). Fit from the SMA analysis is expressed in purple. One of the species analyzed (Chinavia ubica, with genitalia detached) is highlighted with its genital capsule to illustrate the measured traits (light blue bars).
Table 1.
Results of Generalized Least Squares Modeling.
| Model/Predictor Variable | AIC |
|---|---|
| Body size (pure allometric model) | 140.21 |
| Body size + phylogeny | 235.13 |
| Body size + development rate | 141.75 |
| Body size + phylogeny + development rate | 217.09 |
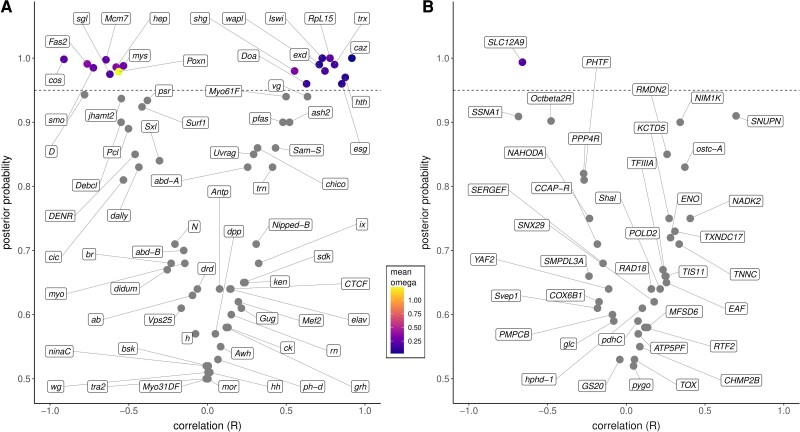
Since body size had a strong effect on genital size, and genital size follows an isometric scaling, we used the ratio between these two variables to test for the genotype–phenotype correlations. This approach allowed us to discount body size, modeling only body-size-free genital variation instead. For the 68 genital genes tested, 18 genes (26.5%) showed a significant correlation between omega (ω) and corrected genital size (posterior probability > 0.95) (fig. 3A). Out of these 18 genes, three exhibited very strong positive correlations (cabeza, escargot and homothorax), and one exhibited a very strong negative correlation (costa) (fig. 3). Seven genes showed strong correlations, while nine genes had moderate correlations (fig. 3). We replicated the same analyses for a set of 40 control, non-developmental, genes. Our of these, a single gene (2.5%), SLC12A9, showed significant correlation (fig. 3B). A full table of the Coevol results is provided as supplementary data S1, Supplementary Material online.
Fig. 3.
Results of phenotype–genotype association (body-size-corrected genital width versus omega [ω]) estimated in Coevol, showing the degree of correlation versus the posterior probability (y axis). Each point represents a gene, and the dashed line is the posterior probability cut-off of 0.95. Omega values for each gene with significant correlation with the phenotype are represented by point colors and were taken from the site-wise model from CodeML (the mean omega of all codons). (A) Genes with predicted genital development functions; (B control, “non-genital” genes.
For the 19 genes with significant association with male genital size, the analyses in CodeML revealed that nine genes had only negatively selected or neutral sites (fig. 4). The majority of sites of the other nine genes showed a prevalence of negative selection and neutral evolution, with only a few sites positively selected (fig. 4). For these genes, the number of positively selected sites varied from two (costa, smoothened and sugarless) to nine (Pox neuro and shotgun). Omega values for the positively selected sites ranged between 1.37 (costa) and 8.09 (Pox neuro).
Fig. 4.
Selective regime estimated in CodeML for each codon (colored points) of all genes that showed significant association with genital size (detected in Coevol). The vertical dashed line approximates the positive selection zone threshold, where dN/dS are equal (ω = 1).
Discussion
In this study, we conducted the largest and most comprehensive analysis of genital size evolution, with samples representing an entire insect order, the Hemiptera. Below, we first discuss the morphological patterns we revealed framed into the phylogenetic perspective, considering how our data fits the predictions of the theory of genital evolution. Second, we discuss our findings of the adaptive molecular evolution analyses, focusing on differences observed in substitution rates for different genes, as well as their implications for male genitalia evo-devo. Lastly, we provide a perspective on how our approach can be expanded to strengthen our understanding of these genotype–phenotype correlations.
Constraints of Body Size and Development Rate in Genital Evolution
We showed that there is great variation in male genital size across the Hemiptera, and the magnitude of among-species variation may vary across clades. This observation is not surprising, and it is consistent with recent studies that revealed heterogeneous patterns of genital change in beetles (Rudoy and Ribera 2016; Genevcius et al. 2020). Nevertheless, we still observed a clear isometric trend, meaning that the evolution of genital size is constrained by body size. In principle, this result may seem contradicting with the theory of genital allometry, which postulates that genital traits develop more independently of body size than other traits (House and Simmons 2007). We emphasize, however, that allometry is a phenomenon that takes place at the populational/individual level, where organ growth may be more or less sensitive to body size variation (Mirth et al. 2016). While most studies that found negative allometry for genital traits have been conducted at the intraspecific level (e.g., Orbach et al. 2018; De-Lima et al. 2019), a few studies at broader scales have found complex scenarios. For example, Galicia-Mendoza et al. (2021) found distinct allometric patterns for the genitalia of aggressive and non-aggressive damselfly species. In spiders, it has been shown that non-intromittent genitalia are evolutionarily correlated with body size, while intromittent genitals are not (Lupše et al. 2016). In our study, the observed isometric scaling results most likely from a physical constraint, given that among-species variation in body size is remarkable (see fig. 1).
Male genital size was found to be secondarily influenced by development rate. The relative weak influence of development rate (compared with body size) was expected due to functional and ontogenetic characteristics of the genitalia. Since immatures do not copulate, genitalia are the latest structures to develop during ontogeny, together with wings. External genitals in hemipterans develop mostly during the fifth and last nymphal instar, and are only completely formed in adults (Singh 1971). Therefore, unlike other structures that are functionally important since early development such as the mouth parts, genitalia are completely absent in the stages where we observe massive body size increase, that is, during the first four molts.
Molecular Signatures of Genital Macro-Evolution
We investigated the evolutionary patterns of 68 genital-related genes and 40 “non-genital” control genes across 92 hemipteran species and demonstrated that, for 19 genes, the ratio of non-synonymous and synonymous mutations (dN/dS or ω) correlated with genital size. None of these genes were master regulators of signaling pathways, like the hox genes that determine segmental identity of posterior body (e.g., abd-a, abd-b, and Antp), nor sex determining genes (e.g., ix and Sxl). As expected, such genes had low correlations with the phenotype, probably because their coding regions are highly conserved (Krumlauf 2018), in contrast with the rapidly evolving genitalia (Genevcius et al. 2017). For these genes, changes in sequence composition would likely yield drastic phenotypic outcomes and be highly deleterious during development (Zhang et al. 2021; Chen et al. 2022). Another group of genes that showed overall low correlation with genital size were the appendage-patterning genes like decapentaplegic, hedgehog, and wingless. Genitalia are commonly thought to be serially homologous to other appendages like legs and antennae due to demonstrated role of these appendage-patterning genes in genital growth in holometabolous (Estrada and Sánchez-Herrero 2001; Macagno and Moczek 2015). In contrast, our results demonstrate that there are no signatures of an adaptive process acting on these genes concerted with genital size. Our data seems more in line with either of the two hypothesis that (i) the male genitalia in Hemiptera may be a highly derived appendage primordia which lost appendage-patterning functions, or (ii) genitalia are modified abdominal segments with absolutely no appendage homology (Aspiras et al. 2011). Note that it is still possible that appendage-patterning genes may correlate with genitalia only in the level of expression (see discussion below).
We found significant phenotype–genotype correlations for genes previously known to be associated with genital size. For example, knockdown in homothorax has caused a reduction in copulatory organs in the milkweed bug (Aspiras et al. 2011); in Drosophila, Mcm7 knockdown has been associated with changes in clasper size (Tanaka et al. 2015), while costa mutants showed increased claspers and reduced hypandrium (Sánchez et al. 1997). To some extent, the concordance between these ontogeny-level studies and our results at the macro-evolutionary scale provides validation to our approach. This is especially important given the paucity of studies linking evolutionary processes measured at the macroscale and direct experimental observations of gene function (Moury and Simon 2011). Interestingly, Mcm7 has been shown important in the development of genital traits with very recent divergence in Drosophila (Tanaka et al. 2015; Hagen et al. 2021). Taken together, these observations and our own results may indicate Mcm7 as a potential candidate involved not only in morphogenesis but also in speciation and lineage diversification.
Another interesting aspect related to these “genital size genes” is that they involve completely different developmental pathways with distinct molecular and biological functions. For example, homothorax is a TALE homeobox transcription factor that forms the homothorax-extradencticle route, promoting cell division and cuticle formation (Rieckhof et al. 1997). In contrast, costa participates in the hedgehog pathway and it is directly involved in growth control (Sánchez et al. 1997). In addition, we also observed significant correlations with genes related to other features of genitalia apart from size: bristle patterning, sclerotization, pigmentation, and the formation of several accessory structures (table 2). These results highlight that genital size is not a “simple trait” whose evolution has been merely facilitated or accompanied by changes in pathways that regulate overall body size. Rather, they may suggest that genital size evolution is only possible with accumulated changes associated with diverse morphogenetic processes, for instance, the formation of cuticle composition, sensory apparatus, and muscle development. Nevertheless, understanding why the evolution of genital size may be linked with the evolution of these other features is not straightforward. A plausible explanation is that the adaptive value of bigger genitals may be indirect. Since we measured the size of the genital capsule, which revolves the remaining genital structures, larger genitals may be associated with increased complexity (Song 2009) by allowing the accommodation of increased number of structures common in insect genitalia, such as hooks, setae, folds, etc. The development and evolution of these structures in more complex genitalia would predict selection in distinct molecular and biological functions, as suggested by our data.
Table 2.
The 18 Genital Genes Whose dN/dS Were Found to Have a Significant Correlation With Genital Size. Because we did not Detect Orthologs for all Species for all Genes, Each Analysis had a Different Number of Species (“sp” column). Final Alignment Size (Align. Size) Used in Coevol Analyses is Denoted in Nucleotides. Gene Function and Their Phenotypic Effects are Described With its Respective Reference.
| Gene | sp. (n) | Align. Size | Function Summary | Effects on Genitalia Development (sp) | Reference |
|---|---|---|---|---|---|
| caz | 42 | 276 | Encodes a chromatin binding protein involved in locomotion, synaptic growth at the neuromuscular junction | Not described (Dmel) | Wang et al. (2011) |
| esg | 23 | 441 | Encodes a snail-type transcription factor with ectodermal expression in embryos that contributes to stem cell maintenance and morphogenesis | Epandrial ventral lobes formation (Dmel) | Vincent et al. (2019) |
| hth | 55 | 480 | Encodes an homeodomain transcription factor mainly responsible for appendages patterning | Control of genital size and formation of phallobase and aedeagus (Ofas) | Aspiras et al. (2011) |
| cos | 30 | 498 | Encodes a kinesin-like protein and negatively regulates the hedgehog pathway that is involved in pattern formation and growth control | Claspers and hypandrium size, fusion of lateral plates (Dmel) | Sánchez et al. (1997) |
| exd | 44 | 948 | Encodes a homeodomain transcription factor that is imported to the nuclueus upon binding the product of hth. Acts in proximal appendages patterning and cell division promotion | Genital bristle patterning (Dmel) and genital sclerotization (Tcas) | Gonzalez-Crespo and Morata (1995) |
| Fas2 | 49 | 684 | Encodes a neuronal recognition molecule, pathway recognition for axons during the development of nerve fascicles | Genital rotation control (Dmel) | Ádám et al. (2003) |
| ISWI | 87 | 2,784 | Energy-transducing component of chromatin-remodeling complexes required for homeotic gene expression, larval blood cell development, X chromosome morphology, ecdysteroid signaling and metamorphosis | Not described (Dmel) | Deuring et al. (2000) |
| RpL15 | 84 | 615 | A structural constituent of ribosome, predicted to be involved in cytoplasmic translation. | Genital rotation and gonad size control (Dmel) | Schulze et al. (2001) |
| smo | 55 | 1,665 | Encodes a critical component of the hedgehog signaling pathway. It is regulated by phosphorylation, dimerization, and cell-surface accumulation upon Hedgehog stimulation. | Determines the correct number of structures such as the genital arc, the claspers and the hypandrium bristle (Dmel) | Gorfinkiel et al. (1999) |
| trx | 48 | 975 | Encodes a chromatin-modifying enzyme involved in gene regulation. This activity antagonizes the epigenetic silencing by Polycomb group proteins. It contributes to axon guidance, eye development and germ cell migration. | Identity of genital segments (Dmel) | Breen (1999) |
| wapl | 48 | 1,575 | Encodes a protein that interacts with the product of pds5 to form the releasin complex that enables sister chromatid separation at mitosis by removing the cohesin ring complex from chromosomes. It also influences gene activation and silencing through interactions with cohesin. | Genital rotation control (Dmel) | Cunningham et al. (2012) |
| Doa | 73 | 1,017 | Encodes an essential Ser/Thr protein kinase with SR proteins. Contributes to somatic sex determination (splicing of dsx transcript) and morphogenesis across the entire body. | Bristles patterning and pigmentation control (Dmel) | Du et al. (1998) |
| MCM7 | 82 | 2,145 | Encodes a component of the MCM2-7 hexamer, which forms part of the CMG complex. The CMG complex is the main DNA helicase that functions during DNA replication. | Controls clasper size and posterior lobe size and shape (Drosophila spp) | Tanaka et al. 2015; Hagen et al. (2021) |
| sgl | 68 | 1,389 | Involved in the biosynthesis of glycosaminoglycans. Required for wingless signaling in different tissues. | Claspers formation (Drosophila spp) | Hagen et al. (2021) |
| hep | 47 | 471 | Encodes a serine/threonine protein kinase involved in the bsk pathway, which mediates immuno response and morphogenesis. Required for epithelial cell sheet movement | Genital positioning and rotation control (Dmel) | Rousset et al. (2010) |
| mys | 80 | 2,112 | Encodes a β subunit of the integrin dimer. The product of mys acts as adhesion/signaling protein regulating cellular adhesion, migration and survival. | Genital rotation control (Dmel) | Fraichard et al. (2010) |
| Poxn | 35 | 225 | Encodes a transcriptional factor that specifies the differences between mono-innervated external sensory organs and poly innervated external sensory organs. Determines the fate to form larval organs and adult chemosensory bristles. | Formation of genital cuticle, penis, claspers, bristles and posterior lobes (Dmel) | Boll and Noll (2002) |
| shg | 61 | 4,077 | Encode calcium-dependent cell adhesion proteins. Functions in cell intercalation in the lateral epidermis during germband extension. Contributes to the determination of body left-right asymmetry by interfering in myosin activities | Genital rotation control (Dmel) | Petzoldt et al. (2012) |
Dmel, Drosophila melanogaster; Ofas, Oncopeltus fasciatus; Tcas, Tribolium castaneum.
For the analyses of control genes, we found a single gene (SLC12A9) with significant (and moderate) correlation with genital size. This result indicate that correlations found between developmental/genital genes and genital size are not mere artifacts, thus, strengthening our approach and conclusions. The gene SLC12A9 is an important solute carrier in mammals, but little is known about its function in insects. However, genes of the solute carrier family super-family are known to determine cell growth in various tissues, including in Drosophila larvae (Velentzas et al. 2018), and may also have a role in sex differentiation and gonad development in vertebrates (Almstrup et al. 2016). It remains to be tested whether this correlation has a functional significance for the Hemiptera.
Although we observed very strong correlations between genital size and ω for some genes, we found that mean cross-species ω values were overall low (fig. 3). This is consistent with a scenario where the majority of changes in genital size have been possible due to a relaxation of purifying selection in developmental genes rather than by persistent positive selection. In fact, this scenario is probably more often the rule then the exception due to the highly pleiotropic nature of the morphogenetic genes (Partha et al. 2017; Womack et al. 2018; Mariano-Martins et al. 2022). From our dataset, illustrative examples are the transcription factors escargot, cabeza, and homothorax. Among different structures, these genes also mediate wing morphogenesis (Fuse et al. 1996; Casares and Mann 2000), which is a phenotype that seems highly conserved across hemipteran groups. Still, for half of the genes that correlated with genital size, we observed at least a few sites under positive selection (fig. 4). This highlights the possibility of heterogeneous evolutionary mechanisms acting in different protein regions, and such differentially affected regions may be interesting targets for future functional studies. It is also important to emphasize that synonymous mutations are not necessarily dissociated with adaptive evolution as they may be advantageous in some contexts like changing splicing patterns and adjusting the speed of RNA translation (Chu and Wei 2019).
Yet regarding the tests of selection regime, the gene Pox neuro stands out for showing some sites with very strong positive selection (fig. 4). Pox neuro encodes transcription factors that are pivotal for the development of genitalia and other organs in Drosophila. While the underlying causes of such strongly selected sites cannot be determined with our data, it is possible that these sites correspond to specific introns whose roles are more determinant for genital development than other structures, such as demonstrated by (Boll and Noll 2002). Another interesting observation is that Pox neuro is not only a morphogene, but also a courtship gene, which is directly involved in exclusive-male courtship behaviors and fertility (Hall 1994; Boll and Noll 2002). Fast evolution via positive selection has been strongly linked with sex-biased expression (Ávila et al. 2018) and courtship behavior divergence (Arbuthnott 2009). Thus, we hypothesize that these two factors may be associated with the high rates of non-synonymous mutations of Pox neuro.
One last key question that emerges from our results is why some genes exhibit positive correlations with the phenotype and others exhibit negative correlations (see fig. 3). While this question can only be precisely answered with deeper understanding of gene functions coupled with the underlying selective mechanism in the phenotype, we hypothesize that genes with negative correlations may represent “moderator genes” (Baker et al. 2021). In this scenario, mutations in protein-coding regions would be thought to consistently decrease phenotypic change. This may be particularly the case of genes that act as negative regulators of developmental pathways such as costa and Fasciclin 2, which are involved in transcription factor sequestering of the hedgehog pathway and the inhibition of epidermal growth, respectively (Mao and Freeman 2009; Li et al. 2016). In addition, genes with marked pleiotropic and epistatic behaviors may also act as moderators due to their global effects on developmental pathways, such as smoothened and sugarless.
Future Directions
Our approach has been shown interesting to reveal the genes and potential loci within these genes involved with the evolution of male genital size. We show that of 68 genital genes analyzed, 18 had positive signatures of genotype–phenotype correlation. We emphasize, however, that associating phenotypes with mutation rates in coding regions, although powerful, only represents partial picture of the adaptive mechanisms underlying (macro)evolution. We thus highlight four directions for future research that may provide further advance on these mechanisms.
The first direction draws from the observation that we cannot completely discard the relevance of the genes without significant correlation because, apart from sequence structure, other features may evolve themselves: expression levels (Romero et al. 2012), expression timing (Keyte and Smith 2014), regulatory sequence composition (Hoekstra and Coyne 2007), structure of gene regulatory networks (Smith et al. 2018), among others. Measuring (co)expression levels of candidate genes in species with different relative genital sizes, and modeling these features across the phylogeny, represent an interesting future avenue yet largely unexplored (Nomura et al. 2021). Secondly, it is important to note that our prior hypotheses of genotype–phenotype associations derive mostly from studies on Drosophila melanogaster. There are certainly other important genes and pathways that can be discovered by conducting transcriptomic and functional studies in non-traditional model insects, such as the Hemiptera. Third, an important characteristic of COEVOL and similar analyses is that they estimate a single omega value for the entire gene. This is not a problem per se, but it may exclude genes for which single sites may coevolve with the phenotype, thus, only genes with a global correlation will be found. To our knowledge, no study has yet used the COEVOL methods to estimate covariances between phenotypes and isolate codons. Even though there will be computational and theoretical challenges, this is another promising approach. Lastly, one crucial aspect of genotype–phenotype modeling is the assumption that this correlation is unidirectional (Smith et al. 2015), for example, increases in omega leading to increases in trait size. However, it is possible that changes in omega rates may implicate heterogeneous phenotypic changes across different clades, making the detection of the correlation more difficult. A very recent method has been developed to account for this issue (Baker et al. 2021), and it may be applied for future studies on genital evolution as well.
Materials and Methods
Phenotypic and Developmental Data
We used the maximum width of the external male genitalia as a proxy for genital size and the maximum abdominal width as a proxy for body size. Genital width was chosen due to its documented role in genital coupling with females (Singh-Pruthi 1925; Moreno-García and Cordero 2008; Wang et al. 2009; Genevcius and Schwertner 2017; Ruschel et al. 2019; and see introduction). The majority of measures were collected directly from scaled images published in the literature or online databases, but a few species were measured from personal figures (supplementary data S2, Supplementary Material online). All measures were taken in the software ImageJ.
We compiled information on the total post-embryonic development duration in number of days from the literature. Because different studies were conducted in different temperature conditions, we used the Campbell’s (1974) developmental model to standardize developmental duration and determine the developmental rate, defined as 1/(developmental time). The minimum temperature for successful development in Hemiptera usually varies between 10 and 16 °C, thus, we used the median 13.5 for the minimum developmental temperature in the model. We then calculated an adjusted developmental rate for a standard temperature of 25 °C. The vast majority of data was obtained from studies with controlled temperatures (supplementary data S2, Supplementary Material online). For the few studies conducted in the field or at room temperature, we used the mean month temperature where the study was conducted, taken either from the respective study (when reported) or from web databases (climate-data.org and gspatial).
Genital Development Genes and Bioinformatics
We searched for genes associated with genital development in the platforms FlyBase and VectorBase and also protein products of arthropods from The Gene Ontology Resource that are assigned with the ontology terms GO:0035112 (genitalia morphogenesis) and GO:0048806 (genitalia development). We compiled an initial set of 148 genes, for which we could obtain 68 ortholog sequences in the genomes of the bed bug (Cimex lectularius) or the kissing bug (Rhodnius prolixus) based on VectorBase annotations. We used protein sequences from these two bugs as reference to search for the orthologs across all publicly available hemipteran transcriptomes. To check if phenotype–genotype correlations are confined to or stronger in these 68 developmental genes, we replicated our comparative analyses in a set of 40 control genes. These genes were sampled randomly from the genome of C. lectularius, our hemipteran reference. To ensure that we were not sampling developmental genes with undiscovered genital morphogenetic roles, we excluded genes that are assigned with any developmental process (GO:0032502). For this, GO terms for the C. lectularius genome, which comprises ∼24,200 coding sequences (NCBI code GCA_000648675.3), were annotated using the online functional annotator eggNOG-mapper (http://eggnog-mapper.embl.de). After pruning developmental genes, we had ∼19,000 genes from where we sampled the random 40 control genes.
We downloaded 84 hemipteran transcriptomes from the TSA database and assembled de novo 8 transcriptomes with raw RNA-seq data obtained from the SRA database (supplementary data S2, Supplementary Material online), totaling 92 species. We used Trinity v. 2.11.0 (Grabherr et al. 2011) for the transcriptome assembly, with minimum contig length of 199 bp and other parameters set to default. To search for the orthologs of the 68 genital and 40 control genes in these transcriptomes, we used blastX in Diamond v. 0.9.21.122 (Buchfink et al. 2014), with a minimum e-value of 0.0001, 40% of query length coverage, and 60% of identity. We further selected the best hit for each species and each gene filtering by bitscore with the python script “blastfilterer.py” (https://github.com/juancrescente/biopyutils/blob/master/blastFilter.py).
To detect protein-coding regions and conduct alignments we used AlignWise v. 0.38 (Evans and Loose 2015) with the muscle algorithm and TranslatorX v. 9.0 (Abascal et al. 2010) for amino acid-guided alignments. We did a manual curation of each alignment to remove misaligned and gap-rich sequences.
Phylogeny
Since there is not a single phylogeny for all the taxa of interest, we used a super-tree approach in CLANN with the average consensus algorithm, which preserves branch lengths (Creevey and McInerney 2005). We inferred the super-tree using 11 molecular phylogenies of hemipterans (Li et al. 2012; Nováková et al. 2013; Zhang et al. 2016; Wang et al. 2017; Wang et al. 2019; Johnson et al. 2018; De Moya et al. 2019; Forthman et al. 2019; Liu et al. 2019; Cao et al. 2020; Genevcius et al. 2021), posteriorly replacing taxa with closely related species to match those with genital and transcriptomic data (supplementary data S2, Supplementary Material online). We dated the phylogeny using the function “chronos” in the R package Ape v. 5.5 (Paradis et al. 2003), with a relaxed model and the same calibration points as in Johnson et al. (2018). The resulting ultrametric tree was used for all comparative analyses.
Comparative Analyses
We started by evaluating the allometric pattern of genital size evolution. We first conducted a GLS analysis where log-transformed genital width was the response variable and the log of the major abdominal width was the predictor variable. Second, we also tested for the effect of development in the evolution of genital size by including in the first model the adjusted developmental rate as a second predictor variable. Third, we also included in these models a phylogenetic correlation matrix estimated under a Brownian motion model to test the effect of phylogenetic non-independence (pGLS). We then compared these models using the AIC.
To distinguish between allometry and isometry, we conducted a standardized major axis analysis (SMA) using the R package smatr 3 (Warton et al. 2012). The SMA analysis tests if the slope of the relationship between log-transformed genital and body sizes is significantly different from 1, which is the expected slope for an isometric correlation.
To investigate the genomic signatures underlying genital size evolution, we modeled the evolution and coevolution of molecular substitution patterns and genital size in Coevol v. 1.5 (Lartillot and Poujol 2011). Briefly, the program models the evolution of synonymous (dS) and non-synonymous substitutions (dN) across a phylogeny, as well as the phenotypic data of interest, using a Brownian motion model in a Markov Chain Monte Carlo (MCMC) approach. Here, we were particularly interested in the ratio dN/dS (i.e., omega, ω), which may inform about adaptation. The association between genital size and ω was estimated using a multivariate Brownian motion model, where the strength of the association was assessed using the partial correlation coefficient R, and the significance was determined by a posterior probability (Lartillot and Poujol 2011). We interpret R > 0.5 as moderate correlations, R > 0.7 as strong correlations and R > 0.85 as very strong correlations. We adopted a significance threshold for the correlations of 0.95 of posterior probability. MCMC burn-in was set to 15%, mixing and convergence were assessed using the module tracecomp within Coevol (Lartillot and Poujol 2011), and the analyses were stopped once a minimum effective sample size of 40 was achieved for the following parameters: log-prior, log-likelihood, tree length, dN/dS and the covariance matrix (Lartillot and Poujol 2011).
We next determined the most likely selection regime acting on the genes that showed significant association with genital size, that is, negative selection, neutral evolution, or positive selection. For this, we fit each alignment to a site model (M2a) in CodeML from PAML v. 4.9 (Yang 2007), which estimates omega independently for each site (i.e., codon). We used the same tree from the previous analysis and the Bayes Empirical Bayes criterion to determine the selection regime.
Supplementary Material
Acknowledgments
We are grateful to Dr David A. Rider for providing many of the articles used to gather morphological and developmental data and Gisele A. Cardoso for valuable comments on the manuscript and coding tips. This work was supported by FAPESP grants numbers 2019/10966-6 and 2020/05636-4 to T.T.T. and a postdoctoral fellowship to B.C.G. (process n. 18/18184-4). This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—with a PhD fellowship to DCC (Finance Code 001).
Contributor Information
Bruno C Genevcius, Department of Genetics and Evolutionary Biology, University of Sao Paulo, Sao Paulo (SP), Brazil.
Denis C Calandriello, Department of Genetics and Evolutionary Biology, University of Sao Paulo, Sao Paulo (SP), Brazil.
Tatiana T Torres, Department of Genetics and Evolutionary Biology, University of Sao Paulo, Sao Paulo (SP), Brazil.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
B.C.G. and T.T.T. designed the research. B.C.G. and D.C.C. collected the data. B.C.G. analyzed the data. B.C.G., D.C.C., and T.T.T. interpreted the results and wrote the paper.
Data availability
We provide as supplementary material the access codes to public transcriptomic data, developmental data with respective references, and morphological measurements.
References
- Abascal F, Zardoya R, Telford MJ. 2010. Translatorx: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ádám G, Perrimon N, Noselli S. 2003. The retinoic-like juvenile hormone controls the looping of left-right asymmetric organs in Drosophila. Development 130:2397–2406. [DOI] [PubMed] [Google Scholar]
- Almstrup K, et al. 2016. Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Sci Rep. 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuthnott D. 2009. The genetic architecture of insect courtship behavior and premating isolation. Heredity 103:15–22. [DOI] [PubMed] [Google Scholar]
- Aspiras AC, Smith FW, Angelini DR. 2011. Sex-specific gene interactions in the patterning of insect genitalia. Dev Biol. 360:369–380. [DOI] [PubMed] [Google Scholar]
- Averof M, Cohen SM. 1995. Wings from ancestral gills. Nature 639:636–639. [DOI] [PubMed] [Google Scholar]
- Ávila V, Campos JL, Charlesworth B. 2018. The effects of sex-biased gene expression and x-linkage on rates of sequence evolution in Drosophila. Mol Biol Evol. 35:655–665. [DOI] [PubMed] [Google Scholar]
- Baker J, Meade A, Venditti C. 2021. Genes underlying the evolution of tetrapod testes size. BMC Biol. 19:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W, Noll M. 2002. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development 129:5667–5681. [DOI] [PubMed] [Google Scholar]
- Breen TR. 1999. Mutant alleles of the Drosophila trithorax gene produce common and unusual homeotic and other developmental phenotypes. Genetics 152:319–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2014. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. [DOI] [PubMed] [Google Scholar]
- Campbell A, Frazer BD, Gilbert NGAP, Gutierrez AP, Mackauer M. 1974. Temperature requirements of some aphids and their parasites. J Appl Ecol. 431–438. [Google Scholar]
- Cao Y, Wu HT, Li M, Chen WT, Yuan ML. 2020. The complete mitochondrial genome of Nysius fuscovittatus (Hemiptera: Lygaeidae). Mitochondrial DNA Part B Resour. 5:3501–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. 2000. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development 127:1499–1508. [DOI] [PubMed] [Google Scholar]
- Chen SJ, Liu XY, Zhang JL, Yang ZN, Xu HJ. 2022. Abdominal-B contributes to abdominal identity in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol Biol. 9:1–10. [DOI] [PubMed] [Google Scholar]
- Chu D, Wei L. 2019. Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC Cancer 19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creevey CJ, McInerney JO. 2005. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics 21:390–392. [DOI] [PubMed] [Google Scholar]
- Cunningham MD, et al. 2012. Wapl antagonizes cohesin binding and promotes polycomb-group silencing in Drosophila. Development. 139:4172–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Lima AKS, Paschoaletto IP, De Oliveira Pinho L, Benmamman P, Klaczko J. 2019. Are hemipenial traits under sexual selection in Tropidurus lizards? Hemipenial development, male and female genital morphology, allometry and coevolution in Tropidurus torquatus (Squamata: Tropiduridae). PLoS One 14:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moya RS, et al. 2019. Deep instability in the phylogenetic backbone of Heteroptera is only partly overcome by transcriptome-based phylogenomics. Insect Syst Divers. 3:1–14. [Google Scholar]
- Deuring R, et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 5:355–365. [DOI] [PubMed] [Google Scholar]
- Dreyer AP, Shingleton AW. 2019. Insulin-insensitivity of male genitalia maintains reproductive success in Drosophila. Biol Lett. 15:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, McGuffin EE, Dauwalder B, Rabinow L, Mattox W. 1998. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol Cell. 2:741–750. [DOI] [PubMed] [Google Scholar]
- Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. 2012. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337:860–864. [DOI] [PubMed] [Google Scholar]
- Estrada B, Sánchez-Herrero E. 2001. The Hox gene abdominal-B antagonizes appendage development in the genital disc of Drosophila. Development 128:331–339. [DOI] [PubMed] [Google Scholar]
- Evans T, Loose M. 2015. Alignwise: a tool for identifying protein-coding sequence and correcting frame-shifts. BMC Bioinformatics. 16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthman M, Miller CW, Kimball RT. 2019. Phylogenomic analysis suggests Coreidae and Alydidae (Hemiptera: Heteroptera) are not monophyletic. Zool Scr. 48:520–534. [Google Scholar]
- Fraichard S, et al. 2010. Tenectin is a novel αPS2βPS integrin ligand required for wing morphogenesis and male genital looping in Drosophila. Dev Biol. 340:504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Sasabe M, Nagata N, Takami Y, Sota T. 2019. Genetic basis of species-specific genitalia reveals role in species diversification. Sci Adv. 5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse N, Hirose S, Hayashi S. 1996. Determination of wing cell fate by the escargot and snail genes in Drosophila. Development 122:1059–1067. [DOI] [PubMed] [Google Scholar]
- Galicia-Mendoza DI, Sanmartín-Villar I, García-Miranda Ó, Cordero-Rivera A. 2021. Territorial damselflies are larger and show negative allometry in their genitalia. Biol J Linn Soc. 134:697–706. [Google Scholar]
- Genevcius BC, et al. 2021. Phylogeny of the stink bug tribe Chlorocorini (Heteroptera, Pentatomidae) based on DNA and morphological data: the evolution of key phenotypic traits. Syst Entomol. 46:327–338. [Google Scholar]
- Genevcius BC, Baker J, Bianchi FM, Marvaldi AE. 2020. Female-driven intersexual coevolution in beetle genitalia. J Evol Biol. 33:957–965. [DOI] [PubMed] [Google Scholar]
- Genevcius BC, Caetano DS, Schwertner CF. 2017. Rapid differentiation and asynchronous coevolution of male and female genitalia in stink bugs. J Evol Biol. 30:461–473. [DOI] [PubMed] [Google Scholar]
- Genevcius BC, Schwertner CF. 2017. Strong functional integration among multiple parts of the complex male and female genitalia of stink bugs. Biol J Linn Soc. 122:774–786. [Google Scholar]
- Gonzalez-Crespo S, Morata G. 1995. Control of Drosophila adult pattern by extradenticle. Development 121:2117–2125. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N, Sánchez L, Guerrero I. 1999. Drosophila terminalia as an appendage-like structure. Mech Dev. 86:113–123. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen JFD, et al. 2021. Unraveling the genetic basis for the rapid diversification of male genitalia between Drosophila species. Mol Biol Evol. 38:437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JC. 1994. The mating of a fly. Science 264:1702–1714. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- House CM, Simmons LW. 2007. No evidence for condition-dependent expression of male genitalia in the dung beetle Onthophagus taurus. J Evol Biol. 20:1322–1332. [DOI] [PubMed] [Google Scholar]
- Johnson KP, et al. 2018. Phylogenomics and the evolution of hemipteroid insects. Proc Natl Acad Sci U S A. 115:12775–12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Youssef N, Susko E, Bielawski JP, Beaulieu J. 2020. A phenotype-genotype codon model for detecting adaptive evolution. Syst Biol. 69:722–738. [DOI] [PubMed] [Google Scholar]
- Keyte AL, Smith KK. 2014. Heterochrony and developmental timing mechanisms: changing ontogenies in evolution. Semin Cell Dev Biol. 34:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. 2018. Hox genes, clusters and collinearity. Int J Dev Biol. 62:659–663. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Poujol R. 2011. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol Biol Evol. 28:729–744. [DOI] [PubMed] [Google Scholar]
- Li T, et al. 2016. Ubr3, a novel modulator of Hh signaling affects the degradation of costal-2 and Kif7 through poly-ubiquitination. PLoS Genet. 12:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tian Y, Zhao Y, Bu W. 2012. Higher level phylogeny and the first divergence time estimation of heteroptera (insecta: Hemiptera) based on multiple genes. PLoS One 7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz DM, Tomoyasu Y. 2018. Dual evolutionary origin of insect wings supported by an investigation of the abdominal wing serial homologs in Tribolium. Proc Natl Acad Sci U S A. 115:E658–E667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. 2019. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera : Heteroptera) inferred from mitochondrial genome sequences. Syst Entomol 4:2–10. [Google Scholar]
- Lupše N, Cheng RC, Kuntner M. 2016. Coevolution of female and male genital components to avoid genital size mismatches in sexually dimorphic spiders. BMC Evol Biol. 16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno ALM, Moczek AP. 2015. Appendage-patterning genes regulate male and female copulatory structures in horned beetles. Evol Dev. 253:248–253. [DOI] [PubMed] [Google Scholar]
- Mallarino R, Abzhanov A. 2012. Paths less traveled: evo-devo approaches to investigating animal morphological evolution. Annu Rev Cell Dev Biol. 28:743–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Freeman M. 2009. Fasciclin 2, the Drosophila orthologue of neural cell-adhesion molecule, inhibits EGF receptor signalling. Development 136:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano-Martins P, Monfardini RD, Lo-Man-Hung N, Torres TT. 2022. Evidence of positive selection on six spider developmental genes. J Exp Zool Part B Mol Dev Evol 338:1–9. [DOI] [PubMed] [Google Scholar]
- Medved V, et al. 2015. Origin and diversification of wings: insights from a neopteran insect. Proc Natl Acad Sci U S A. 112:15946–15951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth CK, Frankino WA, Shingleton AW. 2016. Allometry and size control: what can studies of body size regulation teach us about the evolution of morphological scaling relationships? Curr Opin Insect Sci. 13:93–98. [DOI] [PubMed] [Google Scholar]
- Moreno-García M, Cordero C. 2008. On the function of male genital claspers in Stenomacra marginella (Heteroptera: Largidae). J Ethol. 26:255–260. [Google Scholar]
- Moury B, Simon V. 2011. DN/dS-based methods detect positive selection linked to trade-offs between different fitness traits in the coat protein of potato virus y. Mol Biol Evol. 28:2707–2717. [DOI] [PubMed] [Google Scholar]
- Nomura S, Fujisawa T, Sota T. 2021. Role of sex-concordant gene expression in the coevolution of exaggerated male and female genitalia in a beetle group. Mol Biol Evol. 38:3593–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E, et al. 2013. Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol Phylogenet Evol. 68:42–54. [DOI] [PubMed] [Google Scholar]
- Orbach DN, Hedrick B, Würsig B, Mesnick SL, Brennan PLR. 2018. The evolution of genital shape variation in female cetaceans. Evolution 72:261–273. [DOI] [PubMed] [Google Scholar]
- Organ CL, Cooper LN, Hieronymus TL. 2015. Macroevolutionary developmental biology: embryos, fossils, and phylogenies. Dev Dyn. 244:1184–1192. [DOI] [PubMed] [Google Scholar]
- Paradis E, Strimmer K, Claude J, Noel Y, Bolker B. 2003. Analyses of phylogenetics and evolution: the ape package. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Partha R, et al. 2017. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. Elife 6:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzoldt AG, et al. 2012. DE-Cadherin regulates unconventional Myosin ID and Myosin IC in Drosophila left-right asymmetry establishment. Development 139:1874–1884. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. 1997. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91:171–183. [DOI] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 13:505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset R, et al. 2010. The Drosophila serine protease homologue Scarface regulates JNK signalling in a negative-feedback loop during epithelial morphogenesis. Development 137:2177–2186. [DOI] [PubMed] [Google Scholar]
- Rudoy A, Ribera I. 2016. The macroevolution of size and complexity in insect male genitalia. PeerJ. 4:e1882:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruschel TP, Bianchi FM, Campos LA. 2019. Genital coupling, morphology and evolution of male holding structures in Cicadinae (Hemiptera: Cicadidae). Biol J Linn Soc 128:1–16. [Google Scholar]
- Sánchez L, Casares F, Gorfinkiel N, Guerrero I. 1997. The genital disc of Drosophila melanogaster. II. Role of the genes hedgehog, decapentaplegic and wingless. Dev Genes Evol. 207:229–241. [DOI] [PubMed] [Google Scholar]
- Sanger TJ, Rajakumar R. 2019. How a growing organismal perspective is adding new depth to integrative studies of morphological evolution. Biol Rev. 94:184–198. [DOI] [PubMed] [Google Scholar]
- Schulze S, et al. 2001. Essential genes in proximal 3L heterochromatin of Drosophila melanogaster. Mol Gen Genet. 264:782–789. [DOI] [PubMed] [Google Scholar]
- Singh-Pruthi H. 1925. The morphology of the male genitalia in rhynchota. Trans Entomol Soc London 1:127–267. [Google Scholar]
- Singh M. 1971. Development of male reproductive organs of Chrysocoris stollii. J Kansas Entomol Soc. 44:433–440. [Google Scholar]
- Smith MD, et al. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Davidson LA, Rebeiz M. 2020. Evolutionary expansion of apical extracellular matrix is required for the elongation of cells in a novel structure. Elife 9:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Rebeiz M, Davidson L. 2018. From pattern to process: studies at the interface of gene regulatory networks, morphogenesis, and evolution. Curr Opin Genet Dev. 51:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. 2009. Species-specificity of male genitalia is characterized by shape, size, and complexity. Insect Syst Evol. 40:159–170. [Google Scholar]
- Tanaka KM, et al. 2015. Genetic architecture and functional characterization of genes underlying the rapid diversification of male external genitalia between Drosophila simulans and Drosophila mauritiana. Genetics 200:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KM, Kamimura Y, Takahashi A. 2018. Mechanical incompatibility caused by modifications of multiple male genital structures using genomic introgression in Drosophila. Evolution 72:2406–2418. [DOI] [PubMed] [Google Scholar]
- Tang HY, Smith-Caldas MSB, Driscoll MV, Salhadar S, Shingleton AW. 2011. FOXO Regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Nishimura T, Hirayama A, Takami Y. 2021. Heterochrony and growth rate variation mediate the development of divergent genital morphologies in closely related Ohomopterus ground beetles. Evol Dev. 23:19–27. [DOI] [PubMed] [Google Scholar]
- Usami T, Yokoyama J, Kubota K, Kawata M. 2006. Genital lock-and-key system and premating isolation by mate preference in carabid beetles (Carabus subgenus Ohomopterus). Biol J Linn Soc Lond. 87:145–154. [Google Scholar]
- Velentzas PD, et al. 2018. The proton-coupled monocarboxylate transporter hermes is necessary for autophagy during cell death. Dev Cell. 47:281–293.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BJ, et al. 2019. An atlas of transcription factors expressed in male pupal terminalia of Drosophila melanogaster. G3 Genes, Genomes, Genet 9:3961–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2017. Comparative mitogenomic analysis of mirid bugs (Hemiptera: Miridae) and evaluation of potential DNA barcoding markers. Peer J 5:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, et al. 2019. When did the ancestor of true bugs become stinky? Disentangling the phylogenomics of Hemiptera-Heteroptera. Cladistics 35:42–66. [DOI] [PubMed] [Google Scholar]
- Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. 2011. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 121:4118–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RR, Liang AP, Webb MD. 2009. A new tropiduchid planthopper genus and species from China with descriptions of in copula genitalic structures (Hemiptera: Fulgoromorpha). Syst Entomol. 34:434–442. [Google Scholar]
- Warton DI, Duursma RA, Falster DS, Taskinen S. 2012. Smatr 3- an R package for estimation and inference about allometric lines. Methods Ecol Evol. 3:257–259. [Google Scholar]
- Womack MC, Fiero TS, Hoke KL. 2018. Trait independence primes convergent trait loss. Evolution 72:679–687. [DOI] [PubMed] [Google Scholar]
- Xu J, Yu Y, Chen K, Huang Y. 2019. Intersex regulates female external genital and imaginal disc development in the silkworm. Insect Biochem Mol Biol. 108:1–8. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2016. Evolution of the assassin’s arms: insights from a phylogeny of combined transcriptomic and ribosomal DNA data (Heteroptera: Reduvioidea). Sci Rep. 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Xie YC, Li HJ, Zhuo JC, Zhang CX. 2021. Pleiotropic roles of the orthologue of the Drosophila melanogaster intersex gene in the brown planthopper. Genes (Basel) 12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We provide as supplementary material the access codes to public transcriptomic data, developmental data with respective references, and morphological measurements.