Abstract
Thermoprofundales, formerly Marine Benthic Group D (MBG-D), is a ubiquitous archaeal lineage found in sedimentary environments worldwide. However, its taxonomic classification, metabolic pathways, and evolutionary history are largely unexplored because of its uncultivability and limited number of sequenced genomes. In this study, phylogenomic analysis and average amino acid identity values of a collection of 146 Thermoprofundales genomes revealed five Thermoprofundales subgroups (A–E) with distinct habitat preferences. Most of the microorganisms from Subgroups B and D were thermophiles inhabiting hydrothermal vents and hot spring sediments, whereas those from Subgroup E were adapted to surface environments where sunlight is available. H2 production may be featured in Thermoprofundales as evidenced by a gene cluster encoding the ancient membrane-bound hydrogenase (MBH) complex. Interestingly, a unique structure separating the MBH gene cluster into two modular units was observed exclusively in the genomes of Subgroup E, which included a peripheral arm encoding the [NiFe] hydrogenase domain and a membrane arm encoding the Na+/H+ antiporter domain. These two modular structures were confirmed to function independently by detecting the H2-evolving activity in vitro and salt tolerance to 0.2 M NaCl in vivo, respectively. The peripheral arm of Subgroup E resembles the proposed common ancestral respiratory complex of modern respiratory systems, which plays a key role in the early evolution of life. In addition, molecular dating analysis revealed that Thermoprofundales is an early emerging archaeal lineage among the extant MBH-containing microorganisms, indicating new insights into the evolution of this ubiquitous archaea lineage.
Keywords: archaea, evolution, horizontal gene transfer, hydrogenase, marine benthic group d, metagenomics
Introduction
Archaeal cells are abundant and widespread in diverse habitats on Earth (∼1.1 × 1029 cells), where they are estimated to constitute 37% and 87% of all prokaryotes in the global ocean and deep subsurface marine sediments, respectively (Lipp et al. 2008; Hoshino and Inagaki 2019). However, our understanding of the mechanisms underlying archaeal life is still limited when compared with that of other microorganisms (Baker et al. 2020). With the rapid expansion of high-quality archaeal genomes from pure isolates, metagenome-assembled genomes (MAGs) and single-cell amplified genomes (SAGs), the archaeal tree of life has expanded dramatically to at least 30 archaeal phyla (Tahon et al. 2021).
Thermoprofundales is an archaeal order within the class Thermoplasmata, which is formerly known as MBG-D (Marine Benthic Group D) (Vetriani et al. 1999), DHVEG-1 (Deep-sea Hydrothermal Vent Euryarchaeota Group 1) (Takai and Horikoshi 1999), or Izemarchaea (Adam et al. 2017). Multiple studies based on 16S rRNA gene sequencing indicate that Thermoprofundales is an abundant and cosmopolitan lineage widely distributed in sedimentary habitats. Approximately 70% of the available Thermoprofundales sequences were identified in samples collected from mangrove surface sediments, open ocean, and hydrothermal vents (Zhou et al. 2018), and Thermoprofundales often contributes more than 10% to total archaeal sequences in the community (Lloyd et al. 2013). This lineage is also able to adapt to distinct environmental conditions, including the tolerance of extreme ranges in salinity (freshwater vs. hypersaline) and temperature (polar vs. hydrothermal) (Swan et al. 2010; Zhou et al. 2018). Lokiarchaeota, Hadesarchaea, and anaerobic methanotrophic archaea (ANME) commonly co-occur with Thermoprofundales, suggesting the existence of syntrophic interactions or similar selective pressures in a yet unknown manner (Inagaki et al. 2006; Zhou et al. 2018).
Recently, the sequencing of some Thermoprofundales MAGs and SAGs provided preliminary insights into the metabolic features of these microorganisms. They encode a large number of genes for extracellular peptidases, such as gingipain, clostripain, and collagenase, which are common and active in the sediments from Aarhus Bay (Denmark) (Lloyd et al. 2013), White Oak River estuary (USA) (Lazar et al. 2017), and Shenzhen Futian mangrove (China) (Zhou et al. 2018). Moreover, acetate and ethanol might be produced by Thermoprofundales through anaerobic fermentation processes (Lazar et al. 2017; Zhou et al. 2018). Given the ubiquity and predominance of Thermoprofundales in sedimentary environments, it has been proposed that this archaeal lineage contributes significantly to global carbon cycles (Lloyd et al. 2013; Zhou et al. 2018). However, Thermoprofundales remains one of the least studied archaeal groups, because of the limited number of sequenced genomes and the lack of pure/enriched cultures.
Here, we aim to reveal the phylogenetic diversity, metabolic features, and evolutionary history of Thermoprofundales through the metagenomic mining of 146 genomes obtained in this study or retrieved from public databases. Our analyses included the identification of five monophyletic Thermoprofundales lineages, which inhabit distinct environments and possess versatile metabolic pathways. The evolutionary history of Thermoprofundales was brought to light by mapping events of horizontal gene transfer (HGT) and gene gain and loss at its ancestral node. A unique gene cluster encoding the membrane-bound hydrogenase (MBH) complex stood out as an ancient trait, because the MBH is among the 62 core proteins in the anaerobic prokaryote common ancestor and it represents the simplest form of respiration in extant organisms (Sousa et al. 2016; Yu et al. 2018). The hydrogenase activity of the large subunit protein of the MBH gene cluster was confirmed by measuring H2 production in vitro. Our findings substantially expand understanding of the metabolism and evolution of the cosmopolitan sedimentary archaea Thermoprofundales.
Results and Discussion
An Abundant Archaeal Lineage in Sedimentary Environments Worldwide
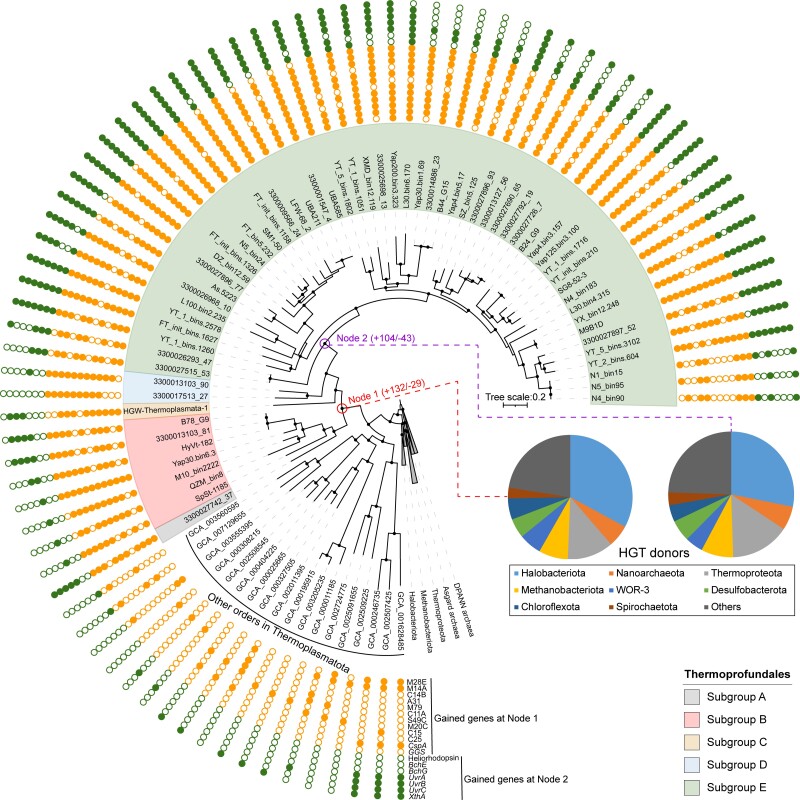
A total of 146 Thermoprofundales genomes (>50% completeness and <5% contamination) were retrieved from metagenomes and single cells sequenced in this study (55 MAGs), and downloaded from the NCBI Assembly database (53 MAGs and 2 SAGs) (Coordinators 2018) and the genomic catalog of Earth’s microbiomes (36 MAGs) (Nayfach et al. 2020). A total of 63 genomes are more than 80% complete (supplementary table S1, Supplementary Material online). The phylogenomic tree based on 55 conserved single-copy ubiquitous archaeal genes (Dombrowski et al. 2020) showed that Thermoprofundales forms a monophyletic lineage in the Candidatus phylum Thermoplasmatota and its ancestral node is adjacent to the orders Methanomassiliicoccales, Thermoplasmatales, Acidiprofundales, Poseidoniales, and MGIII (fig. 1) (Rinke et al. 2021). The average amino acid identity (AAI) value between each genome in Thermoprofundales and that from its adjacent orders was <50% (supplementary fig. S1 and table S2, Supplementary Material online), confirming that Thermoprofundales is an order-level lineage (Zhou et al. 2018) according to the proposed standards for high-quality taxa descriptions of uncultivated microorganisms (Konstantinidis et al. 2017; Rinke et al. 2021).
Fig. 1.
Phylogenomic tree of 146 Thermoprofundales genomes. Different colors covering the tree branches indicate the five Thermoprofundales subgroups. The filled or hollow star at each genome ID indicates the full or partial MBH subunit genes, respectively. The colors of the inner ring indicate the availability of light. The colors of the middle ring indicate the temperature range. The colors of the outer ring indicate the environmental source of each genome. Ultrafast bootstrapping was used to estimate the reliability of each branch with 1,000 times resampling, and the nodes with a bootstrap value >80 are marked with black dots.
Global distribution analysis as revealed by the 16S rRNA genes and MAGs both showed that Thermoprofundales is distributed in various sedimentary environments with wide temperature and salinity ranges (supplementary fig. S2, Supplementary Material online), presumably contributing to global carbon biogeochemical cycles as reported previously (Lloyd et al. 2013; Zhou et al. 2018). Based on the 16S rRNA gene, the relative abundances of Thermoprofundales were 0.1–6.3% in freshwater sediment, 0.3–8.6% in saline lake sediment, 0.1–17.1% in intertidal sediment, and 0.1–4.3% in marine sediment, which was consistent with those based on the MAGs abundance (supplementary table S3, Supplementary Material online). The highest relative abundances of Thermoprofundales as revealed by 16S rRNA genes and MAGs were found in sedimentary environments, such as an intertidal sediment from the southern coast of the United States (17.1%) and a hydrothermal sediment from the Guaymas Basin in the Gulf of California (24.5%), respectively. When compared with the upper water column which is dominated by free-living microorganisms, sedimentary habitats generally have low dissolved oxygen levels and high organic matter content, where abundant anaerobic heterotrophs adopt nonmotile or particle-attached lifestyles (Orcutt et al. 2011; Zhou et al. 2018).
Five Subgroups with Distinct Habitat Preferences and Metabolic Features
The Thermoprofundales MAGs were classified into five subgroups according to their phylogenetic positions (fig. 1) and AAI values (supplementary fig. S1, Supplementary Material online), whose average values between each pair of genomes in the same subgroup were higher than 63.0% (supplementary table S2, Supplementary Material online). Genomes from Subgroups B, D, and E showed distinct habitat preferences (fig. 1). For example, 76.0% and 100% of the genomes from Subgroup B and Subgroup D were identified in sediments from deep-sea hydrothermal vents, which are characterized by extremely high temperatures for microorganisms (e.g., >60 °C) (Dombrowski et al. 2018). This is further supported by the identification of a reverse gyrase gene (the hallmark gene of some hyperthermophiles and/or moderate thermophiles (Campbell et al. 2009; Lipscomb et al. 2017; Feng et al. 2019)) exclusively in genomes of the Subgroups B and D (supplementary fig. S3 and table S4, Supplementary Material online). Therefore, these two subgroups seem to comprise thermophiles, and the result is consistent with a recent report in which Subgroups B and D were assigned as JdFR-43 and HyVt, respectively (Liu et al. 2022). However, the preferred habitats for Subgroups A and C were not revealed because of the underrepresentation of genomes in these two subgroups (two for Subgroup A and one for Subgroup C) (fig. 1).
A survey combining the 16S rRNA gene sequences identified in the MAGs of this study with those from a previous report (Zhou et al. 2018) showed that the sequences of Subgroup E clustered in a monophyletic lineage, which included clades from 1 to 11 as classified previously (Zhou et al. 2018) (supplementary fig. S4, Supplementary Material online). The lineage of Subgroup E had the highest diversity among the five subgroups based on the MAGs. A high percentage (84.2%) of the MAGs were obtained from terrestrial or marine surface environments (e.g., freshwater river, intertidal zone, shrimp pond, saline lake, and seawater), which are characterized by high sunlight incidence and mild temperatures (15–40 °C) (fig. 1; supplementary table S1, Supplementary Material online). Interestingly, a gene encoding the light-sensing heliorhodopsin was identified exclusively in Subgroup E (supplementary fig. S5, Supplementary Material online). However, phototrophy might not be supported in Thermoprofundales because heliorhodopsin has not yet to be confirmed to display proton-pumping activity (Pushkarev et al. 2018), and the genomes of Thermoprofundales lack the key genes for photosynthesis (PsaAB and PsbAD). Nevertheless, the heliorhodopsin gene in Thermoprofundales was always accompanied by several protein-encoding genes involved in carotenoid biosynthesis (CrtZ, CrtB, CrtD, CruA, and CrtU) and repair of DNA damage caused by light-induced oxidative stress (PhrB and UvsE) (Meulenbroek et al. 2013; Zhang et al. 2017) in the same gene cluster (supplementary fig. S6, Supplementary Material online). Carotenoid production occurs often in phototrophic microorganisms, such as photosynthetic bacteria (e.g., Arthrospira, Rhodobacter, and Rhodosporidium), extremely halophilic archaea (Halobacteria), microalgae, and some yeast (Mussagy et al. 2019). In Subgroup E, some genomes possess complete gene sets for the de novo biosynthesis of bacterioruberin and zeaxanthin (fig. 2), and key genes such as bisanhydrobacterioruberin hydratase (CruF) and beta-carotene 3-hydroxylase (CrtZ) were first detected in the Candidatus phylum Thermoplasmatota (Rinke et al. 2021) (supplementary figs. S7 and S8, Supplementary Material online). Carotenoids produced by these pathways might act as antioxidants and protect cells against oxidative damage, such as that resulting from exposure to sunlight, UV radiation, and/or H2O2 (Giani et al. 2019). Due to the adjacent gene arrangement among heliorhodopsin, carotenoid biosynthesis, and DNA damage repair, we speculate that the heliorhodopsin in Subgroup E functions as a light sensor to mitigate light-induced oxidative stress, probably by regulating the expression of genes for carotenoid biosynthesis and light-induced DNA damage repair (Pushkarev et al. 2018; Bulzu et al. 2021).
Fig. 2.
Reconstructed metabolic pathways of Thermoprofundales. A full list of genes labeled with different letters is provided in supplementary table S4, Supplementary Material online.
Evolutionary History of Thermoprofundales
To shed light on the evolutionary history of Thermoprofundales, gene gain and loss events were predicted by mapping all the orthologous genes found in high-quality genomes (>80% completeness and <5% contamination) to a phylogenomic tree. In general, 132 and 29 genes were gained and lost at the ancestral node of Thermoprofundales, respectively (Node 1 in fig. 3). For example, multiple genes involved in protein degradation were gained at this node (supplementary table S5, Supplementary Material online), making Thermoprofundales an important protein degrader in global sedimentary environments (Lloyd et al. 2013; Lazar et al. 2017; Zhou et al. 2018). Two genes related to survival in fluctuating environmental conditions were gained: a gene for cold shock protein CspA and a gene for glucosylglycerate synthase (GGS). These two genes might help Thermoprofundales to cope with multiple stressful conditions, such as harsh temperatures, osmotic pressure, oxidative stress, starvation, and pH extremes (Keto-Timonen et al. 2016; Nunes-Costa et al. 2017). Of note, the evolution of Subgroup E genomes had unique features when compared with other subgroups (104 gains and 43 losses; Node 2 in fig. 3). Among the 104 gained genes at the ancestral node of Subgroup E, a heliorhodopsin gene and two genes involved in chlorophyll synthesis (BchE and BchG) possibly facilitate the utilization of sunlight (supplementary table S6, Supplementary Material online). Meanwhile, four DNA repair genes (UvrABC and XthA), which might participate in the removal of damaged bases caused by strong sunlight (Verhoeven et al. 2002), were also gained at Node 2. This suggests that microorganisms in Subgroup E evolved strategies to both use light energy and prevent its damage, which may be driving forces to separate this subgroup from others during the evolution of Thermoprofundales.
Fig. 3.
Ancestral genome content reconstruction and HGT donor predictions of Thermoprofundales. The solid or open circles outside the tree represent the presence or absence of the genes, respectively. Node 1 and Node 2 indicate the ancestral node of all Thermoprofundales genomes and Subgroup E, respectively. The values in parentheses indicate the number of gained (+) and lost (−) genes, and full lists of the genes are provided in supplementary table S5 and S6, Supplementary Material online. The pie charts show the ratios of HGT donors. The phylogenomic tree based on 55 conserved single-copy ubiquitous archaeal genes was constructed using IQ-TREE with ModelFinder. Ultrafast bootstrapping was used to estimate the reliability of each branch with 1,000 times resampling, and the nodes with a bootstrap value >80 are marked with black dots.
HGT is the process of acquiring foreign DNA from other organisms through transformation, transduction, and conjugation (e.g., conjugative plasmids) (Douglas and Langille 2019). It is a major driving force for microbial evolution and plays important roles in archaeal speciation, metabolism, and adaptation (Wagner et al. 2017; Brito 2021). On average, 3.5% of the genes in a Thermoprofundales genome were identified as HGT genes, and 70.4% of these HGTs were derived from other distantly related archaeal genomes rather than bacterial ones (fig. 3; supplementary table S7, Supplementary Material online). Halobacteriota and Thermoproteota (GTDB taxonomy (Rinke et al. 2021), formerly a group of archaea including Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota) were the major HGT donors. This result is in contrast with the hypothesis that most of the HGTs in some mesophilic archaea originated from bacteria (López-García et al. 2015), and previous reports showed that gene transfer from bacteria to archaea occurred five times more frequently than from archaea to bacteria (Nelson-Sathi et al. 2015). The functional patterns between archaeal and bacterial-derived HGTs were further compared using the Thermoprofundales genomes at Nodes 1 and 2 in figure 3, respectively. The result revealed that archaeal donors contributed more HGTs in the processes of transcription, and carbohydrate transport and metabolism. However, bacterial donors contributed more HGTs related to energy production and conversion, cell wall/membrane/envelope biogenesis, and translation, ribosomal structure, and biogenesis (supplementary fig. S9, Supplementary Material online). In addition, we examined the HGT donors in genomes from other orders in the Candidatus phylum Thermoplasmatota as proposed elsewhere (Rinke et al. 2021), including Acidiprofundales, Thermoplasmatales, Methanomassiliicoccales, Poseidoniales, and MGIII (supplementary fig. S10, Supplementary Material online). More than 53% of the HGTs identified in Acidiprofundales, Thermoplasmatales, and Methanomassiliicoccales were derived from other archaeal genomes. However, more than 92% of the HGTs in Poseidoniales and MGIII were from bacterial genomes (supplementary fig. S10, Supplementary Material online). This result minimizes the potential biases caused by the overall dominance of bacterial genomes in the reference database (191,527 bacterial vs. 3,073 archaeal genomes in GTDB Release 95; Rinke et al. 2021), and it further confirms that Thermoprofundales acquired most of its HGTs from other distantly related archaeal species.
In addition, 437 sequences from the Thermoprofundales genomes were predicted as plasmids using PlasFlow (Krawczyk et al. 2018) and PlasClass (Pellow et al. 2020) (supplementary table S8, Supplementary Material online). Plasmids are mobile genetic elements (MGEs), and they contain nonchromosomal fragments of DNA and facilitate the fast evolution and adaptation of microorganisms to diverse environments (Heuer and Smalla 2012). They typically lack essential genes for microbial growth, but possess genes involved in response to changes in environmental conditions or exposure to pollutants (e.g., heavy metals and antibiotics) (Carattoli 2013). Interestingly, the gene encoding an extracellular protein-degrading enzyme for gingipain (Merops family C25) was located in the plasmids of 27 genomes. This enzyme, which participates in the protein remineralization process, is abundant and active in samples from Aarhus Bay sediments of Denmark (Lloyd et al. 2013). Some structural genes encoding the archaellum were located in plasmids from 19 Thermoprofundales genomes, including some key genes for archaellum rotation and assembly (FlaI, FlaJ, and FlaB). As a motility structure peculiar to the Archaea domain (Albers and Jarrell 2018), the archaellum may enable Thermoprofundales to move toward favorable micro-niches (Herzog and Wirth 2012), and/or mediate its surface attachment, cell–cell communication, and extracellular electron exchange (Näther et al. 2006; Walker et al. 2019). Six Thermoprofundales plasmids also encoded the complete gene sets for riboflavin biosynthesis (RibBHE), a precursor of the coenzymes flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD). These two coenzymes are major electron carriers in multiple and ubiquitous redox reactions involved in energy conservation pathways and cellular processes (Thakur et al. 2017).
H2 Production by an Ancient MBH Complex
Phylogenetic reconstruction revealed that 71.2% of the genomes from the five Thermoprofundales subgroups encoded a gene of the Group 4d [NiFe] hydrogenase, which is an MBH-type H2-producing hydrogenase (Vignais and Billoud 2007) (fig. 4a; supplementary table S4, Supplementary Material online). The MBH complex is composed of 14 subunits, including a membrane arm and a peripheral arm (Schut et al. 2013). The membrane arm is formed by ten subunits (MbhABCDEFGHIM) located in the cell membrane, where it functions as an Na+/H+ antiporter domain (fig. 2). The peripheral arm is formed by four subunits (MbhJKLN) exposed to the cytoplasm, where it functions as a [NiFe] hydrogenase domain (Yu et al. 2018). The cryo-EM structure of this enzyme from a hyperthermophilic archaeon, Pyrococcus furiosus, was recently obtained. It is reported that the H2-evolving catalytic site is located at the E21 and the [NiFe] center (C68, C71, C374, and C377) of the large subunit MbhL (Yu et al. 2018). Sequence alignment and 3D structure reconstruction of the MbhL coding genes revealed that all the H2-evolving catalytic sites are conserved in Thermoprofundales, when compared with P. furiosus (supplementary fig. S11, Supplementary Material online). Furthermore, the MbhL gene from Thermoprofundales FT_bin5.232 was expressed in Escherichia coli BL21 strain to test its activity. Consistent with previous reports (Sapra et al. 2000; McTernan et al. 2015), the purified enzyme showed catalytic activity for H2 production as assayed by gas chromatography (fig. 4b).
Fig. 4.
(a) Phylogenetic tree of Thermoprofundales based on the MbhL gene. The shaded area indicates the position of the MbhL gene, and the red branches indicate the MbhL genes of Thermoprofundales. The label at each clade indicates a hydrogenase group. (b) H2 production by the recombinant MbhL protein from Thermoprofundales FT_bin5.232. Hydrogenase activity was measured in a reaction medium containing the recombinant MbhL protein or inactivated MbhL protein. A blank assay was run without any protein. The dashed line marks the H2 peak by gas chromatography.
Hydrogen metabolism (including H2 oxidation and production) is one of the most ancient and widespread metabolic traits, whose metabolite (i.e., H2) is widely used to generate energy to support microbial growth (Lane et al. 2010; Greening et al. 2016). Microorganisms produce H2 by using the [NiFe]-hydrogenase (Groups 3b and 4d) or [FeFe]-hydrogenase (Greening et al. 2016; Søndergaard et al. 2016). These H2 producers contribute to the major biogenic pool of H2 on Earth (e.g., anoxic sediments and gastrointestinal tracts) (Greening and Boyd 2020). In Thermoprofundales, the production of H2 is catalyzed by the MBH-type [NiFe] hydrogenase, and reduced ferredoxin is the natural electron donor of this enzyme (Yu et al. 2018). Various genes coding for the production of reduced ferredoxin were detected in the Thermoprofundales genomes, showing that other components needed for hydrogen production are not a limitation in this microorganism (fig. 2). For example, identified pathways that lead to the generation of reduced ferredoxin include pyruvate and 2-oxoglutarate oxidation (pyruvate ferredoxin oxidoreductase, 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase) in the tricarboxylic acid cycle, and glyceraldehyde oxidation (aldehyde ferredoxin oxidoreductase) in the glycolysis pathway.
New Insights into the Evolution of Modern Respiratory Systems
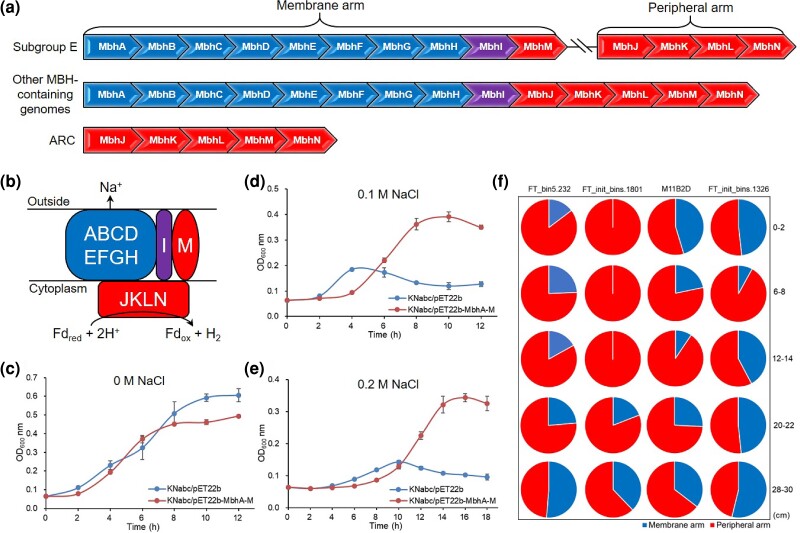
In Subgroup E, gene arrangement of the 14 subunits of MBH differed from those in subgroups A, B, C, D, and P. furiosus (Yu et al. 2018) (fig. 5a). In the genomes of Subgroup E, the peripheral arm genes of the [NiFe] hydrogenase domain were separated from the membrane arm genes of the Na+/H+ antiporter domain. A detailed survey of all the genomes in the NCBI GenBank database (January 25, 2021) further revealed that the discontinuous gene arrangement of MBH was unique to Thermoprofundales Subgroup E in Bacteria and Archaea (fig. 6). Phylogenetic analysis of the concatenated 4 peripheral arm genes (MbhJKLN), the 10 membrane arm genes (MbhABCDEFGHIM), and the complete 14 genes (MbhA–N) showed a consistent placement of Thermoprofundales as a monophyletic lineage adjacent to some Aciduliprofundum genomes (fig. 6, supplementary fig. S12, Supplementary Material online). These results are also coherent with the genome tree of all the MBH-containing genomes, revealing that the genes for the peripheral and membrane arms in Thermoprofundales are evolutionary conserved.
Fig. 5.
(a) Gene arrangement of the MBH complex in Thermoprofundales. ARC, the proposed common ARC. (b) Illustration of the MBH complex. (c–e) Growth performances of Escherichia coli KNabc strains cultured in 0, 0.1, and 0.2 M NaCl, respectively. KNabc/pET-22b-MbhA-M, a KNabc strain containing a gene cluster MbhABCDEFGHIM inserted into a pET-22b(+) plasmid. KNabc/pET-22b, a KNabc strain containing an empty pET-22b(+) plasmid. Three replicates are performed for each treatment. (f) Relative transcriptional abundances of the peripheral and membrane arms in four MAGs from the FT mangrove sediments. The numbers at the right of each row indicate sediment depth intervals of the FT metatranscriptomes.
Fig. 6.
(a) Phylogenomic tree of the MBH-containing archaeal genomes in the NCBI GenBank database. The tree based on 55 archaeal marker genes is constructed using IQ-TREE with ModelFinder. (b) Phylogenetic tree of the concatenated 14-subunits MBH gene in the NCBI GenBank database. The colors covering the tree branches indicate different archaeal lineages. The nodes with a bootstrap value >80 are marked with black dots.
Metatranscriptomic analysis of the FT mangrove sediments revealed distinct transcriptional patterns between the genes of the peripheral arm and the membrane arm in some MAGs from Subgroup E (fig. 5f), suggesting that these two arms could function independently. To further confirm this, a recombinant plasmid containing the membrane arm gene cluster MbhABCDEFGHIM from FT_bin5.232 was introduced into an antiporter-deficient strain E. coli KNabc, which is commonly used to test the function of Na+/H+ antiporters (Shao et al. 2021). In general, when cultured in LBK medium containing either 0.1 or 0.2 M NaCl, a much better growth performance was observed for KNabc transformed with the membrane arm plasmid than that for the same strain with an empty plasmid (fig. 5c–e). This showed that the membrane arm protein of Subgroup E partially compensated the deficiency of KNabc for growing at high salt concentrations, confirming its function as an Na+ transporter.
Molecular dating analysis revealed that Subgroup E was an early diverged lineage among all the MBH-containing microorganisms in Thermoplasmatota (fig. 7). The ancestor of Subgroup E might have emerged from a habitat that resembles the sediments of terrestrial surface (fig. 1), which are characterized by mild temperature (<40 °C), low salinity (<2%), low oxygen availability, and intense sunlight incidence, when compared with the deep sea. The ancestor of Subgroup E possibly experienced extensive gene duplication, rearrangement, and HGT events throughout its evolutionary history, including changes in the region of the MBH gene cluster. The separation of MBH into two modules, as observed in Subgroup E, might facilitate the adaption to the aquatic environments on the terrestrial surface. This would be achieved through a more efficient but less energy-consuming processes for hydrogen production and proton pumping, respectively. For example, 1) the synthesis of a 4-subunits (MbhJKLN) instead of a 14-subunits (MbhA–N) hydrogenase complex is functional for hydrogen production; 2) Subgroup E microorganisms may have a reduced need to pump out excess sodium when compared with other subgroups, since most of the genomes in Subgroup E were retrieved from terrestrial habitats with low salinity. These speculations are supported by the much higher transcriptional abundances of the peripheral arm than those of the membrane arm in four MAGs from the FT mangrove sediments (fig. 5f), showing that hydrogen production is more transcriptionally active than proton pumping in Subgroup E.
Fig. 7.
Evolutionary timeline of the MBH-containing archaeal lineages. The values in parentheses are the posterior 95% confidence intervals.
Interestingly, the gene structure of the peripheral arm in Subgroup E resembles that of the proposed common ancestral respiratory complex (ARC) of modern respiratory systems such as MBH and complex I (fig. 5a). The ARC, encoded by 5-subunits genes (MbhJKLMN), is a core protein module that can be traced back to the Last Universal Common Ancestor (LUCA) of Bacteria and Archaea; it is thought to function independently as a hydrogen-producing enzyme, which plays a key role in the early evolution of life (Schut et al. 2016). It is hypothesized that MBH shares a common ancestor with Complex I, and both of them possibly evolve from the Mrp (multiple resistance and pH adaptation) antiporter by acquiring a membrane-anchored hydrogenase module (Yu et al. 2018). The Mrp antiporter is an Na+ (or K+)/H+ exchanger representing an ancestor of many essential redox-driven proton pumps including MBH and Complex I (Steiner and Sazanov 2020). The conserved structural features in the two complexes indicate a similar energy conservation mechanism between the peripheral and membrane arms. However, a major difference between the two complexes is the absence of a proposed sodium ion translocation unit in the membrane arm of Complex I, but it is shared between MBH and Mrp antiporter (Yu et al. 2018). This indicates that the membrane arm of MBH evolves earlier than that of Complex I. Meanwhile, the phylogenetic tree of representative [NiFe]-hydrogenase groups also showed that the MbhL gene in the peripheral arm of MBH emerged earlier than the NuoD gene of complex I (fig. 4a). A primary driving force in the evolution of ARC to modern respiratory complexes may be the availability of oxygen and associated high redox potential compounds (Schut et al. 2016). However, when compared with ARC, the lack of an MbhM subunit in the peripheral arm of Subgroup E does not appear to impact its function as a hydrogenase, because the MbhM is reported to be a membrane anchor for the peripheral arm and it did not show any ion translocation activity in P. furiosus (Yu et al. 2018).
Conclusion
Our study provides a comprehensive insight into the abundant and widespread sedimentary archaeal group Thermoprofundales, and it expands the knowledge of Thermoprofundales’ diversity, evolution, and ecological roles in global biogeochemical cycles. The genomic evolution of Thermoprofundales is driven by HGT events, of which ∼70.5% are derived from other distantly related archaeal lineages. Thermoprofundales possesses a separated two modular gene structure of MBH with high similarity to the ARC system, and the two modules were confirmed to be functionally independent and active. Our findings uncover an unprecedented role of Thermoprofundales as an H2 producer in global sedimentary environments, and Thermoprofundales may fuel other H2 consumers and facilitate extensive microbe-microbe interactions in the community.
Materials and Methods
Sample Collection, Nucleic Acid Extraction and Sequencing
The seawater samples of the Yap metagenomes were obtained in the Yap Trench area of the western Pacific during the 37th Dayang cruise in 2016. Seawater samples were collected by using CTD SBE911plus (Sea-Bird Electronics, USA) at depth intervals of 4, 30, 75, 125, 200, and 2,000 mbsl. Detailed information is described elsewhere (Zhang et al. 2018; Liu et al. 2021). Briefly, 8 l of each seawater sample was filtered through a 0.22 μm-mesh membrane filter immediately after recovery onboard. The membrane was cut into ∼0.2 cm2 pieces with flame-sterilized scissors, and the DNA was extracted using PowerSoil® DNA Isolation Kit (MoBio, USA) following the manufacturer’s protocol. Sequencing was performed on Illumina Hiseq X Ten platform using 2 × 150 bp pair-end technology.
The sediment samples of DZ, XMD, YX, and FT were obtained from four representative mangrove nature reserves (China) in 2017, including Danzhou Xinyinggang Nature Reserve (DZ), Ximendao National Marine Reserve (XMD), Yunxiao Zhangjiangkou National Nature Reserve (YX), and Shenzhen Futian National Nature Reserve (FT). The DZ, XMD, and YX sediment samples were collected at the top 10 cm. The FT sediment samples were collected at depth intervals of 0–2, 6–8, 12–14, 20–22, and 28–30 cm. Detailed information on the processing of samples is described elsewhere (Zhang et al. 2019). Briefly, genomic DNA was extracted from 0.3 g of each sediment using a DNeasy PowerSoil kit (Qiagen, Germany) according to the manufacturer’s instructions. Total RNA was extracted from 2 g of each FT sediment using an RNeasy PowerSoil Total RNA Kit (Qiagen). A DNase Max Kit (Qiagen) and an RNeasy PowerClean Pro Cleanup Kit (Qiagen) were used to remove genomic DNA and purify the remaining RNA, respectively. Sequencing was performed on Illumina NovaSeq 6000 platform using 2 × 150 bp pair-end technology.
The YT samples were obtained from the Rongcheng Swan Lake Nature Reserve (China) in 2018. Details are described elsewhere (Liu et al. 2021). The sediment cores were collected using columnar samplers at depth intervals of 0–2, 21–26, and 36–41 cm in a seagrass meadow (YT_C) and a nonseagrass-covered site nearby (YT_FC), respectively. DNA was extracted from 10 g sediment of each sample by using PowerMax® Soil DNA Isolation Kit (MoBio, Germany) according to the manufacturer’s protocol. Sequencing was performed on Illumina HiSeq 2500 platform using 2 × 150 bp pair-end technology.
The JLS1 sample was collected from the surface sediment of Jiulong River estuary (China) in 2018. Details are described elsewhere (Zou et al. 2020). DNA was extracted from 10 g sediment of each sample using PowerMax® Soil DNA Isolation Kit following the manufacturer’s instructions. Sequencing was performed on Illumina HiSeq 2000 platform using 2 × 150 bp pair-end technology.
The L30 and L100 samples were collected from the drainage ditch of Ningbo Chunlin shrimp aquafarm (China) in 2019. The sediments of the top 2–10 cm were sampled at 30 m (L30) and 100 m (L100) from the outlet discharge point, respectively. Details are described elsewhere (Lu et al. 2021). Genomic DNA was extracted from 0.5 g of each sediment with a FastDNA® SPIN Kit for Soil (MP Biomedicals, USA) according to the manufacturer’s protocol. Sequencing was performed on Illumina HiSeq 2500 platform using 2 × 150 bp pair-end technology.
The QZM sample was obtained from a hot spring sediment at Tibet (China) in 2015, and its temperature was 62 °C. Genomic DNA was extracted from 0.3 g of the sediments using a PowerSoil® DNA Isolation Kit (MoBio, Germany) according to the manufacturer’s instructions. Sequencing was performed on Illumina Hiseq X Ten platform using 2 × 150 bp pair-end technology.
The N1, N4, and N5 samples were collected from the surface sediments of three adjacent locations in Qinghai Lake (China) in 2018. Approximately 10 g of each sample was applied for DNA extraction with the procedure as described elsewhere (Chen et al. 2016; Natarajan et al. 2016). Sequencing was performed on Illumina HiSeq 2500 platform using 2 × 250 bp pair-end technology.
Metagenomic Assembly and Binning
The Yap4, Yap30, Yap75, Yap125, Yap 200, Yap2000, DZ, XMD, YX, JLS1, L30, and L100 sets of metagenomes were assembled and binned using the same method (Zhang et al. 2022). Briefly, raw reads of each sample were trimmed using Sickle (v.1.33) (https://github.com/najoshi/sickle) with default settings, and the reads were then de novo assembled into contigs using IDBA-UD (v.1.1.1) (Peng et al. 2012) with the parameters “-mink 65 -maxk 145 -step 10.” Metagenomic binning was performed using MetaBAT2 (v.2.12.1) (Kang et al. 2019) with nine different combinations of specificity and sensitivity parameters (-m 2,000, 2,500, or 3,000; –maxP 85, 90, or 95), and the bins were merged and refined by DAS Tool (v.1.1) (Sieber et al. 2018).
The FT and YT sets of metagenomes were assembled and binned using the same method. Raw reads were trimmed with the read_qc module from metaWRAP (v.1.1) (Uritskiy et al. 2018). Clean reads from the same sampling site were pooled together before de novo assemble to one co-assembly, and they were then assembled using MEGAHIT (v.1.1.2) (Li et al. 2015) with the flag “–presets meta-large.” Metagenomic binning was performed using MetaBAT2 (v.2.12.1) with eight different combinations of specificity and sensitivity parameters (–maxP 60 or 95; –minS 60 or 95; –maxEdges 200 or 500), and the bins were merged and refined by DAS Tool (v.1.1) (Sieber et al. 2018).
The N1, N4, N5, and QZM sets of metagenomes were assembled and binned using the same method. Raw reads were trimmed and filtered using fastp (v.0.20.0) with default parameters (Chen et al. 2018). Clean reads of each sample were separately assembled into contigs using SPAdes (v.3.13.0) (Prjibelski et al. 2020) with the parameters “-meta -only-assembler -k 21, 31, 55, 77, 99, 121.” Metagenomic binning was performed using MetaBAT2 (v.2.12.1) and MaxBin2 (v.2.2.5) (Wu et al. 2016) modules in the metaWRAP pipeline (v.1.0.3) (Uritskiy et al. 2018), and the bins were merged and refined by Binning_refiner (v.1.2) (Song and Thomas 2017).
The completeness, contamination, and heterogeneity of the MAGs were determined based on lineage-specific conserved marker gene sets in each genome by CheckM (v.1.0.7) (Parks et al. 2015).
Retrieval of Thermoprofundales Genomes
The collection of Thermoprofundales genomes was obtained from three sources, which included 55 MAGs retrieved from the metagenomes of this study, 55 MAGs/SAGs downloaded from the NCBI GenBank database (Coordinators 2018) (January 25, 2021), and 36 MAGs from the genomic catalog of Earth’s microbiomes (Nayfach et al. 2020). Taxonomic assignment of the genomes was performed by using “classify” workflow in the GTDB-Tk software (v.1.3.0) (Chaumeil et al. 2020), and candidate Thermoprofundales genomes were assigned to “c__E2” in the GTDB taxonomy. A phylogenomic tree based on 55 conserved single-copy ubiquitous archaeal genes (Dombrowski et al. 2020) was then constructed using IQ-TREE (v.1.6.3) (Nguyen et al. 2015) with ModelFinder (Kalyaanamoorthy et al. 2017), and the Thermoprofundales genomes formed a monophyletic clade adjacent to other orders from the Candidate phylum Thermoplasmatota (e.g., Methanomassiliicoccales, Thermoplasmatales, Acidiprofundales, Poseidoniales, and MGIII). The tree is visualized using the iTOL online tool (v.5) (Letunic and Bork 2021). The AAI value shared by any two genomes was calculated using CompareM software with default parameters (v.0.1.2) (https://github.com/dparks1134/CompareM), and the heatmap was drawn using TBtools (v.1.075) (Chen et al. 2020).
Gene Annotation, Metabolic Reconstruction, and Metatranscriptomic Activity
The protein-coding genes of each genome were predicted by Prodigal (v.2.6.3) (Hyatt et al. 2010) using “-p meta” option. Orthologous gene families were identified using OrthoFinder (v.2.2.1) (Emms and Kelly 2019) with the parameters “-S diamond -M msa.” Gene annotation was performed against multiple web servers and protein databases and using the BLASTP program (E-value cutoff ≤1 × 10–5) (Altschul et al. 1990), including the Rapid Annotations using Subsystems Technology (RAST) server (Brettin et al. 2015), the Kyoto Encyclopedia of Genes and Genomes (KEGG) server (Kanehisa et al. 2021), the Clusters of Orthologous Genes (COG) (Galperin et al. 2021), the archaeal Clusters of Orthologous Genes (arCOG) (Makarova et al. 2015), the Protein families database (Pfam) (Mistry et al. 2021), the Conserved Domain Database (CDD) (Lu et al. 2020), the Protein Clusters (PRK) (https://www.ncbi.nlm.nih.gov/proteinclusters/), The Institute for Genomic Research’s database of protein FAMilies (TIGRFAM) (Haft et al. 2013), the Carbohydrate-Active enZymes database (CAZy) (Lombard et al. 2014), and the MEROPS database (Rawlings et al. 2017). Signal peptides and extracellular peptidases were predicted using SignalP (v.5.0) (Almagro Armenteros et al. 2019) and PSORTb (v.3.0.3) (Yu et al. 2010), respectively. Potential metabolic pathways were reconstructed based on the above predicted annotations and reference pathways depicted in KEGG (Kanehisa et al. 2021) and MetaCyc (Caspi et al. 2020). In FT mangrove sediments, transcriptomic abundances of the separated peripheral and membrane arms from Subgroup E were determined by mapping non-rRNA reads to the corresponding sequences using Bowtie 2 (v.2.3.4.1) (Langmead and Salzberg 2012) with default settings, and they were calculated using the RPKM (reads per kilobase per million sequenced reads) method (Robinson and Oshlack 2010; Zhang et al. 2018).
Construction of Phylogenetic Trees
Phylogenetic trees of all the key genes in this study (i.e., MbhL, reverse gyrase, 16S rRNA, CruF, CrtZ, and the concatenated 14-subunits, 10-subunits, and 4-subunits MBH) were constructed using IQ-TREE (v.1.6.3) with ModelFinder, and ultrafast bootstrapping was used to estimate the reliability of each branch with 1,000 times resampling. The trees were visualized using iTOL (v.5).
Sequence Alignment and 3D Structure Reconstruction
The protein sequences of the MbhL gene were aligned using free end gaps with the Blosum62 cost matrix in Geneious Prime® software (v.2020.2.4) (https://www.geneious.com). Three-dimensional structures of the MbhL protein were reconstructed using the C-I-TASSER online tool with default options (Zheng et al. 2021).
Expression, Purification, and Enzymatic Assay of MbhL
The MbhL gene from Thermoprofundales FT_bin5.232 was fused with a PelB signal peptide and a maltose-binding protein (MBP) tag. The nucleotide sequences were then synthesized after codon optimization for E. coli, inserted into the NdeI–XhoI site of plasmid pET-22b(+), and transformed into an E. coli BL21(DE3) strain. Meanwhile, an empty plasmid pET-22b(+) was transformed into an E. coli BL21(DE3) as the negative control. The expression was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in a total volume of 2.0 l at 37 °C for 4 h when the OD600 was 0.6. The cells were harvested by centrifugation at 8,000 rpm for 5 min and then lysed by sonication in 1×PBS buffer (1.8 mM KH2PO4, 8.1 mM Na2HPO4, 136.9 mM NaCl, 2.7 mM KCl, pH = 7.4). The cell lysate was centrifuged at 12,000 rpm for 10 min to remove debris, and the supernatant was applied to an MBP column. After washing with 1×PBS buffer, the fusion protein was eluted with 1×PBS buffer containing 10 mM maltose, and then dialyzed into a storage buffer (1.8 mM KH2PO4, 8.1 mM Na2HPO4, 300 mM NaCl, 2.7 mM KCl, 10% glycerol, pH = 7.4).
Hydrogenase activity was examined by measuring H2 production at 37 °C over a period of 15 min in a 10 ml anaerobic culture tube, as described elsewhere (Adams and Mortenson 1984) with few modifications. Briefly, the air in the headspace of each tube was replaced with Argon before the experiment. The 2 ml assay mixture contained 25 μg/ml fusion protein, 1 mM methyl viologen, and 20 mM sodium dithionite in 50 mM EPPS buffer (pH = 8.0). The inactivated MbhL protein was prepared by boiling the recombinant MbhL at 100 °C for 30 min. The H2 concentration was measured by gas chromatography (Agilent 7890B, USA).
NaCl Tolerance Test
Nucleotide sequences of the gene cluster MbhABCDEFGHIM from Thermoprofundales FT_bin5.232 were synthesized after codon optimization for E. coli, inserted into the NdeI–XhoI site of plasmid pET-22b(+), and transformed into an antiporter-deficient strain E. coli KNabc. Meanwhile, an empty plasmid pET-22b(+) was transformed into an E. coli KNabc as the negative control. They were separately incubated in an LBK medium (10 g/l tryptone, 5 g/l yeast extract, 87 mM KCl, pH = 7.0) with 100 μg/ml ampicillin at 37 °C and 200 rpm. When the OD600 (optical density at 600 nm) of the culture liquid was ∼0.5, a total of 50 μl was inoculated into 5 ml of LBK medium with 100 μg/ml ampicillin and 0.25 mM IPTG at NaCl concentrations of 0, 0.1, and 0.2 M, respectively. The growth curve was determined by measuring the OD600 of culture liquid at an interval of 2 h at 37 °C and 200 rpm. Triplicate samples were conducted for each treatment.
Molecular Dating Analysis
Estimation of the species divergence time was performed using the MCMCTree tool (v.4.9j) (Yang 2007) with several temporal calibrations as described elsewhere (Ren et al. 2019). Briefly, the root of Archaea is about 4,380–3,460 Mya (Wolfe and Fournier 2018), Thermoproteales and Sulfolobales both originate after the GOE (about 2,330 Mya) (Blank 2009; Luo et al. 2016), and MRCA of Sulfolobus solfataricus and Sulfolobus islandicus originate after 475 Mya (Blank 2011). The parameters in MCMCTree were used as below: first, the codeml module was used to calculate the overall substitution rate for each gene family. Then, the mcmctree module (with usedata = 3) was used to calculate the maximum likelihood estimation of branch lengths, gradient, and Hessian (Reis and Yang 2011). Finally, the mcmctree module (with usedata = 2) was used to estimate the divergence times. The estimation has been repeated for two times, and the two replicates had a consistent performance as shown in supplementary figure S13, Supplementary Material online. The parameters of the mcmctree module were “burnin = 20,000, sampfreq = 10, nsample = 200,000.”
Prediction of HGT Events and Plasmid Sequences
HGT events and their potential donors in Thermoprofundales genomes were predicted by HGTector (v.2.0b3) (Zhu et al. 2014) using default parameters and the “–donor-name” option. Plasmid sequences were predicted using PlasFlow (Krawczyk et al. 2018) and PlasClass (Pellow et al. 2020) with default parameters, and the results from the two software were combined with the criteria “probability threshold >0.7 and sequence length >5Kbps.”
Global Distribution and Relative Abundance
For 16S rRNA genes, the relative abundance of Thermoprofundales was calculated by searching a representative sequence from each subgroup against the Earth Microbiome Project (Thompson et al. 2017) and eight intertidal sediment samples along the southeast coast of China (Zhang et al. 2019). Detailed information for data processing is described elsewhere (Zhang et al. 2019). Briefly, filtered reads after chimeras removal were de-multiplexed and clustered into operational taxonomic units at 97% sequence similarity cutoff using VSEARCH (v.2.13.3) (Rognes et al. 2016). Taxonomic assignments were performed using “assign_taxonomy.py” in QIIME software (v.1.9.0) (Caporaso et al. 2010) against the SILVA132 database (Quast et al. 2013). In addition, a representative sequence from each subgroup was also submitted to the Integrated Microbial Next Generation Sequencing (IMNGS) web tool (Lagkouvardos et al. 2016) to search for Thermoprofundales in the NCBI SRA database. For metagenomes, the 146 Thermoprofundales genomes were dereplicated at 95% identity with CoverM software (v.0.6.1) (https://github.com/wwood/CoverM) to avoid arbitrary mapping among highly similar genomes, and the relative abundance of the dereplicated genomes in each metagenome was calculated using CoverM with the program “coverm genome.”
Gene Gain and Loss Analysis
High-quality (>80% completeness and <5% contamination) Thermoprofundales and reference genomes were selected (including the ancestral nodes, sister lineages and outgroups) to reconstruct the genome contents and phylogenomic tree using OrthoFinder (v.2.2.1) and IQ-TREE (v.1.6.3) (based on 55 conserved marker genes; Dombrowski et al. 2020), respectively. Processes governing the evolution of the five Thermoprofundales subgroups were inferred from predicted gene gain and loss events using BadiRate (v.1.35) (Librado et al. 2012) with options “-anc -bmodel FR -rmodel BDI -ep CWP.”
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant nos 91951102, 32225003, 31970105, 32061133009), the Innovation Team Project of Universities in Guangdong Province (grant no. 2020KCXTD023), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB42000000), and the Science and Technology Innovation Committee of Shenzhen (grant nos JCYJ20190808152403587, JCYJ20200109105010363). We thank Qianjun He, Chao Xia, and Zhaokui Jin for help with the measurement of H2. We thank Lin Meng for preparing the E. coli KNabc strain. We thank Wan Liu for the genome submission to eLMSG.
Contributor Information
Xinxu Zhang, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Yuhan Huang, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Yang Liu, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Wei Xu, Key Laboratory of Marine Biogenetic Resources, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, Fujian, China.
Jie Pan, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Xiaowei Zheng, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Huan Du, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Cuijing Zhang, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Zhongyi Lu, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Dayu Zou, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Zongbao Liu, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Mingwei Cai, Institute of Chemical Biology, Shenzhen Bay Laboratory, Shenzhen, Guangdong, China.
Jinbo Xiong, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, Ningbo, Zhejiang, China.
Yaxin Zhu, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Zhiyang Dong, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Hongchen Jiang, State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan, Hubei, China.
Hailiang Dong, State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Beijing, China.
Juquan Jiang, Department of Microbiology and Biotechnology, College of Life Sciences, Northeast Agricultural University, Harbin, Heilongjiang, China.
Zhuhua Luo, Key Laboratory of Marine Biogenetic Resources, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, Fujian, China.
Li Huang, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Meng Li, Archaeal Biology Center, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China; Shenzhen Key Laboratory of Marine Microbiome Engineering, Institute for Advanced Study, Shenzhen University, Shenzhen, Guangdong, China.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The genome sequences from the current study have been deposited in eLMSG (an eLibrary of Microbial Systematics and Genomics, https://www.biosino.org/elmsg/) under accession numbers LMSG_G000004137.1–LMSG_G000004191.1. The accession numbers of the metagenomic raw reads of each sample used in this study are provided in supplementary table S1, Supplementary Material online. Metatranscriptomic raw reads of the five FT samples have been deposited in NODE (National Omics Data Encyclopedia, https://www.biosino.org/node/) with accession numbers OES078050–OES078054.
References
- Coordinators NR . 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S. 2017. The growing tree of archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11:2407–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MWW, Mortenson LE. 1984. The purification of hydrogenase II (uptake hydrogenase) from the anaerobic N2-fixing bacterium Clostridium pasteurianum. Biochim Biophys Acta Bioenergy. 766:51–61. [Google Scholar]
- Albers S-V, Jarrell KF. 2018. The archaellum: an update on the unique archaeal motility structure. Trends Microbiol. 26:351–362. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37:420–423. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Baker BJ, De Anda V, Seitz KW, Dombrowski N, Santoro AE, Lloyd KG. 2020. Diversity, ecology and evolution of archaea. Nat Microbiol. 5:887–900. [DOI] [PubMed] [Google Scholar]
- Blank CE. 2009. Phylogenomic dating-a method of constraining the age of microbial taxa that lack a conventional fossil record. Astrobiology. 9:173–191. [DOI] [PubMed] [Google Scholar]
- Blank CE. 2011. An expansion of age constraints for microbial clades that lack a conventional fossil record using phylogenomic dating. J Mol Evol. 73:188–208. [DOI] [PubMed] [Google Scholar]
- Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, et al. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 5:8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito IL. 2021. Examining horizontal gene transfer in microbial communities. Nat Rev Microbiol. 19:442–453. [DOI] [PubMed] [Google Scholar]
- Bulzu P-A, Kavagutti Vinicius S, Chiriac M-C, Vavourakis Charlotte D, Inoue K, Kandori H, Andrei A-S, Ghai R, Hallam Steven J. 2021. Heliorhodopsin evolution is driven by photosensory promiscuity in monoderms. mSphere. 6:e00661-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK, Wu D, Robinson JM, Khouri HM, Eisen JA, et al. 2009. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 5:e1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol. 303:298–304. [DOI] [PubMed] [Google Scholar]
- Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD. 2020. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 48:D445–D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2020. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 13:1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen P, Zhang L, Guo X, Dai X, Liu L, Xi L, Wang J, Song L, Wang Y, Zhu Y, et al. 2016. Diversity, biogeography, and biodegradation potential of Actinobacteria in the deep-sea sediments along the southwest Indian Ridge. Front Microbiol. 7:1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N, Teske AP, Baker BJ. 2018. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun. 9:4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N, Williams TA, Sun J, Woodcroft BJ, Lee J-H, Minh BQ, Rinke C, Spang A. 2020. Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution. Nat Commun. 11:3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas GM, Langille MGI. 2019. Current and promising approaches to identify horizontal gene transfer events in metagenomes. Genome Biol Evol. 11:2750–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wang Y, Zubin R, Wang F. 2019. Core metabolic features and hot origin of Bathyarchaeota. Engineering 5:498–504. [Google Scholar]
- Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49:D274–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani M, Garbayo I, Vílchez C, Martínez-Espinosa RM. 2019. Haloarchaeal carotenoids: healthy novel compounds from extreme environments. Mar Drugs. 17:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10:761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Boyd E. 2020. Editorial: microbial hydrogen metabolism. Front Microbiol. 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41:D387–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog B, Wirth R. 2012. Swimming behavior of selected species of archaea. Appl Environ Microbiol. 78:1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H, Smalla K. 2012. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev. 36:1083–1104. [DOI] [PubMed] [Google Scholar]
- Hoshino T, Inagaki F. 2019. Abundance and distribution of archaea in the subseafloor sedimentary biosphere. ISME J. 13:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F, Kuypers MMM, Tsunogai U, Ishibashi J, Nakamura K, Treude T, Ohkubo S, Nakaseama M, Gena K, Chiba H, et al. 2006. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system. Proc Natl Acad Sci U S A. 103:14164–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. 2021. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keto-Timonen R, Hietala N, Palonen E, Hakakorpi A, Lindström M, Korkeala H. 2016. Cold shock proteins: a minireview with special emphasis on csp-family of enteropathogenic Yersinia. Front Microbiol. 7:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Rosselló-Móra R, Amann R. 2017. Uncultivated microbes in need of their own taxonomy. ISME J. 11:2399–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PS, Lipinski L, Dziembowski A. 2018. Plasflow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 46:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, Clavel T. 2016. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 6:33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Allen JF, Martin W. 2010. How did LUCA make a living? Chemiosmosis in the origin of life. Bioessays 32:271–280. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar CS, Baker BJ, Seitz KW, Teske AP. 2017. Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. ISME J. 11:1118–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. [DOI] [PubMed] [Google Scholar]
- Librado P, Vieira FG, Rozas J. 2012. Badirate: estimating family turnover rates by likelihood-based methods. Bioinformatics 28:279–281. [DOI] [PubMed] [Google Scholar]
- Lipp JS, Morono Y, Inagaki F, Hinrichs K-U. 2008. Significant contribution of archaea to extant biomass in marine subsurface sediments. Nature 454:991–994. [DOI] [PubMed] [Google Scholar]
- Lipscomb GL, Hahn EM, Crowley AT, Adams MWW. 2017. Reverse gyrase is essential for microbial growth at 95 °C. Extremophiles 21:603–608. [DOI] [PubMed] [Google Scholar]
- Liu Y, Makarova KS, Huang W-C, Wolf YI, Nikolskaya AN, Zhang X, Cai M, Zhang C-J, Xu W, Luo Z, et al. 2021. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 593:553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-F, Yang L, Liu Z-L, Chen J, Fang B, Zhou L, Liu J-F, Yang S-Z, Gu J-D, Mu B-Z. 2022. Discovery of the non-cosmopolitan lineages in Candidatus thermoprofundales. Environ Microbiol. 24:3063–3080. [DOI] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, et al. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Zivanovic Y, Deschamps P, Moreira D. 2015. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 13:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48:D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhang X, Wang C, Li M, Chen J, Xiong J. 2021. Responses of sediment resistome, virulence factors and potential pathogens to decades of antibiotics pollution in a shrimp aquafarm. Sci Total Environ. 794:148760. [DOI] [PubMed] [Google Scholar]
- Luo G, Ono S, Beukes Nicolas J, Wang David T, Xie S, Summons Roger E. 2016. Rapid oxygenation of Earth’s atmosphere 2.33 billion years ago. Sci Adv. 2:e1600134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. 2015. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life 5:818–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTernan PM, Chandrayan SK, Wu C-H, Vaccaro BJ, Lancaster WA, Adams MWW. 2015. Engineering the respiratory membrane-bound hydrogenase of the hyperthermophilic archaeon Pyrococcus furiosus and characterization of the catalytically active cytoplasmic subcomplex. Protein Eng Des Sel. 28:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenbroek EM, Peron Cane C, Jala I, Iwai S, Moolenaar GF, Goosen N, Pannu NS. 2013. UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions. Nucleic Acids Res. 41:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar Gustavo A, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res. 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussagy CU, Winterburn J, Santos-Ebinuma VC, Pereira JFB. 2019. Production and extraction of carotenoids produced by microorganisms. Appl Microbiol Biotechnol. 103:1095–1114. [DOI] [PubMed] [Google Scholar]
- Natarajan VP, Zhang X, Morono Y, Inagaki F, Wang F. 2016. A modified SDS-based DNA extraction method for high quality environmental DNA from seafloor environments. Front Microbiol. 7:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther DJ, Rachel R, Wanner G, Wirth R. 2006. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J Bacteriol. 188:6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F, Wu D, Paez-Espino D, Chen IM, Huntemann M, et al. 2020. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 39:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Costa D, Maranha A, Costa M, Alarico S, Empadinhas N. 2017. Glucosylglycerate metabolism, bioversatility and mycobacterial survival. Glycobiology 27:213–227. [DOI] [PubMed] [Google Scholar]
- Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. 2011. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev. 75:361–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. Checkm: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellow D, Mizrahi I, Shamir R. 2020. Plasclass improves plasmid sequence classification. PLoS Comp Biol. 16:e1007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. [DOI] [PubMed] [Google Scholar]
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 70:e102. [DOI] [PubMed] [Google Scholar]
- Pushkarev A, Inoue K, Larom S, Flores-Uribe J, Singh M, Konno M, Tomida S, Ito S, Nakamura R, Tsunoda SP, et al. 2018. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. 2017. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46:D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis Md, Yang Z. 2011. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 28:2161–2172. [DOI] [PubMed] [Google Scholar]
- Ren M, Feng X, Huang Y, Wang H, Hu Z, Clingenpeel S, Swan BK, Fonseca MM, Posada D, Stepanauskas R, et al. 2019. Phylogenomics suggests oxygen availability as a driving force in Thaumarchaeota evolution. ISME J. 13:2150–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Chuvochina M, Mussig AJ, Chaumeil P-A, Davín AA, Waite DW, Whitman WB, Parks DH, Hugenholtz P. 2021. A standardized archaeal taxonomy for the genome taxonomy database. Nat Microbiol. 6:946–959. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra R, Verhagen MFJM, Adams MWW. 2000. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 182:3423–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut GJ, Boyd ES, Peters JW, Adams MWW. 2013. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol Rev. 37:182–203. [DOI] [PubMed] [Google Scholar]
- Schut GJ, Zadvornyy O, Wu C-H, Peters JW, Boyd ES, Adams MWW. 2016. The role of geochemistry and energetics in the evolution of modern respiratory complexes from a proton-reducing ancestor. Biochim Biophys Acta Bioenergy 1857:958–970. [DOI] [PubMed] [Google Scholar]
- Shao L, Xu T, Zheng X, Shao D, Zhang H, Chen H, Zhang Z, Yan M, Abdel-Motaal H, Jiang J. 2021. A novel three-TMH Na+/H+ antiporter and the functional role of its oligomerization. J Mol Biol. 433:166730. [DOI] [PubMed] [Google Scholar]
- Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. 2018. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 3:836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søndergaard D, Pedersen CNS, Greening C. 2016. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 6:34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-Z, Thomas T. 2017. Binning_refiner: improving genome bins through the combination of different binning programs. Bioinformatics 33:1873–1875. [DOI] [PubMed] [Google Scholar]
- Sousa FL, Nelson-Sathi S, Martin WF. 2016. One step beyond a ribosome: the ancient anaerobic core. Biochim Biophys Acta Bioenergy. 1857:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J, Sazanov L. 2020. Structure and mechanism of the Mrp complex, an ancient cation/proton antiporter. eLife 9:e59407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. 2010. Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea. Appl Environ Microbiol. 76:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahon G, Geesink P, Ettema TJG. 2021. Expanding archaeal diversity and phylogeny: past, present, and future. Annu Rev Microbiol. 75:359–381. [DOI] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur K, Tomar SK, Singh AK, Mandal S, Arora S. 2017. Riboflavin and health: a review of recent human research. Crit Rev Food Sci Nutr. 57:3650–3660. [DOI] [PubMed] [Google Scholar]
- Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, et al. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritskiy GV, DiRuggiero J, Taylor J. 2018. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven EEA, Wyman C, Moolenaar GF, Goosen N. 2002. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 21:4196–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol. 65:4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Billoud B. 2007. Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev. 107:4206–4272. [DOI] [PubMed] [Google Scholar]
- Wagner A, Whitaker RJ, Krause DJ, Heilers J-H, van Wolferen M, van der Does C, Albers S-V. 2017. Mechanisms of gene flow in archaea. Nat Rev Microbiol. 15:492–501. [DOI] [PubMed] [Google Scholar]
- Walker DJF, Martz E, Holmes DE, Zhou Z, Nonnenmann SS, Lovley DR. 2019. The archaellum of Methanospirillum hungatei is electrically conductive. Mbio 10:e00579–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe JM, Fournier GP. 2018. Horizontal gene transfer constrains the timing of methanogen evolution. Nat Ecol Evol. 2:897–903. [DOI] [PubMed] [Google Scholar]
- Wu Y-W, Simmons BA, Singer SW. 2016. Maxbin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wu C-H, Schut GJ, Haja DK, Zhao G, Peters JW, Adams MWW, Li H. 2018. Structure of an ancient respiratory system. Cell 173:1636–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Z, Xu W, Pan J, Huang Y, Cai M, Luo Z, Li M. 2022. Genomic insights into versatile lifestyle of three new bacterial candidate phyla. Sci China Life Sci. 65:1547–1562. [DOI] [PubMed] [Google Scholar]
- Zhang C-J, Pan J, Duan C-H, Wang Y-M, Liu Y, Sun J, Zhou H-C, Song X, Li M. 2019. Prokaryotic diversity in mangrove sediments across southeastern China fundamentally differs from that in other biomes. mSystems 4:e00442-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang L, Zhong D. 2017. Photolyase: dynamics and electron-transfer mechanisms of DNA repair. Arch Biochem Biophys. 632:158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu W, Liu Y, Cai M, Luo Z, Li M. 2018. Metagenomics reveals microbial diversity and metabolic potentials of seawater and surface sediment from a hadal biosphere at the Yap Trench. Front Microbiol. 9:2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y. 2021. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods. 1:100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Liu Y, Lloyd KG, Pan J, Yang Y, Gu J-D, Li M. 2018. Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, thermoprofundales (MBG-D archaea). ISME J. 13:885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Kosoy M, Dittmar K. 2014. HGTector: an automated method facilitating genome-wide discovery of putative horizontal gene transfers. BMC Genomics. 15:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Wan R, Han L, Xu Min N, Liu Y, Liu H, Kao S-J, Li M, Atomi H. 2020. Genomic characteristics of a novel species of ammonia-oxidizing archaea from the Jiulong river estuary. Appl Environ Microbiol. 86:e00736-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences from the current study have been deposited in eLMSG (an eLibrary of Microbial Systematics and Genomics, https://www.biosino.org/elmsg/) under accession numbers LMSG_G000004137.1–LMSG_G000004191.1. The accession numbers of the metagenomic raw reads of each sample used in this study are provided in supplementary table S1, Supplementary Material online. Metatranscriptomic raw reads of the five FT samples have been deposited in NODE (National Omics Data Encyclopedia, https://www.biosino.org/node/) with accession numbers OES078050–OES078054.