Abstract
The fish oil constituent docosahexaenoic acid (DHA, 22:6 n-3) is a signaling lipid with anti-inflammatory properties. The molecular mechanisms underlying the biological effect of DHA are poorly understood. Here, we report the design, synthesis, and application of a complementary pair of bio-orthogonal, photoreactive probes based on the polyunsaturated scaffold DHA and its oxidative metabolite 17-hydroxydocosahexaenoic acid (17-HDHA). In these probes, an alkyne serves as a handle to introduce a fluorescent reporter group or a biotin-affinity tag via copper(I)-catalyzed azide-alkyne cycloaddition. This pair of chemical probes was used to map specific targets of the omega-3 signaling lipids in primary human macrophages. Prostaglandin reductase 1 (PTGR1) was identified as an interaction partner that metabolizes 17-oxo-DHA, an oxidative metabolite of 17-HDHA. 17-oxo-DHA reduced the formation of pro-inflammatory lipids 5-HETE and LTB4 in human macrophages and neutrophils. Our results demonstrate the potential of comparative photoaffinity protein profiling for the discovery of metabolic enzymes of bioactive lipids and highlight the power of chemical proteomics to uncover new biological insights.
Introduction
Dietary omega-3 polyunsaturated fatty acids (PUFAs) are generally considered to be beneficial for human health.1−3 For example, the fish oil constituent docosahexaenoic acid (DHA, 22:6 n-3, Scheme 1A) is important for brain development, cardiovascular function, and the immune system.2−7 Mechanistic studies indicate that many of the favorable effects of omega-3 fatty acids are due to their interaction with immunological processes,1,2 which has been reiterated by the discovery of their oxidized metabolites involved in the resolution of inflammation.8,9 Malfunction of this resolution phase of inflammation is hypothesized to contribute to many chronic inflammatory diseases, such as rheumatoid arthritis and asthma.10,11 A better molecular understanding of the biological role of these lipids in the resolution phase of inflammation is required to develop therapeutics for these diseases.
Scheme 1. Oxidative Metabolism of DHA (1) and Synthetic Strategy of Photoaffinity Probes 4 and 5.
(A) Metabolic pathway of DHA to 17-oxo-DHA, resolvins, and protectins. (B) Structures of photoaffinity-click (pac-)probes based on DHA (1) and 17-HDHA (2). The synthetic strategy included the use of soybean lipoxygenase (sLOX) to introduce the hydroxyl and the use of Wittig reactions to join building blocks 6, 8, and 9. COX, cyclooxygenase; DHA, docosahexaenoic acid; GST, glutathione S-transferase; HDHA, hydroxydocosahexaenoic acid; LOX, lipoxygenase; P450, cytochrome P450; sHE, soluble epoxide hydrolase.
The resolution of inflammation requires the intricate orchestration of cells of the innate immune system via soluble mediators. Among these, oxidative metabolites of DHA play a central role.7,12 Biochemical studies have shown that DHA is oxidized into a central precursor, 17-hydroxydocosahexaenoic acid (17-HDHA, 2) via multiple pathways, including 15-lipoxygenase and cyclooxygenase acetylated by aspirin (Scheme 1A).13 Oxidation by 15-lipoxygenase results in the formation of 17(S)-HDHA, while acetylated COX generates 17(R)-HDHA.14 Importantly, 17-HDHA has protective effects in various animal models of colitis, arthritis, and renal reperfusion.15−18 In vitro, human macrophages produce less TNFα and more IL-10 following exposure to 17-HDHA.19,20 Treatment with 17-HDHA also reduces LTB4 production in both isolated murine and human neutrophils.20,21 Moreover, 17-HDHA can be further metabolized to specialized pro-resolving lipid mediators (SPMs), such as D-series resolvins and protectins (Scheme 1A), a process involving several enzymes. Resolvins and protectins are bioactive lipids with potent pro-resolving activities, including halting the infiltration of neutrophils and enhancing the non-phlogistic clearance of apoptotic cells, cellular debris, and microbes by macrophages, thereby stimulating the resolution of inflammation, while promoting tissue regeneration.8,22 On the other hand, 17-HDHA can be metabolized to 17-oxo-docosahexaenoic acid (17-oxo-DHA, 3), which may limit the formation of SPMs. Insight into its protein interaction partners in human immune cells would be of great benefit in obtaining a better molecular understanding of the biological role and metabolism of DHA and its oxidative metabolites.
Lipid photoaffinity probes have been successfully used to map protein–lipid interactions on a global scale in their native environment.23 Bio-orthogonal photoaffinity lipid probes consist of a lipid modified with a photoreactive group and bio-orthogonal ligation handle.24 The photoreactive group is activated by irradiation with light, generating a reactive species that may form a covalent and irreversible bond with the interacting protein. The ligation handle is used to attach a reporter group via bio-orthogonal chemistry, which allows for visualization or isolation of the probe-bound protein in a complex biological sample. This affinity-based protein profiling (AfBPP) approach has been reported for multiple lipid classes, including phospholipids,25,26 fatty acids,23,27 sphingolipids,28,29 and sterols,24,30 but has not been applied to omega-3 PUFAs, arguably due to the synthetic challenges associated with their preparation.31
An important drawback of photoreactive lipid-based probes is their inherent high lipophilicity and nonspecific binding to proteins. Significant overlap between protein targets of lipid probes has been documented, making it difficult to assign specific interaction partners to a given probe and to study their biological role.32,33 Competition experiments with non-labeled lipids have been applied to identify specific binding partners but were not always successful, possibly due to the accumulation of both probe and competitor lipids in the cellular membrane.34,35
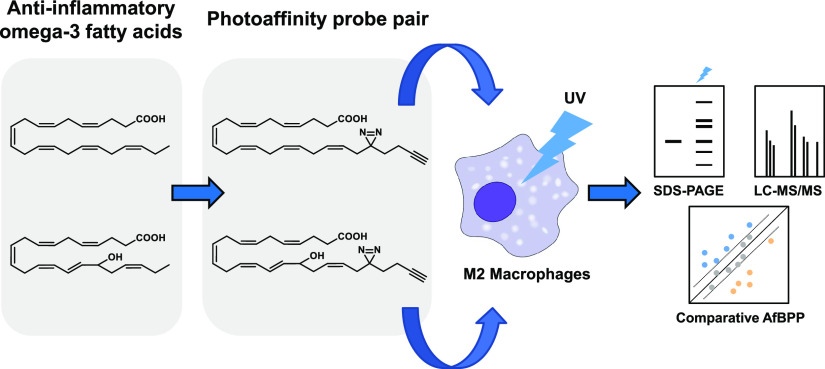
Here, we describe the design, synthesis, and application of a pair of complementary photoaffinity probes (4 and 5) based on the structure of DHA and 17-HDHA, respectively (Scheme 1B). Our main aim was to map specific lipid targets of 17-HDHA in primary human macrophages by comparative AfBPP (Figure 1). Probe 5 retained the anti-inflammatory properties of the parent lipid in human M2 macrophages. Using chemical proteomics, we identified prostaglandin reductase 1 (PTGR1) as a lipid-binding partner. Subsequent, biochemical and genetic studies revealed that PTGR1 metabolizes 17-oxo-DHA, an oxidative metabolite of 17-HDHA. 17-oxo-DHA reduced the biosynthesis of the pro-inflammatory lipid mediators 5-HETE and LTB4 in human macrophages and neutrophils. Thus, comparative photoaffinity labeling revealed PTGR1 as a metabolic hub in bioactive lipid signaling in human macrophages.
Figure 1.
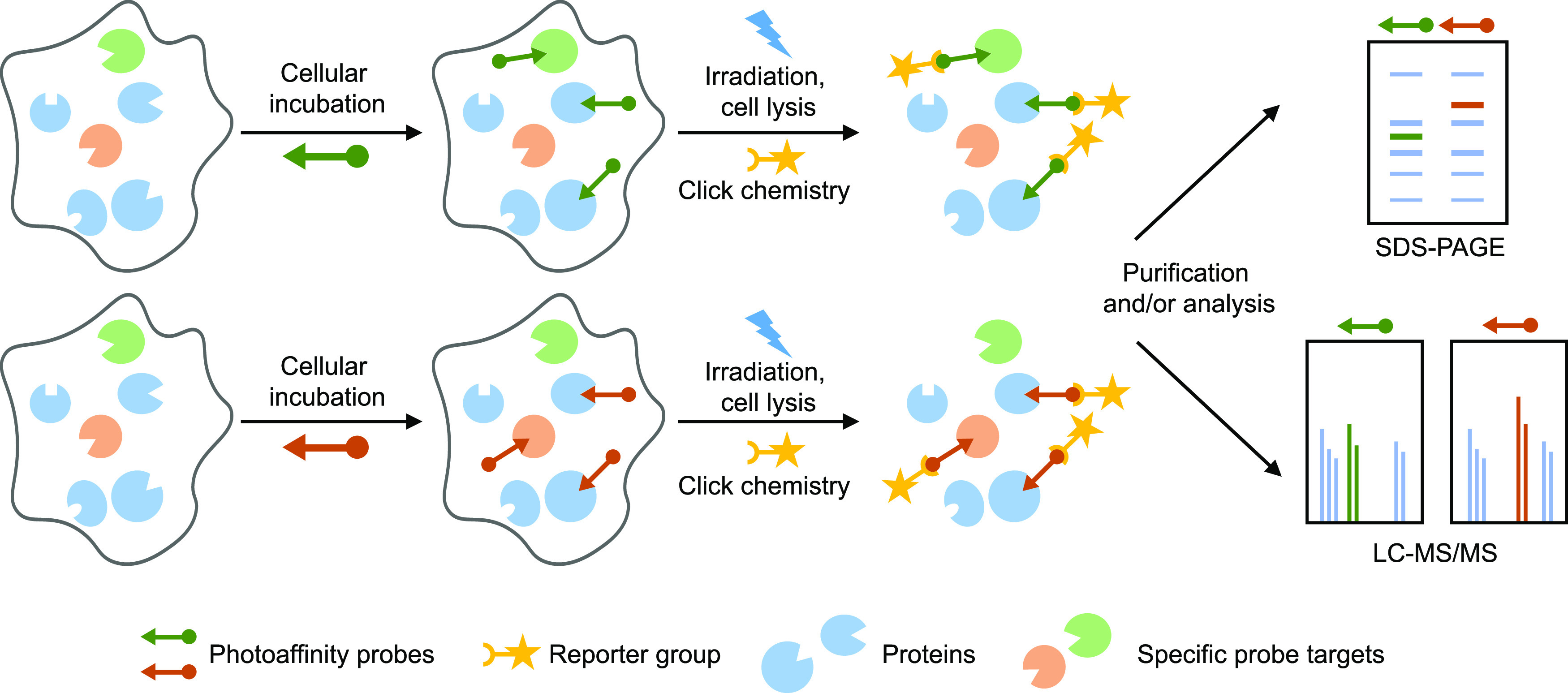
Schematic overview of the comparative photoaffinity-based protein profiling (AfBPP) experiment. Two highly similar, bifunctional photoreactive probes are used to identify probe-interacting proteins using chemical proteomics. Cells are supplied with either probe, and after an incubation period, the photoreactive group is activated using UV light, which results in the formation of a covalent and irreversible bond with the probe-interacting protein. The probe-bound protein can then be investigated by ligation to a fluorescent group and SDS-PAGE separation or ligation to an affinity handle, enriched, and identified using LC–MS/MS. Probe-specific targets can be identified by comparing the interaction landscapes of the individual probes.
Results
Design and Synthesis of Photoaffinity Probes 4 and 5
In the design of photoaffinity probes 4 and 5 (Scheme 1B) and to ensure they closely resemble the signaling lipids DHA and 17-HDHA, respectively, we decided to keep the PUFA scaffold intact and substitute the omega carbon with a diazirine- and alkyne-containing minimalistic bifunctional group.36 Diazirines are small photoreactive groups with short reactive half-lives upon activation, thereby minimizing the interference and reducing non-specific labeling. The alkyne is the smallest bio-orthogonal tag available and has physio-chemical properties similar to the alkyl chain of fatty acids.37,38
To synthesize the probes in an efficient manner, we made use of a chemoenzymatic approach. Probe 5 was produced by soybean lipoxygenase using probe 4 as the substrate (Scheme 1B). Probe 4 was synthesized by combining two strategic building blocks, the minimalistic bifunctional photoreactive linker 6 and the PUFA scaffold (7), using a Wittig reaction. Building block 7 was generated by combining the two dienes 8 and 9, also via a Wittig reaction. This synthetic strategy avoids the reduction of six skipped (non-conjugated) alkynes at once, which would lead to a complex mixture of partially hydrogenated products.39 Moreover, this strategy did not require the assembly of large, skipped polyalkyne structures, which are tremendously unstable.40,41 Moreover, this strategy allowed the installation of the labile diazirine at a late stage in the synthesis.
First, minimalist linker derivative 10 was synthesized following a previously reported route36 with minor modifications (Scheme S1). To enable its use in a Wittig reaction, the iodide was substituted with triphenylphosphine at 70 °C to obtain phosphonium salt 6 in quantitative yield (Scheme 2). To obtain fragments 8 and 9, skipped alkynes were assembled and partially hydrogenated using P-2 nickel. To this end, butynol (11) was protected with a tert-butyl(dimethyl)silyl (TBS) group to afford alkyne 12 in 98% yield. This compound was deprotonated using n-butyllithium and reacted with paraformaldehyde to afford alcohol 13, which was tosylated to obtain 14. This intermediate was used without further purification for a copper(I)-mediated coupling to butynol to afford skipped alkyne 15 in 71% yield over three steps. The skipped alkyne was partially hydrogenated using P-2 nickel catalyst to furnish alcohol 9 in 64% yield.
Scheme 2. Synthesis of Building Blocks 6, 8, and 9.
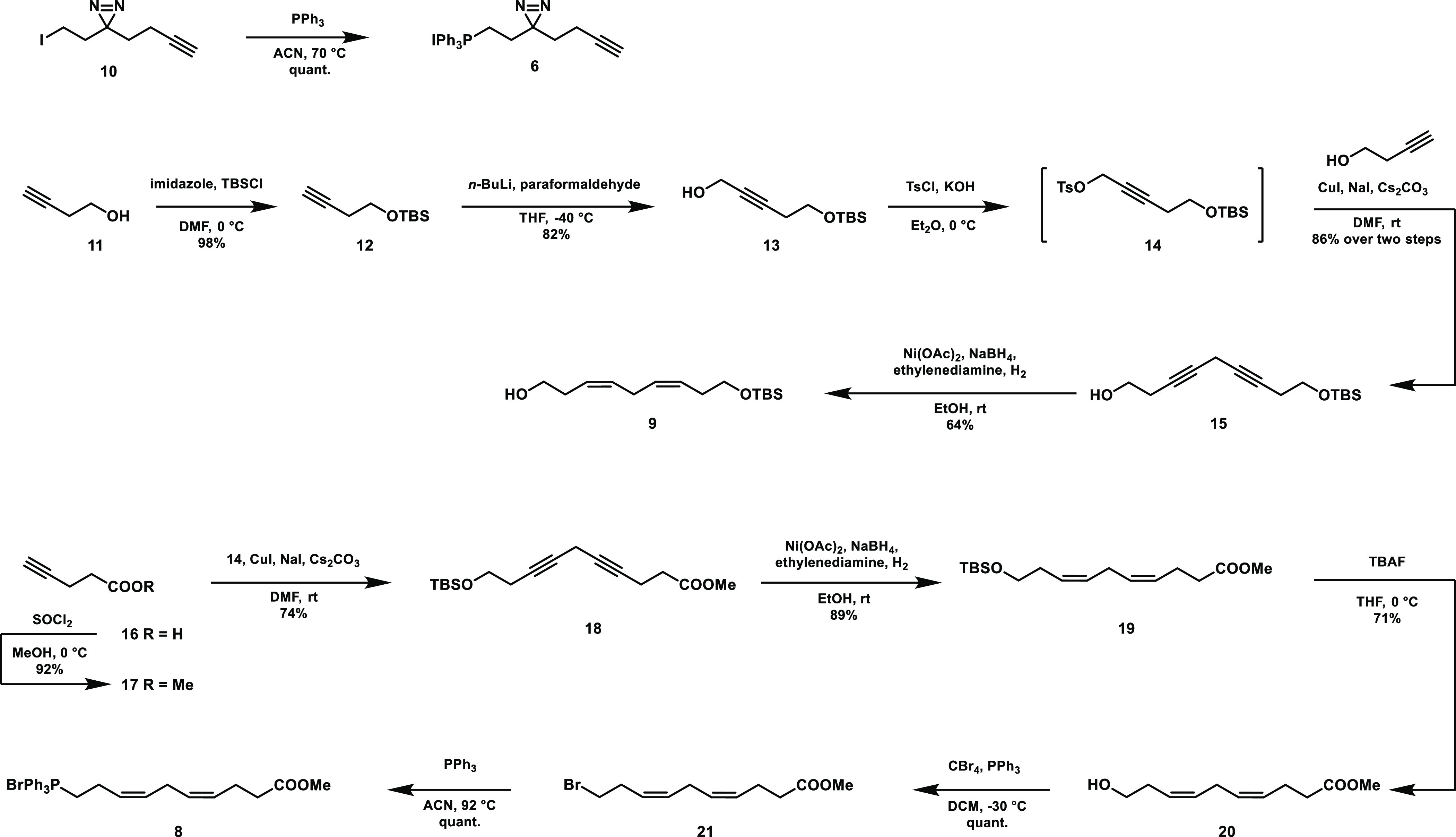
For fragment 8, pentynoic acid (16) was protected by esterification in methanol to obtain 17, which was reacted with sulfonate ester 14 in a second copper(I)-mediated coupling to afford skipped alkyne 18 (Scheme 2). Skipped alkyne 18 was partially hydrogenated to afford silylated 19. The TBS group was removed using tetrabutylammonium fluoride (TBAF) to afford alcohol 20, which was halogenated to form 21. This was then substituted with triphenylphosphine to afford phosphonium salt 8 in 43% yield from pentynoic acid.
To join intermediates 8 and 9, a Wittig reaction was performed by deprotonation of 8 to form ylid 22 using lithium bis(trimethylsilyl)amide (LiHMDS) at a reduced temperature, and the addition of freshly prepared aldehyde 23 at −100 °C (Scheme 3). Aldehyde 23 was prepared by oxidation of alcohol 9 using DMP directly before use due to the instability of β,γ-unsaturated aldehydes.42,43 The Wittig reaction afforded with high preference the Z-isomer, which could be separated from the undesired E-isomer by column chromatography to afford 24 in 57% yield. 24 was in turn deprotected using TBAF to obtain alcohol 7 in 87% yield.
Scheme 3. Formation of Probes 4 and 5 by Assembly of Building Blocks 6, 8, and 9.
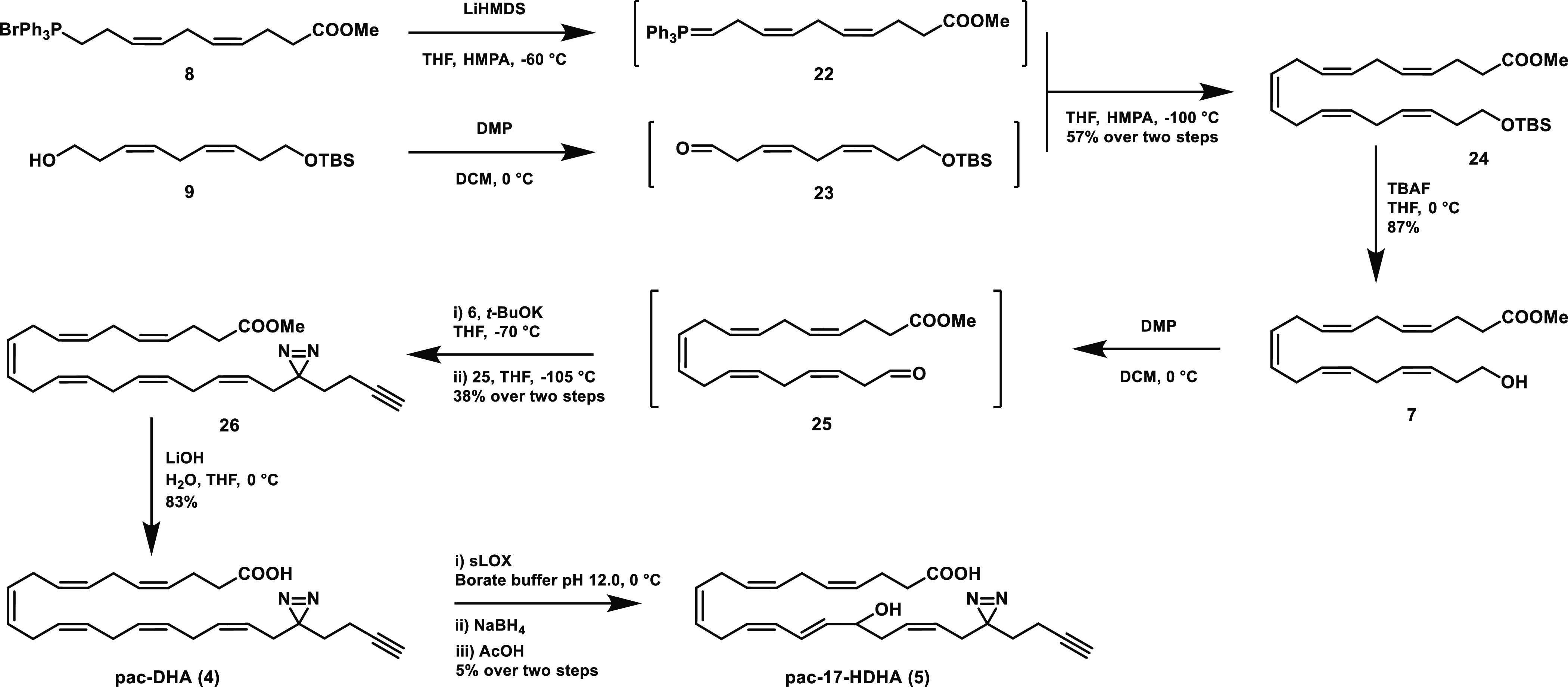
Building block 6 was installed in probe 4 by generating the final double bond using another Wittig reaction, for which alcohol 7 was oxidized with DMP, while simultaneously treating phosphonium salt 6 with KOtBu at a reduced temperature. Higher temperatures or stronger bases resulted in the loss of the diazirine. The freshly generated aldehyde was added to the ylid at −105 °C, which afforded methyl ester 26 in 38% yield after workup and purification by column chromatography, which was capable of removing the E-isomer. The Z-configuration of the last double bond was checked by NMR, where a Z-characteristic coupling constant of 10.4 Hz was found. Saponification of the ester in 26 yielded photoaffinity probe 4 in 83% yield.
To generate the desired photoaffinity-click (pac)-17-HDHA (5), commercially available soybean lipoxygenase (sLOX) was used, which catalyzes the oxidation of DHA (1) to 17-hydroperoxy-DHA, which can be reduced to 17-HDHA (2).44,45 While DHA (1) was fully converted into 17-HDHA by sLOX using previously reported conditions,46 this resulted in the complete loss of the diazirine when using probe 4 as a substrate. To this end, the reaction conditions with probe 4 were optimized for enzyme loading, temperature, and incubation time. This resulted in the formation of probe 5, which could be obtained after high-performance liquid chromatography (HPLC) purification. The position of the hydroxyl group was confirmed by fragmentation on LC–MS after hydrogenation of the alkenes using previously reported conditions.47 Of note, the final enzymatic step yielded only a small amount of probe 5, which was sufficient for subsequent biological experiments. Further optimization of reaction conditions or modification of the lipoxygenase enzyme may improve the yield, although the latter strategy comes with risk, as changing the lipoxygenase structure is known to change its positional specificity.48,49
Mapping Protein Interaction Partners of Probes 4 and 5
Since our main aim was to identify specific binding partners of 17-HDHA, probe 5 was tested in a cellular assay using primary human M2 macrophages to confirm it was biologically equivalent to 17-HDHA in reducing the inflammatory response. M2 macrophages were differentiated from monocytes of healthy donors in the presence of macrophage colony-stimulating factor (M-CSF). Upon stimulation with Ca2+ ionophore, M2 macrophages rapidly synthesize the proinflammatory eicosanoid 5-HETE from arachidonic acid (AA) by 5-lipoxygenase (Figure 2A). A liquid chromatography–mass spectrometry (LC–MS) method was developed to quantify 5-HETE in these human macrophages. Pre-incubation with 17-HDHA lipid (1 μM) or probe 5 (1 μM) significantly reduced the conversion of AA into 5-HETE. DHA, which was taken along as a negative control compound, was less effective (Figure 2A), thereby confirming the specificity of the 17-HDHA response. In conclusion, probe 5 mimics the anti-inflammatory signaling capacity of 17-HDHA in human macrophages and can be used to investigate the binding partners of its parent lipid.
Figure 2.

Comparative ABPP in human M2 macrophages using pac-17-HDHA and pac-DHA. (A) 5-HETE formation (LC–MS/MS) produced by ionophore-stimulated M2 macrophages. Data represent means ± SD of cells obtained from a representative donor (n = 3). **p < 0.01, ****p < 0.0001 in comparison to ionophore-treated control using a one-way ANOVA with Dunnett’s multiple comparisons correction. n.d.: not detected. (B) Probe targets of pac-17-HDHA (5) and pac-DHA (4) conjugated to Cy5-N3 using CuAAC chemistry and analyzed by SDS-PAGE and in-gel fluorescent scanning show UV-dependent and probe-specific labeling. Coomassie (CM) served as a protein loading control. Asterisks indicate probe-specific bands. (C) Waterfall plot of proteins identified using probe 5 in M2 macrophages. UV enrichment is capped at 20-fold. (D) UV enrichment of probe 5-enriched targets by probe 4 and probe 5 is shown, with a 2-fold cutoff indicating probe-specific targets, with probe 4-specific targets in blue and probe 5-specific targets in orange. Promiscuous lipid probe binders33 are indicated in red. Data represent means ± SD of cells obtained from a representative donor (n = 3). (E) Cellular component analysis of probe 5-interacting proteins by gene ontology (GO). (F) PTGR1 amino acid sequence starting at tryptophan W275. One of the identified tryptic peptides is underlined and bold. (G, H) Gel-based AfBPP of GFP- or PTGR1-overexpressing HEK-293-T cells using probes 4 and 5 and indicated a lipid competitor. Expression of PTGR1 was checked by anti-FLAG western blot.
Next, we investigated the protein interaction landscape of probe 5 in human M2 macrophages using two-step AfBPP. To identify specific targets of 17-HDHA, we also employed probe 4 to map general, promiscuous lipid binding proteins binding with equal preference to both probes. To this end, cells were incubated with probe 4 or 5 in serum-free medium for 30 min to allow for sufficient uptake. Crosslinking was effected by UV-irradiation (λ = 350 nm, 10 min) using a CaproBox,50 which irradiated the cells with simultaneous cooling at 4 °C to counteract the heat induced by the irradiation. Next, the cells were harvested, lysed, and subjected to copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC, “click”-reaction)51 conditions utilizing Cy5-N3 to enable the visualization of the probe-bound proteins by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis and in-gel fluorescence scanning (Figure 2B). This resulted in the visualization of many fluorescent bands for both probes, which were absent in the non-irradiated samples, demonstrating that the probes do not covalently interact with proteins without UV irradiation. Although a large overlap in fluorescent bands was revealed after labeling by either probe 4 or 5, there were also several probe-specific bands observed (Figure 2B).
To identify the probe-interacting proteins, a chemical proteomics experiment was performed. Label-free quantification was used in order to compare the relative abundance of probe-bound proteins in the samples.52 Briefly, the tagged proteins were ligated to biotin-N3 via click chemistry, and the resulting biotin-labeled proteins were enriched using avidin-coated agarose beads and digested using trypsin and the resulting tryptic peptides were analyzed by LC–MS using a Synapt G2-Si instrument. The data processing was performed with the commercial software Progenesis, which resulted in the identification and quantification of 179 proteins after deselection based on identified unique peptides and appearance in the CRAPome.53 Of these, 34 were significantly UV-enriched by probe 5 (Figure 2C,D). To identify probe 5-specific interacting proteins, the UV enrichment profiles of both probes were compared (Figure 2D). Proteins that were previously identified as promiscuous lipid probe binders33 are indicated in red. All promiscuous lipid probe binders, with the singular exception of voltage-dependent anion channel 2 (VDAC2), were equally enriched by both probes. Eight and ten proteins were specifically labeled by probes 4 and 5, respectively. This demonstrated that comparative AfBPP is capable of discerning probe-specific interactions.
Gene ontology (GO) enrichment analysis revealed that the proteins significantly UV-enriched by probe 5 are mainly found in organelles in lipid metabolism, such as the endoplasmic reticulum and mitochondria (Table 1). On the other hand, probe 4-specific targets are associated with lipid particles and transport vesicles, such as perilipin-2 and -3, which are involved in lipid droplet formation. This may suggest DHA is incorporated in lipid droplets. Of note, probe-5-specific targets were significantly associated with the extracellular exosomes GO term (Figure 2E)54 and predominantly annotated as oxidoreductase enzymes, for example, aldehyde dehydrogenase 1A1 (ALDH1A1) and prostaglandin reductase-1 (PTGR1). The main activity of ALDH1A1 consists of producing the signaling lipid retinoic acid by oxidation of retinaldehyde, but it can also react with other aldehydes such as 4-hydroxy-2-nonenal (4-HNE).55,56 Specific binding of probe 5 indicates ALDH1A1 could be involved 17-HDHA metabolism, possibly via a reactive aldehyde metabolite analogous to 4-hydroxy-2-nonenal. PTGR1 is known for its role in eicosoanoid and 4-HNE metabolism.57,58 PTGR1 expression is increased during inflammation and is involved in the resolution of inflammation through modulation of the HMGB1-miR522-3P-PTGR1 axis.59 PTGR1 functions as 15-oxo-prostaglandin 13-reductase and acts on 15-oxo-PGE1 and 15-oxo-PGE2.57,59 It also catalyzes the conversion LTB4 into its biologically less active metabolite, 12-oxo-LTB4, which is an initial and key step of metabolic inactivation of LTB4.60
Table 1. List of all Proteins Significantly UV-Enriched by Probe 5 and Their Probe Specificity.
| Uniprot accession | Gene name | Unique peptides | Description | Specific |
|---|---|---|---|---|
| Q9H2F3 | HSD3B7 | 3 | 3 beta-hydroxysteroid dehydrogenase type 7 | probe 5 |
| Q9NZ08 | ERAP1 | 20 | endoplasmic reticulum aminopeptidase 1 | probe 5 |
| Q14914 | PTGR1 | 11 | prostaglandin reductase 1 | probe 5 |
| Q9BTZ2 | DHRS4 | 10 | dehydrogenase/reductase SDR family member 4 | probe 5 |
| Q16698 | DECR1 | 4 | 2,4-dienoyl-CoA reductase, mitochondrial | probe 5 |
| Q01469 | FABP5 | 2 | fatty acid-binding protein, epidermal | probe 5 |
| O75874 | IDH1 | 8 | isocitrate dehydrogenase [NADP] cytoplasmic | probe 5 |
| Q9Y6N5 | SQOR | 6 | sulfide:quinone oxidoreductase, mitochondrial | probe 5 |
| P00352 | ALDH1A1 | 17 | retinal dehydrogenase 1 | probe 5 |
| P05091 | ALDH2 | 16 | aldehyde dehydrogenase, mitochondrial | probe 5 |
| Q92928 | RAB1C | 2 | putative Ras-related protein Rab-1C | probe 4 |
| Q99541 | PLIN2 | 11 | perilipin-2 | probe 4 |
| Q9BQE5 | APOL2 | 4 | apolipoprotein L2 | probe 4 |
| O60664 | PLIN3 | 2 | perilipin-3 | probe 4 |
| P45880 | VDAC2 | 7 | voltage-dependent anion-selective channel protein 2 | probe 4 |
| O15127 | SCAMP2 | 2 | secretory carrier-associated membrane protein 2 | probe 4 |
| Q8NBQ5 | HSD17B11 | 2 | estradiol 17-beta-dehydrogenase 11 | probe 4 |
| Q9H8H3 | METTL7A | 2 | methyltransferase-like protein 7A | probe 4 |
Validation of Probe 5-Specific Targets and Role of PTGR1 in 17-HDHA Metabolism
Two probe 5-enriched proteins, PTGR1 (isoform 1, Figure 2F) and ALDH1A1, were chosen as representative targets for validation as 17-HDHA interaction partners (Figure 2F). Both proteins were recombinantly expressed with a FLAG-tag in HEK-293T cells. Cells were treated with either probe 4 or 5, irradiated, and lysed, and probe-labeled proteins were ligated to Cy5-N3 using click chemistry. The proteins were resolved on SDS-PAGE gel and visualized with a fluorescent scanner (Figures 2G and S1). A fluorescent band was observed in a UV-dependent manner at the expected molecular weight for PTGR1 and ALDH1A1 in the samples treated with probe 5, but not by probe 4. The fluorescent band overlapped with a FLAG-tag signal in the western blot. Next, we investigated whether the labeling of PTGR1 could be outcompeted by 17-HDHA or DHA. Both lipids did not prevent the labeling of PTGR1 by probe 5 (Figure 2H). Inherent high lipophilicity, low aqueous solubility, and partitioning into the cellular membrane may explain why target occupancy studies are challenging with lipids.34,35 Alternatively, it is conceivable that probe 5 was converted into an active metabolite, such as a 17-oxo-derivative, that labels PTGR1. To investigate this hypothesis, we first tested if primary human M2 macrophages and human THP-1 cells are capable of forming 17-oxo-DHA using 17-HDHA as a substrate. To this end, a targeted lipidomics method was set up to quantify 17-oxo-DHA. Upon incubation of M2 macrophages and THP-1 cells with 17-HDHA, we could detect the formation of 17-oxo-DHA in a concentration-dependent manner (Figure 3A). To study whether PTGR1 uses 17-HDHA or 17-oxo-DHA as a substrate, we genetically deleted PTGR1 from THP-1 cells by CRISPR/Cas9. Two different KO clones were generated and treated with 17-HDHA or 17-oxo-DHA (Figures 3B and S3). It was found 17-oxo-DHA levels increased in the KO cells compared to wildtype cells, whereas 17-HDHA levels remained the same (Figure 3C,D). Conversely, overexpressing PTGR1 in U2OS cells resulted in lower 17-oxo-DHA levels, but 17-HDHA concentrations were not affected upon incubation with 17-HDHA (Figure 3E). To determine whether 17-oxo-DHA directly interacted with PTGR1, we performed a competition experiment with probe 5 on recombinant PTGR1 expressed in HEK293T cells. It was observed that 17-oxo-DHA could reduce the PTGR1 labeling by probe 5 (Figure 3F). Next, it was tested if PTGR1 could use 17-oxo-DHA as a substrate. Incubation of 17-oxo-DHA with purified, recombinant PTGR1 led to the conversion of the lipid into a metabolite with a mass of 17-oxo-DHA plus two hydrogen atoms in a temperature- and protein-dependent manner (Figure S5). Likely, the 15-cis double bond was reduced by the analogy of the action of PTGR1 on prostaglandins. Taken together, these results indicate that 17-oxo-DHA is a substrate of PTGR1.
Figure 3.

Investigation of 17-HDHA to 17-oxo-DHA metabolism and anti-inflammatory signaling. (A) 17-Oxo-DHA produced by M2 macrophages. Cells were either pretreated for 2 h with 17-HDHA or vehicle control. Data represent means ± SD of cells obtained from a representative donor (n = 3). (B) Characterization of PTGR1 CRISPR/Cas9 knockout THP1 cells. Single-cell clones were generated of which two were further cultured. (C) 17-Oxo-DHA produced by WT or PTGR1 KO THP1 cells treated with 1 μM 17-HDHA for 4 h. (D) 17-Oxo-DHA remaining after treating WT or PTGR1 KO THP1 cells with 1 μM 17-oxo-DHA for 3 h. (E) 17-HDHA and 17-oxo-DHA levels of U2OS cells overexpressing PTGR1 or control. Cells were treated for 4 h with 300 nM 17-HDHA. (F) Gel-based AfBPP of control or PTGR1-overexpressing HEK-293-T cells using probes 4 and 5 and indicated a lipid competitor. Expression of PTGR1 was checked by anti-FLAG western blot. Asterisk indicates PTGR1 band. (G) 5-HETE/AA ratio (LC–MS/MS) produced by ionophore-stimulated M2 macrophages. Cells were either pretreated for 15 min with 17-oxo-DHA or vehicle control. Data from THP-1 and U2OS cells represent means ± SD (n = 3–4). *p < 0.05, **p < 0.01, ***p < 0.001 in comparison to control (dotted line) using a one-way ANOVA with Dunnett’s multiple comparisons correction. ##p < 0.01 using a Student’s t-test. n.d.: not detected.
Finally, to investigate whether 17-oxo-DHA has anti-inflammatory effects, primary human M2 macrophages and neutrophils were stimulated with an ionophore with or without 17-oxo-DHA present. Notably, dose-dependent inhibition of the pro-inflammatory markers in both macrophages (Figure 3G) and neutrophils (Figure S4) was observed, thereby indicating that 17-oxo-DHA may limit the inflammatory response.
Conclusions
Here, we developed a comparative chemical proteomic approach with two complementary bifunctional photoreactive probes 4 and 5, which were based on the scaffold of the PUFA DHA and its oxidative metabolite 17-HDHA. Our aim was to identify specific targets of 17-HDHA. The probes were synthesized by forming building blocks containing four of the alkenes of DHA by partial hydrogenation and combining them by Wittig reactions, forming the last two double bonds. The final step in the synthesis of probe 5 was performed by employing an enzymatic reaction using soybean lipoxygenase.
Using both probes in primary human macrophages from healthy human donors, we identified several probe-specific lipid–protein interactions. PTGR1 was selected as a representative example for validation as a protein target of 17-HDHA. PTGR1 is known for its role in eicosoanoid and 4-HNE metabolism but was so far not linked to omega-3 lipid metabolism.57,58 PTGR1 is involved in immune signaling by inactivation of pro-inflammatory eicosanoids such as prostaglandins and LTB4. Here, we have shown that PTGR1 also uses 17-oxo-DHA as a substrate, likely by acting as a 15-reductase. Since 17-oxo-DHA reduces the formation of pro-inflammatory markers 5-HETE and LTB4, its metabolism by PTGR1 may limit this anti-inflammatory effect. Thus, PTGR1 is simultaneously metabolizing pro- and anti-inflammatory lipids. Further studies may shed light on the biological role of the novel metabolite generated by PGTR1.
Altogether, our study extends previous reports, suggesting that PTGR1 serves as a metabolic hub that inactivates proinflammatory LTB4 and metabolizes anti-inflammatory 17-oxo-DHA, thereby modulating the cellular levels of these important signaling lipids that act in concert to modulate the human macrophage–neutrophil axis. Finally, our results highlight the use of complementary bifunctional, photoreactive probes to identify specific protein interaction partners of promiscuous, lipophilic signaling molecules and also showcase the power of chemical proteomics in guiding the discovery of novel biological insights in primary human cells.
Acknowledgments
This work was supported by grant ICI-00016 from the Institute for Chemical Immunology. M.v.d.S. received base-funding from Oncode Institute. We thank Caprotec for kindly providing the Caprobox. Dr. Richard van den Berg is acknowledged for feedback on the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c06827.
Author Contributions
⊥ B.G. and J.H.H. contributed equally to this work.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE61 partner repository with the dataset identifier PXD027600.
Supplementary Material
References
- Calder P. C. Omega-3 Fatty Acids and Inflammatory Processes: From Molecules to Man. Biochem. Soc. Trans. 2017, 45, 1105–1115. 10.1042/BST20160474. [DOI] [PubMed] [Google Scholar]
- Layé S.; Nadjar A.; Joffre C.; Bazinet R. P. Anti-Inflammatory Effects of Omega-3 Fatty Acids in the Brain: Physiological Mechanisms and Relevance to Pharmacology. Pharmacol. Rev. 2018, 70, 12–38. 10.1124/pr.117.014092. [DOI] [PubMed] [Google Scholar]
- Larrieu T.; Layé S. Food for Mood: Relevance of Nutritional Omega-3 Fatty Acids for Depression and Anxiety. Front. Physiol. 2018, 9, 1047. 10.3389/fphys.2018.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder P. C. Marine Omega-3 Fatty Acids and Inflammatory Processes: Effects, Mechanisms and Clinical Relevance. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2015, 1851, 469–484. 10.1016/j.bbalip.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Proudman S. M.; James M. J.; Spargo L. D.; Metcalf R. G.; Sullivan T. R.; Rischmueller M.; Flabouris K.; Wechalekar M. D.; Lee A. T.; Cleland L. G. Fish Oil in Recent Onset Rheumatoid Arthritis: A Randomised, Double-Blind Controlled Trial within Algorithm-Based Drug Use. Ann. Rheum. Dis. 2015, 74, 89–95. 10.1136/annrheumdis-2013-204145. [DOI] [PubMed] [Google Scholar]
- Gioxari A.; Kaliora A. C.; Marantidou F.; Panagiotakos D. P. Intake of ω-3 Polyunsaturated Fatty Acids in Patients with Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. Nutrition 2018, 45, 114–124.e4. 10.1016/j.nut.2017.06.023. [DOI] [PubMed] [Google Scholar]
- Navarini L.; Afeltra A.; Gallo Afflitto G.; Margiotta D. P. E. Polyunsaturated Fatty Acids: Any Role in Rheumatoid Arthritis?. Lipids Health Dis. 2017, 16, 197. 10.1186/s12944-017-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C. N. Resolution Phase of Inflammation: Novel Endogenous Anti-Inflammatory and Proresolving Lipid Mediators and Pathways. Annu. Rev. Immunol. 2007, 25, 101–137. 10.1146/annurev.immunol.25.022106.141647. [DOI] [PubMed] [Google Scholar]
- Serhan C. N. Pro-Resolving Lipid Mediators Are Leads for Resolution Physiology. Nature 2014, 510, 92–101. 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnig C.; Bezema T.; Calder P. C.; Charloux A.; Frossard N.; Garssen J.; Haworth O.; Dilevskaya K.; Levi-Schaffer F.; Lonsdorfer E.; et al. Activation of Resolution Pathways to Prevent and Fight Chronic Inflammation: Lessons From Asthma and Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 1699. 10.3389/fimmu.2019.01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G.; Neurath M. F. Resolution of Chronic Inflammatory Disease: Universal and Tissue-Specific Concepts. Nat. Commun. 2018, 9, 3261. 10.1038/s41467-018-05800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I.; Glass C. K. Anti-Inflammatory Therapy in Chronic Disease: Challenges and Opportunities. Science 2013, 339, 166–172. 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C. N.; Fredman G.; Yang R.; Karamnov S.; Belayev L. S.; Bazan N. G.; Zhu M.; Winkler J. W.; Petasis N. A. Novel Proresolving Aspirin-Triggered DHA Pathway. Chem. Biol. 2011, 18, 976–987. 10.1016/j.chembiol.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weylandt K. H.; Chiu C.-Y.; Gomolka B.; Waechter S. F.; Wiedenmann B. Omega-3 Fatty Acids and Their Lipid Mediators: Towards an Understanding of Resolvin and Protectin Formation. Prostaglandins Other Lipid Mediators 2012, 97, 73–82. 10.1016/j.prostaglandins.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Valdes A. M.; Ravipati S.; Menni C.; Abhishek A.; Metrustry S.; Harris J.; Nessa A.; Williams F. M. K.; Spector T. D.; Doherty M.; et al. Association of the Resolvin Precursor 17-HDHA, but Not D- or E- Series Resolvins, with Heat Pain Sensitivity and Osteoarthritis Pain in Humans. Sci. Rep. 2017, 7, 10748. 10.1038/s41598-017-09516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A. F.; Claudino R. F.; Dutra R. C.; Marcon R.; Calixto J. B. Omega-3 Fatty Acid-Derived Mediators 17(R)-Hydroxy Docosahexaenoic Acid, Aspirin-Triggered Resolvin D1 and Resolvin D2 Prevent Experimental Colitis in Mice. J. Immunol. 2011, 187, 1957–1969. 10.4049/jimmunol.1101305. [DOI] [PubMed] [Google Scholar]
- Duffield J. S.; Hong S.; Vaidya V. S.; Lu Y.; Fredman G.; Serhan C. N.; Bonventre J. V. Resolvin D Series and Protectin D1 Mitigate Acute Kidney Injury. J. Immunol. 2006, 177, 5902–5911. 10.4049/jimmunol.177.9.5902. [DOI] [PubMed] [Google Scholar]
- Lima-Garcia J. F.; Dutra R. C.; da Silva K.; Motta E. M.; Campos M. M.; Calixto J. B. The Precursor of Resolvin D Series and Aspirin-Triggered Resolvin D1 Display Anti-Hyperalgesic Properties in Adjuvant-Induced Arthritis in Rats. Br. J. Pharmacol. 2011, 164, 278–293. 10.1111/j.1476-5381.2011.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Périz A.; Planagumà A.; Gronert K.; Miquel R.; López-Parra M.; Titos E.; Horrillo R.; Ferré N.; Deulofeu R.; Arroyo V.; et al. Docosahexaenoic Acid (DHA) Blunts Liver Injury by Conversion to Protective Lipid Mediators: Protectin D1 and 17S-Hydroxy-DHA. FASEB J. 2006, 20, 2537–2539. 10.1096/fj.06-6250fje. [DOI] [PubMed] [Google Scholar]
- von Hegedus J. H.; Kahnt A. S.; Ebert R.; Heijink M.; Toes R. E. M.; Giera M.; Ioan-Facsinay A. Toll-like Receptor Signaling Induces a Temporal Switch towards a Resolving Lipid Profile in Monocyte-Derived Macrophages. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2020, 1865, 158740 10.1016/j.bbalip.2020.158740. [DOI] [PubMed] [Google Scholar]
- Chiu C.-Y.; Gomolka B.; Dierkes C.; Huang N. R.; Schroeder M.; Purschke M.; Manstein D.; Dangi B.; Weylandt K. H. Omega-6 Docosapentaenoic Acid-Derived Resolvins and 17-Hydroxydocosahexaenoic Acid Modulate Macrophage Function and Alleviate Experimental Colitis. Inflammation Res. 2012, 61, 967–976. 10.1007/s00011-012-0489-8. [DOI] [PubMed] [Google Scholar]
- Dalli J.; Winkler J. W.; Colas R. A.; Arnardottir H.; Cheng C.-Y. C.; Chiang N.; Petasis N. A.; Serhan C. N. Resolvin D3 and Aspirin-Triggered Resolvin D3 Are Potent Immunoresolvents. Chem. Biol. 2013, 20, 188–201. 10.1016/j.chembiol.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niphakis M. J.; Lum K. M.; Cognetta A. B. III; Correia B. E.; Ichu T.-A.; Olucha J.; Brown S. J.; Kundu S.; Piscitelli F.; Rosen H.; et al. A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells. Cell 2015, 161, 1668–1680. 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce J. J.; Cognetta A. B.; Niphakis M. J.; Tully S. E.; Cravatt B. F. Proteome-Wide Mapping of Cholesterol-Interacting Proteins in Mammalian Cells. Nat. Methods 2013, 10, 259–264. 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland M. M.; Bostic H. E.; Gong D.; Speers A. E.; Lucas N.; Cho W.; Cravatt B. F.; Best M. D. Phosphatidylinositol 3,4,5-Trisphosphate Activity Probes for the Labeling and Proteomic Characterization of Protein Binding Partners. Biochemistry 2011, 50, 11143–11161. 10.1021/bi201636s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Du S.; Cazenave-Gassiot A.; Ge J.; Lee J.-S.; Wenk M. R.; Yao S. Q. Global Mapping of Protein–Lipid Interactions by Using Modified Choline-Containing Phospholipids Metabolically Synthesized in Live Cells. Angew. Chem. 2017, 129, 5923–5927. 10.1002/ange.201702509. [DOI] [PubMed] [Google Scholar]
- Haberkant P.; Raijmakers R.; Wildwater M.; Sachsenheimer T.; Brügger B.; Maeda K.; Houweling M.; Gavin A.-C.; Schultz C.; van Meer G.; et al. In Vivo Profiling and Visualization of Cellular Protein–Lipid Interactions Using Bifunctional Fatty Acids. Angew. Chem., Int. Ed. 2013, 52, 4033–4038. 10.1002/anie.201210178. [DOI] [PubMed] [Google Scholar]
- Höglinger D.; Nadler A.; Haberkant P.; Kirkpatrick J.; Schifferer M.; Stein F.; Hauke S.; Porter F. D.; Schultz C. Trifunctional Lipid Probes for Comprehensive Studies of Single Lipid Species in Living Cells. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 1566–1571. 10.1073/pnas.1611096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberkant P.; Stein F.; Höglinger D.; Gerl M. J.; Brügger B.; Van Veldhoven P. P.; Krijgsveld J.; Gavin A.-C.; Schultz C. Bifunctional Sphingosine for Cell-Based Analysis of Protein-Sphingolipid Interactions. ACS Chem. Biol. 2016, 11, 222–230. 10.1021/acschembio.5b00810. [DOI] [PubMed] [Google Scholar]
- Zhuang S.; Li Q.; Cai L.; Wang C.; Lei X. Chemoproteomic Profiling of Bile Acid Interacting Proteins. ACS Cent. Sci. 2017, 3, 501–509. 10.1021/acscentsci.7b00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin T. K.; Micalizio G. C. Convergent and Stereospecific Synthesis of Complex Skipped Polyenes and Polyunsaturated Fatty Acids. Nat. Chem. 2010, 2, 638–643. 10.1038/nchem.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum K. M.; Sato Y.; Beyer B. A.; Plaisted W. C.; Anglin J. L.; Lairson L. L.; Cravatt B. F. Mapping Protein Targets of Bioactive Small Molecules Using Lipid-Based Chemical Proteomics. ACS Chem. Biol. 2017, 12, 2671–2681. 10.1021/acschembio.7b00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenders S. T. A.; Gagestein B.; van der Stelt M.. Opportunities for Lipid-Based Probes in the Field of Immunology. In Activity-Based Protein Profiling; Cravatt B. F., Hsu K.-L., Weerapana E., Eds.; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, 2018; pp 283–319. [DOI] [PubMed] [Google Scholar]
- Bockelmann S.; Mina J. G. M.; Korneev S.; Hassan D. G.; Müller D.; Hilderink A.; Vlieg H. C.; Raijmakers R.; Heck A. J. R.; Haberkant P.; et al. A Search for Ceramide Binding Proteins Using Bifunctional Lipid Analogs Yields CERT-Related Protein StarD7. J. Lipid Res. 2018, 59, 515–530. 10.1194/jlr.M082354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarik M.; Biam R. S.; Robichaud P.-P.; Surette M. E. The Impact of PUFA on Cell Responses: Caution Should Be Exercised When Selecting PUFA Concentrations in Cell Culture. Prostaglandins Leukot. Essent. Fatty Acids 2020, 155, 102083 10.1016/j.plefa.2020.102083. [DOI] [PubMed] [Google Scholar]
- Li Z.; Hao P.; Li L.; Tan C. Y. J.; Cheng X.; Chen G. Y. J.; Sze S. K.; Shen H.-M.; Yao S. Q. Design and Synthesis of Minimalist Terminal Alkyne-Containing Diazirine Photo-Crosslinkers and Their Incorporation into Kinase Inhibitors for Cell- and Tissue-Based Proteome Profiling. Angew. Chem. 2013, 125, 8713–8718. 10.1002/ange.201300683. [DOI] [PubMed] [Google Scholar]
- Kleiner P.; Heydenreuter W.; Stahl M.; Korotkov V. S.; Sieber S. A. A Whole Proteome Inventory of Background Photocrosslinker Binding. Angew. Chem., Int. Ed. 2017, 56, 1396–1401. 10.1002/anie.201605993. [DOI] [PubMed] [Google Scholar]
- Sakurai K. Photoaffinity Probes for Identification of Carbohydrate-Binding Proteins. Asian J. Org. Chem. 2015, 4, 116–126. 10.1002/ajoc.201402209. [DOI] [Google Scholar]
- Vik A.; Hansen T. V. Synthetic Manipulations of Polyunsaturated Fatty Acids as a Convenient Strategy for the Synthesis of Bioactive Compounds. Org. Biomol. Chem. 2018, 16, 9319–9333. 10.1039/C8OB02586J. [DOI] [PubMed] [Google Scholar]
- Balas L.; Durand T.; Saha S.; Johnson I.; Mukhopadhyay S. Total Synthesis of Photoactivatable or Fluorescent Anandamide Probes: Novel Bioactive Compounds with Angiogenic Activity. J. Med. Chem. 2009, 52, 1005–1017. 10.1021/jm8011382. [DOI] [PubMed] [Google Scholar]
- Heitz M. P.; Wagner A.; Mioskowski C.; Noel J. P.; Beaucourt J. P. Synthesis of All-Cis-1-Bromo-4,7,10,13-Nonadecatetraene: A Precursor to C-1-Labeled Arachidonic Acid. J. Org. Chem. 1989, 54, 500–503. 10.1021/jo00263a049. [DOI] [Google Scholar]
- Saha G.; Basu M. K.; Kim S.; Jung Y.-J.; Adiyaman Y.; Adiyaman M.; Powell W. S.; FitzGerald G. A.; Rokach J. A Convenient Strategy for the Synthesis of β,γ-Unsaturated Aldehydes and Acids. A Construction of Skipped Dienes. Tetrahedron Lett. 1999, 40, 7179–7183. 10.1016/S0040-4039(99)01498-7. [DOI] [Google Scholar]
- Rosell M.; Villa M.; Durand T.; Galano J.-M.; Vercauteren J.; Crauste C. Total Syntheses of Two Bis-Allylic-Deuterated DHA Analogues. Asian J. Org. Chem. 2017, 6, 322–334. 10.1002/ajoc.201600565. [DOI] [Google Scholar]
- Coffa G.; Imber A. N.; Maguire B. C.; Laxmikanthan G.; Schneider C.; Gaffney B. J.; Brash A. R. On the Relationships of Substrate Orientation, Hydrogen Abstraction, and Product Stereochemistry in Single and Double Dioxygenations by Soybean Lipoxygenase-1 and Its Ala542Gly Mutant. J. Biol. Chem. 2005, 280, 38756–38766. 10.1074/jbc.M504870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechetkin I. R.; Osipova E. V.; Tarasova N. B.; Mukhitova F. K.; Hamberg M.; Gogolev Y. V.; Grechkin A. N. Specificity of Oxidation of Linoleic Acid Homologs by Plant Lipoxygenases. Biochemistry 2009, 74, 855–861. 10.1134/S0006297909080069. [DOI] [PubMed] [Google Scholar]
- Dobson E. P.; Barrow C. J.; Kralovec J. A.; Adcock J. L. Controlled Formation of Mono- and Dihydroxy-Resolvins from EPA and DHA Using Soybean 15-Lipoxygenase. J. Lipid Res. 2013, 54, 1439–1447. 10.1194/jlr.M036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovich I. A. A One-Step Method of 10,17-Dihydro(Pero)Xydocosahexa-4Z,7Z,11E,13Z,15E,19Z-Enoic Acid Synthesis by Soybean Lipoxygenase. J. Lipid Res. 2006, 47, 854–863. 10.1194/jlr.D500042-JLR200. [DOI] [PubMed] [Google Scholar]
- Walther M.; Ivanov I.; Myagkova G.; Kuhn H. Alterations of Lipoxygenase Specificity by Targeted Substrate Modification and Site-Directed Mutagenesis. Chem. Biol. 2001, 8, 779–790. 10.1016/S1074-5521(01)00050-3. [DOI] [PubMed] [Google Scholar]
- Ivanov I.; Golovanov A. B.; Ferretti C.; Canyelles-Niño M.; Heydeck D.; Stehling S.; Lluch J. M.; González-Lafont À.; Kühn H. Mutations of Triad Determinants Changes the Substrate Alignment at the Catalytic Center of Human ALOX5. ACS Chem. Biol. 2019, 14, 2768–2782. 10.1021/acschembio.9b00674. [DOI] [PubMed] [Google Scholar]
- Luo Y.; Blex C.; Baessler O.; Glinski M.; Dreger M.; Sefkow M.; Köster H. The CAMP Capture Compound Mass Spectrometry as a Novel Tool for Targeting CAMP-Binding Proteins: From Protein Kinase A To Potassium/Sodium Hyperpolarization-Activated Cyclic Nucleotide-Gated Channels. Mol. Cell. Proteomics 2009, 8, 2843–2856. 10.1074/mcp.M900110-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himo F.; Lovell T.; Hilgraf R.; Rostovtsev V. V.; Noodleman L.; Sharpless K. B.; Fokin V. V. Copper(I)-Catalyzed Synthesis of Azoles. DFT Study Predicts Unprecedented Reactivity and Intermediates. J. Am. Chem. Soc. 2005, 127, 210–216. 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- van Rooden E. J.; Florea B. I.; Deng H.; Baggelaar M. P.; van Esbroeck A. C. M.; Zhou J.; Overkleeft H. S.; van der Stelt M. Mapping in Vivo Target Interaction Profiles of Covalent Inhibitors Using Chemical Proteomics with Label-Free Quantification. Nat. Protoc. 2018, 13, 752–767. 10.1038/nprot.2017.159. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D.; Wright Z.; Couzens A. L.; Lambert J. P.; St-Denis N. A.; Li T.; Miteva Y. V.; Hauri S.; Sardiu M. E.; Low T. Y.; et al. The CRAPome: A Contaminant Repository for Affinity Purification-Mass Spectrometry Data. Nat. Methods 2013, 10, 730–736. 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Yoval-Sánchez B.; Rodríguez-Zavala J. S. Differences in Susceptibility to Inactivation of Human Aldehyde Dehydrogenases by Lipid Peroxidation Byproducts. Chem. Res. Toxicol. 2012, 25, 722–729. 10.1021/tx2005184. [DOI] [PubMed] [Google Scholar]
- Vasiliou V.; Pappa A.; Petersen D. R. Role of Aldehyde Dehydrogenases in Endogenous and Xenobiotic Metabolism. Chem.-Biol. Interact. 2000, 129, 1–19. 10.1016/S0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Mesa J.; Alsina C.; Oppermann U.; Parés X.; Farrés J.; Porté S. Human Prostaglandin Reductase 1 (PGR1): Substrate Specificity, Inhibitor Analysis and Site-Directed Mutagenesis. Chem.-Biol. Interact. 2015, 234, 105–113. 10.1016/j.cbi.2015.01.021. [DOI] [PubMed] [Google Scholar]
- Sánchez-Rodríguez R.; Torres-Mena J. E.; Quintanar-Jurado V.; Chagoya-Hazas V.; Rojas del Castillo E.; del Pozo Yauner L.; Villa-Treviño S.; Pérez-Carreón J. I. Ptgr1 Expression Is Regulated by NRF2 in Rat Hepatocarcinogenesis and Promotes Cell Proliferation and Resistance to Oxidative Stress. Free Radical Biol. Med. 2017, 102, 87–99. 10.1016/j.freeradbiomed.2016.11.027. [DOI] [PubMed] [Google Scholar]
- Kang G.-J.; Lee H.-J.; Byun H. J.; Kim E. J.; Kim H. J.; Park M. K.; Lee C.-H. Novel Involvement of MiR-522-3p in High-Mobility Group Box 1-Induced Prostaglandin Reductase 1 Expression and Reduction of Phagocytosis. Biochim. Biophys. Acta, Mol. Cell Res. 2017, 1864, 625–633. 10.1016/j.bbamcr.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Yamamoto T.; Yokomizo T.; Nakao A.; Izumi T.; Shimizu T. Immunohistochemical Localization of Guinea-Pig Leukotriene B4 12-Hydroxydehydrogenase/15-Ketoprostaglandin 13-Reductase. Eur. J. Biochem. 2001, 268, 6105–6113. 10.1046/j.0014-2956.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Bai J.; Bandla C.; García-Seisdedos D.; Hewapathirana S.; Kamatchinathan S.; Kundu D. J.; Prakash A.; Frericks-Zipper A.; Eisenacher M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.