Abstract
Advances in genomics have led to the identification of many risk loci with hundreds of genes and thousands of DNA variants associated with neuropsychiatric disorders. A significant barrier to understanding the genetic underpinnings of complex diseases is the lack of functional characterization of risk genes and variants in biological systems relevant to human health and connecting disease-associated variants to pathological phenotypes. Characterizing gene and DNA variant functions requires genetic perturbations followed by molecular and cellular assays of neurobiological phenotypes. However, generating null or mutant alleles is low throughput, making it impossible to characterize disease-associated variants in large quantities efficiently. CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) screens can be leveraged to dissect the biological consequences of the tested genes and variants in their native context. Nevertheless, testing non-coding variants associated with complex diseases remains non-trivial. In this review, we first discuss the current challenges of interpreting the function of the non-coding genome and approaches to prioritizing disease-associated variants in the context of the 3D epigenome. Second, we provide a brief overview of high-throughput CRISPRi and CRISPRa screening strategies applicable for characterizing non-coding sequences in appropriate biological systems. Lastly, we discuss the promising prospects of using CRISPR-based technologies to dissect DNA sequences associated with neuropsychiatric diseases.
Introduction
Although neuropsychiatric disorders, such as intellectual disabilities, autism and schizophrenia, exhibit genetic heritability, the exact genetic underpinnings remain elusive. Large-scale studies such as the ENCODE and Epigenome Roadmap projects have annotated millions of candidate cis-regulatory elements (cCREs) on the basis of their chromatin accessibility, associations to histone modifications, transcription factor (TF) binding, DNA hypomethylation, etc. One of the most striking findings from these annotations is that putative regulatory sequences harbor a disproportionally large number of sequence variants associated with diverse human traits and diseases, leading to the theory that genetic variations in regulatory elements contribute substantially to common complex human diseases (1,2). However, the reliability of these descriptive maps to capture the functional CREs remains, for the most part, to be demonstrated. In this review, we will discuss how to prioritize non-coding variants by leveraging 3D epigenome annotation and how to characterize their functional impact with CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) screens.
3D Epigenome Annotation and Prioritization of Non-Coding Variants Associated with Neuropsychiatric Diseases
Genome-wide association studies (GWAS) have revealed thousands of non-coding DNA variants associated with neuropsychiatric disorders. However, it has been challenging to prioritize risk variants, identify the cell types in which variants function or the target genes they regulate and measure the effect sizes of disease-associated variants on their target gene expression due to the following reasons: first, genetic loci are themselves hard to decipher, as the majority are located in poorly annotated non-coding regions, whose impact is governed by cis-regulatory mechanisms that remain to be understood; second, regulatory sequences often interact with their target genes over long genomic distances, precluding a straightforward identification of disease risk genes and limiting the interpretation of non-coding variants from GWAS. Typically, neighboring genes are assigned as risk loci for non-coding variants. However, this nearest gene model is challenged by both experimental and computational evidence (3,4); third, the complexity of brain tissues, with numerous heterogeneous cell types (5), makes any direct interpretation of the genetic information burdensome, as genetic loci can affect only a subset of cell types during specific cellular and pathophysiological states. Therefore, to understand the functional impact of non-coding variants, it is vital to go beyond genetic associations and integrate diverse data types to elucidate the target genes and pathways of the genetic variants in a cell-type-specific manner.
To overcome these challenges, epigenomic annotations can be leveraged to prioritize risk variants that affect the gene regulatory functions of cCREs. For example, open chromatin regions, chromatin states associated with active histone marks and TF binding sites can be used for prioritizing variants contributing to disease by disrupting the regulatory functions of enhancers (2). One model for temporal–spatial control of cell-type-specific gene expression from distal elements is that cCREs, such as enhancers, can loop to their target genes and activate gene transcription. Thus, information on 3D chromatin structure, in particular, high-resolution chromatin loops between cCREs and their gene targets, can further facilitate the identification of risk genes and biological processes affected by non-coding variants (6–9). The physical interactions between cCREs and gene promoters can be mapped using several chromatin conformation capture methods, including Hi-C (10), Micro-C (11), HiChIP (12,13) and Genome Architecture Mapping (14). Such information can pinpoint causal variants to their linked genes, and their dysregulation can be one of the mechanisms of disease pathogenesis via impacting transcriptional regulation. Thus, integrative approaches combining chromatin spatial organization studies with medical genetic information, along with other multi-omics data, can facilitate the identification of putative risk variants and genes. For example, microdissected human brain regions were used for genome-wide mapping of long-range chromatin interactions to prioritize neuropsychiatric variants in cCREs and their target genes (7,15).
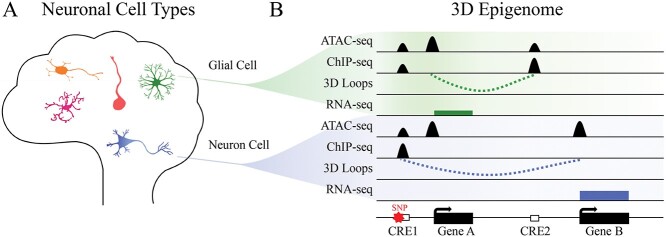
Complex diseases often involve multiple dysregulated loci with cell-type-specific patterns of activity. This presents unique challenges for deciphering disease etiology when attempting to distinguish causative mechanisms from secondary phenotypes within heterogeneous samples. For these reasons, comprehensive annotation of regulatory relationships in specific, well-characterized cell populations, should enable in-depth insights into complex disease biology (Fig. 1). To this end, several studies used specific cell types sorted from human brain tissues for the identification of high-resolution chromatin loops between promoters and distal cis-regulatory elements (6,9,16). In one study, the authors isolated excitatory neurons, oligodendrocytes, microglia and astrocytes from the adult brain and revealed that Alzheimer’s disease variants are significantly enriched in microglia-specific enhancers (6). In another study, the authors isolated radial glia, intermediate progenitor cells, excitatory neurons and interneurons from the developing human cortex, and identified tens of thousands of cell-type-specific chromatin interactions. Further analysis discovered that lineage-specific genes are enriched for promoters with unusually high interactives, termed super interactive promoters (SIPs) (9). SIPs represent a unique mechanism for fine-tuning gene expression by having multiple distal regulatory elements essential for cellular identity and function. In addition, human-induced pluripotent stem cell (iPSC) differentiation models remain attractive for 3D epigenome studies for cell types that cannot be readily isolated from the human brain (17–19). For example, iPSC neuronal and glia differentiation have been used for interrogating cell-type-specific 3D chromatin interactions (8,20). These studies provided an excellent resource for annotating disease variants and target gene pairs. Moreover, iPSC differentiation offers convenient models for testing the functionalities of disease variant regions through genetic perturbation.
Figure 1.
Characterizing human neuronal cells using 3D epigenome. (A) The human brain contains diverse heterogeneous neuronal cell types, which include neuons, astrocytes, oligodendrocytes and microglia et al. (B) Different cell types show distinct 3D epigenome landscape and gene regulatory network. In the example, besides the shared open chromatin regions, the neuronal and glial cells have unique open chromatin regions marked by ATAC-seq signal, which indicate potential cell-type-specific regulatory function of CREs. Active chromatin marks, 3D chromatin contacts and RNA-seq data indicate that CRE1 can regulate Gene B in the neuronal cells and that CRE2 can regulate Gene A in glial cells. Due to the cell-type-specific transcriptional regulation of CREs, functional validation of disease-related SNP within CRE1 would be best performed in neuronal cells.
High-Throughput CRISPRi and CRISPRa Approaches to Characterize Disease-Associated Non-Coding Loci
Clustered regularly interspaced short palindromic repeats (CRISPR), originally derived from bacteria adaptive immune systems (21), is a versatile genome-editing tool. The original CRISPR was constructed through two key components, a single-guide RNA (gRNA) and a Cas9 endonuclease from Streptococcus pyogenes (22). Cas9 can be directed to the specific target site by a gRNA with complementary endogenous sequences and generate double strand breaks (DSBs) that can make indels through non-homologous DNA end joining (22–24). CRISPR can also be used for precision genome editing. In particular, homology-directed repair of CRIPSR-induced DSBs enables precise editing with defined donor DNA sequences, including base substitutions, sequence insertions and deletions (25). In this review, we will focus on CRISPRi and CRISPRa epigenome editing tools, which are more suitable for high-throughput genetic screens due to their higher efficiency in perturbing both alleles, thus yielding more consistent and robust results compared with CRISPR indel and CRISPR deletion approaches. CRISPRi and CRISPRa are achieved by using the catalytically dead Cas9 (dCas9), which binds to the target site without inducing DNA cleavage. CRISPRi can block the transcription initiation or elongation by dCas9 alone (26,27). In addition, CRISPRi can effectively repress transcription by fusing dCas9 with transcriptional repressor domains, such as KRAB (28), to induce gene silencing and enhancer inhibition more efficiently than dCas9 alone (28,29). CRISPRa can activate gene expression by fusing dCas9 with one or more activation domains, including VP64 (28), VP64-p65-Rta (30), synergistic activation mediator (31) or SunTag (32). CRISPRi and CRISPRa both result in reversible epigenome editing, as the systems must be constitutively expressed to perturb function. This is no longer the case with CRISPRoff (33), a system that establishes epigenomic silencing by fusing KRAB, Dnmt3A and Dnmt3L to dCas9. CRSIPRoff recruits histone deacetylases, histone methyltransferases and heterochromatin protein 1, leading to the formation of heterochromatin (28,29) and DNA methylation and establishing a heritable repressive state throughout cell proliferation and differentiation (33,34).
Although CRISPRi and CRISPRa can be effectively used for interrogating genome function for both genes and cCREs (28,35–37), genes, rather than non-coding regions containing risk variants, are more extensively studied by CRISPR approaches. In the following section, we will discuss a few genetic screening strategies that are applicable to advance our understanding of cCREs and disease-associated non-coding variants.
Pooled CRISPR screens with survival/proliferation phenotypes
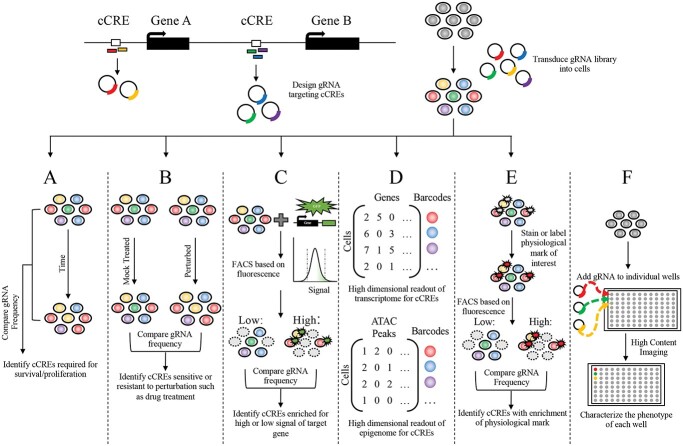
Survival and proliferation are classical phenotypes that have been widely used in the context of CRISPR screens. In this approach, gRNA libraries can be delivered together with CRISPRi and CRISPRa machinery to induce silencing or activation of regions of interest in cells. By quantifying the relative depletion or enrichment of gRNAs between two time points, loci that are essential for cell survival, differentiation or proliferation can be identified (Fig. 2A). This technique has been used to identify genes essential to the survival/differentiation of neurons with both CRISPRi and CRISPRa perturbations (38,39). Similarly, CRISPRi and CRISPRa screens on the basis of survival and proliferation phenotypes can be applied to identify genomic loci that confer sensitivity or resistance to toxic conditions, including drug treatment or cellular stress, by analyzing guide distribution before and after treatment (Fig. 2B). For example, the GPX4 gene, which encodes a glutathione peroxidase, was only found to be essential for neuronal survival under oxidative stress conditions (39). In principle, survival phenotype can identify cCREs regulating cell growth and survival genes. However, most disease-associated variants are not necessarily linked to the survival and proliferation phenotypes, and cell survival may not be sensitive enough to identify enhancers with GWAS-identified variants that only exert subtle effects on essential gene expression. Thus, CRISPR screen strategies directly link to transcriptional output and disease-relevant cellular function are more desirable.
Figure 2.
Strategies of CRISPR screens. Experiment overview for high-throughput CRISPR screening, where gRNAs designed to target cCREs are cloned and then delivered into the cell type of interest containing CRISPR machinery. After delivery, the transduce cells containing the pooled gRNAs can be characterized by several different techniques with varying readouts. (A) Comparing the distribution of pooled gRNAs between two time points can identify loci essential for proliferation and survivability. (B) Assessing the relative depletion or enrichment of gRNAs between a perturbed condition such as drug treatment compared with a mock treatment at the same time point or the time point before treatment, connects loci that confer sensitivity of resistance to the perturbation. (C) Quantifying the expression changes induced by cCREs of an endogenous gene tagged with GFP allows FACS to separate cells with high or low levels of fluorescence/gene expression and identify loci that regulate the expression of the tagged gene. (D) Conducting scRNA-seq or scATAC-seq identifies high-dimensional transcriptional or epigenomic phenotypes associated with perturbation of cCREs. (E) Separating pooled cells, for example by FACS, with a labeled physiological biomarker such as protein levels, allows for the gRNA distribution of cells with and without the marker to be compared with identify loci associated with a biological process. (F) Complex phenotypes that cannot be measured in pooled screens can be investigated in arrayed screens where gRNAs are introduced and characterized in individual wells.
Pooled CRISPR screens on the basis of gene expression
The 3D genome annotation has identified many cCREs physically interacting with gene promoters, suggesting their functionality in regulating gene expression. Several studies have used CRISPRi and CRISPRa in a massively parallel fashion to screen for enhancers of a gene of interest (Fig. 2C). Genes expression levels can be either detected by an endogenously tagged fluorescent reporter (36,40,41) or quantified signals with fluorescence in situ hybridization (42,43) using fluorescence-activated cell sorting (FACS) after perturbing cCREs. Although these studies typically focus on down regulation of target genes (36,42,43), upregulation through CRISPRa experiments can also identify potential CREs (44).
Pooled CRISPR screens coupled with single-cell RNA sequencing (scRNA-seq), e.g. Mosaic-Seq (45), can identify genes, pathways and genetic interactions in cells with specific perturbations (46–48) (Fig. 2). Moreover, scATAC-seq can also be coupled with CRISPR screens to identify how perturbations alter the epigenomic landscape and gene regulatory networks (49–51). Furthermore, genes identified as targets of perturbed regions can be used for gene ontology analysis to identify shared and unique pathways under different CRISPR conditions. However, due to the limited power and high-sequencing cost, they are often utilized to follow-up prioritized targets (52,53). These limitations are further exacerbated when screening for CREs, as they are more abundant than expressed genes and often have smaller effect sizes on transcription. To overcome these limitations, targeted quantification of select genes (54,55), or increasing the multiplicity of infection (56), can lead to increased power to detect transcriptional changes following CRISPR perturbation.
Pooled CRISPR screens on the basis of cellular physiological functions
Currently, there is a shortage of tractable biological models to test cCREs in the human genome at scale. In addition, there is a big gap between observing transcription effects caused by perturbation at cCREs and such changes in transcription that contribute to disease phenotypes. As mentioned in the gene reporter-based strategy, CRISPR screens for interrogating cCREs are low throughput, generally focusing on cCREs of one gene at a time (36,40,41). CRISPRpath, a scalable screening strategy can be applied for parallelly characterizing CREs from multiple genomic loci by leveraging genes converging on phenotypes linked via a specific biological pathway (57). Similarly, cellular assays can be used to screen for cCREs of multiple genes that affect the same physiological functions associated with disease states. For example, Liu et al. identified factors upregulated by CRISPRa that facilitate neuronal cell fate in mouse embryonic stem cells by sorting cells that express the neuronal marker TUBB3 upon activation (58). Future studies focusing on additional disease-relevant phenotypes that can be selected by FACS, including active mitochondria and calcium signaling in neurons or phagocytosis assay and cytokine production in immune response in microglia, should yield novel insights into the genetic basis of complex neuropsychiatric diseases (Fig. 2E). Certain phenotypes, such as complex morphologies, rely on high-content imaging analysis. After characterizing pooled cells via microscopy, multiple methods exist to identify the guides enriched for the phenotype. First, targeted in situ sequencing can be used to identify the gRNA expressed in each cell (59). Second, targeted photoactivation of a fluorescent protein in an automated manner, allows for separation via FACS (60). Lastly, a recent publication developed image-enabled cell sorting, which simultaneously combines fluorescence microscopy and flow cytometry techniques in a novel manner that allows for screens of cell morphology and localization of labeled proteins (61). This method holds great promise in performing genetic screens using complex phenotypes.
Array-based screens for complex phenotypes
Although all previous screens focused on pooled libraries, complex phenotypes, such as the excitability of neurons, or the secretion of specific factors, generally cannot be disentangled through pooled formats and must be investigated in array screens. The key principle of these screens is to deliver gRNA to separate wells and subsequently evaluate the phenotype of interest associated with each well (Fig. 2F). Compared with the above methods, array screens are relatively low throughput and normally rely on automation to discern phenotypes.
CRISPR screens in biologically relevant systems
The majority of large-scale CRISPR screens have been conducted in transformed cell lines with little relevance to human health and diseases. Although screens in non-neuronal systems have been able to discover hits that subsequently reproducible results in neuronal cell types (62,63), it is highly likely that neuronal-specific functional sequences would be missed in a different cell type. For example, a study screening for essential genes in iPSC-derived neurons identified genes specific to neurons, including MAP3K12, MAPK8 and CDKN1C (38). Recently, a few studies have embarked on functional characterization in iPSC differentiation models (20,38,39), primary cells isolated from the human brain (9) and mouse brain tissues (52). For example, CRISPRview combines CRISPR technology with single molecule FISH and immunostaining to interrogate DNA sequences at a single-cell level, allowing the very first test of brain enhancers using cells directly isolated from the human brain (9). In another study, a total of 35 autism spectrum disorder/neurodevelopmental delay risk genes were evaluated by perturb-seq using the developing mouse brain in utero (52). Similar strategies could be considered for testing cCREs and non-coding variants in vivo.
Alternatively, brain organoids can also be used for functional characterization of disease-associated variants with the following advantages. First, organoids consist of a diverse collection of cell types communicating with each other, allowing interrogation of gene function in diverse cell types, including those that are hard to isolate from in vivo or iPSC differentiation models. Moreover, cell lines are typically grown in 2D, rather than 3D, which can lead to varying results in screens, as shown by recent work in cancer organoids (64). Although organoids provide many advantages, challenges of this model include that they do not fully recapitulate in vivo tissues, still lack certain cell types such as microglia (65,66), and take extensive time to mature.
Finally, although this review focused on how CRISPR technologies are applied to characterize DNA variants and elements associated with neuropsychiatric disorders, these methods are readily applicable to other complex diseases, such as cancer. For example, CRISPRi screens identified iso-form specific loss of function in gastric cancer (67), highlighted prostate cancer risk loci involved in 3D genome architecture (68), as well as mapped regulatory networks in estrogen receptor-positive breast cancer (53). In addition, large-scale CRISPRa and CRISPRi screens are used to identify essential genes of cytokine production with stimulation in human primary T cells (69).
Conclusions/Future Directions
Regulatory elements function in a cell and content-dependent manner. We are still in the infancy of a comprehensive understanding of how chromatin architecture contributes to genome function in the human brain. Recent publications leveraging single-cell sequencing have highlighted the heterogeneity of the developing human cortex (5,70). Each cell type can also exhibit different cell states depending on environment and signaling cues, underscoring the necessity of annotating the epigenomic regulation in a cell-type-specific manner. Future studies that generate reference and disease-specific maps of genome architecture in brain cell types across different time points and cell states, can aid in prioritizing genes and genetic variants that confer an individual’s risk for disease. Once putative risk genes and variants are identified via 3D epigenome annotation, CRISPRi and CRSIPRa screens can be leveraged for further functional characterization. Although results from existing methods of characterizing cCREs are helpful for learning the rules of gene regulation, there is an astronomical gap between the large number of cCREs and disease-associated variants and how exactly they contribute to human disease and traits. The majority of functional characterization studies of cis-regulatory sequences have been done using transformed cell lines, so their biological relevance to human health and disease is debatable; most genetic perturbations of cCREs depend on transcriptional output phenotypes and fail to address whether such gene expression changes are biologically significant for cellular function; the combinatorial effects of cCREs on target gene expression remain largely unexplored. Thus, future studies of testing cCREs and DNA variants using disease-relevant phenotypes in biologically relevant systems should offer novel insights into the genetic basis of neuropsychiatric diseases. In addition, current CRISPR screens failed to reveal the exact single nucleotide that is causal for disease, making it challenging to identify the causal variants among thousands of candidate variants associated with diseases and traits. Applying prime editing (71) to CRISPR screens will allow for rigorous evaluation of disease-associated variants at the nucleotide level, an essential step toward personalized prediction, diagnosis and treatment of otherwise intractable neuropsychiatric diseases.
Conflicts of Interest statement. None to declare.
Funding
US National Institute of Health (NIH) grants (R01 AG057497, R01 EY027789, UM1 HG009402 and U01DA052713 to Y.S).
Contributor Information
Ian R Jones, Institute for Human Genetics, University of California, San Francisco, CA, USA.
Xingjie Ren, Institute for Human Genetics, University of California, San Francisco, CA, USA.
Yin Shen, Institute for Human Genetics, University of California, San Francisco, CA, USA; Department of Neurology, University of California, San Francisco, CA, USA.
References
- 1. Roadmap Epigenomics Consortium, Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., Kheradpour, P., Zhang, Z., Wang, J. et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The ENCODE Project Consortium, Abascal, F., Acosta, R., Addleman, N.J., Adrian, J., Afzal, V., Ai, R., Aken, B., Akiyama, J.A., Jammal, O.A. et al. (2020) Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature, 583, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mumbach, M.R., Satpathy, A.T., Boyle, E.A., Dai, C., Gowen, B.G., Cho, S.W., Nguyen, M.L., Rubin, A.J., Granja, J.M., Kazane, K.R. et al. (2017) Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet., 49, 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu, Z., Zhang, F., Hu, H., Bakshi, A., Robinson, M.R., Powell, J.E., Montgomery, G.W., Goddard, M.E., Wray, N.R., Visscher, P.M. and Yang, J. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet., 48, 481–487. [DOI] [PubMed] [Google Scholar]
- 5. Nowakowski, T.J., Bhaduri, A., Pollen, A.A., Alvarado, B., Mostajo-Radji, M.A., Di Lullo, E., Haeussler, M., Sandoval-Espinosa, C., Liu, S.J., Velmeshev, D. et al. (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science, 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nott, A., Holtman, I.R., Coufal, N.G., Schlachetzki, J.C.M., Yu, M., Hu, R., Han, C.Z., Pena, M., Xiao, J., Wu, Y. et al. (2019) Brain cell type-specific enhancer–promoter interactome maps and disease – risk association. Science, 366, 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Won, H., de la Torre-Ubieta, L., Stein, J.L., Parikshak, N.N., Huang, J., Opland, C.K., Gandal, M.J., Sutton, G.J., Hormozdiari, F., Lu, D. et al. (2016) Chromosome conformation elucidates regulatory relationships in developing human brain. Nature, 538, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song, M., Yang, X., Ren, X., Maliskova, L., Li, B., Jones, I.R., Wang, C., Jacob, F., Wu, K., Traglia, M. et al. (2019) Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat. Genet., 51, 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song, M., Pebworth, M.-P., Yang, X., Abnousi, A., Fan, C., Wen, J., Rosen, J.D., Choudhary, M.N.K., Cui, X., Jones, I.R. et al. (2020) Cell-type-specific 3D epigenomes in the developing human cortex. Nature, 587, 644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rao, S.S.P., Huntley, M.H., Durand, N.C., Stamenova, E.K., Bochkov, I.D., Robinson, J.T., Sanborn, A.L., Machol, I., Omer, A.D., Lander, E.S. and Aiden, E.L. (2014) A 3D map of the human genome at Kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsieh, T.-H.S., Weiner, A., Lajoie, B., Dekker, J., Friedman, N. and Rando, O.J. (2015) Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell, 162, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mumbach, M.R., Rubin, A.J., Flynn, R.A., Dai, C., Khavari, P.A., Greenleaf, W.J. and Chang, H.Y. (2016) HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods, 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang, R., Yu, M., Li, G., Chee, S., Liu, T., Schmitt, A.D. and Ren, B. (2016) Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res., 26, 1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beagrie, R.A., Scialdone, A., Schueler, M., Kraemer, D.C.A., Chotalia, M., Xie, S.Q., Barbieri, M., de Santiago, I., Lavitas, L.-M., Branco, M.R. et al. (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature, 543, 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu, L., Liu, X., Huang, W.-K., Giusti-Rodríguez, P., Cui, J., Zhang, S., Xu, W., Wen, Z., Ma, S., Rosen, J.D. et al. (2020) Robust hi-C maps of enhancer-promoter interactions reveal the function of non-coding genome in neural development and diseases. Mol. Cell, 79, 521–534.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu, B., Won, H., Mah, W., Park, R.B., Kassim, B., Spiess, K., Kozlenkov, A., Crowley, C.A., Pochareddy, S., PsychENCODE Consortium et al. (2021) Neuronal and glial 3D chromatin architecture informs the cellular etiology of brain disorders. Nat. Commun., 12, 3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang, C., Ward, M.E., Chen, R., Liu, K., Tracy, T.E., Chen, X., Xie, M., Sohn, P.D., Ludwig, C., Meyer-Franke, A. et al. (2017) Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Reports, 9, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The Pharmacogenomics of Bipolar Disorder Study, Mertens, J., Wang, Q.-W., Kim, Y., Yu, D.X., Pham, S., Yang, B., Zheng, Y., Diffenderfer, K.E., Zhang, J. et al. (2015) Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature, 527, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fernandopulle, M.S., Prestil, R., Grunseich, C., Wang, C., Gan, L. and Ward, M.E. (2018) Transcription factor-mediated differentiation of human iPSCs into neurons: rapid differentiation of iPSCs into neurons. Curr. Protoc. Cell Biol., 79, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajarajan, P., Borrman, T., Liao, W., Schrode, N., Flaherty, E., Casiño, C., Powell, S., Yashaswini, C., LaMarca, E.A., Kassim, B. et al. (2018) Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science, 362, eaat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D.A. and Horvath, P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science, 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- 22. Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A. and Charpentier, E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mali, P., Yang, L., Esvelt, K.M., Aach, J., Guell, M., DiCarlo, J.E., Norville, J.E. and Church, G.M. (2013) RNA-guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A. and Zhang, F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu, P.D., Lander, E.S. and Zhang, F. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larson, M.H., Gilbert, L.A., Wang, X., Lim, W.A., Weissman, J.S. and Qi, L.S. (2013) CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc., 8, 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qi, L.S., Larson, M.H., Gilbert, L.A., Doudna, J.A., Weissman, J.S., Arkin, A.P. and Lim, W.A. (2021) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 184, 844. [DOI] [PubMed] [Google Scholar]
- 28. Gilbert, L.A., Larson, M.H., Morsut, L., Liu, Z., Brar, G.A., Torres, S.E., Stern-Ginossar, N., Brandman, O., Whitehead, E.H., Doudna, J.A. et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedman, J.R., Fredericks, W.J., Jensen, D.E., Speicher, D.W., Huang, X.P., Neilson, E.G. and Rauscher, F.J. (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev., 10, 2067–2078. [DOI] [PubMed] [Google Scholar]
- 30. Chavez, A., Scheiman, J., Vora, S., Pruitt, B.W., Tuttle, M., P R Iyer, E., Lin, S., Kiani, S., Guzman, C.D., Wiegand, D.J. et al. (2015) Highly efficient Cas9-mediated transcriptional programming. Nat. Methods, 12, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konermann, S., Brigham, M.D., Trevino, A.E., Joung, J., Abudayyeh, O.O., Barcena, C., Hsu, P.D., Habib, N., Gootenberg, J.S., Nishimasu, H. et al. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature, 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanenbaum, M.E., Gilbert, L.A., Qi, L.S., Weissman, J.S. and Vale, R.D. (2014) A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell, 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nuñez, J.K., Chen, J., Pommier, G.C., Cogan, J.Z., Replogle, J.M., Adriaens, C., Ramadoss, G.N., Shi, Q., Hung, K.L., Samelson, A.J. et al. (2021) Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell, 184, 2503–2519.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amabile, A., Migliara, A., Capasso, P., Biffi, M., Cittaro, D., Naldini, L. and Lombardo, A. (2016) Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell, 167, 219–232.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hilton, I.B., D’Ippolito, A.M., Vockley, C.M., Thakore, P.I., Crawford, G.E., Reddy, T.E. and Gersbach, C.A. (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol., 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fulco, C.P., Munschauer, M., Anyoha, R., Munson, G., Grossman, S.R., Perez, E.M., Kane, M., Cleary, B., Lander, E.S. and Engreitz, J.M. (2016) Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science, 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thakore, P.I., D’Ippolito, A.M., Song, L., Safi, A., Shivakumar, N.K., Kabadi, A.M., Reddy, T.E., Crawford, G.E. and Gersbach, C.A. (2015) Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods, 12, 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian, R., Gachechiladze, M.A., Ludwig, C.H., Laurie, M.T., Hong, J.Y., Nathaniel, D., Prabhu, A.V., Fernandopulle, M.S., Patel, R., Abshari, M. et al. (2019) CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron, 104, 239–255.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian, R., Abarientos, A., Hong, J., Hashemi, S.H., Yan, R., Dräger, N., Leng, K., Nalls, M.A., Singleton, A.B., Xu, K. et al. (2021) Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat. Neurosci., 24, 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Diao, Y., Li, B., Meng, Z., Jung, I., Lee, A.Y., Dixon, J., Maliskova, L., Guan, K., Shen, Y. and Ren, B. (2016) A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res., 26, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diao, Y., Fang, R., Li, B., Meng, Z., Yu, J., Qiu, Y., Lin, K.C., Huang, H., Liu, T., Marina, R.J. et al. (2017) A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods, 14, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fulco, C.P., Nasser, J., Jones, T.R., Munson, G., Bergman, D.T., Subramanian, V., Grossman, S.R., Anyoha, R., Doughty, B.R., Patwardhan, T.A. et al. (2019) Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet., 51, 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reilly, S.K., Gosai, S.J., Gutierrez, A., Mackay-Smith, A., Ulirsch, J.C., Kanai, M., Mouri, K., Berenzy, D., Kales, S., Butler, G.M. et al. (2021) Direct characterization of cis-regulatory elements and functional dissection of complex genetic associations using HCR–FlowFISH. Nat. Genet., 53, 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simeonov, D.R., Gowen, B.G., Boontanrart, M., Roth, T.L., Gagnon, J.D., Mumbach, M.R., Satpathy, A.T., Lee, Y., Bray, N.L., Chan, A.Y. et al. (2017) Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature, 549, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie, S., Duan, J., Li, B., Zhou, P. and Hon, G.C. (2017) Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell, 66, 285–299.e5. [DOI] [PubMed] [Google Scholar]
- 46. Dixit, A., Parnas, O., Li, B., Chen, J., Fulco, C.P., Jerby-Arnon, L., Marjanovic, N.D., Dionne, D., Burks, T., Raychowdhury, R. et al. (2016) Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell, 167, 1853–1866.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adamson, B., Norman, T.M., Jost, M., Cho, M.Y., Nuñez, J.K., Chen, Y., Villalta, J.E., Gilbert, L.A., Horlbeck, M.A., Hein, M.Y. et al. (2016) A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell, 167, 1867–1882.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Datlinger, P., Rendeiro, A.F., Schmidl, C., Krausgruber, T., Traxler, P., Klughammer, J., Schuster, L.C., Kuchler, A., Alpar, D. and Bock, C. (2017) Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods, 14, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rubin, A.J., Parker, K.R., Satpathy, A.T., Qi, Y., Wu, B., Ong, A.J., Mumbach, M.R., Ji, A.L., Kim, D.S., Cho, S.W. et al. (2019) Coupled single-cell CRISPR screening and Epigenomic profiling reveals causal gene regulatory networks. Cell, 176, 361–376.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pierce, S.E., Granja, J.M. and Greenleaf, W.J. (2021) High-throughput single-cell chromatin accessibility CRISPR screens enable unbiased identification of regulatory networks in cancer. Nat. Commun., 12, 2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liscovitch-Brauer, N., Montalbano, A., Deng, J., Méndez-Mancilla, A., Wessels, H.-H., Moss, N.G., Kung, C.-Y., Sookdeo, A., Guo, X., Geller, E. et al. (2021) Profiling the genetic determinants of chromatin accessibility with scalable single-cell CRISPR screens. Nat. Biotechnol., 39, 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jin, X., Simmons, S.K., Guo, A., Shetty, A.S., Ko, M., Nguyen, L., Jokhi, V., Robinson, E., Oyler, P., Curry, N. et al. (2020) In vivo perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science, 370, eaaz6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lopes, R., Sprouffske, K., Sheng, C., Uijttewaal, E.C.H., Wesdorp, A.E., Dahinden, J., Wengert, S., Diaz-Miyar, J., Yildiz, U., Bleu, M. et al. (2021) Systematic dissection of transcriptional regulatory networks by genome-scale and single-cell CRISPR screens. Sci. Adv., 7, eabf5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marshall, J.L., Doughty, B.R., Subramanian, V., Guckelberger, P., Wang, Q., Chen, L.M., Rodriques, S.G., Zhang, K., Fulco, C.P., Nasser, J. et al. (2020) HyPR-seq: single-cell quantification of chosen RNAs via hybridization and sequencing of DNA probes. Proc. Natl. Acad. Sci. U. S. A., 117, 33404–33413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schraivogel, D., Gschwind, A.R., Milbank, J.H., Leonce, D.R., Jakob, P., Mathur, L., Korbel, J.O., Merten, C.A., Velten, L. and Steinmetz, L.M. (2020) Targeted perturb-seq enables genome-scale genetic screens in single cells. Nat. Methods, 17, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gasperini, M., Hill, A.J., McFaline-Figueroa, J.L., Martin, B., Kim, S., Zhang, M.D., Jackson, D., Leith, A., Schreiber, J., Noble, W.S. et al. (2019) A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell, 176, 377–390.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ren, X., Wang, M., Li, B., Jamieson, K., Zheng, L., Jones, I.R., Li, B., Takagi, M.A., Lee, J., Maliskova, L. et al. (2021) Parallel characterization of cis-regulatory elements for multiple genes using CRISPRpath. Sci. Adv., 7, eabi4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu, Y., Yu, C., Daley, T.P., Wang, F., Cao, W.S., Bhate, S., Lin, X., Still, C., Liu, H., Zhao, D. et al. (2018) CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell, 23, 758–771.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feldman, D., Singh, A., Schmid-Burgk, J.L., Carlson, R.J., Mezger, A., Garrity, A.J., Zhang, F. and Blainey, P.C. (2019) Optical pooled screens in human cells. Cell, 179, 787–799.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yan, X., Stuurman, N., Ribeiro, S.A., Tanenbaum, M.E., Horlbeck, M.A., Liem, C.R., Jost, M., Weissman, J.S. and Vale, R.D. (2021) High-content imaging-based pooled CRISPR screens in mammalian cells. J. Cell Biol., 220, e202008158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schraivogel, D., Kuhn, T.M., Rauscher, B., Rodríguez-Martínez, M., Paulsen, M., Owsley, K., Middlebrook, A., Tischer, C., Ramasz, B., Ordoñez-Rueda, D. et al. (2022) High-speed fluorescence image–enabled cell sorting. Science, 375, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kramer, N.J., Haney, M.S., Morgens, D.W., Jovičić, A., Couthouis, J., Li, A., Ousey, J., Ma, R., Bieri, G., Tsui, C.K. et al. (2018) CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet., 50, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rousseaux, M.W.C., Vázquez-Vélez, G.E., Al-Ramahi, I., Jeong, H.-H., Bajić, A., Revelli, J.-P., Ye, H., Phan, E.T., Deger, J.M., Perez, A.M. et al. (2018) A druggable genome screen identifies modifiers of α-synuclein levels via a tiered cross-species validation approach. J. Neurosci., 38, 9286–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han, K., Pierce, S.E., Li, A., Spees, K., Anderson, G.R., Seoane, J.A., Lo, Y.-H., Dubreuil, M., Olivas, M., Kamber, R.A. et al. (2020) CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature, 580, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bhaduri, A., Andrews, M.G., Mancia Leon, W., Jung, D., Shin, D., Allen, D., Jung, D., Schmunk, G., Haeussler, M., Salma, J. et al. (2020) Cell stress in cortical organoids impairs molecular subtype specification. Nature, 578, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pollen, A.A., Bhaduri, A., Andrews, M.G., Nowakowski, T.J., Meyerson, O.S., Mostajo-Radji, M.A., Di Lullo, E., Alvarado, B., Bedolli, M., Dougherty, M.L. et al. (2019) Establishing cerebral organoids as models of human-specific brain evolution. Cell, 176, 743–756.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Davies, R., Liu, L., Taotao, S., Tuano, N., Chaturvedi, R., Huang, K.K., Itman, C., Mandoli, A., Qamra, A., Hu, C. et al. (2021) CRISPRi enables isoform-specific loss-of-function screens and identification of gastric cancer-specific isoform dependencies. Genome Biol., 22, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ahmed, M., Soares, F., Xia, J.-H., Yang, Y., Li, J., Guo, H., Su, P., Tian, Y., Lee, H.J., Wang, M. et al. (2021) CRISPRi screens reveal a DNA methylation-mediated 3D genome dependent causal mechanism in prostate cancer. Nat. Commun., 12, 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schmidt, R., Steinhart, Z., Layeghi, M., Freimer, J.W., Bueno, R., Nguyen, V.Q., Blaeschke, F., Ye, C.J. and Marson, A. (2022) CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science, 375, eabj4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Allen, D.E., Donohue, K.C., Cadwell, C.R., Shin, D., Keefe, M.G., Sohal, V.S. and Nowakowski, T.J. (2022) Fate mapping of neural stem cell niches reveals distinct origins of human cortical astrocytes. Science, 376, 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Scholefield, J. and Harrison, P.T. (2021) Prime editing – an update on the field. Gene Ther., 28, 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]