Abstract
The 17-kDa antigen of Bartonella henselae has previously been shown to elicit a strong humoral immune response in patients with cat scratch disease (CSD) and to be useful in screening human serum samples for CSD. In this study, PCR amplification of genes homologous to the 17-kDa antigen gene of B. henselae was performed using genomic DNAs from several species of Bartonella, including the currently recognized human pathogens. Amplicons of similar size were demonstrated using the following chromosomal DNA templates: B. henselae (two strains), B. quintana (two strains), B. elizabethae, B. clarridgeiae, B. vinsonii subsp. vinsonii, and B. vinsonii subsp. berkhoffii. No evidence of a B. bacilliformis homolog of the 17-kDa antigen gene was obtained using multiple primer pairs. DNA sequencing revealed open reading frames capable of coding for proteins with sizes similar to that of the 17-kDa antigen of B. henselae in all of the amplicons; however, extensive sequence divergence across the genus was noted. Cloning of the amplified products into pUC19 resulted in recombinants that directed synthesis of homologs of the 17-kDa protein. Immunoblot analysis using human sera from CSD cases demonstrated very little cross-reactivity among different species for this protein. In contrast, immunoblots using rabbit serum raised to the recombinant B. henselae antigen showed extensive cross-reactivity with the proteins of other Bartonella species. The data suggest that the use of the 17-kDa antigen as a serologic reagent may allow the development of more specific diagnostic assays. Furthermore, the nucleotide sequences from the various versions of the 17-kDa antigen gene should be useful for rapid identification of Bartonella at the species level.
The genus Bartonella consists of several recognized species that were reclassified by merging the genera Rochalimaea and Grahamella with Bartonella (11, 14). All species are oxidase-negative, fastidious gram-negative bacilli (14). Presently there are four species that are established human pathogens: B. bacilliformis, B. henselae, B. quintana, and B. elizabethae (6). Recently, a newly described species, B. clarridgeiae, has been associated with cat scratch disease (CSD) in humans (15, 28, 30). Bartonella species that have not yet been linked to human disease include, B. vinsonii, isolated from a Canadian vole (39); B. vinsonii subsp. berkhoffii, isolated from dogs (13, 29); and several species isolated from rodents (11, 20, 21). However, a new subspecies, B. vinsonii subsp. arupensis, was recently isolated from the blood of a single patient (40). Specific and practical diagnostic tests have not yet been developed for most of these species, and hence the tools necessary to associate them with human disease do not exist.
The disease spectrum among the human pathogens ranges from severe life-threatening infections such as the hemolytic anemia caused by B. bacilliformis, the agent of Carrion's disease and Oroya fever (1), to the relatively benign but common CSD caused by B. henselae. It is estimated that annually in the United States there are 22,000 cases of CSD contracted from domestic cats (26). In addition, both B. henselae and B. quintana have been shown to cause more severe disease syndromes, including fever with bacteremia, endocarditis, bacillary angiomatosis, and peliosis hepatis, in both immunocompromised and immunocompetent patients (6, 37). In one case a novel bacterium, B. elizabethae, was isolated from a 31-year-old man with aortic tissue vegetation (17); however, additional reports linking this organism to human disease have not been published.
Diagnosis of Bartonella infection is most frequently accomplished by serology. Isolation is possible but requires extended incubation periods far greater than are needed for most bacteria. Extended incubation times and fastidious growth requirements result in low sensitivity associated with isolation and problems with contamination of primary plating media (6). PCR is an option in laboratories with the proper equipment and expertise but has not yet gained widespread use in clinical diagnostic laboratories (7). For these reasons as well as convenience, serology remains the most frequently utilized means of diagnosis. Serologic assays are simple and easy to perform, but currently used methods only confirm exposure and do not conclusively indicate an acute infection (6). The indirect fluorescent-antibody assay (IFA) is the most common and thoroughly evaluated serologic test (2, 9, 16, 33, 35, 41). However, cross-reactivity among Bartonella species (23, 27) and variable sensitivities observed for the IFA in different laboratories have led some investigators to question the usefulness of this test (2, 9, 33). To address these concerns, we have focused on identifying and characterizing protein antigens of the various Bartonella species that may be of value as diagnostic reagents. One such protein, the 17-kDa antigen, was identified from B. henselae (5). The reactivity of a recombinant fusion protein derived from the 17-kDa antigen of B. henselae in a Western blot format was shown to correlate well with IFA results and diagnosis of CSD (5). The focus of this study is to identify homologs of the 17-kDa protein in other species of Bartonella. Recombinant versions of these proteins should prove useful for serodiagnosis, and the corresponding genes may be of value as targets for species-specific amplification.
MATERIALS AND METHODS
Bacterial strains and preparation of genomic DNA.
The sources and designations of the various isolates, representing six species of Bartonella, used in this study are summarized in Table 1. Bacteria were cultivated on heart infusion agar supplemented with 5% defibrinated rabbit blood at 37°C in 5% CO2. Cultures were incubated for 3 to 5 days until growth was sufficient. B. bacilliformis was cultivated at 28°C without supplemental CO2 for 7 to 10 days. Colony morphology and staining of bacterial cells by the Gimenez procedure (18) were used to monitor cell growth and purity. DNA was extracted using a procedure previously described (3). Briefly, cell growth was harvested into sterile TE buffer (10 mM Tris [pH 8.0] and 1 mM EDTA). Sodium lauryl sarcosinate was added to a final concentration of 1.0%, and proteinase K was added to a final concentration of 100 μg/ml. After 2 h of incubation at 65°C, the bacterial lysate was repeatedly extracted with an equal volume of buffer-saturated phenol and chloroform. DNA was precipitated by the addition of 1/10 volume of 3 M sodium acetate and 2.5 volumes of cold ethanol. The yield and size of the genomic DNA were assessed by agarose gel electrophoresis.
TABLE 1.
Properties of the 17-kDa antigen homologs from different Bartonella spp.
| Strain | ATCC no. (reference) | Species | Molecular mass (kDa) | No. of amino acid residues |
|---|---|---|---|---|
| Houston-1 | 49882 (34) | B. henselae | 16.9 | 148 |
| San Antonio-1 | —a (31) | B. henselae | 16.9 | 148 |
| 94-F40 | 700095 (30) | B. clarridgeiae | 16.9 | 148 |
| Fuller | VR-358 | B. quintana | 16.9 | 148 |
| U.Mass. | — | B. quintana | 16.9 | 148 |
| F9251 | 49927 (17) | B. elizabethae | 20.3 | 177 |
| B. vinsonii subsp. berkhoffii | 51672 (29) | B. vinsonii | 17.8 | 155 |
| Vole agent (B. vinsonii subsp. vinsonii) | VR-152 (39) | B. vinsonii | 26.2 | 234 |
| KC583 | 35685 | B. bacilliformis | Not found | |
| KC584 | 35686 | B. bacilliformis | Not found |
—, no ATCC number.
PCR amplification of the 17-kDa antigen gene homologs.
Genomic DNAs from the species listed in Table 1 were used as a template for PCR. Multiple primer pair combinations were constructed from regions surrounding the B. henselae (Houston-1) 17-kDa antigen gene; however, four different primer pair combinations were shown to be optimal and were used to amplify each template as follows. B. clarridgeiae was amplified with primer pair 17KAF (5′ GGAATGAATGATGAGATCGC 3′) and 17KBR (5′ GTTGAGAAGACTATTCATCG-3′). B. quintana and B. henselae, were amplified with primer pair 240 (5′ GCTCTAGACAGGGACAAAGTTCCGTTGTTGC 3′) and 241 (5′-CGGGGTACCGCCATTGTCGTCACAATGACG 3′). B. elizabethae and B. vinsonii subsp. vinsonii were amplified with primer pair 17KAF and R2 (5′ TGAAAAGAGGTCCAAGACCT 3′). B. vinsonii subsp. berkhoffii was amplified with primer pair 17KBR (5′ CTGAGCGAGAATTTGAGCTG 3′) and 17KAR (5′ CCAGAAATGCTCTCAAACGG 3′). The positions of these primers are indicated in Fig. 1. An additional primer pair derived from highly conserved sequences internal to the 17-kDa antigen gene, consisting of IntF (5′ GAAAAAATATAGCTTAGTCAC 3′) and IntR (5′CTAAAGTCGGACATCAGATT 3′), was also used to confirm the presence of a 17-kDa antigen gene homolog in all of the Bartonella species that tested positive. Amplification was performed using the following conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 50°C for 2 min, and 70°C for 2 min. The last cycle was followed by incubation at 70°C for 7 min to ensure the adenylation of the 3′ end. PCR amplifications were performed in a DNA thermocycler (MJ Research, Watertown, Mass.) using EasyStart 100 prealiquoted tubes (Molecular BioProducts, Inc., San Diego, Calif.).
FIG. 1.
Line diagram showing relative positions of oligonucleotide primers used for PCR amplification of the 17-kDa antigen gene from Bartonella species. The positions of the oligonucleotides are with respect to the sequence of the 17-kDa antigen gene and flanking sequences of B. henselae Houston-1 (accession number U23447). Nucleotide positions: 17KBF, 2921 to 2940; 240, 2986 to 3008; 17KAF, 3035 to 3054; 17KAR, 3609 to 3629; 17KBR, 3772 to 3791; 241, 3806 to 3826; R2, 4012 to 4031; Intf, 3158 to 3178; and IntR, 3583 to 3601.
Cloning.
Amplicons were cloned directly from the PCR mixture or following gel extraction into pCR2.1-TOPO according to the directions of the manufacturer (Invitrogen, Carlsbad, Calif.). The ligation junction of pCR2.1 is located between two EcoRI cleavage sites. The resulting ligation mixture was transformed into One Shot cells (Invitrogen) and plated on Luria-Bertani (LB) agar containing ampicillin (100 μg/ml) and 80 μl of X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 20 mg/ml. White colonies were selected and cultured overnight in LB broth with ampicillin. Plasmid DNA was isolated by alkaline lysis and cleaved with EcoRI, and the insert size was confirmed by agarose gel electrophoresis.
DNA sequencing.
Plasmid DNA was isolated using the standard protocol from a QIAprep Spin Plasmid Kit (Qiagen Inc., Valencia, Calif.). Clones representing each strain were manually sequenced by a modification (42) of the dideoxy chain termination method of Sanger et al. (36). The resulting denatured double-stranded plasmid was sequenced using a Sequenase Quick-Denature Plasmid Sequencing Kit according to the directions of the manufacturer (Amersham Life Science, Cleveland, Ohio). 35S-dATP-labeled sequencing reaction mixtures were electrophoresed on a 6% acrylamide gel. The dried gel was exposed to X-ray film, and the sequence was recorded. Analysis of DNA sequences was performed using DNAsis version 2.5 for Windows (Hitachi, San Bruno, Calif.).
PCR amplification for ligation into pUC19.
Specific oligonucleotide pairs derived from the sequence obtained from each species were synthesized in order to amplify the entire 17-kDa gene with the putative ribosome binding site from each individual Bartonella strain. Each oligonucleotide primer derived from the 5′ end of the gene was designed with an XbaI site (-TCTAGA-) near the 5′ end, and the primer derived from the 3′ end of the gene contained a BamHI site (-GGATCC-) near the 5′ end to allow directional cloning. Amplification was achieved through initial denaturation at 94°C for 4 min; 3 cycles of 94°C for 1 min, 42°C for 2 min, and 67°C for 2 min; and 30 cycles of 94°C for 1 min, 50°C for 2 min, and 70°C for 1 min 30 s. Each amplicon was digested with XbaI and BamHI and ligated into pUC 19 cleaved with the same two enzymes so that the 17-kDa antigen gene homologs were immediately downstream of the inducible lacZ α-peptide promoter. The ligation mixtures were transformed into E. coli JM109 as previously described (19), and clones to be used for expression were identified by restriction endonuclease analysis and agarose gel electrophoresis. All recombinants were sequenced again using fluorescent-dye-labeled primers with a Thermosequenase cycle sequencing kit (Amersham) and an automated DNA sequencing and genetic analysis system (Li-Cor Inc., Lincoln, Nebr.).
SDS-PAGE and immunoblotting.
Clones were grown to early log phase at 37°C in 5 ml of LB broth containing ampicillin (100 μg/ml) and induced with 1 mM isopropyl thio-β-d-galactopyranoside (IPTG) for an additional 3.5 h. Bacterial cells were harvested by centrifugation and resuspended in one-third of the original culture volume of 1× sample buffer (63 mM Tris [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate [SDS], 0.0025% bromophenol blue) (Novex, San Diego, Calif.), and β-mercaptoethanol was added to a final concentration of 1%. The samples were boiled for 5 min and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a 4 to 20% gradient minigel (Novex). MultiMark multicolored standards were used to determine the approximate molecular weight (Novex). The proteins were transferred to nitrocellulose and blocked overnight in Tris-buffered saline (TBS) with 5% skim milk. The resulting membrane filter was incubated with serum diluted 1:200 (human sera) or 1:300 (rabbit serum) in TBS–5% skim milk for 2 h. The membrane filter was washed in TBS with 0.05% Tween 20 four times and exposed to either peroxidase-labeled goat anti-rabbit affinity-purified antibodies (Kirkegaard and Perry, Gaithersburg, Md.; diluted 1:7,000 in TBS with 5% skim milk) or peroxidase-labeled goat anti-human affinity purified antibodies (Kirkegaard and Perry; diluted 1:5,000 in TBS with 5% skim milk). The filter was then washed, and bound antibody was detected with TMB membrane substrate according to directions of the manufacturer (Kirkegaard and Perry).
The human sera used for immunoblots were collected from human immunodeficiency virus (HIV)-infected patients attending the HIV Clinic at the Bay Pines Veterans Affairs Medical Center, Bay Pines, Fla., after informed consent was obtained using a protocol approved by the Institutional Review Board. Serum samples were tested for antibodies to Bartonella by IFA as previously described (35). Only sera with IFA titers of 128 or greater were included, and these were tested individually (at a 1:200 dilution) or pooled (and then diluted 1:200). In another experiment, an individual serum sample obtained from a patient diagnosed with CSD with an IFA titer of 2,048 was provided by Patricia Emmanuel, Department of Pediatrics, University of South Florida College of Medicine. In other experiments, polyclonal hyperimmune rabbit serum raised to a fusion protein of the B. henselae (Houston-1) 17-kDa antigen was used at a 1:300 dilution. To produce this serum, a New Zealand White rabbit was immunized with 500 μg of the fusion protein, which has been previously described (5). The rabbit was boosted with an additional 500 μg after 4 weeks, and the animal was bled 1 week later and the serum was collected. The B. henselae (Houston-1) 17-kDa antigen expressed in Escherichia coli was used as a positive control, and E. coli JM109 harboring pUC19 with no insert was used as a negative control.
In vitro transcription-translation.
Plasmid template was used in an in vitro transcription-translation reaction designed for circular prokaryotic templates (Promega, Madison, Wis.). Plasmid-encoded proteins were labeled with [35S]methionine and resolved on a 4 to 20% gradient gel (Novex). The resulting gel was exposed to Enhance autoradiography enhancer (NEN Life Science Products, Boston, Mass.), dried, and exposed to X-ray film.
Nucleotide sequence accession numbers.
The nucleotide sequence for the 17-kDa antigen gene of B. henselae has been previously published (5) and deposited in the GenBank database under accession number U23447. The accession numbers for the genes from other strains of Bartonella are as follows: B. henselae (San Antonio-1), AF199503; B. quintana (Fuller), AF199006; B. quintana (U.Mass), AF199007; B. elizabethae, AF195504; B. clarridgeiae, AF195506; B. vinsonii subsp. vinsonii, AF195505; and B. vinsonii subsp. berkhoffii, AF200337.
RESULTS
PCR amplification.
PCR amplification of templates from various Bartonella species resulted in products of the approximate predicted size from B. henselae (San Antonio-1 strain), B. clarridgeiae, B. quintana (Fuller strain), B. quintana (U.Mass strain), B. vinsonii subsp. vinsonii, B. vinsonii subsp. berkhoffii, and B. elizabethae with one or more primer pairs. The primer pairs which provided the best amplification for subsequent cloning and sequencing are described in Materials and Methods. B. bacilliformis produced small amounts of a PCR product that was much larger than predicted for a homolog of the 17-kDa antigen gene using primer pairs 17KAF-17KAR, 17KAF-17KBR, and 240-241. Sequencing of clones harboring these amplicons resulted in the identification of an open reading frame capable of coding for a protein of 18 kDa. However, despite the similarity in predicted size with the 17-kDa antigen, no obvious amino acid sequence identity was observed. In an additional experiment utilizing primers internal to the most highly conserved coding regions of the 17-kDa antigen gene (IntF and IntR), product was amplified from all Bartonella species tested except B. bacilliformis (data not shown). Thus, despite the use of multiple primer pairs for amplification, no evidence of a 17-kDa antigen gene was found for either strain of B. bacilliformis.
Sequence analysis.
DNA sequencing revealed open reading frames capable of coding for proteins with deduced sizes similar to that previously described for the B. henselae 17-kDa antigen. Proteins of similar sizes were predicted from the sequence obtained from B. henselae (San Antonio-1 strain). B. quintana (Fuller strain and U.Mass strain), B. clarridgeiae, and B. vinsonii subsp. berkhoffii (Table 1). However, the B. vinsonii subsp. vinsonii and B. elizabethae versions of the gene were substantially larger (Table 1). Thus, there seems to be some discontinuity in predicted size among the various species, including both a human pathogen (B. elizabethae) and a strain currently thought to be nonpathogenic for humans (B. vinsonii subsp. vinsonii).
All homologs of the gene exhibited certain characteristics of prokaryotic gene structure. The antigen genes from B. henselae (San Antonio), B. clarridgeiae, B. quintana (Fuller), B. quintana (U.Mass.), B. vinsonii subsp. vinsonii, B. vinsonii subsp. berkhoffii, and B. elizabethae contained the identical polypurine-rich sequence (AGGAG) immediately upstream of the presumed ATG initiator methionine codon. These sequences presumably serve as ribosome binding sites for the antigen genes from the various species and strains. Similar sequences have been found immediately upstream of other B. henselae genes (4, 5, 12; A.W.O. Burgess, J.-Y. Paquet, and J.-J. Letesson, and B. Anderson, submitted for publication).
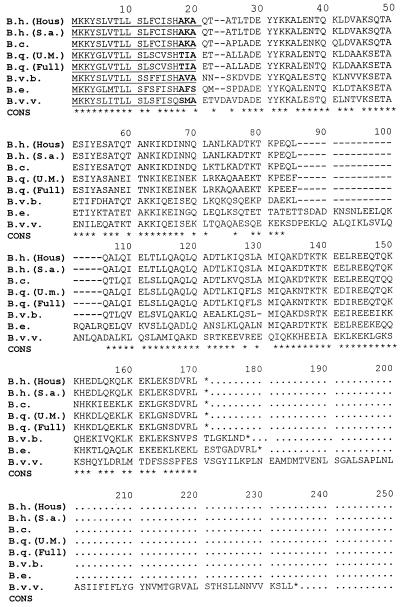
The putative initiator methionine is followed by a stretch of 18 to 25 residues that define a hydrophobic domain in all of the species that were analyzed using the algorithm of Hopp and Woods (24). In addition, two lysine codons follow the methionine start codon in all species. These properties are strongly predictive of bacterial signal peptides involved in targeting proteins for translocation across the cytoplasmic membrane (25). The deduced amino acid sequence alignment for B. henselae (San Antonio-1), B. vinsonii subsp. berkhoffii, and B. clarridgeiae indicates potential A-X-A peptidase cleavage sites (Fig. 2). Identical sequences have been shown on two other proteins of B. henselae that are processed and cleaved by signal peptidase before insertion into the outer membrane (12; Burgess and Anderson, submitted). Likewise, similar or identical peptide sequences have been shown to function as signal peptidase cleavage sites in E. coli (25). The other species revealed potential cleavage site variations that are similar to sites reported for E. coli, differing at one position from the consensus sequence of A-X-A (25): T-I-A, B. quintana (Fuller); T-I-A, B. quintana (U.Mass); A-F-S, B. elizabethae; and S-M-A, B. vinsonii subsp. vinsonii.
FIG. 2.
Multiple-sequence alignment of the deduced amino acid sequences of the 17-kDa antigen gene homologs from various Bartonella strains. The alignment was generated using the alignment routine of Higgins and Sharp (22) with a gap penalty of 5 and a window size of 5. The start codon (Met) is indicated at position 1, and stop codons are shown with asterisks. Putative signal sequences are underlined at residues 1 to 17, and potential signal peptidase cleavage sites are shown in boldface and underlined at positions 17 to 20. At positions where there is agreement between five or more of the different strains, a consensus sequence (CONS) is indicated by an asterisk. The sequences are identified on the left as follows. B. henselae Houston-1 strain, B.h. (Hous); B. henselae San Antonio-1 strain, B.h. (S.a.); B. clarridgeiae, B.c.; B. quintana U.Mass strain, B.q. (U.M.); B. quintana Fuller strain, B.q. (Full); B. vinsonii subsp. berkhoffii, B.v.b.; B. elizabethae, B.e.; and B. vinsonii subsp. vinsonii, B.v.v.
When the deduced amino acid sequence for the 17-kDa antigen gene is used to study phylogenetic relationships among the genus Bartonella, a number of interesting observations are noted (Fig. 3). First, no sequence variation was observed between the two different strains of B. henselae. Likewise, no amino acid variation was noted among the two strains of B. quintana sequenced, even though over 30 years separates their isolation. Second, B. henselae appears to be more closely related to B. clarridgeiae (approximately 15% amino acid sequence divergence) than to B. quintana (approximately 30% sequence divergence). Third, the remaining species, including B. elizabethae, B. vinsonii subsp. vinsonii, and B. vinsonii subsp. berkhoffii, appear to be only remotely related to the B. henselae, B. quintana, and B. clarridgeiae group. Finally, the two subspecies of B. vinsonii have homologs of the 17-kDa antigen that exhibit extensive sequence divergence (50%) (Fig. 3).
FIG. 3.
Dendrogram of the deduced amino acid sequences from the 17-kDa antigen genes from each of the Bartonella species or subspecies. The multiple-sequence alignment was generated by the CLUSTAL package, with the branching order and matching percentages indicated at each branch point (22).
Expression and antigenicity of the 17-kDa antigen homologs.
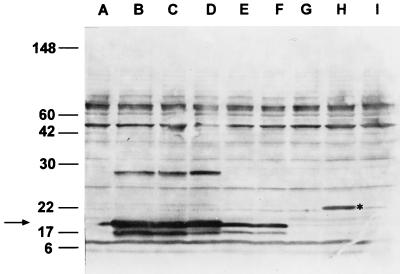
Immunoblot analysis with polyclonal anti-17-kDa rabbit serum demonstrated reactivity with clones of the antigen genes from B. henselae (Fig. 4, lanes B and C), B. clarridgeiae (lane D), and B. quintana (lanes E and F). These clones produce a doublet band with a size identical to that of the 17-kDa antigen from the Houston-1 strain of B. henselae. It is likely that the sources of the two bands seen in the doublet are proteins that are cleaved to various degrees by E. coli signal peptidase. An additional band is seen with B. henselae and B. clarridgeiae at approximately 28 kDa (Fig. 4, lanes B to D); it is possible that this protein is either incompletely solubilized (denatured) 17-kDa antigen or a dimer of the protein. It should be noted that only the B. henselae and B. clarridgeiae genes produced the 28-kDa band in E. coli, suggesting that some intrinsic variation of these versions of the protein results in the slower-migrating form of the antigen. Unlike those of the other Bartonella species, the B. elizabethae version of the antigen migrated at approximately 21 kDa (Fig. 4, lane H). The two subspecies of B. vinsonii failed to show any obvious immunoreactive bands at any size (Fig. 4, lanes G and I). When the DNAs from the recombinant E. coli strains harboring the genes derived from the two subspecies of B. vinsonii were used as templates in in vitro transcription-translation reactions, both plasmids directed synthesis of proteins of the predicted size (data not shown). Thus, these proteins are expressed in E. coli but are not reactive with the rabbit anti-B. henselae 17-kDa antigen serum. Cross-reactive rabbit antibodies are observed to react with bands at other sizes but are also reactive with the E. coli-plus-vector control (Fig. 4, lane A), indicating they are E. coli proteins. These data indicate that epitope(s) recognized by the immunized rabbit is conserved in some but not all species of Bartonella.
FIG. 4.
Immunoblot of E. coli strains containing plasmids encoding homologs of the 17-kDa antigen gene from different Bartonella species reacted with polyclonal rabbit serum raised to the 17-kDa antigen of B. henselae. Total proteins from E. coli strains were resolved by SDS-PAGE and transferred to nitrocellulose. The serum sample used was raised to recombinant B. henselae 17-kDa antigen as described in the text. The sources of the antigen genes in the recombinants are as follows: lane B, B. henselae (Houston-1); lane C, B. henselae (San Antonio-1); lane D, B. clarridgeiae; lane E, B. quintana (Fuller); lane F, B. quintana (U.Mass); lane G, B. vinsonii subsp. berkhoffii; lane H, B. elizabethae; and lane I, B. vinsonii subsp. vinsonii. Lane A, E. coli host strain without cloned Bartonella DNA. The positions of molecular mass standards are indicated at the left in kilodaltons, and the position of the 17-kDa antigenic proteins is indicated with an arrow. The position of the larger B. elizabethae homolog is marked with an asterisk.
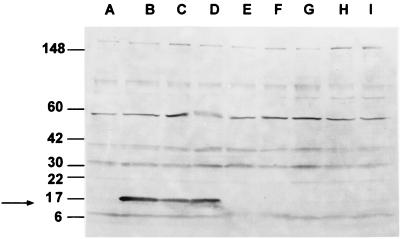
The immunoblot shown in Fig. 5 was reacted with a representative human serum specimen from a patient diagnosed with CSD who was shown to have a positive IFA titer to B. henselae. Reactivity with a protein of approximately 17 kDa was noted only with the two strains of B. henselae (lanes B and C) and B. clarridgeiae (lanes D). However, unlike the immunoblot with the rabbit serum, there was no cross-reactivity with either strain of B. quintana (lanes E and F), B. elizabethae (lane H), or the two strains of B. vinsonii (lanes G and I). A pool of three other human sera gave identical results, reacting only with the B. henselae and B. clarridgeiae clones (data not shown). Additional bands are observed at other sizes but are reactive with the E. coli-plus-vector control, indicating that they are E. coli proteins. These results suggest that infection in humans may elicit antibodies that are not broadly reactive across the genus.
FIG. 5.
Immunoblot of E. coli strains containing plasmids encoding homologs of the 17-kDa antigen gene from different Bartonella species reacted with human serum. Total proteins from E. coli strains were resolved by SDS-PAGE and transferred to nitrocellulose. The serum sample used was from a patient clinically diagnosed with CSD and having an IFA titer to B. henselae of 2,048. The sources of the antigen genes in the recombinants are as follows: lane B, B. henselae (Houston-1); lane C, B. henselae (San Antonio-1); lane D, B. clarridgeiae; lane E, B. quintana (Fuller); lane F, B. quintana (U.Mass); lane G, B. vinsonii subsp. berkhoffii; lane H, B. elizabethae; and lane I, B. vinsonii subsp. vinsonii. Lane A, E. coli host strain without cloned Bartonella DNA. The positions of molecular mass standards are indicated at the left in kilodaltons. The positions of the 17-kDa antigen homologs is indicated with an arrow.
DISCUSSION
Human infection by B. henselae and B. quintana results in a diverse array of clinical symptoms. Laboratory diagnosis of infections caused by these two agents requires isolation and identification, PCR amplification of bacterial DNA, or the presence of specific antibodies detected by serology. Only serologic testing is widely available, and it remains the most common means of diagnosing infections caused by Bartonella species. Several serologic assays have been described for detecting specific antibodies, including enzyme-linked immunosorbent assay (8, 23, 38) and IFA (35). Of these the IFA is the most widely used and thoroughly evaluated test (16). However, recent reports have indicated that the predictive value of the IFA may vary among different investigators (2, 9, 16, 33, 41). Variable results and sensitivities of this test have resulted in reports questioning the use of the IFA for diagnosis (9). The cause of such variability is not known, but it may be due to different methods of antigen preparation. Likewise, cross-reactivity between Bartonella and Coxiella, Chlamydia, and other bacteria has been well documented (23, 27). It is also known that the IFA is not specific for individual Bartonella species, with a patient's serum usually being reactive with antigens from one or more Bartonella species by IFA (6).
B. quintana appears to be a common pathogen both in the United States and abroad, and B. elizabethae and B. clarridgeiae have been associated with human disease (6). A number of other Bartonella species, subspecies, and strain variants have been associated with rodents, providing a common potential reservoir for transmission. In addition, B. vinsonii subsp. vinsonii and B. vinsonii subsp. berkhoffii have been identified as being capable of causing diseases of veterinary importance. Standardized antigens for serologic testing to detect specific antibodies to many of these Bartonella species have not been described. By characterizing individual protein antigens of various Bartonella species, we may be able to identify antigens or epitopes specific for each of the Bartonella species that are more specific than current serologic tests.
To investigate the humoral immune response to Bartonella infection, we have focused on individual protein antigens. One such antigen, the 17-kDa antigen of B. henselae, has been expressed as a fusion protein and shown to be reactive with sera from 92% of the CSD cases tested, suggesting that it may be of value as a diagnostic reagent (5). A homologous version of this antigen was found in all species of Bartonella tested except B. bacilliformis. The nucleotide sequence and the corresponding deduced amino acid sequence shared various levels of homology among the species tested (Fig. 2). However, the first 4 amino acids (MKKY) were identical for all species, and the first 20 residues were similar with regard to overall hydrophobicity and charge. The hydrophobicity and presence of lysine residues at the immediate amino terminus are characteristic of translocated proteins of bacteria. The lysines are thought to interact with the phosphate groups of the membrane phospholipids, and the hydrophobic core domain is thought to interact with the lipid moieties (25). The hydrophobic core is followed by a potential signal peptidase cleavage site. Alanine is frequently found at the −3 and −1 positions upstream of the cleavage site in E. coli (25) and was found in the B. henselae and B. elizabethae antigens in this study. Other small neutral side chains, such as glycine, serine, and threonine, are often seen in these positions (25), which was consistent with findings for B. quintana (Fuller and U.Mass), B. elizabethae, and B. vinsonii subsp. vinsonii. The signal sequence of A-X-A has been shown by our laboratory to be a signal peptidase cleavage site for two other outer membrane proteins of B. henselae (12; Burgess and Anderson, submitted). The presence of doublet bands on the immunoblots is also consistent with the role of signal peptidase in processing this antigenic protein. Thus, it is likely that the proteins from each of the Bartonella species included in this study are translocated across the cytoplasmic membrane.
The deduced amino acid sequences were used to construct a dendrogram of phylogenetic relationships. Surprisingly, the two species that were the most closely related were B. henselae and B. clarridgeiae (84.5%). The B. henselae and B. quintana versions of the 17-kDa antigen showed extensive sequence divergence (>30%), and no variation for different strains within B. henselae (Houston-1 and San Antonio-1 strains) and B. quintana (Fuller and U.Mass strains) was noted. In contrast, the two subspecies of B. vinsonii produced vastly different deduced amino acid sequences with extensive sequence divergence for the antigen (45.6%). It is possible that these two subspecies are more remotely related than previously thought (29), or, alternatively, that genetic exchange involving the 17-kDa antigen gene has hastened the evolutionary process.
When the cloned versions of the 17-kDa antigen from each of the species were reacted with human serum samples from patients with Bartonella infections, only B. henselae and the B. clarridgeiae were reactive. These data are also consistent with the phylogenetic relationship showing that the antigen is more closely related between B. henselae and B. clarridgeiae than any two other species. These results also suggest that the epitope(s) recognized by human sera from patients infected with B. henselae is not cross-reactive with B. quintana; however, since isolation from the patients whose sera were used in this study was not attempted, this observation cannot be confirmed. If this lack of cross-reactivity is confirmed upon further evaluation, recombinant 17-kDa antigens could be used to differentiate infections caused by B. henselae and B. clarridgeiae from those caused by B. quintana. Since B. henselae and B. clarridgeiae are associated with CSD and B. quintana is more frequently associated with severe systemic disease, identification of B. quintana as a causative agent may indicate the need for more aggressive treatment with antibiotics that are not always prescribed for CSD. The rabbit serum raised to purified recombinant B. henselae 17-kDa protein was broadly cross-reactive with all species except B. vinsonii, showing that at least one epitope recognized by the rabbit serum is well conserved.
The identification and sequencing of homologs of the 17-kDa antigen should facilitate the development of both serologic and genetic tools for the diagnosis of Bartonella infections. The use of recombinant protein derived from each of the pathogenic species of Bartonella as an antigen for enzyme-linked immunosorbent assay should permit rapid serologic testing that discriminates infections caused by B. henselae from those caused by B. quintana. If the need arises for serologic assays that are specific for other Bartonella species, then it should be possible to utilize the appropriate 17-kDa antigen homolog as an antigen. Additionally, the use of conserved antigen genes as targets for gene probes and PCR primers has proven useful for rapid detection and identification of bacteria directly in clinical specimens or clinical isolates. Currently, the species level identification of Bartonella isolates is often based on PCR amplification of conserved genes followed by the use of specific probes (7) or restriction fragment length polymorphism analysis (10, 32). The use of PCR primers derived from segments of the 17-kDa antigen gene which are unique to each species could easily shorten and simplify this process.
ACKNOWLEDGMENTS
We thank Dorsey Kordick of North Carolina State University for providing the strains of B. clarridgeiae and B. vinsonii and Gary Litman, All Children's Hospital, for automated sequencing services. We are also greatly appreciative of Pat Emmanuel, USF Department of Pediatrics, for providing the serum sample from a clinically diagnosed case of CSD.
This research was supported by Public Health Service grant R29-AI38178 from the National Institutes of Health.
REFERENCES
- 1.Alexander B. A review of Bartonellosis in Ecuador and Columbia. Am J Trop Med Hyg. 1995;52:354–359. doi: 10.4269/ajtmh.1995.52.354. [DOI] [PubMed] [Google Scholar]
- 2.Amerein M P, De Breil D, Jaulhac B, Meyer P, Monteil H, Piemont Y. Diagnostic value of the indirect immunofluorescence assay in cat scratch disease with Bartonella henselae and Afipia felis antigens. Clin Diagn Lab Immunol. 1996;3:200–204. doi: 10.1128/cdli.3.2.200-204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson B, Goldsmith C, Johnson A, Padmalayam I, Baumstark B. Bacteriophage-like particle of Rochalimaea henselae. Mol Microbiol. 1994;13:67–73. doi: 10.1111/j.1365-2958.1994.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson B, Jones D, Burgess A. Cloning, expression and sequence analysis of the Bartonella henselae gene encoding the HtrA stress-response protein. Gene. 1996;178:35–38. doi: 10.1016/0378-1119(96)00330-7. [DOI] [PubMed] [Google Scholar]
- 5.Anderson B, Lu E, Jones D, Regnery R. Characterization of a 17-kilodalton antigen of Bartonella henselae reactive with sera from patients with cat scratch disease. J Clin Microbiol. 1995;33:2358–2365. doi: 10.1128/jcm.33.9.2358-2365.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson B, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson B, Sims K, Regnery R, Robinson L, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barka N E, Hadfield T, Patnaik M, Schwartzman W A, Peter J B. EIA for detection of Rochalimaea henselae-reactive IgG, IgM, and IgA antibodies in patients with suspected cat-scratch disease. J Infect Dis. 1993;167:1503–1504. doi: 10.1093/infdis/167.6.1503. [DOI] [PubMed] [Google Scholar]
- 9.Bergmans A M, Peeters M F, Schellekens J F, Vos M C, Sabbe L J, Ossewaarde J M, Verbakel H, Hooft H J, Schouls L M. Pitfalls and fallacies of cat scratch disease serology: evaluation of Bartonella henselae-based indirect fluorescence assay and enzyme-linked immunoassay J. Clin Microbiol. 1997;35:1931–1937. doi: 10.1128/jcm.35.8.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birtles R J. Differentiation of Bartonella species using restriction endonuclease analysis of PCR-amplified 16S rRNA genes. FEMS Microbiol Lett. 1995;129:261–265. doi: 10.1111/j.1574-6968.1995.tb07590.x. [DOI] [PubMed] [Google Scholar]
- 11.Birtles R J, Harrison T G, Saunders N A, Molyneux D H. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov. Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 12.Bowers T J, Sweger D, Jue D, Anderson B. Isolation, sequencing, and expression of the gene encoding a major protein from the bacteriophage associated with Bartonella henselae. Gene. 1998;206:49–52. doi: 10.1016/s0378-1119(97)00580-5. [DOI] [PubMed] [Google Scholar]
- 13.Breitschwerdt E B, Kordick D L, Malarkey D E, Keene B, Hadfield T L, Wilson K. Endocarditis in a dog due to infection with a novel Bartonella subspecies. J Clin Microbiol. 1995;33:154–160. doi: 10.1128/jcm.33.1.154-160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner D J, O'Connor S P, Winkler H H, Steigerwalt A G. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov. Bartonella vinsonii comb. nov., Bartonella henselae comb. nov., and Bartonella elizabethae comb. nov., and to remove the family Bartonellaceae from the order Rickettsiales. Int J Syst Bacteriol. 1993;43:777–786. doi: 10.1099/00207713-43-4-777. [DOI] [PubMed] [Google Scholar]
- 15.Clarridge J E, Raich T J, Pirwani D, Simon B, Tsai L, Rodriguez-Barradas M C, Regenery R, Zollo A, Jones D C, Rambo C. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J Clin Microbiol. 1995;33:2107–2113. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalton M J, Robinson L E, Cooper J, Regnery R L, Olson J G, Childs J E. Use of Bartonella antigens for serologic diagnosis of cat-scratch disease at a national reference center. Arch Intern Med. 1995;155:1670–1676. [PubMed] [Google Scholar]
- 17.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gimenez D F. Staining rickettsiae in yolk-sac cultures. Stain Technol. 1964;39:135–140. doi: 10.3109/10520296409061219. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 1. Oxford, United Kingdom: IRL Press; 1985. pp. 109–136. [Google Scholar]
- 20.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piemont Y. Bartonella tricoborum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 21.Heller R, Kubina M, Mariet P, Riegel P, Delacour G, Dehio C, Kasten R, Boulouis H J, Monteil H, Chomel B, Piemont Y. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int J Syst Bacteriol. 1999;49:283–288. doi: 10.1099/00207713-49-1-283. [DOI] [PubMed] [Google Scholar]
- 22.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 23.Hollingdale M R, Herrmann J E, Vinson J W. Enzyme immunoassay of antibody to Rochalimaea quintana: diagnosis of trench fever and serologic cross-reactions among other rickettsiae. J Infect Dis. 1978;137:578–582. doi: 10.1093/infdis/137.5.578. [DOI] [PubMed] [Google Scholar]
- 24.Hopp T P, Woods K R. Prediction of antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izard J W, Kendall D A. Signal peptides: exquisitely designed transport promoters. Mol Microbiol. 1994;13:765–773. doi: 10.1111/j.1365-2958.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 26.Jackson L S, Perkins B A, Wenger J D. Cat scratch disease in the United States: an analysis of three national databases. Am J Public Health. 1993;83:1707–1711. doi: 10.2105/ajph.83.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knobloch J, Bialek R, Muller G, Asmus P. Common surface epitope of Bartonella bacilliformis and Chlamydia psittaci. Am J Trop Med Hyg. 1988;39:427–433. doi: 10.4269/ajtmh.1988.39.427. [DOI] [PubMed] [Google Scholar]
- 28.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwalt A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kordick D L, Swaminathan B, Greene C E, Wilson K H, Whitney A M, O'Connor S, Hollis D G, Matar G M, Steigerwalt A G, Malcolm G B, Hayes P S, Hadfield T L, Breitschwerdt E B, Brenner D J. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int J Syst Bacteriol. 1996;46:704–709. doi: 10.1099/00207713-46-3-704. [DOI] [PubMed] [Google Scholar]
- 30.Lawson P A, Collins M D. Description of Bartonella clarridgeiae sp. nov. isolated from the cat of a patient with Bartonella henselae septicemia. Med Microbiol Lett. 1996;5:64–73. [Google Scholar]
- 31.Lucey D, Dolan M J, Moss C W, Garcia M, Hollis D G, Wenger S, Morgan G, Almeida R, Leong D, Greisen K S, Welch D F, Slater L N. Relapsing illness due to Rochalimaea henselae in immunocompetent hosts: implication for therapy and new epidemiological associations. Clin Infect Dis. 1992;14:683–688. doi: 10.1093/clinids/14.3.683. [DOI] [PubMed] [Google Scholar]
- 32.Norman A F, Regnery R, Jameson P, Greene C, Krause D C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raoult D, Dupont H T, Enea-Mutillod M. Positive predictive value of Rochalimaea henselae antibodies in the diagnosis of cat-scratch disease. Clin Infect Dis. 1994;19:335. doi: 10.1093/clinids/19.2.355. [DOI] [PubMed] [Google Scholar]
- 34.Regnery R L, Anderson B E, Clarridge III J E, Rodriquez-Barradas M, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regnery R L, Olson J G, Perkins B A, Bibb W. Serologic response to “Rochalimaea henselae” antigen in suspected cat scratch disease. Lancet. 1992;339:1443–1445. doi: 10.1016/0140-6736(92)92032-b. [DOI] [PubMed] [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;76:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slater L N, Welch D F, Min K-W. Rochalimaea henselae causes bacillary angiomatosis and peliosis hepatis. Arch Intern Med. 1992;152:602–606. [PubMed] [Google Scholar]
- 38.Szelc-Kelly C M, Goral S, Perez-Perez G I, Perkins B, Regnery R L, Edwards K M. Serologic response to Bartonella and Afipia antigens in patients with cat scratch disease. Pediatrics. 1995;96:1137–1142. [PubMed] [Google Scholar]
- 39.Weiss E, Dasch G A. Differential characteristics of strains of Rochalimaea: Rochalimaea vinsonii sp. nov., the Canadian vole agent. Int J Syst Bacteriol. 1982;32:305–314. [Google Scholar]
- 40.Welch D F, Carroll K C, Hofmeister E K, Persing D H, Robison D A, Steigerwalt A G, Brenner D J. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zangwill K M, Hamilton D H, Perkins B A, Regnery R L, Plikaytis B D, Hadler J L, Cartter M L, Wenger J D. Cat scratch disease in Connecticut: epidemiology, risk factors and evaluation of a new diagnostic test. N Engl J Med. 1993;329:8–13. doi: 10.1056/NEJM199307013290102. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Scholl R, Browse J, Somerville C. Double stranded DNA as a choice for DNA sequencing. Nucleic Acids Res. 1998;16:1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]