Abstract
Modified thermal cycling conditions were explored in an effort to improve the reproducibility and resolving power of repetitive-element PCR (rep-PCR) fingerprinting. Assay performance was rigorously evaluated under standard and modified cycling conditions, using as a test set 12 strains putatively representing 12 serovars of Salmonella enterica. For all three fingerprint types (ERIC2, BOXA1R, and composite fingerprints), the use of extremely elevated annealing temperatures plus an initial “touchdown” cycling routine yielded significant improvements in day-to-day reproducibility and discriminating power despite the somewhat sparser appearance of the fingerprints. Modified cycling conditions markedly reduced the variability of fingerprints between cyclers, allowing fingerprints from different cyclers to be analyzed together without the degradation of assay performance that occurred with between-cycler analyses under standard cycling conditions. With modified cycling, composite fingerprints exhibited the lowest reproducibility but the highest net discriminating power of the three fingerprint types. rep-PCR fingerprints led to the discovery of a serotyping error involving one of the 12 test strains. These data demonstrate that modified cycling regimens that incorporate elevated annealing temperatures (with or without an initial touchdown routine) may markedly improve the performance of rep-PCR fingerprinting as a bacterial typing tool.
Bacterial strain typing at the subspecific level is an essential tool for contemporary public health and hospital infection control efforts (8, 18, 27, 33), as well as for basic research involving the molecular epidemiology and evolutionary biology of pathogenic bacteria (2, 6, 10, 21–24). Traditional subspecific typing methods include serotyping, phage typing, biotyping, plasmid profiling, multilocus enzyme electrophoresis, conventional restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis (PFGE). Their strengths notwithstanding, all of these methods have one or more significant drawbacks, including being slow or cumbersome; requiring highly specialized equipment, skills, and/or reagents; relying on variable or unstable traits; and yielding uninterpretable results for some strains (8, 18, 23, 33).
PCR-based fingerprinting is a simple, rapid, and broadly applicable typing method that is potentially available to any laboratory with PCR capability. Fingerprints are generated using either arbitrary primers (random amplified polymorphic DNA [35], arbitrarily primed PCR [34], or DNA amplification fingerprinting [5]) or repetitive-element-based primers (rep-PCR) (32). PCR fingerprinting has been reported to be useful in a variety of infection control and molecular epidemiological applications involving diverse bacterial types (11–13, 25, 27, 29, 31, 36). However, concerns have been raised regarding the irreproducibility of PCR-generated fingerprints (1, 20, 27; E. M. Jutras, P. Rochelle, R. de Leon, M. Stewart, and R. Wolfe, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. Q-109, p. 439, 1998). Claims of reproducibility from proponents of PCR fingerprinting generally have not been supported with specific data. On the contrary, reports of manipulations which ostensibly improve the reproducibility of PCR fingerprinting (7, 9, 13, 32; G. Lisby, D. L. Baggesen, and U. Skibsted, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. L-6, p. 354, 1998) suggest that irreproducibility is a greater problem with this method than is generally acknowledged.
Although our preliminary experience with rep-PCR typing of E. coli (14–16) and Salmonella enterica (J. R. Johnson, unpublished data) confirmed the method's speed and simplicity, we also found that with the published cycling conditions (32) the assay's reproducibility and strain discrimination on a day-to-day basis were inadequate, even with the use of kit-purified genomic DNA instead of boiled lysates and commercial PCR beads instead of hand-compounded master mixes (M. Saluta, W.-T. E. Ting, M. Koonge, and C. Tseng, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. H-124, p. 297, 1998).
The comparatively high GC content of the ERIC and BOXA1R primers used in rep-PCR (32) suggested to us that at the recommended annealing temperature (52°C) these primers might bind to and initiate DNA synthesis from partially mismatched recognition sites. Such mismatched annealing would be expected to be unstable and highly temperature dependent and hence sensitive to small temperature shifts as might occur from run to run or from day to day on a given thermal cycler or between cyclers. We hypothesized that the use of higher annealing temperatures would yield more specific (and hence more temperature-stable) priming, resulting in greater day-to-day and cycler-to-cycler reproducibility of fingerprints. (This is similar to the principle underlying “touchdown” [TD] cycling, in which to increase the specificity of primer binding in early cycles, annealing temperatures are initially set higher than the ultimate annealing temperature and then are decreased in a stepwise fashion with each cycle until the ultimate annealing temperature is reached [7, 9].) In the present study we sought to rigorously and quantitatively evaluate the impact of extremely elevated annealing temperatures on the reproducibility and discriminating power of rep-PCR fingerprints, using as the test substrate strains representing different serovars of S. enterica.
MATERIALS AND METHODS
Strains.
Twelve strains putatively representing 12 different serovars of S. enterica were selected from the Minneapolis VA Medical Center clinical microbiology laboratory's freezer bank of Salmonella isolates. Both common and uncommon serovars were included (see Fig. 1). The isolates had been identified as Salmonella according to standard methods (17), and serotypes had been determined by the Minnesota Department of Health reference laboratory. Strains were stored at −70°C until ready for use.
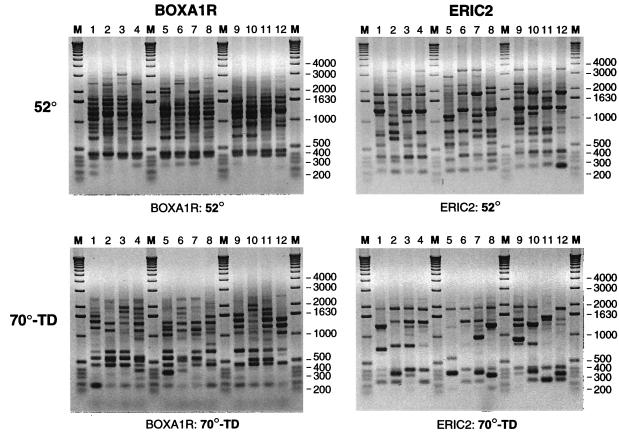
FIG. 1.
BOXA1R (left) and ERIC2 (right) rep-PCR fingerprints of 12 S. enterica isolates, as generated using either standard 52°C cycling (top panels) or 70-TD cycling (bottom panels). Lanes: 1, serovar Infantis; 2, serovar Newport; 3, serovar Enteritidis; 4, serovar Tennessee; 5, serovar St. Paul; 6, serovar “Mbandaka”; 7, serovar Havana; 8, serovar Heidelberg; 9, serovar Hadar; 10, serovar Typhimurium; 11, serovar Ohio; 12, serovar London; M, molecular weight marker.
Template DNA and primers.
Template DNA was extracted from three separate colonies of each of the 12 Salmonella strains using a commercial genomic DNA purification kit (Qiagen, Valencia, Calif.). Samples were stored at 4°C. Primers evaluated included ERIC1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′), ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′), and BOXA1R (5′-CTACGGCAAGGCGACGCTGACG-3′) (32). In preliminary experiments in which the three primers were tested singly and in all combinations ERIC2 alone and BOXA1R alone yielded the most diverse fingerprints and therefore, were selected for use in the remainder of the study.
PCR conditions.
Amplifications were done using Ready to Go PCR beads (Pharmacia), with 50 ng of template DNA and 20 pmol of primer in a 25-μl reaction volume. The two thermal cyclers used (cycler A [MTC-200 dual block] and cycler B [MTC-100 single block]; both from MJ Research, Watertown, Mass.) had been purchased 4 years apart and were kept in different laboratories on different floors of the building.
The study was designed to compare standard with modified cycling conditions. The standard cycling routine was as previously described, i.e., a preliminary denaturation step of 7 min at 95°C; 30 cycles of denaturation for 30 s at 90°C, annealing for 1 min at 52°C, and extension for 8 min at 65°C; and then a final extension step for 16 min at 65°C (32). The modified cycling routines incorporated elevated annealing temperatures (up to 72°C), with or without the addition of an initial 10-cycle, 5°C TD routine (7, 9). The preliminary denaturation step was for 2 min at 94°C. If a TD routine was used, it included denaturation for 30 s at 94°C, ramping at 1.5°C per s to the TD annealing temperature (which for the first cycle was set at 5°C above the ultimate annealing temperature and then in subsequent cycles was decreased by 0.5°C per cycle until the ultimate annealing temperature was reached), annealing for 1 min, ramping at 0.1°C per s to 72°C (extension temperature), and extension for 4.5 min at 72°C. This was followed by 25 cycles (35 cycles if no initial TD routine was used) of denaturation for 30 s at 94°C, ramping at 1.5°C per s to the ultimate annealing temperature, annealing for 1 min, ramping at 0.1°C per s to 72°C, and extension for 4.5 min at 72°C, with a final extension step of 1 min at 72°C. For most of the study, the modified cycling routine had an ultimate annealing temperature of 70°C following an initial 10-cycle TD routine from 75°C (70-TD cycling).
PCR products were electrophoresed in 1.0% agarose gels, stained with ethidium bromide, and visualized using a UV transilluminator and a digital image capture system (Gel Doc; Bio-Rad, Hercules, Calif.). Electrophoresis and gel analysis were done in a laboratory different from either of the PCR laboratories, and after tubes had been opened, PCR products were not carried back from this laboratory into either of the PCR laboratories.
The ERIC2 and BOXA1R primers were each used separately to generate fingerprints from DNA samples extracted in triplicate from each of the 12 Salmonella strains, with both standard and 70-TD cycling conditions used to amplify each DNA sample with each primer on two different thermal cyclers. In addition, the paired ERIC2 and BOXA1R fingerprints generated for each sample on a particular cycler with a particular cycling routine were digitally combined head-to-tail to create a “virtual” composite fingerprint, which then was analyzed in the same way as the individual ERIC2 and BOXA1R fingerprints.
Fingerprint analysis.
Images were manipulated and analyzed using the Multi-Analyst and Molecular Analyst software applications (Bio-Rad). Lanes were scanned densitometrically, and their densitometric tracks were normalized with respect to a molecular size standard (1-kb ladder; Gibco/BRL, Gaithersburg, Md.) which was included in four lanes on every gel. Densitometric tracks from each sample lane were then compared in a pairwise fashion with those of other lanes from the same gel or different gels. Pearson's correlation coefficient was used to calculate the degree of overall similarity between pairs of tracks. Since overall densitometric tracks were analyzed, neither the operator nor the computer defined the number or position of discrete bands within each track, and hence no operator judgment was involved in the analyses. Preliminary experiments indicated that reproducibility and discriminating power were generally better with this approach than with band-based analyses, which required subjective judgments by the operator (data not shown).
Analysis of dendrograms and performance indices.
For visual comparison of standard versus 70-TD cycling both on individual cyclers and across cyclers, similarity dendrograms were constructed using the unpaired group method of analysis (26). Differences between standard and 70-TD cycling dendrograms with respect to the proportion of strains having all replicate fingerprints clustered together, without interposition of fingerprints from other strains, were evaluated using McNemar's test. Comparisons between cycling regimens also were analyzed statistically by using the correlation coefficients to calculate indices for same-strain reproducibility, different-strain differentiation, and net discriminating power as obtained under different conditions. A strain's similarity index under a particular set of conditions was calculated as the mean of the similarity coefficients for all pairwise combinations between different replicates of that strain as tested under those conditions (high values = better same-strain reproducibility). A strain's differentiation index under a particular set of conditions was calculated by determining for each replicate of that strain the highest similarity coefficient between it and any replicate of one of the 11 other strains as tested under the same conditions and averaging these values for all replicates of the strain (high values = poor different-strain differentiation). A strain's net discriminating power under a particular set of conditions was the difference between its similarity index and its differentiation index. Means for these three indices were calculated across the 12 strains for each set of conditions, and a paired t test was used to compare these indices between conditions, with individual strains serving as the unit of analysis. The threshold for statistical significance was a P value of <0.05.
RESULTS
Appearance of fingerprints.
Robust fingerprints could still be generated with the ERIC2 and BOXA1R primers at annealing temperatures as high as 72°C (i.e., 20°C higher than the standard annealing temperature), even with the addition of an initial 10-cycle TD ramp that began 5°C higher than the final annealing temperature. Fingerprints generated on different days at annealing temperatures of 65°C (with TD), 70°C (without TD), and 72°C (with TD) were quite similar (data not shown), suggesting that day-to-day reproducibility might be high at any annealing temperature within this range. Thus, 70-TD cycling was selected for comparison with standard cycling.
With both the ERIC2 and BOXA1R primers, fingerprints generated using 70-TD cycling consistently differed substantially from those generated using standard cycling (Fig. 1). Although they contained fewer bands overall, 70-TD fingerprints also exhibited new strain-specific bands. Background shadowing, which under standard conditions was especially marked with the BOXA1R primer, was substantially reduced.
Dendrograms.
In dendrograms based on fingerprints from a single cycler, with either standard cycling or 70-TD cycling most strains' replicate fingerprints clustered together, separated from the fingerprints of other strains. However, the 70-TD dendrograms were more deeply forked between serovars and had shorter terminal branches connecting the replicate fingerprints of each isolate (data not shown). Furthermore, in one or more of the six standard cycling dendrograms (ERIC2, BOXA1R, and composite fingerprints from each cycler), six strains had at least one replicate fingerprint clustered with a different strain's fingerprints rather than with the rest of the index strain's fingerprints, whereas in the 70-TD cycling dendrograms these same strains' replicate fingerprints consistently clustered together, apart from the fingerprints of other strains. The reverse pattern (i.e., better same-strain resolution with standard cycling) occurred only once with a single strain (not shown).
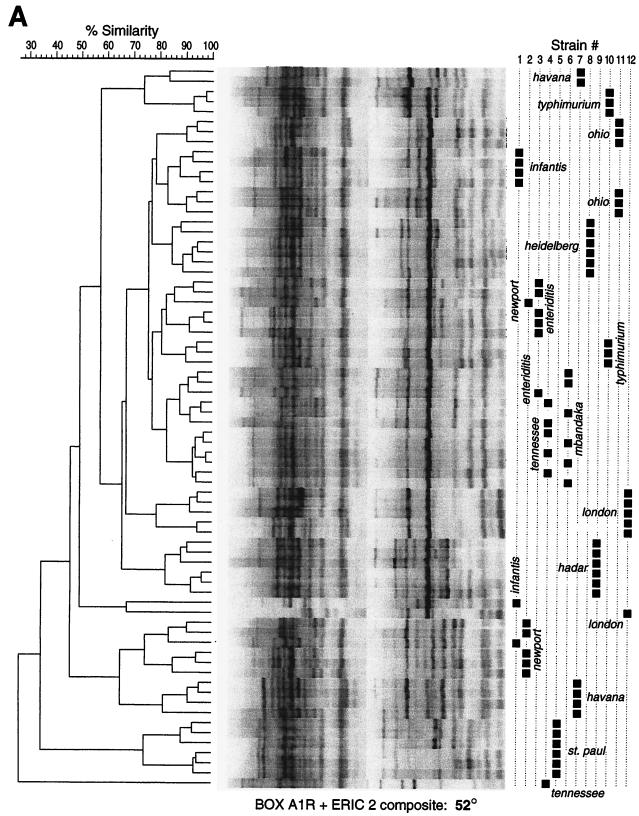
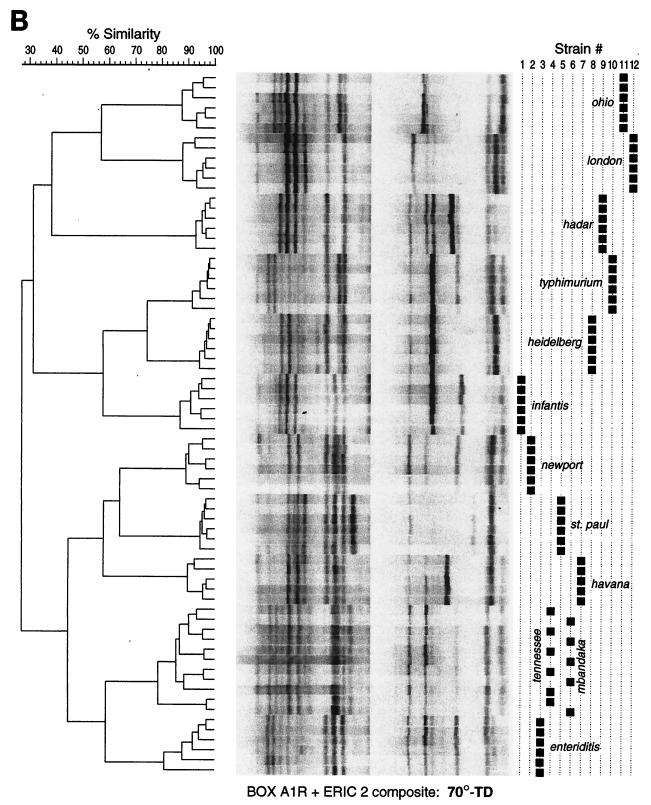
When fingerprints from both cyclers were combined into a single dendrogram for each primer type and cycling routine (combined-cycler dendrograms), the superior resolving power provided by 70-TD cycling was even more apparent (Fig. 2). With 70-TD cycling, replicate fingerprints from all serovars except Mbandaka and Tennessee clustered distinctly by serovar, well separated from other serovars. In contrast, with standard cycling, 8 of the 12 strains were incompletely resolved in one or more of the three combined-cycler dendrograms (P < 0.05 by McNemar's test) (Fig. 2). Further investigation of the Mbandaka and Tennessee isolates revealed that both strains had been isolated from the same patient within a 1-year interval. When newly serotyped by the Minnesota Department of Health, both isolates were unambiguously identified as S. enterica serovar Tennessee, confirming their identity and providing phenotypic validation of the genotyping results.
FIG. 2.
Combined-cycler dendrograms based on standard 52°C cycling (A) or 70-TD cycling (B), with BOXA1R-ERIC2 composite fingerprints of 12 S. enterica isolates (six replicate fingerprints per isolate). Strains: 1, serovar Infantis; 2, serovar Newport; 3, serovar Enteritidis; 4, serovar Tennessee; 5, serovar St. Paul; 6, serovar “Mbandaka”; 7, serovar Havana; 8, serovar Heidelberg; 9, serovar Hadar; 10, serovar Typhimurium; 11, serovar Ohio; 12, serovar London.
Performance indices.
An explanation for the superior resolving capability of 70-TD cycling as observed in dendrograms was suggested by statistical analysis of the underlying correlation coefficients. With all three types of fingerprints (ERIC2, BOXA1R, and composite), on average the same-strain reproducibility (Table 1), different-strain differentiation (Table 2), and net discriminating power (Table 3) were consistently better with 70-TD cycling than with standard cycling, whether data from the two cyclers were analyzed separately or in combination. The use of 70-TD cycling all but eliminated the marked decrease in reproducibility (Table 1) and net discriminating power (Table 3) that occurred with standard cycling when fingerprints were combined across cyclers.
TABLE 1.
Reproducibility of rep-PCR fingerprints from 12 isolates of S. enterica in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprints from cyclers A and B analyzed separately or combined | Cycling regimen | Similarity index (%)
|
||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| Separate | Standard | 92.5ab | 89.3ef | 89.7ij |
| 70-TD | 95.1ac | 93.8eg | 91.6ik | |
| Combined | Standard | 86.5bd | 81.4fh | 83.2jl |
| 70-TD | 93.7cd | 93.8gh | 90.8kl | |
P < 0.001.
P = 0.01.
P = 0.01.
P = 0.02.
P > 0.10.
P < 0.001.
P > 0.10.
P = 0.005.
P > 0.10.
P < 0.001.
P = 0.04.
P = 0.003.
TABLE 2.
Differentiation capability of rep-PCR fingerprints from 12 isolates of S. enterica in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprints from cyclers A and B analyzed separately or combined | Cycling regimen | Differentiation index (%)a
|
||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| Separate | Standard | 79.1bc | 87.2fg | 78.5jk |
| 70-TD | 74.0bd | 78.7fh | 64.8jl | |
| Combined | Standard | 79.9ce | 85.9gi | 78.0km |
| 70-TD | 74.3de | 78.2hi | 68.1lm | |
Smaller values indicate better differentiation.
P > 0.10.
P > 0.10.
P > 0.10.
P > 0.10.
P < 0.002.
P > 0.10.
P > 0.10.
P = 0.003.
P = 0.03.
P > 0.10.
P > 0.10.
P = 0.02.
TABLE 3.
Net discrimination power of rep-PCR fingerprints from 12 isolates of S. enterica in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprints from cyclers A and B analyzed separately or combined | Cycling regimen | Net discrimination power (%)a
|
||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| Separate | Standard | 14.3bc | 8.0fg | 11.8jk |
| 70-TD | 21.5bd | 16.0fh | 23.4jl | |
| Combined | Standard | 7.2ce | 8.3gi | 6.0km |
| 70-TD | 19.3de | 15.6hi | 22.7lm | |
Larger values for net discrimination power (similarity index − differentiation index) indicate better discrimination.
P > 0.10.
P = 0.01.
P = 0.05.
P = 0.03.
P > 0.10.
P > 0.10.
P > 0.10.
P = 0.04.
P = 0.01.
P = 0.003.
P > 0.10.
P < 0.001.
We next compared the performance characteristics of ERIC2, BOXA1R, and composite fingerprints (as generated under 70-TD cycling), to determine whether one of the individual primers gave better reproducibility, differentiation, or net discrimination than the other and whether composite fingerprints provided any advantage over individual primer fingerprints (Tables 4 to 6). Whether the two cyclers were analyzed separately or in combination, ERIC2 and BOXA1R fingerprints did not differ significantly with respect to any of the three performance measures, although ERIC2 fingerprints consistently did somewhat better. In contrast, composite fingerprints gave significantly poorer same-strain reproducibility than did fingerprints from either primer alone (Table 4), yet they more than compensated for this by giving significantly better different-strain differentiation (Table 5) and so on balance delivered better net discriminating power than did either of the individual primers, particularly BOXA1R (Table 6).
TABLE 4.
Comparison of reproducibility of ERIC2, BOXA1R, and composite fingerprints with 70-TD cycling
| Fingerprints from cycler A, B, or both A and B | Similarity index (%)
|
P valuea
|
|||
|---|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ERIC vs composite | BOX vs composite | |
| A alone | 95.6 | 94.9 | 93.5 | 0.004 | 0.005 |
| B alone | 94.6 | 92.7 | 89.7 | 0.01 | 0.001 |
| A and B combined | 93.7 | 93.8 | 90.8 | <0.001 | <0.001 |
For all comparisons of ERIC2 and BOXA1R, P > 0.10.
TABLE 6.
Comparison of net discrimination power of ERIC2, BOXA1R, and composite fingerprints with 70-TD cycling
| Fingerprints from cycler A, B, or both A and B | Net discrimination power (%)a
|
P value, BOX vs compositeb | ||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| A alone | 22.0 | 15.5 | 25.0 | 0.02 |
| B alone | 20.3 | 14.9 | 22.1 | 0.10 |
| A and B combined | 19.3 | 15.6 | 22.7 | 0.07 |
Larger values for net discriminating power (similarity index − differentiation index) indicate better discrimination.
For comparisons of ERIC2 versus either BOXA1R or composite, P > 0.10.
TABLE 5.
Comparison of differentiation capabilities of ERIC2, BOXA1R, and composite fingerprints with 70-TD cycling
| Fingerprints from cycler A, B, or both A and B | Differentiation index (%)a
|
P valueb
|
|||
|---|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ERIC vs composite | BOX vs composite | |
| A alone | 69.7 | 76.9 | 64.6 | 0.07 | 0.02 |
| B alone | 74.0 | 78.1 | 68.7 | 0.08 | 0.01 |
| A and B combined | 74.3 | 78.2 | 68.1 | 0.02 | 0.02 |
Smaller values indicate better differentiation.
For all comparisons of ERIC2 and BOXA1R, P > 0.10.
DISCUSSION
In the present study we rigorously evaluated the impact of radically modified cycling parameters on the performance of rep-PCR fingerprinting. We found that ultra-high annealing temperatures (in combination with TD cycling plus modified cycle times and ramp speeds) yielded significantly better same-strain reproducibility, different-strain differentiation, and net discriminating power, particularly across thermal cyclers, than did published rep-PCR conditions. In striking contrast to standard conditions, the modified cycling parameters allowed complete segregation of what proved to be 11 different serovars of S. enterica even when fingerprints from different days, different DNA preparations, and different thermal cyclers located in different laboratories were pooled for analysis using an operator-independent analysis system.
Although the mechanisms underlying the improved performance of rep-PCR that we observed under modified cycling conditions are unknown, increased specificity of primer binding is probably important (7, 9). We suspect that at annealing temperatures sufficiently high to restrict primer binding to precisely complementary loci in the target DNA, small variations in annealing temperature would be unlikely to substantially alter the distribution of primer sites occupied, which should result in considerable temperature stability of amplification fingerprints. In contrast, at lower (mismatch-tolerant) annealing temperatures, including probably the standard 52°C recommended for use with the ERIC2 and BOXA1R primers (32), the number and distribution of potential primer binding sites would be expected to vary continuously with the temperature, resulting in much greater temperature dependence of amplification fingerprints. Since it is likely that there are minute differences from day to day or during a single PCR run in the precise temperatures maintained by a cycler at each step in a routine, and all the more so between cyclers, some intrinsic variability of fingerprints would be expected, which should be greater at lower, mismatch-tolerant annealing temperatures than at higher, more stringent annealing temperatures.
TD cycling is designed to achieve specific primer binding in the crucial early cycles of PCR even in the absence of precise knowledge of the optimal annealing temperature range (7, 9). Whether the abbreviated TD routine that we incorporated into our modified cycling regimen contributed substantially to improved assay performance is unknown. Since in preliminary experiments fingerprints generated at an annealing temperature of 70°C without an initial TD routine matched closely those generated with annealing temperatures of either 65 or 72°C with an initial 5°C TD routine, it is likely that the most important component of our modified cycling regimen was the markedly elevated plateau annealing temperature per se. The importance of the modified ramp speeds in the elevated-temperature regimens (9) is unknown.
The high degree of same-strain reproducibility of fingerprints achievable with modified cycling conditions suggests that it may be possible for rep-PCR, when performed appropriately, to be used reliably not just for same-day screening of small groups of isolates but for construction of a database of fingerprints against which subsequently generated fingerprints could be compared. Furthermore, the stability of fingerprints across cyclers observed with modified cycling suggests that rep-PCR fingerprints are not necessarily cycler specific. This raises the possibility of cross comparisons of PCR fingerprints between laboratories, analogous to the approach used by the Centers for Disease Control and Prevention PulseNet system for comparing PFGE fingerprints between different public health laboratories around the United States (J. Besser, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., session 164/Y, p. 27, 1998; B. Swanimathan, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., session 164/Y, p. 27, 1998).
One unexpected discovery of the present study, the misidentification of serovar Tennessee as serovar Mbandaka, illustrates how PCR-based fingerprinting can in some instances supersede traditional Salmonella serotyping. The surprisingly indistinguishable PCR fingerprints of two isolates putatively representing these two dissimilar serovars prompted further epidemiological and serological investigations, which revealed that both isolates probably represented of a single strain of S. enterica serovar Tennessee. Whether the resolving power within S. enterica of optimized rep-PCR fingerprinting on balance is equivalent or superior to that of conventional serotyping remains to be determined through examination of representatives of other serovars and of additional representatives of the serovars studied here, ideally with validation from a “gold standard” comparison method such as PFGE for discrimination below the serovar level.
Although PCR technology is rapidly becoming a standard component of contemporary public health and research microbiology laboratories, the same cannot be said for the sophisticated gel analysis and dendrogram construction applications used in the present study, which are highly specialized and costly. However, it should be noted that this software system was in no way responsible for the improved performance of rep-PCR fingerprinting that we achieved, which instead was due strictly to modified cycling conditions. On the other hand, the software system was useful for our rigorous quantitative assessment of assay performance under standard and modified PCR conditions and would be valuable to users of PCR-based fingerprinting for computer-assisted database construction and searching. Nonetheless, any laboratory with PCR capability presumably could achieve improved reproducibility of rep-PCR fingerprints by adopting modified cycling conditions such as described here and could continue to analyze fingerprints visually or with an alternative computerized system.
To our knowledge the present study provides the most rigorous, best controlled, and most quantitative assessment to date of the reproducibility and discriminating power of a PCR-based fingerprinting system (3, 4, 13, 19, 20, 28, 34, 35). It also differs from much of the prior work in the field by relying on judgment-free methods for fingerprint definition and analysis which preclude the introduction of observer bias.
Since the present study included only Salmonella, the applicability of its findings to other genera is uncertain. Our preliminary experience suggests that although rep-PCR assay performance improves at higher temperature with Escherichia coli as well, with E. coli annealing temperatures cannot be elevated to the same extent as with Salmonella without fading of fingerprints (14a)). Thus, rep-PCR conditions may need to be optimized for each organism type.
In summary, we found that with S. enterica the use of extremely elevated annealing temperatures for rep-PCR fingerprinting yielded markedly improved same-strain reproducibility and different-strain differentiation, particularly across thermal cyclers, as compared with standard rep-PCR conditions. These findings invite a more extensive evaluation of rep-PCR with modified cycling conditions for typing of Salmonella and an exploration of its use with other genera.
ACKNOWLEDGMENTS
Miguel Azar, Carol Shanholtzer, and the technologists of the VA Medical Center Clinical Microbiology Laboratory provided the Salmonella isolates. The Minnesota Department of Health Microbiology Laboratory did the confirmatory serotyping, Dave Prentiss prepared the figures, and Diana Owensby assisted with manuscript preparation.
Grant support was from VA Merit Review and National Institutes of Health grant DK 47504 (to J.R.J.).
REFERENCES
- 1.Arbeit R D, Maslow J N, Mulligan M E. Polymerase chain reaction-mediated genotyping in microbial epidemiology. Clin Infect Dis. 1994;18:1018–1019. doi: 10.1093/clinids/18.6.1017. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Arbeit R D, Kim C, Beltran P, Crowe H, Steinback S, Campanelli C, Wilson R A, Selander R K, Goldstein R. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: pap-related sequences compared with rrn operons. Infect Immun. 1990;58:471–479. doi: 10.1128/iai.58.2.471-479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassam B J, Caetano-Anolles G, Gresshoff P M. DNA amplification fingerprinting of bacteria. Appl Microbiol Biotechnol. 1992;38:70–76. doi: 10.1007/BF00169422. [DOI] [PubMed] [Google Scholar]
- 4.Birch M, Denning D W, Law D. Rapid genotyping of Escherichia coli O157 isolates by random amplification of polymorphic DNA. Eur J Clin Microbiol Infect Dis. 1996;15:297–302. doi: 10.1007/BF01695661. [DOI] [PubMed] [Google Scholar]
- 5.Caetano-Anolles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–94. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 6.Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- 7.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenstein B I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis. 1990;161:595–602. doi: 10.1093/infdis/161.4.595. [DOI] [PubMed] [Google Scholar]
- 9.Gallego F J, Martinez I. Method to improve reliability of random-amplified polymorphic DNA markers. BioTechniques. 1997;23:663–664. doi: 10.2144/97234bm27. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Martinez J, Martinez-Murcia A J, Rodriguez-Valera F, Zorraquino A. Molecular evidence supporting the existence of two major groups in uropathogenic Escherichia coli. FEMS Immunol Med Microbiol. 1996;14:231–244. doi: 10.1111/j.1574-695X.1996.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 11.Georghiou P R, Hamill R J, Wright C E, Versalovic J, Koeuth T, Watson D A, Lupski R. Molecular epidemiology of infections due to Enterobacter aerogenes: identification of hospital outbreak-associated strains by molecular techniques. Clin Infect Dis. 1995;20:84–94. doi: 10.1093/clinids/20.1.84. [DOI] [PubMed] [Google Scholar]
- 12.Giesendorf B A J, van Belkum A, Koeken A, Stegeman H, Henkens M H C, van der Plas J. Development of species-specific DNA probes for Campylobacter jejuni, Campylobacter coli, and Campylobacter lari by polymerase chain reaction fingerprinting. J Clin Microbiol. 1993;31:1541–1546. doi: 10.1128/jcm.31.6.1541-1546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iriarte M P, Owen R J. Repetitive and arbitrary primer DNA sequences in PCR-medicated fingerprinting of outbreak and sporadic isolates of Campylobacter jejuni. FEMS Immunol Med Microbiol. 1996;15:17–22. doi: 10.1111/j.1574-695X.1996.tb00353.x. [DOI] [PubMed] [Google Scholar]
- 14.Johnson J R, Brown J J. Colonization with and acquisition of uropathogenic Escherichia coli strains as revealed by polymerase chain reaction-based detection. J Infect Dis. 1998;177:1120–1124. doi: 10.1086/517409. [DOI] [PubMed] [Google Scholar]
- 14a.Johnson J R, O'Bryan T T. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin Diagn Lab Immunol. 2000;7:265–273. doi: 10.1128/cdli.7.2.265-273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson J R, Russo T A, Scheutz F, Brown J J, Zhang L, Palin K, Rode C, Bloch C, Marrs C F, Foxman B. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapGJ96 (“Class I”) and PrsGJ96 (“Class III”) Gal(α1-4)Gal-binding adhesins. J Infect Dis. 1997;175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 16.Johnson J R, Stapleton A E, Russo T A, Scheutz F S, Brown J J, Maslow J N. Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4:H5 containing the class I and class III alleles of papG. Infect Immun. 1997;65:2153–2159. doi: 10.1128/iai.65.6.2153-2159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennette E H, Balows A, Hausler W J, Shadomy H J, editors. Manual of clinical microbiology. 4th ed. Washington, D.C.: American Society for Microbiology; 1985. [Google Scholar]
- 18.Lupski J R. Molecular epidemiology and its clinical application. JAMA. 1993;270:1363–1364. [PubMed] [Google Scholar]
- 19.Madico G, Akopyants N S, Berg D E. Arbitrarily primed PCR DNA fingerprinting of Escherichia coli O157:H7 strains by using templates from boiled cultures. J Clin Microbiol. 1995;33:1534–1536. doi: 10.1128/jcm.33.6.1534-1536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M C, Gonzalez-Hevia M A, Moro I, Mendoza M C. Genetic typing methods applied to the differentiation of clonal lines among Salmonella enterica serogroup G strains causing human salmonellosis. FEMS Immunol Med Microbiol. 1997;19:215–221. doi: 10.1111/j.1574-695X.1997.tb01090.x. [DOI] [PubMed] [Google Scholar]
- 21.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picard B, Sevali Garcia J, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, Ingraham K L, Magasanik B, Low K B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 24.Selander R K, Korhonen T K, Väisänen-Rhen V, Williams P H, Pattison P E, Caugant D A. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986;52:213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snelling A M, Gerner-Smidt P, Hawkey P M, Heritage J, Parnell P, Porter C, Bodenham A R, Inglis T. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (REP-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J Clin Microbiol. 1996;34:1193–1202. doi: 10.1128/jcm.34.5.1193-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokal R R, Sneath P H A. Principles of numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1963. [Google Scholar]
- 27.Swaminathan B, Barrett T J. Amplification methods for epidemiologic investigations of infectious diseases. J Microbiol Methods. 1995;23:129–139. [Google Scholar]
- 28.Telenius H, Carter N P, Bebb C E, Nordenskjold M, Ponder B A, Tunnacliffe A. Degenerative oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 29.van Belkum A, Bax R, Peerbooms P, Goessens W H F, van Leeuwen N, Quint W G V. Comparison of phage typing and DNA fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1993;31:798–803. doi: 10.1128/jcm.31.4.798-803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Belkum A, Meis J. Polymerase chain reaction-mediated genotyping in microbial epidemiology. Clin Infect Dis. 1994;18:1017–1018. doi: 10.1093/clinids/18.6.1017. [DOI] [PubMed] [Google Scholar]
- 31.van Belkum A, Struelens M, Quint W. Typing of Legionella pneumophila strains by polymerase chain reaction-mediated DNA fingerprinting. J Clin Microbiol. 1993;31:2198–2200. doi: 10.1128/jcm.31.8.2198-2200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versalovic J, Schneid M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 33.Wachsmuth K. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis. 1986;8:682–692. doi: 10.1093/clinids/8.5.682. [DOI] [PubMed] [Google Scholar]
- 34.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woods C R, Versalovic J, Koeuth T, Lupski J R. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J Clin Microbiol. 1992;30:2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]