Abstract
Repetitive-element PCR (rep-PCR) fingerprinting is a promising molecular typing tool for Escherichia coli, including for discriminating between pathogenic and nonpathogenic clones, but is plagued by irreproducibility. Using the ERIC2 and BOXA1R primers and 15 E. coli strains from the ECOR reference collection (three from each phylogenetic group, as defined by multilocus enzyme electrophoresis [MLEE], including virulence-associated group B2), we rigorously assessed the effect of extremely elevated annealing temperatures on rep-PCR's reproducibility, discriminating power, and ability to reveal MLEE-defined phylogenetic relationships. Modified cycling conditions significantly improved assay reproducibility and discriminating power, allowing fingerprints from different cyclers to be analyzed together with minimal loss of resolution. The correspondence of rep-PCR with MLEE with respect to tree structure and regression analysis of distances was substantially better with modified than with standard cycling conditions. Nonetheless, rep-PCR was only a fair surrogate for MLEE, and when fingerprints from different days were compared, it failed to distinguish between different clones within all-important phylogenetic group B2. These findings indicate that although the performance and phylogenetic fidelity of rep-PCR fingerprinting can be improved substantially with modified assay conditions, even when so improved rep-PCR cannot fully substitute for MLEE as a phylogenetic typing method for pathogenic E. coli.
Escherichia coli, the most frequent cause of urinary tract infections, neonatal sepsis and meningitis, and bacterial infectious diarrhea, is responsible for an enormous burden of morbidity, mortality, and health care costs (14, 15, 25, 33, 40, 41, 46, 48). Paradoxically, as the predominant facultative member of the normal human colonic flora, E. coli is present in most individuals as a harmless commensal (48). Pathogenic and commensal strains of E. coli to a large extent derive from separate evolutionary groups within the highly clonal E. coli population (8, 42, 45). Strains from lineages associated with pathogenecity typically possess specific virulence traits which confer the ability to cause disease in intact hosts (6, 24, 27, 37, 42, 46). These virulence traits are inherited vertically within the resulting virulent clones (27, 37, 46, 51) but also can be transmitted horizontally to other lineages (2, 27, 36, 37, 43), sometimes as part of blocks of virulence genes known as pathogenicity-associated islands (4, 5, 21, 22, 31, 50).
Investigation of E. coli virulence in relation to population structure requires a genotyping method that can reveal underlying genetic relationships between different E. coli strains. Traditional O, K, and H serotypes, plasmid profiles, and biotypes in general are unreliable indicators of clonal relationships (9, 13, 45, 54). In contrast, electrophoretic mobility patterns for multiple metabolic enzymes (multilocus enzyme electrophoresis [MLEE]), DNA sequence analysis of such housekeeping genes, and restriction fragment length polymorphisms in and around genomic ribosomal DNA loci (ribotyping) give largely concordant and reproducible assessments of the E. coli population structure (2, 11, 20, 32, 37, 45).
However, these established clonotyping methods are technically cumbersome or costly. Hence, there has been considerable recent interest in exploiting the simplicity and versatility of PCR technology to develop an alternative clonotyping method for E. coli. Amplification fingerprinting using arbitrary oligonucleotides as primers (3), which has been described by its developers as random amplified polymorphic DNA (RAPD) (58), arbitrarily primed PCR (56), and DNA amplification fingerprinting (7), yielded somewhat encouraging results in several studies that evaluated it as an evolutionary typing tool for diarrheagenic E. coli (55) or for E. coli in general (11, 17, 18). However, RAPD fingerprinting may have poor day-to-day reproducibility (1, 49) and a limited ability to reproduce evolutionary relationships as defined by MLEE (18; J. R. Johnson, unpublished data).
An alternative approach to PCR-based fingerprinting, repetitive-element PCR (rep-PCR), uses as primers oligonucleotides homologous to defined sequences which are present in multiple copies in the bacterial genome (35, 52, 53). rep-PCR has been predicted to yield more reproducible fingerprints than arbitrarily primed PCR because it relies on defined target sequences and thus can be used under stringent amplification conditions (1). Indeed, rep-PCR's same-day reproducibility and discriminating power have sufficed for small-scale epidemiological and phylogenetic studies involving wild-type E. coli strains (26, 29, 30). However, in our experience, day-to-day reproducibility has been as problematical with rep-PCR as with RAPD (Johnson, unpublished data). The marked improvement in reproducibility of rep-PCR fingerprints of Salmonella that resulted from the use of extremely elevated annealing temperatures (27a) prompted us to evaluate modified amplification conditions also with E. coli. In the present study we compared the performance characteristics of rep-PCR fingerprinting under standard versus modified amplification conditions and evaluated the ability of rep-PCR to assess genetic relationships between different strains of E. coli from the well-characterized ECOR reference collection, which represents the range of genetic diversity present in the species as a whole.
MATERIALS AND METHODS
Strains.
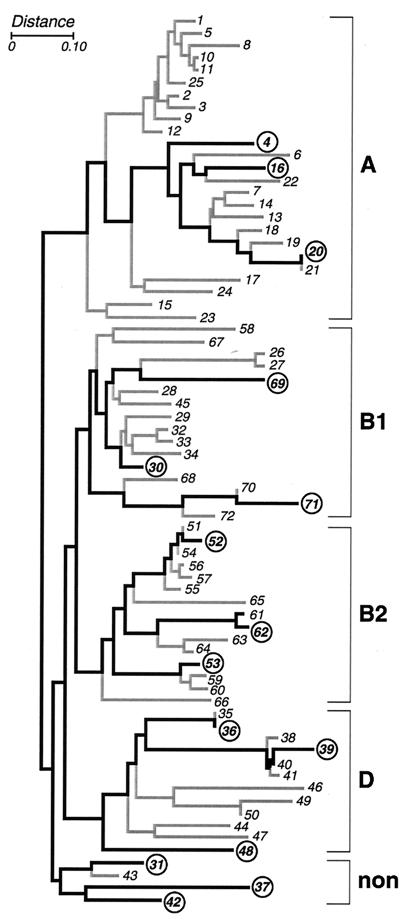
Three representative E. coli strains from each of the four major phylogenetic groups of the ECOR reference collection (groups A, B1, B2, and D) and from the remaining nonaligned strains, as defined by Herzer et al. using MLEE with 38 metabolic enzymes, were used as the test substrate (15 strains total) (Fig. 1) (23, 39). Strains were stored at −70°C until ready for use.
FIG. 1.
MLEE-based dendrogram for the ECOR reference collection of E. coli as derived by Herzer et al., using the NJ method to compare electrophoretic polymorphisms for 38 metabolic enzymes (23). Brackets at the right demarcate the five phylogenetic groups (A, B1, B, D, and nonaligned [non]). Heavy lines connect the 15 strains used in the present study (circles).
Template DNA and primers.
Template DNA was extracted from a pure culture of each of the 15 ECOR strains using a commercial kit (Pharmacia, Piscataway, N.J.). Primers evaluated included ERIC1R, ERIC2, BOXA1R, and MBO-REP (53). In preliminary experiments in which the primers were tested singly or in combination, ERIC2 alone and BOXA1R alone yielded the clearest and most diverse fingerprints (data not shown) and therefore were used for the remainder of the study.
PCR conditions.
Amplifications were done using Ready to Go PCR beads (Pharmacia), with 50 ng of template DNA and 20 pmol of primer in a 25-μl reaction volume. The two thermal cyclers used (cycler A [MTC-100 single block] and cycler B [MTC-200 dual block]; both from MJ Research, Watertown, Mass.) had been purchased 4 years apart and were kept in different laboratories on different floors of the building.
Standard and modified cycling conditions were compared. The standard cycling routine was as previously described (53), including the recommended 52°C annealing temperature. The modified cycling routines incorporated elevated annealing temperatures (up to 70°C), with or without the addition of an initial 10-cycle, 5°C “touchdown” (TD) routine (12, 16, 27a). The preliminary denaturation step was for 2 min at 94°C. The TD routine included denaturation for 30 s at 94°C, ramping at 1.5°C per s to the TD annealing temperature (which for the first cycle was set at 5°C above the plateau annealing temperature and then in subsequent cycles was decreased by 0.5°C per cycle until the plateau annealing temperature was reached), annealing for 1 min, ramping at 0.1°C per s to 72°C (extension temperature), and extension for 4.5 min at 72°C. The plateau portion consisted of 25 cycles of denaturation for 30 s at 94°C, ramping at 1.5°C per s to the plateau annealing temperature, annealing for 1 min, ramping at 0.1°C per s to 72°C, and extension for 4.5 min at 72°C, with a final extension step of 1 min at 72°C.
PCR products were electrophoresed in 1.0% agarose gels, stained with ethidium bromide, and visualized using a UV transilluminator and a digital image capture system (Gel Doc; Bio-Rad, Hercules, Calif.). In preliminary experiments ERIC2 and BOXA1R fingerprints were quite stable over annealing temperatures ranging from 60 to 66°C. Since at higher annealing temperatures the fingerprints abruptly shifted and then faded or disappeared (particularly with the ERIC2 primer), for the bulk of the study a plateau annealing temperature of 65°C was used, preceded by an initial TD routine beginning at 70°C (65-TD cycling).
DNA samples from each of the 15 ECOR strains were amplified with each primer separately (ERIC2 and BOXA1R) on each of the two thermal cyclers under both standard and 65-TD cycling conditions in three separate runs each, for a total of 360 amplifications. In addition, the paired ERIC2 and BOXA1R fingerprints generated for each sample on a particular cycler with a particular cycling routine were digitally combined head-to-tail to create a virtual composite fingerprint, which then was analyzed in the same manner as the individual ERIC2 and BOXA1R fingerprints.
Fingerprint analysis.
Images were analyzed using Multi-Analyst and Molecular Analyst (Bio-Rad). Densitometric tracks from each lane were normalized with respect to a molecular size standard (250-bp ladder; Gibco/BRL, Gaithersburg, Md.), which was included in four lanes on every gel, and then were compared in a pairwise fashion with tracks from other lanes from the same gel or different gels. Only the portion of each lane from just above the level of the 3,500-bp marker to just below the level of the 250-bp marker was analyzed, since almost all bands occurred within this size range (see, e.g., Fig. 2), and higher bands were noticeably irreproducible (data not shown). Pearson's correlation coefficient was used to calculate the degree of overall similarity between pairs of tracks. Neither the operator nor the computer defined the number or position of discrete bands within each track, and no operator judgement was involved in the analyses. Preliminary experiments indicated that reproducibility and discriminating power were generally better with this approach than with band-based analyses, which required subjective judgements by the operator (data not shown).
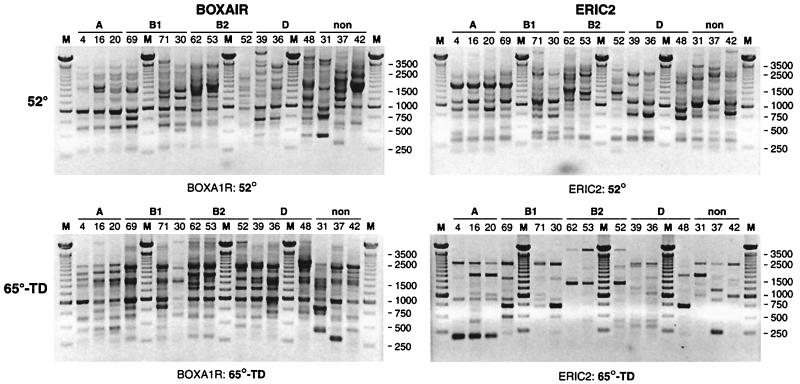
FIG. 2.
Representative rep-PCR gels for the 15 ECOR strains, showing fingerprints as generated using the BOXA1R primer (left) or the ERIC2 primer (right), with standard cycling (top) or 65-TD cycling (bottom). Lanes are labeled with strain numbers and bracketed according to ECOR group (A, B1, B2, D, and nonaligned [non]). Lanes M, 250-bp molecular size marker. Note that fingerprints for a given strain as generated with the same primer differ considerably between standard cycling and 65-TD cycling.
Performance indices.
Comparisons of assay performance between cycling regimens and fingerprint types were analyzed by using pairwise similarity coefficients to derive three different performance indices for each set of conditions. A strain's similarity index was the mean of the similarity coefficients for all pairwise comparisons between different replicates of that strain (high values = better same-strain reproducibility). A strain's differentiation index was the mean of the highest similarity coefficients between each replicate of the strain and any replicate of a strain from a different ECOR group (high values = poor different-strain differentiation). A strain's net discriminating power was the difference between the strain's similarity index and its differentiation index. Means for these three indices were calculated for the 15 strains for each set of conditions, and a paired t test was used to compare indices between conditions, with individual strains serving as the unit of analysis. Throughout, the threshold for statistical significance was a P value of <0.05.
Dendrogram analysis.
Assay performance also was evaluated by analysis of dendrograms, which were constructed from matrices of similarity coefficients by using the unpaired group method of analysis (UPGMA) (47). Dendrograms were assessed qualitatively for their structural similarity to the MLEE-based dendrogram for the ECOR collection as derived by Herzer et al. using the neighbor-joining (NJ) method (Fig. 1) (23). Dendrograms also were assessed for the degree to which individual strains (or phylogenetic groups) were fully resolved, i.e., had all replicate fingerprints from that strain (or phylogenetic group) in a single cluster that included only fingerprints from that strain (or members of the same phylogenetic group).
Regression comparison of rep-PCR versus MLEE.
To directly assess the ability of rep-PCR to reproduce phylogenetic relationships as defined by MLEE without interference from the use of different tree construction algorithms, pairwise similarity coefficients for the 15 test strains as derived from rep-PCR were directly compared by simple regression with MLEE-derived pairwise distances for the same 15 strains (55; T. L. Whittam, laboratory website [http://www.bio.psu.edu/People/Faculty/Whittam/Lab]). First, all 105 pairwise comparisons of the 15 strains from each typing method were analyzed. Next, repeated reanalysis was done after exclusion of each ECOR phylogenetic group in turn. Finally, analysis was repeated after exclusion of the two phylogenetic groups whose exclusion individually yielded the greatest improvement in correspondence of rep-PCR and MLEE.
RESULTS
Appearance of fingerprints.
65-TD fingerprints differed substantially from standard cycling fingerprints for all strains with both primers (Fig. 2). Compared with standard cycling fingerprints, 65-TD fingerprints were somewhat sparser but also exhibited unique bands in both the high- and low-molecular-weight ranges.
Performance indices.
65-TD cycling yielded dramatic improvements in reproducibility for each fingerprint type (Table 1). Its impact on differentiating power was variable, depending on the fingerprint type (Table 2). The net effect of these changes was a modest (BOXA1R) or major (ERIC2 and composite fingerprints) improvement in net discriminating power with 65-TD cycling (Table 3).
TABLE 1.
Reproducibility of rep-PCR fingerprints from 15 ECOR strains in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprints from cyclers A and B analyzed separately or combined | Cycling regimen | Similarity index (%)a
|
||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| Separate | Standardbd | 71.8 | 66.7 | 67.3 |
| 65-TDbe | 80.3 | 76.7 | 73.0 | |
| Combined | Standardcd | 63.5 | 54.8 | 57.8 |
| 65-TDce | 80.4 | 73.1 | 71.9 | |
Higher values indicate better reproducibility. Six replicate fingerprints per strain were included in each analysis.
With A separate from B, for standard versus 65-TD cycling, P = 0.02 (ERIC2 and BOXA1R) and P = 0.06 (composite).
With A and B combined, for standard versus 65-TD cycling, P < 0.001 for all comparisons.
With standard cycling, for A separate from B versus A and B combined, P < 0.001 for all comparisons.
With 65-TD cycling, for A separate from B versus A and B combined, P > 0.10 (ERIC2 and composite) and P = 0.01 (BOXA1R).
TABLE 2.
Differentiating power of rep-PCR fingerprints from 15 ECOR strains in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprints from cyclers A and B analyzed separately or combined | Cycling regimen | Differentiation index (%)a
|
||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| Separate | Standardbd | 64.4 | 61.0 | 54.4 |
| 65-TDbe | 53.6 | 71.8 | 53.4 | |
| Combined | Standardcd | 68.6 | 60.9 | 58.9 |
| 65-TDce | 55.4 | 73.3 | 52.2 | |
Smaller values indicate better differentiation. Six replicate fingerprints per strain were included in each analysis.
With A separate from B, for standard versus 65-TD cycling, P = 0.03 (ERIC2), P = 0.01 (BOXA1R), and P > 0.10 (composite).
With A and B combined, for standard versus 65-TD cycling, P = 0.01 (ERIC2), P = 0.003 (BOXA1R), and P = 0.06 (composite).
With standard cycling, for A separate from B versus A and B combined, P = 0.01 (ERIC2), P > 0.10 (BOXA1R), and P < 0.001 (composite).
With 65-TD cycling, for A separate from B versus A and B combined, P > 0.10 for all comparisons.
TABLE 3.
Net discrimination power of rep-PCR fingerprints from 15 ECOR strains in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprints from cyclers A and B analyzed separately or combined | Cycling regimen | Net discrimination power (%)a
|
||
|---|---|---|---|---|
| ERIC2 fingerprints | BOXA1R fingerprints | Composite fingerprints | ||
| Separate | Standardbd | 7.4 | 5.7 | 13.0 |
| 65-TDbe | 26.7 | 11.9 | 19.7 | |
| Combined | Standardcd | −5.2 | −6.1 | −1.1 |
| 65-TDce | 25.1 | −0.2 | 19.6 | |
Larger values for net discriminating power (similarity index − differentiation index) indicate better differentiation. Six replicate fingerprints per strain were included in each analysis.
With A separate from B, for standard versus 65-TD cycling, P < 0.001 (ERIC2), P > 0.10 (BOXA1R), and P = 0.03 (composite).
With A and B combined, for standard versus 65-TD cycling, P < 0.001 (ERIC2 and composite) and P = 0.08 (BOXA1R).
With standard cycling, for A separate from B versus A and B combined, P = 0.04 (ERIC2) and P < 0.001 (BOXA1R and composite).
With 65-TD cycling, for A separate from B versus A and B combined, P > 0.10 (ERIC2 and composite) and P = 0.002 (BOXA1R).
The positive effect of 65-TD cycling on assay performance was most striking in combined cycler analyses (Tables 1 to 3). 65-TD cycling essentially eliminated the marked decline in reproducibility and net discriminating power that was observed with standard cycling when fingerprints were combined across cyclers (Tables 1 to 3).
Under both sets of cycling conditions, reproducibility was best with ERIC2 fingerprints (Table 1). Differentiation and net discriminating power were best with composite fingerprints under standard cycling conditions and with both ERIC2 and composite fingerprints with 65-TD cycling (Tables 2 and 3).
Strain resolution in dendrograms.
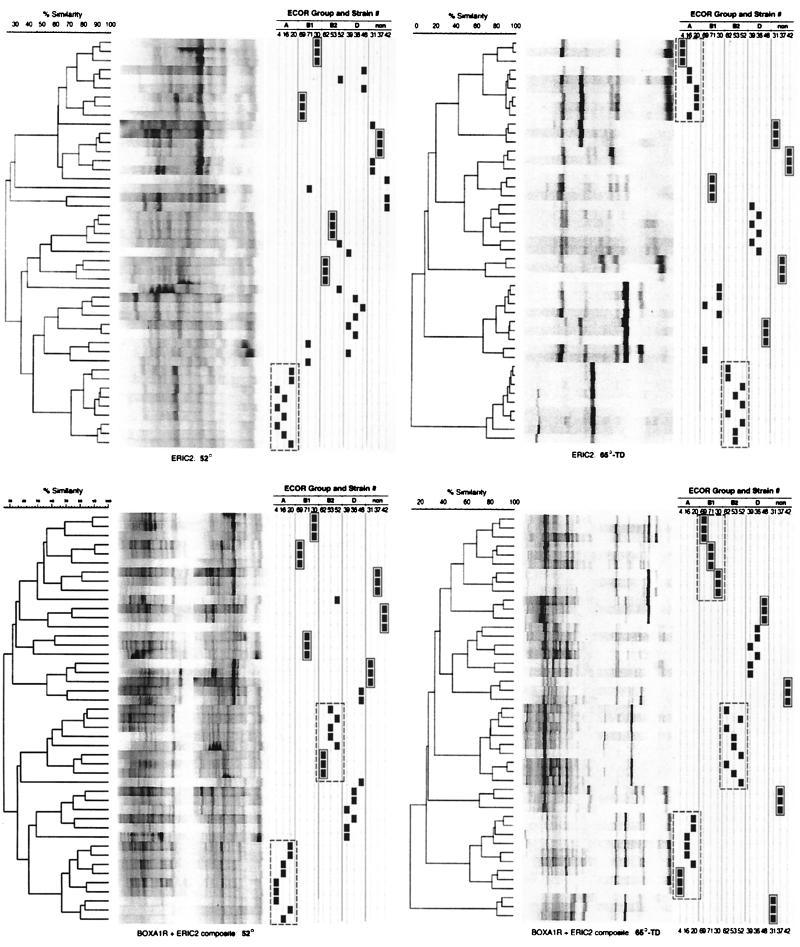
To further assess the ability of rep-PCR to resolve genetically distinct strains with standard versus 65-TD cycling, clustering of each of the 15 ECOR strains' fingerprints in UPGMA-based dendrograms was evaluated. Dendrograms comprising fingerprints from a single cycler run (irrespective of cycling conditions) consistently showed complete separation of the 15 strains (Fig. 3). However, this apparent level of discrimination was lost when replicate runs were incorporated into a single dendrogram (Fig. 4), as would be required if rep-PCR were to be used as a tool for preparation of longitudinal databases. Scrambling of strains in multiple-run dendrograms occurred even when replicate fingerprints from a single cycler were combined (Fig. 4) and was particularly problematic when fingerprints from different cyclers were combined (Table 4), as would occur with between-laboratory comparisons of fingerprints. 65-TD cycling had no consistent impact on strain resolution in single-cycler dendrograms (Table 4; Fig. 4, upper left panel versus upper right panel or lower left panel versus lower right panel), but yielded greatly improved strain resolution in combined-cycler dendrograms, particularly with ERIC2 and composite fingerprints (Table 4).
FIG. 3.
Single-run dendrograms for the 15 ECOR strains based on ERIC2 fingerprints (left) or BOXA1R-ERIC2 composite fingerprints (right), with 65-TD cycling. In both dendrograms, each strain is separated from all other strains. (BOXA1R fingerprints yielded similar results [data not shown].) The ERIC2 dendrogram includes both the largest (B2 strains versus others) and the smallest (B2 strains versus one another) between-strain distances observed in any of the dendrograms. non, nonaligned strains.
FIG. 4.
Triplicate-run dendrograms for the 15 ECOR strains based on ERIC2 fingerprints (upper panels) or BOXA1R-ERIC2 composite fingerprints (lower panels), as generated using standard cycling (left panels) or 65-TD cycling (right panels) on cycler B. The black squares at the right of each dendrogram mark the position in the dendrogram of replicate fingerprints of each strain, as identified at the top. Solid boxes enclose fully resolved strains. Dashed boxes enclose fully resolved phylogenetic groups. (Gel strips are reconstructions and hence underestimate the clarity of the actual gel images.) Summary data for the numbers of strains and phylogenetic groups resolved are shown in Tables 4 and 5, respectively. non, nonaligned strains.
TABLE 4.
Resolution of strains in rep-PCR dendrograms in relation to cycling regimen and use of single versus multiple cyclers
Phylogenetic fidelity.
The ability of rep-PCR fingerprints to accurately reproduce MLEE-defined phylogenetic relationships between the 15 ECOR strains was initially assessed by visual comparison of dendrograms based on rep-PCR (Fig. 4) versus MLEE (Fig. 1). In dendrograms based on triplicate rep-PCR runs from a single cycler, with standard cycling no more than a single phylogenetic group (always either group A or B2) was fully resolved per dendrogram, although in several instances one or two additional ECOR groups approached full resolution (Table 5; Fig. 4, left panels). In contrast, with 65-TD cycling as many as three of the five phylogenetic groups (again usually groups A and B2) were fully resolved per dendrogram (Table 5; Fig. 4, upper right and lower left panels). In combined-cycler dendrograms (not shown), standard cycling failed to fully resolve any phylogenetic groups, and 65-TD cycling fully resolved no more than groups A and/or B2 (Table 5). Complete (or near-complete) resolution of one or more phylogenetic groups occurred only with 65-TD cycling and only with ERIC2 and composite fingerprints (Table 5).
TABLE 5.
Resolution of phylogenetic groups in rep-PCR dendrograms in relation to cycling regimen and use of single versus multiple cyclers
| Fingerprint type | Phylogenetic group(s) fully (partiallya) resolvedb
|
|||||
|---|---|---|---|---|---|---|
| Single-cycler dendrograms
|
Combined-cycler dendrograms
|
|||||
| Standard cycling
|
65-TD cycling
|
|||||
| Cycler A | Cycler B | Cycler A | Cycler B | Standard cycling | 65-TD cycling | |
| BOXA1R | None | A (B1) | None | None (A, B2) | None | None |
| ERIC2 | B2 | B2c | B2 (A) | A, B2d | None (A, B2) | A, B2 |
| Composite | B2 | A (B2)e | A, B2 | A, B1, B2f | None (A, B2) | B2 (A) |
Partially resolved, one or two (single-cycler dendrograms) or one to three (combined-cycler dendrograms) scrambled fingerprints per phylogenetic group.
Nine fingerprints per phylogenetic group in single-cycler dendrograms and 18 fingerprints per group in combined-cycler dendrograms.
Shown in Fig. 4, upper left panel.
Shown in Fig. 4, upper right panel.
Shown in Fig. 4, lower left panel.
Shown in Fig. 4, lower right panel.
In both single-cycler and combined-cycler dendrograms, even within the fully resolved phylogenetic groups the constituent strains themselves were usually scrambled (Fig. 4). This was consistently the case for the group B2 strains (Fig. 4). Exceptions included strain 4 (group A) and the three group B1 strains, which were individually resolved within their respective phylogenetic clusters in one or more dendrograms each (Fig. 4, right panels).
Different clustering methods, such as UPGMA (as used in the present study) and NJ (as used by Herzer et al. [23] (Fig. 1) can generate different tree structures from the same data set (32, 44). Therefore, correspondence of rep-PCR with MLEE also was assessed independently by direct regression analysis of pairwise similarity coefficients and distances between the 15 ECOR strains as derived by rep-PCR and MLEE, respectively. In the total population, although the correspondence of rep-PCR with MLEE was weak regardless of cycling conditions, it was best (and reached statistical significance only) with 65-TD cycling (Table 6).
TABLE 6.
Correspondence of rep-PCR and MLEE distances
| ECOR groups analyzed | Linear regression, rep-PCRa vs MLEEb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ERIC2 fingerprints
|
Composite fingerprints
|
|||||||
| Standard cycling
|
65-TD cycling
|
Standard cycling
|
65-TD cycling
|
|||||
| r | P value | r | P value | r | P value | r | P value | |
| All | 0.12 | 0.24 | 0.23 | 0.03 | 0.12 | 0.24 | 0.27 | 0.006 |
| All but A | 0.06 | 0.61 | 0.14 | 0.25 | 0.14 | 0.25 | 0.10 | 0.42 |
| All but B1 | 0.26 | 0.03 | 0.42 | <0.001 | 0.28 | 0.02 | 0.43 | <0.001 |
| All but B2 | 0.26 | 0.03 | 0.19 | 0.14 | 0.19 | 0.12 | 0.17 | 0.17 |
| All but D | 0.14 | 0.25 | 0.31 | 0.01 | 0.07 | 0.56 | 0.33 | 0.008 |
| All but nonaligned | 0.23 | 0.07 | 0.41 | <0.001 | 0.26 | 0.04 | 0.53 | <0.001 |
| A, B2, D | 0.51 | 0.001 | 0.69 | <0.001 | 0.54 | <0.001 | 0.79 | <0.001 |
Pearson correlation coefficients from pairwise comparisons of rep-PCR fingerprints between strains.
m/n for pairwise comparisons of enzyme electrophoretic mobility polymorphisms between strains, where m is the number of mismatched loci and n is the total number of loci evaluated by MLEE using 38 metabolic enzymes (23). Data are from the Thomas Whittam laboratory website (http://www.bio.psu.edu /People/Faculty/Whittam/Lab).
Independent MLEE analyses of the ECOR collection have assigned individual isolates to different phylogenetic groups (10). To determine the impact of particular groups in our study, the data were reanalyzed after removal of individual phylogenetic groups from the data set. After removal of individual groups, 65-TD cycling usually yielded higher r values and lower P values than did standard cycling (Table 6), with the only exceptions being group B2 (both fingerprint types) and group A (composite fingerprints only). Irrespective of cycling conditions, removal of the B1 and/or the nonaligned strains from the data set improved the correspondence of rep-PCR with MLEE. The highest r values obtained, which reflected quite good correspondence of rep-PCR with MLEE, were with 65-TD cycling in the analyses limited to groups A, B2, and D (Table 6).
DISCUSSION
In the present study we rigorously evaluated the impact of radically modified thermal cycling conditions on the reproducibility, resolving power, and phylogenetic validity of rep-PCR fingerprinting for E. coli, using as a test substrate a panel of genetically well-characterized strains representing all major divisions of the ECOR reference collection (23, 39). Our findings suggest four main conclusions. First, extremely elevated annealing temperatures yield significantly improved overall assay performance compared with standard cycling conditions. Second, with modified assay conditions rep-PCR is able to resolve differences between (and in some instances within) several major phylogenetic groups within the E. coli population. Third, the use of modified cycling conditions generally improves the correspondence of rep-PCR with MLEE. Finally, despite these strengths even modified rep-PCR has significant limitations as a general phylogenetic typing tool for E. coli, since it fails to adequately resolve all major phylogenetic groups; corresponds well quantitatively with MLEE only for strains of ECOR groups A, B2, and D; and (except within an individual PCR run) does not discriminate reliably within ECOR group B2, the source of most extraintestinal pathogenic E. coli (6, 9, 42, 45, 46).
The improved reproducibility of rep-PCR fingerprints that we observed with 65-TD cycling may have been due to more specific primer binding at these temperatures, as compared with the mismatched priming that probably occurred at lower, less stringent annealing temperatures (1, 12, 16, 49). Minor shifts in annealing temperature or reaction conditions (such as are likely occur from run to run or within each PCR run despite standardization efforts) would be predicted to have less effect on the distribution of occupied priming sites, and hence on the resulting amplification fingerprint, at annealing temperatures high enough to require complete complementarity for primer binding than at lower temperatures which might permit occupation of a continuum of variably mismatched priming sites. We still did encounter some degree of irreproducibility of fingerprints even with 65-TD cycling, particularly with the BOXA1R primer. Whether this could be reduced further with other manipulations, such as the use of a “hot-start” routine, remains to be determined. Our disappointing experience with the use of a thermally activated polymerase (which in effect provides a hot start) (Johnson, unpublished data) suggests that most of the irreproducibility observed with PCR fingerprinting is due to factors other than nonspecific primer binding at low temperatures during the first PCR cycle and hence is unlikely to be eliminated by alternative hot-start methods.
To our knowledge, the present study provides the most rigorous evaluation to date of the reproducibility of rep-PCR fingerprinting for E. coli. Same-strain reproducibility, both in the same run and, particularly, in different runs, is of paramount importance to any typing technique, since it directly determines the confidence with which observed similarities or differences can be interpreted as real rather than artifactual. Previous performance evaluations of PCR fingerprinting have included some assessment of reproducibility. However, in most instances this has involved same-day amplification and side-by-side (same-gel) electrophoresis of products, using template DNAs extracted from multiple different subcultures of a single strain (10, 55), whereas the greatest threat to the reproducibility of PCR fingerprinting comes with separate amplifications and gel runs, even when the same template DNA is used (1, 49). Furthermore, in no previous study of which we are aware has assay reproducibility been assessed quantitatively or in a manner that excludes observer bias. In contrast, the present study provides objectively derived quantitative assessments, with statistical analysis of condition-specific differences in assay performance.
That reproducibility and discriminating power are interdependent was demonstrated in the present study by our finding that whereas all 15 strains appeared to be clearly differentiated in each day's PCR run under all conditions, when replicate runs from the same cycler were analyzed together certain groups of strains clearly could not be confidently resolved. Furthermore, when fingerprints from the two different cyclers were combined, even more noise was introduced, further obscuring what had appeared to be clear-cut strain-strain differences. It should be noted that previous studies of PCR fingerprinting for E. coli typically have analyzed discriminating power independently of reproducibility (11, 17, 18, 55).
With respect to reproduction of the MLEE-derived tree, we found that ERIC2 fingerprints by themselves allowed clear separation of groups A and B2 from all others, with large intercluster distances. BOXA1R fingerprints alone were not particularly useful, but when combined with ERIC2 fingerprints they allowed the resolution also of group B1 strains while retaining groups A and B2 in separate clusters. Thus, rep-PCR using one or two primers appeared to provide a facsimile of the MLEE-derived phylogenetic tree as good as or better than that provided by RAPD fingerprinting with the use of a single primer (17) or a combination of five primers (11).
The modest correspondence we found between rep-PCR and MLEE by regression analysis confirms the impression from comparisons of dendrograms that rep-PCR is a mediocre surrogate for MLEE in defining genetic distances between strains. Higher correlation coefficients have been reported by others for regression of RAPD and MLEE (18, 55), although since different strain sets were used in those studies, direct comparisons with the present study may not be valid. This is particularly likely to be true in view of our finding that the correspondence of rep-PCR with MLEE varied substantially depending on the mix of phylogenetic groups included in the analysis. Interestingly, the greatest discrepancies between rep-PCR and MLEE were with the B1 and nonaligned strains, several of which have been reassigned to different phylogenetic groups in successive MLEE-based resortings of the ECOR collection (23, 39, 45). Since rep-PCR appears to approximate MLEE well for group A strains and poorly for nonaligned strains, our study may have been biased against rep-PCR by our inclusion of disproportionately few group A strains and an excess of nonaligned strains, compared with these groups' relative prevalence in the ECOR collection (35 and 6%, respectively) (23).
It should be noted that even the best correlation of rep-PCR with MLEE obtained in the present study after elimination of “problem” phylogenetic groups, i.e., r = 0.79 (when only groups A, B2, and D were included, with 65-TD cycling and composite fingerprints), corresponds to r2 = 0.62, which can be interpreted as indicating that only 62% of the variance of the rep-PCR data is attributable to phylogenetic variation as resolved by MLEE. This value, although disappointingly low, is remarkably similar to values reported for the correlation of MLEE-derived genetic distances with estimates of polynucleotide sequence divergence as obtained by the ultimate comparison standard, hybridization of total cellular DNA (r ∼ 0.8; hence, r2 = 0.64) (45). This observation points out the limitations even of MLEE and suggests that at its best rep-PCR may be no worse a proxy for MLEE than MLEE is for analysis of total cellular DNA. However, to be useful as a general typing method, rep-PCR cannot be selectively applied only to “best-case” organisms. Furthermore, since the 15 strains selected for use in the present study represent a minimal subset of the larger E. coli population, it is likely that inclusion of additional isolates would further complicate, if not confound, attempts to correlate rep-PCR with MLEE for phylogenetic grouping.
If the goal of a phylotyping method for E. coli is primarily to sort strains dichotomously into two groups, one comprising the most virulent lineages (e.g., ECOR group B2 [6, 23, 42], carboxylesterase B2 strains [11, 19, 28], or RAPD or ribo-PCR group a [17]) and the other comprising all remaining strains, then a single RAPD primer may suffice (17), as did the ERIC2 primer alone in the present study. This primer has the added advantages of demonstrated reproducibility and the ability to resolve also ECOR group A. However, none of the published PCR-based fingerprinting methods has been shown to have sufficient discriminating power or evolutionary fidelity to reproducibly resolve separate pathogenic clones within all-important ECOR group B2. Thus, a more discriminating and reproducible yet still phylogenetically valid molecular analysis of E. coli population structure may require either alternative approaches to PCR fingerprinting (32, 38, 57) or the use of non-PCR-based methods (S. D. Reid, C. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. D/B-144, p. 237, 1999).
ACKNOWLEDGMENTS
Howard Ochman provided the ECOR strains, Thomas Whittam provided the MLEE data, Dave Prentiss prepared the figures, and Diana Owensby helped prepare the manuscript.
Grant support was from VA Merit Review and National Institutes of Health grant DK-47504.
REFERENCES
- 1.Arbeit R D, Maslow J N, Mulligan M E. Polymerase chain reaction-mediated genotyping in microbial epidemiology. Clin Infect Dis. 1994;18:1018–1019. doi: 10.1093/clinids/18.6.1017. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Arbeit R D, Kim C, Beltran P, Crowe H, Steinback S, Campanelli C, Wilson R A, Selander R K, Goldstein R. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: pap-related sequences compared with rrn operons. Infect Immun. 1990;58:471–479. doi: 10.1128/iai.58.2.471-479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg D E, Akopyants N S, Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol. 1994;5:13–24. [Google Scholar]
- 4.Bloch C A, Rode C K. Pathogenicity island evaluation in Escherichia coli K1 by crossing with laboratory strain K-12. Infect Immun. 1996;64:3218–3223. doi: 10.1128/iai.64.8.3218-3223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caetano-Anolles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–94. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 8.Caugant D A, Levin B R, Lidin-Janson G, Whittam T S, Svanborg Edén C, Selander R K. Genetic diversity and relationships among strains of Escherichia coli in the intestine and those causing urinary tract infections. Prog Allergy. 1983;33:203–227. doi: 10.1159/000318331. [DOI] [PubMed] [Google Scholar]
- 9.Caugant D A, Levin B R, Orskov I, Orskov F, Svanborg Eden C, Selander R K. Genetic diversity in relation to serotype in Escherichia coli. Infect Immun. 1985;49:407–413. doi: 10.1128/iai.49.2.407-413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cave H, Bingen E, Elion J, Denamur E. Differentiation of Escherichia coli strains using randomly amplified polymorphic DNA analysis. Res Microbiol. 1994;145:141–150. doi: 10.1016/0923-2508(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 11.Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- 12.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenstein B I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis. 1990;161:595–602. doi: 10.1093/infdis/161.4.595. [DOI] [PubMed] [Google Scholar]
- 14.Eisenstein B I, Jones G W. The spectrum of infections and pathogenic mechanisms of Escherichia coli. Adv Intern Med. 1988;33:231–252. [PubMed] [Google Scholar]
- 15.Foxman B. Recurring urinary tract infection: incidence and risk factors. Am J Public Health. 1990;80:331–333. doi: 10.2105/ajph.80.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego F J, Martinez I. Method to improve reliability of random-amplified polymorphic DNA markers. BioTechniques. 1997;23:663–664. doi: 10.2144/97234bm27. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Martinez J, Martinez-Murcia A J, Rodriguez-Valera F, Zorraquino A. Molecular evidence supporting the existence of two major groups in uropathogenic Escherichia coli. FEMS Immunol Med Microbiol. 1996;14:231–244. doi: 10.1111/j.1574-695X.1996.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 18.Gordon D M. The genetic structure of Escherichia coli populations in feral house mice. Microbiology. 1997;143:2039–2046. doi: 10.1099/00221287-143-6-2039. [DOI] [PubMed] [Google Scholar]
- 19.Goullet P, Picard B. Electrophoretic type B2 of carboxylesterase B for characterization of highly pathogenic Escherichia coli strains from extra-intestinal infections. J Gen Microbiol. 1990;33:1–6. doi: 10.1099/00222615-33-1-11. [DOI] [PubMed] [Google Scholar]
- 20.Grimont F, Grimont P A D. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur/Microbiol. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 21.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 22.Guyer D M, Kao J-S, Mobley H L T. Genomic analysis of a pathogenecity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNS-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson J R. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J R. Treatment and prevention of urinary tract infections. In: Mobley H L T, Warren J W, editors. Urinary tract infections: molecular pathogenesis and clinical management. Washington, D.C.: ASM Press; 1996. pp. 95–118. [Google Scholar]
- 26.Johnson J R, Brown J J. Colonization with and acquisition of uropathogenic Escherichia coli strains as revealed by polymerase chain reaction-based detection. J Infect Dis. 1998;177:1120–1124. doi: 10.1086/517409. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J R, Brown J J, Maslow J N. Clonal distribution of the three alleles of the Gal(α1-4)Gal-specific adhesin gene papG among Escherichia coli strains from patients with bacteremia. J Infect Dis. 1998;177:651–661. doi: 10.1086/514230. [DOI] [PubMed] [Google Scholar]
- 27a.Johnson J R, Clabots C. Improved repetitive-element PCR fingerprinting of Salmonella enterica with the use of extremely elevated annealing temperatures. Clin Diagn Lab Immunol. 2000;7:258–264. doi: 10.1128/cdli.7.2.258-264.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson J R, Goullet P H, Picard B, Moseley S L, Roberts P L, Stamm W E. Association of carboxylesterase B electrophoretic pattern with presence and expression of urovirulence factor determinants and antimicrobial resistance among strains of Escherichia coli causing urosepsis. Infect Immun. 1991;59:2311–2315. doi: 10.1128/iai.59.7.2311-2315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J R, Russo T A, Scheutz F, Brown J J, Zhang L, Palin K, Rode C, Bloch C, Marrs C F, Foxman B. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapGJ96 (“Class I”) and PrsGJ96 (“Class III”) Gal(α1-4)Gal-binding adhesins. J Infect Dis. 1997;175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 30.Johnson J R, Stapleton A E, Russo T A, Scheutz F S, Brown J J, Maslow J N. Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4:H5 containing the class I and class III alleles of papG. Infect Immun. 1997;65:2153–2159. doi: 10.1128/iai.65.6.2153-2159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao J, Stucker D M, W. W J, Mobley H L T. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- 33.Levine M M. Escherichia coli infections. N Engl J Med. 1985;313:445–447. doi: 10.1056/NEJM198508153130710. [DOI] [PubMed] [Google Scholar]
- 34.Lin J-J, Kuo J, Ma J. A PCR-based DNA fingerprinting technique: AFLP for molecular typing of bacteria. Nucleic Acids Res. 1996;24:3649–3650. doi: 10.1093/nar/24.18.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupski J L. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marklund B I, Tennent J M, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 37.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milkman R, Bridges M M. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990;126:505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orskov I, Orskov F. Escherichia coli in extra-intestinal infections. J Hyg. 1985;95:551–575. doi: 10.1017/s0022172400060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patton J P, Nash D B, Abrutyn E. Urinary tract infection: economic considerations. Med Clin N Am. 1991;75:495–513. doi: 10.1016/s0025-7125(16)30466-7. [DOI] [PubMed] [Google Scholar]
- 42.Picard B, Sevali Garcia J, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plos K, Hull S I, Hull R A, Levin B R, Orskov I, Orskov F, Svanborg-Edén C. Distribution of the P-associated-pilus (pap) region among Escherichia coli from natural sources: evidence for horizontal gene transfer. Infect Immun. 1989;57:1604–1611. doi: 10.1128/iai.57.5.1604-1611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 45.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, Ingraham K L, Magasanik B, Low K B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 46.Selander R K, Korhonen T K, Väisänen-Rhen V, Williams P H, Pattison P E, Caugant D A. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986;52:213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sokal R R, Sneath P H A. Principles of numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1963. [Google Scholar]
- 48.Sussman M. Escherichia coli and human disease. In: Sussman M, editor. Escherichia coli. Mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 3–48. [Google Scholar]
- 49.Swaminathan B, Barrett T J. Amplification methods for epidemiologic investigations of infectious diseases. J Microbiol Methods. 1995;23:129–139. [Google Scholar]
- 50.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaisanen-Rhen V, Elo J, Vaisanen E, Siitonen A, Orskov I, Orskov F, Svenson S B, Makela P H, Korhonen T. P-fimbriated clones among uropathogenic Escherichia coli strains. Infect Immun. 1984;43:149–155. doi: 10.1128/iai.43.1.149-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Versalovic J, Schneid M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 54.Wachsmuth K. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis. 1986;8:682–692. doi: 10.1093/clinids/8.5.682. [DOI] [PubMed] [Google Scholar]
- 55.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Widjojoatmodjo M N, Fluit A C, Verhoef J. Molecular identification of bacteria by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1995;33:2601–2606. doi: 10.1128/jcm.33.10.2601-2606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]