Abstract
The TP53 gene is unarguably one of the most studied human genes. Its encoded protein, p53, is a tumor suppressor and is often called the “guardian of the genome” due to its pivotal role in maintaining genome stability. Historically, most studies of p53 have focused on its roles in somatic cells and tissues, but in the last 2 decades, its functions in embryonic stem cells (ESCs) and induced pluripotent stem cells have attracted increasing attention. Recent studies have identified p53 as a critical regulator of pluripotency, self-renewal, differentiation, proliferation, and genome stability in mouse and human embryonic stem cells. In this article, we systematically review the studies on the functions of p53 in ESCs, provide an updated overview, attempt to reconcile controversial results described in the literature, and discuss the relevance of these cellular functions of p53 to its roles in tumor suppression.
Keywords: embryonic stem cells, p53, tumor suppressor, self-renewal, differentiation, genomic stability
Graphical Abstract
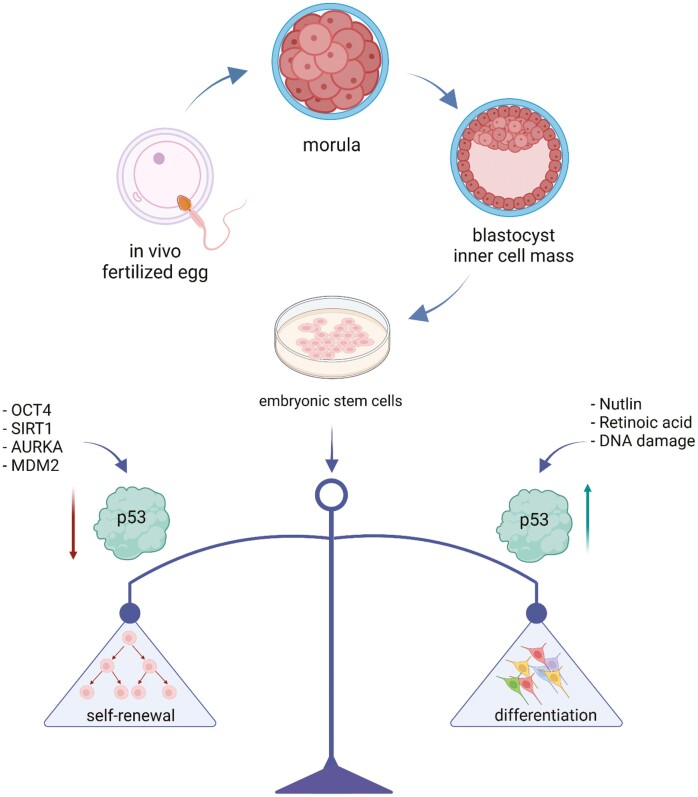
p53 is involved in sustaining the balance between self-renewal and differentiation in embryonic cells for proper development and homeostasis. Created with BioRender.com
Significance Statement.
Embryonic stem cells (ESCs) are capable of self-renewal and differentiate into most cell types. They have great potential in cell therapy and are an excellent model for early embryonic development. p53 is one of the critical proteins that sustain the balance between self-renewal, differentiation, and genomic integrity in ESCs for proper development and homeostasis. This review article discusses the studies on the roles of p53 in ESCs by highlighting the possible underlying mechanisms and emphasizing the controversies in the niche field of p53 research to provide more insights into future studies of p53 functions in cancer and development.
Introduction
Embryonic stem cells (ESCs) are capable of unlimited self-renewal and retain pluripotency to differentiate into all 3 germ layers in the body during gastrulation, which ultimately gives rise to an entire organism.1,2 ESCs are isolated from the inner cell mass (ICM) of blastocysts, an early stage of embryo development.3 The self-renewal capacity of embryonic stem cells is achieved by maintaining a delicate balance between proliferation, self-renewal, and the prevention of apoptosis or senescence.4
The remarkable ability of ESCs to differentiate into all cell types necessitates unique and robust mechanisms to mitigate insults that jeopardize genome stability. For example, ESCs are highly sensitive to DNA damage stressors.5 This hypersensitivity to DNA damage is postulated as one of the mechanisms to minimize the risk of passing unrepaired genomic alterations to daughter cells, as ESCs with damaged DNA are removed from the population, a process of maintaining a low DNA mutation burden.6 This theory is supported by the observation that the mutation frequency is 100 times lower in ESCs than in differentiated cells.6 However, hypersensitivity to DNA damage is a double-edged sword. On the one hand, ESCs maintain a population with a pristine genome by eliminating rogue cells. On the other hand, this mutation-proof mechanism could easily lead to the loss of the ESC population, which could cause developmental abnormalities and cancer.7 It remains unclear how the organism strikes a fine balance between genome stability and cell population stability to ensure normal development. As the “guardian of the genome,” p53 may be a pivotal player in this delicate balancing act.
p53 is a well-known sequence-specific transcription factor that regulates the expression of genes involved in apoptosis, cell cycle arrest, DNA repair, and senescence in response to various genomic and oncogenic insults, thereby contributing to the maintenance of genome stability.8-10 Apart from its roles in sustaining homeostasis in somatic cells, the importance of p53 in stem cell biology has also received significant interest. This review will summarize the roles of p53 in embryonic stem cells, provide an update on recently published studies, and outline controversies in this niche field of p53 research.
An Overview of p53
p53 was first discovered in 1979 as a binding partner of the SV40 large T-antigen.11 It was initially thought to be an oncogene because the mutated TP53 cDNA was cloned from transformed cells. Later studies, however, indicated that p53 is a tumor suppressor and that almost half of the human tumors have either TP53 mutations or deletions.12 Shortly after, germline mutations in the TP53 gene were found to be associated with an inherited disorder, Li-Fraumeni syndrome (LFS). Individuals with LFS have a high risk of developing different types of cancer at a young age.13
Human p53 protein consists of 393 amino acids. Structural and functional analysis has demonstrated that p53 consists of both a nuclear localization signal (NLS) and nuclear export signal (NES), which facilitate the shuttling of p53 between the nucleus and cytoplasm (Fig. 1).14 Without stress, p53 protein levels are low, and the protein is primarily cytoplasmic. Upon various stresses, p53 is stabilized and translocates to the nucleus, where it binds to chromatin and activates its target genes. p53 has 2 amino-terminal transactivation domains (TADs) followed by a conserved proline-rich domain, a central DNA-binding domain, a tetramerization domain, and a carboxy-terminal regulatory domain (CTD).15
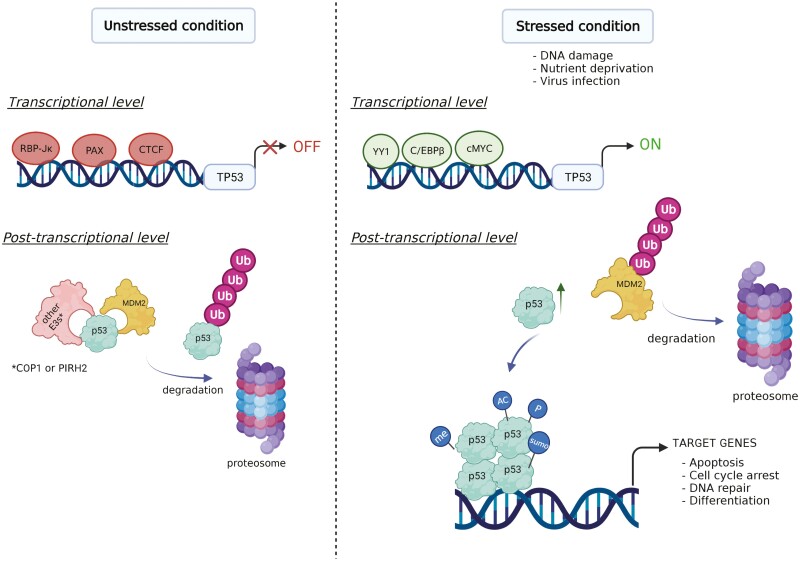
Figure 1.
(A) Domain architecture of p53. The major functional domains of p53 are shown including the N-terminal transactivation domain (TAD), proline-rich domain, DNA-binding domain, carboxy-terminus domain (CTD), as well as the nuclear localization signal (NLS) and nuclear export signal (NES), which are involved in transportation of p53 between the nucleus and cytoplasm. “Hotspot” mutations are also depicted in the DNA-binding domain. Created with IBS (http://ibs.biocuckoo.org/). (B) A schematic showing both transcriptional and post-transcriptional regulation of TP53 (or p53) under varying conditions. In unstressed conditions, TP53 is downregulated by transcription factors such as RBP-Jκ, PAX, and CTCF. At the post-transcriptional level, p53 is ubiquitylated by MDM2 or other E3 ligases and subsequently degraded by the proteasome. Under stressed conditions, transcription factors such as c-MYC, YY1, and C/EBPβ enhance TP53 level. p53 is activated and acquires post-translational modifications such as phosphorylation (P), acetylation (AC), methylation (me), or sumoylation (sumo). Activated p53 then binds to chromatin as a tetramer and regulates its target genes to modulate various cellular functions. Created with BioRender.com.
Most TP53 mutations are missense mutations that cluster in a region encoding the central DNA-binding domain. Among those mutations, 8 amino acid substitutions (R175H, Y220C, G245S, R248Q, R248W, R249S, R273H, and R282W), called “hotspot mutations,” are the most frequently identified in humans (Fig. 1A).16,17 These hotspot mutations are categorized into 2 groups: “contact mutations” (R248 and R273) that affect the ability of p53 to interact with DNA and “conformational mutations” (R175, Y220, G245, R249, and R282) that disrupt the folding of p53.16,17 Some mutations result in the loss of p53’s tumor suppressor activity and therefore are called loss-of-function (LOF) mutations. Others produce p53 mutants that have dominant negative effects or gain new oncogenic functions that promote tumor progression, and these mutations are gain-of-function (GOF).17 The GOF of p53 mutant provides therapeutic opportunities for cancer treatment.
The expression and activity of p53 is tightly regulated. Under normal conditions, p53 protein levels are kept low by E3 ligases such as MDM2,18 COP1,19 and PIRH220 (Fig. 1B). MDM4 (also known as MDMX), a homolog of MDM2, is another negative regulator of p53. Unlike MDM2, MDM4 has no E3 ligase activity and functions by directly inhibiting p53 activity rather than promoting its degradation.21 Various stressors, such as DNA damage, virus infection, oncogene activation, or nutrient deprivation, trigger the release of p53 from its negative regulators and lead to the stabilization and activation of p53.22 Activated p53, in turn, binds to specific sequences of the genome and alters the expression of many genes.23 This process is regulated by several cofactors and numerous post-translational modifications (PTMs), such as phosphorylation, acetylation, sumoylation, and methylation.24 The combinatorial effects of these cofactors and PTMs fine-tune p53 stabilization and activation (Fig. 1B).
The TP53 (Trp53 in mice) gene is also subject to transcriptional and epigenetic regulation, and the mechanisms of this regulation in somatic cells have been reviewed by others.25,26 The transcription factors regulating TP53 are c-MYC, CTCF, YY1, PAX, C/EBPβ, and RBP-Jκ under varying conditions (Fig. 1B).26 Interestingly, the mRNA levels of Trp53 are high in mouse ESCs (mESCs) and downregulated upon differentiation,27 suggesting that transcriptional regulation may be a critical step in regulating the expression of Trp53 in mESCs. In support of this concept, key transcription factors of ESCs, such as TCF3, NANOG, OCT4, have been shown to regulate Trp53 transcription.27,28
One of the primary cellular functions of p53 is to regulate downstream transcriptional targets involved in apoptosis, cell cycle, senescence, DNA repair, ferroptosis, metabolism, autophagy, lysosomal function, and numerous other critical processes.8 In addition to these nuclear functions, p53 has been shown to have cytoplasmic roles. A well-documented cytoplasmic role of p53 is to regulate mitochondrial function and cell death.29 Despite the discovery of all these cellular pathways regulated by p53, it remains unclear which pathway(s) contributes to the tumor-suppressive function of p53.30,31
In addition to its role as a tumor suppressor, p53 has been implicated as a critical factor regulating development. This area has long been overlooked because p53 knockout mice are outwardly “normal” in development. However, there is a resurgence of interest in this topic, which coincides with the studies of p53 in ESCs and induced pluripotency stem cells (iPSCs).32-35 This review aims to comprehensively summarize the studies on the functions of p53 in ESCs and highlight recent studies of p53 in ESCs.
The Multiple Functions of p53 in ESCs
In addition to the functions described for p53 in somatic cells, p53 also modulates self-renewal, differentiation, and pluripotency in ESCs. We note that there is no strict boundary between ESC-specific and non-ESC-specific functions of p53 because some of the non-ESC-specific functions of p53, such as cell cycle regulation, are tightly intertwined with the pluripotent and lineage choice pathways in ESCs.36 The precise outcome of p53 activation in ESCs depends on several factors, including stress types, cell types, differentiation stages, specific post-translational modifications on p53, and associated co-regulatory factors.37 The overall role of p53 is to sustain the balance between self-renewal, differentiation, and genomic integrity in ESCs for proper development and homeostasis.38 These potential roles of p53, in the context of the differences between human and mouse ESCs (hESCs and mESCs), will be discussed below.
A Summary of Comparative p53 Functions in Human and Mouse ESCs
Both hESCs and mESCs have the ability to replicate (self-renewal) and differentiate into most cell types (pluripotency).3,39 The undifferentiated state of hESCs and mESCs is regulated by several core transcription factors, such as OCT4, SOX2, NANOG, KLF4, and other epigenetic modifiers. These factors cooperatively maintain the pluripotency of stem cells while suppressing differentiation.40 Although the major self-renewal and pluripotency networks in hESCs and mESCs are similar, there are some notable differences. For example, the classical media for culturing hESCs and mESCs are different. Leukemia inhibitor factor (LIF) is commonly used in the conventional culture medium for mESCs, while fibroblast growth factor (FGF) is needed for culturing hESCs. In addition, although both hESCs and mESCs are derived from the ICM of blastocysts, hESC gene expression patterns, signaling responses, appearance, and culture requirements are more similar to pluripotent epiblast-derived stem cells (EpiSCs) than mESCs.41,42
The overall role of p53 is similar in hESCs and mESCs. For example, activated p53 induces the differentiation of hESCs and mESCs.34,43-45 However, the molecular mechanisms underlying the functions and the modes of action of p53 differ between hESC and mESCs. How the intrinsic differences between hESCs and mESCs shape p53 functions will be reviewed in the subsequent sections (Fig. 2).
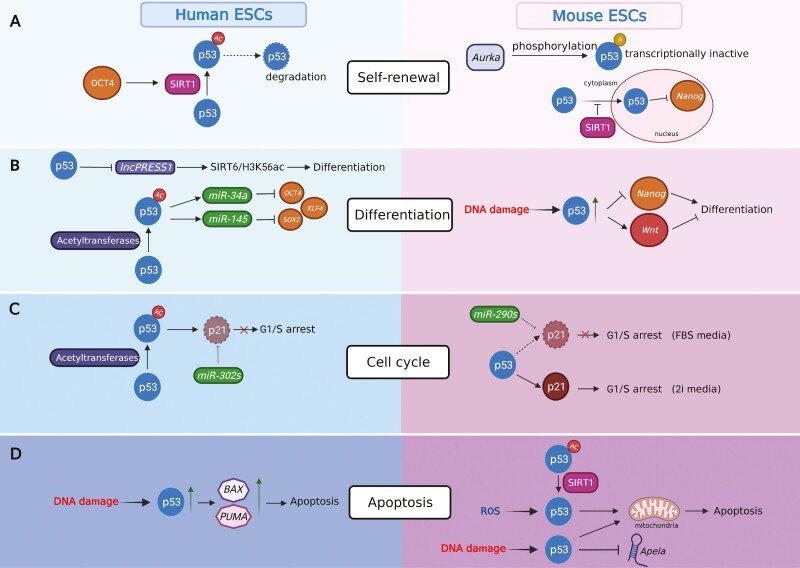
Figure 2.
A comparative view of p53 functions in self-renewal, differentiation, cell cycle, and apoptosis in human and mouse ESCs. Created with BioRender.com.
Roles of p53 in Differentiation and Self-Renewal of ESCs
p53 plays a significant role in maintaining the balance between self-renewal and differentiation through different mechanisms (Fig. 2A and 2B). In mESCs, p53 induces differentiation by transcriptionally repressing master pluripotency regulators such as Nanog, Oct4, and Sox2 (Table 1).34,45 In hESCs, activated p53 induces microRNAs (miR-34 and miR-145), which in turn suppress the expression of master regulators,46 or repress the hESC-specific lncRNA, lncPRESS1, to regulate the SIRT6/H3K56ac axis (Fig. 2B; Table 1).44 Therefore, the effect of p53 on the expression of master regulators in hESCs appears to be indirect, ie, the repression is mediated by intermediatory factors, such as miRs and lncRNAs.
Table 1.
Main p53-regulated targets in ESCs.
| Cell type | Target name | Regulation | Effect | Reference |
|---|---|---|---|---|
| mESCs | Nanog | Repression | Induces differentiation | 34,45 |
| mESCs | Oct4 | Repression | Induces differentiation | 34,45 |
| mESCs | Sox2 | Repression | Induces differentiation | 34,45 |
| hESCs | miR34 (microRNA) | Activation | Induces differentiation | 46 |
| hESCs | miR145 (microRNA) | Activation | Induces differentiation | 46 |
| hESCs | lncPRESS1 (lncRNA) | Repression | Regulation of SIRT6/h3k56ac axis | 44 |
| mESCs | Wnt ligands | Activation | In response to DNA damage, induces anti-differentiation | 47 |
| hESCs | BAX | Activation | Induces apoptosis | 48 |
| hESCs | PUMA | Activation | Induces apoptosis | 48 |
| mESCs | Apela (a regulatory RNA) | Activation | Induces apoptosis | 49 |
In response to DNA damaging agents, p53 promotes the transcription of Wnt genes, which have an anti-differentiation role in mESCs (Fig. 2B; Table 1).50 It has been postulated that p53-induced Wnt ligands released from damaged mESCs may act on neighboring cells to prevent their differentiation, presumable acting as a compensatory mechanism to maintain the cell number in the population.47 This hypothesis assumes that biologically relevant DNA damage only affects a proportion of mESCs and has not been experimentally tested. Interestingly, in hESCs, the induction of WNTs by p53, p63, and p73 in response to differentiation cues is required for mesendodermal differentiation.51
Recent studies found that ESCs with high levels of p53 lost the competition to those with low levels of p53.52,53 Mathematical modeling showed that the competition is dependent on close cell-to-cell contact.52 Given that Wnts mediate short-distance cell–cell communication, it will be interesting to study in the future whether the p53-Wnt axis mediates ESCs competition.
Cell Cycle Regulation of ESCs by p53
In hESCs, the p53-p21 (encoded by the CDKN1A gene) axis is largely deficient in the G1/S checkpoint.54 Upon DNA damage, CDKN1A mRNA is induced while the p21 protein is undetectable, partially due to the repression by miR-302s.55
In mESCs, whether p53 plays a role in cell cycle regulation, particularly G1/S arrest, depends on the culture medium. Naïve-state mESCs cultured in a conventional medium, which contains fetal bovine serum (FBS) and LIF, lack the p53-regulated G1/S checkpoint.33,56 These cells have a very short G1 phase, presumably due to high levels of cyclin A-CDK2 and cyclin E-CDK2.57 Activated p53 does not promote G1/S arrest. Instead, it induces apoptosis and/or differentiation. However, if mESCs are maintained in the ground state by 2i medium, which contains inhibitors of mitogen-activated protein kinase kinase (Mek, PD0325901) and glycogen synthase kinase-3 (Gsk3, CHIR99021), the p53-p21 axis is required for G1/S arrest (Fig. 2C).58 The differential level of p21 in mESCs grown in serum-containing medium and 2i medium is one factor that may explain these seemingly contradictory results. In the serum-containing medium, p53, upon activation, induces the Cdkn1a mRNA; however, the p21 protein is difficult to detect59 (Fig. 2C). Similar to hESCs, the absence of p21 protein partially results from the repression by miRs, such as members of the miR-290 family.59 In the 2i medium, however, the p21 protein is readily detectable.60 Therefore, the transcriptome and/or epigenome, affected by culture conditions, of ESCs can dramatically alter the function of p53. It remains to be determined how the different pathways for differentiation and self-renewal of hESCs and mESCs, as mentioned above, affect the modes of action of p53 in cell cycle regulation of hESCs and mESCs.
The Controversial Roles of p53 in Regulating the Apoptosis of ESCs
In differentiated cells, prolonged or high-dose DNA damage leads to elevated p53 in the nucleus and mitochondria, which in turn activates its targets, such as BAX, PUMA, and NOXA. These pro-apoptotic factors initiate cell death.61 In hESCs, p53 induces rapid apoptosis through several mechanisms. A study by Grandela et al demonstrated that etoposide-induced apoptosis dramatically changes the subcellular localization of p53, which shuttles from the cytosol to the nucleus and alters the expression of the key regulators BAX and PUMA (Fig. 2D; Table 1).48 Others have shown that p53 regulates the translocation of BAX to the mitochondria and mitochondrial priming.62 Interestingly, p53 regulates mitochondria-mediated apoptosis in mESCs, and this role is mediated by a regulatory RNA, Apela (Fig. 2D; Table 1).49 These results suggest that p53 has a convergent role in regulating mitochondria-mediated apoptosis in hESCs and mESCs. Interestingly, studies have shown that ESCs typically have structurally immature and functionally deficient mitochondria.63 As a result, ESCs generally rely on glycolysis for energy production. These observations suggest that p53-regulated pro-apoptotic function of mitochondria is independent from the energy production pathway.
However, compared to hESCs, whether p53 induces apoptosis in mESCs remains highly contentious. Some have shown that genotoxic stress-induced apoptosis of mESCs is dependent on p53,49,64,65 while others have described p53-independent apoptosis under similar conditions.33,66 The factors that cause this discrepancy are unclear but might include the culture and experimental conditions. For example, the issues of different cell confluency (low vs high) and cleaved caspase-3 antibodies for performing apoptosis assays were raised.49,66 It is worth discussing the “optimal” confluence for performing apoptosis assays in mESCs. On the one hand, most in vitro studies, particularly those in cancer, are performed under low confluence conditions, as cells growing under exponential phase are preferred. On the other hand, the high density of ESCs culture mimics the compact sphere-like structure of the ICM in blastocysts, from which ESCs are derived. In addition, as discussed in the cell cycle regulation by p53 in ESCs, cell culture media composition must also be considered. It is also possible that different batches of FBS and LIF could contribute to inconsistent findings reported by different groups. Thus, to address the discrepancy in the literature, future studies need to pay extra attention to these variables.
Ultimately, the value of determining whether p53 promotes apoptosis of ESCs largely depends on whether the in vitro system can model the in vivo function of p53. It is worth noting that an in vivo study showed that p53 is transcriptionally active in mESCs and capable of regulating cell cycle and apoptotic genes in the ICM of blastocysts.67 Given that mESCs are derived from the ICM, this study suggests that p53 has the ability to activate the pro-apoptotic program in mESCs. In another study, p53+/+ and p53−/− embryos were exposed to X-ray irradiation at embryonic days 3.5 (E3.5) and 9.5 (E9.5). Although the survival rate of p53−/− embryos was better than that of p53+/+ due to the lack of apoptosis, p53−/− embryos had more developmental anomalies.7 The implications of this study are 2-fold. First, DNA damage induces p53-dependent apoptosis in E3.5 blastocysts, from which mESCs are derived. Second, in response to DNA damage, p53-dependent apoptosis in early embryonic stages minimizes developmental anomalies in a population by canceling the development of damaged embryos. These in vivo studies serve as valuable guidance for interpreting results from in vitro studies of the apoptotic role of p53 in ESCs.
Metabolic Regulation of ESCs by p53
The role of p53 in the metabolic regulation of ESCs is relatively understudied compared with its role in processes such as differentiation and apoptosis. Since culture conditions undoubtedly affect the metabolism of ESCs, differences in culture techniques and changes in media composition make studying the roles of p53 in the metabolic regulation of ESCs difficult. Recently, p53 was found to regulates glucose and choline metabolism in hESCs, for its disruption was associated with upregulation of phosphatidylcholine and glycolysis and delayed mesendoderm differentiation.68 Common metabolic p53 targets, such as TIGAR and SCO2, were not involved, suggesting that p53 may have different metabolic targets in ESCs compared to differentiated cells. Therefore, the mechanisms by which p53 regulates glucose and choline metabolism in hESCs remain unclear.
Negative Regulators of p53 in ESCs
Although the TP53 gene is highly expressed in ESCs, negative regulators of p53 restrain its activity in the absence of activating cues. For example, ChIP-seq analysis of chromatin bound by phosphorylated p53 (serine 18, S18P; S15P in human) revealed that many p53-bound loci are poised for activation, but p53 activity is suppressed by negative regulators.45 In the absence of stress, p53 activity must be repressed to maintain the undifferentiated status and survival of ESCs. A small-molecule inhibitor of MDM2, Nutlin, prevents the binding of MDM2 and p53 and therefore activates p53. In hESCs, Nutlin treatment leads to rapid differentiation by blocking cell cycle progression via the p53-p21 axis. Since p21 is a cell cycle factor in differentiated cells, this suggests that the differentiation and cell cycle progression processes of hESCs are intertwined. Another study demonstrated that MDM4 (MDMX) is a key negative regulator of p53 in mESCs that directly inhibits p53 activity without affecting p53 protein levels.69
Through a short hairpin RNA (shRNA) screen, Aurora kinase A (Aurka), which is a serine/threonine kinase with key mitotic regulatory functions was identified as a negative regulator of p53 in mESCs, possibly by phosphorylating serine residues 212 and 312 in p53 (Fig. 2A).70 Inhibition of Aurka led to p53 activation and subsequent ectoderm and mesoderm differentiation, overall suggesting that Aurka-p53 signaling in the regulation of self-renewal and differentiation is critical.
OCT4 is a key pluripotency factor involved in maintaining the self-renewal and pluripotency of ESCs. The anti-differentiation role of OCT4 has been previously attributed to its ability to maintain the expression of self-renewal factors while also suppressing the expression of lineage genes. Another mechanism underlying the anti-differentiation function of OCT4 has been reported in hESCs, where OCT4 inhibits p53 activity by inducing the expression of the histone deacetylase SIRT1, a known negative regulator of p53 and deacetylates p53 at lysine 120 and 164 in hESCs (Fig. 2A).71 In mESCs, SIRT1 controls p53 activity by a different mechanism (Fig. 2B); deacetylation of p53 by SIRT1 blocks the translocation of p53 from the cytoplasm to the nucleus in response to endogenous reactive oxygen species (ROS). The retaining of p53 in the cytoplasm triggers mitochondria-dependent apoptosis in mESCs and reduces p53-mediated Nanog repression in the nucleus (Fig. 2D).72 Thus, the balance between pro-apoptotic function and pro-differentiation roles of p53 in mESCs is regulated by SIRT1.
The Relevance of P53’s Roles in ESCs to Its Tumor Suppressive Functions
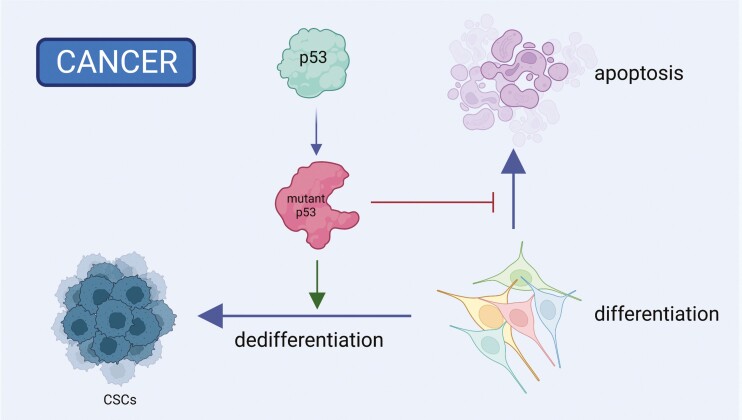
Cancer stem cells (CSCs) or cancer-initiating cells (CICs), which are transformed and stem-like cells, can initiate tumors, persist after cancer therapy, and gain more aggressive features.73 Since embryonic stem cells and cancer stem cells (CSCs) display similarities based on their high plasticity and overlapping gene signatures, studying the roles of p53 in ESCs may shed new light on the role of p53 in preventing the establishment of CSCs in somatic tissues (Fig. 3). The potential role of p53 as a barrier to CSC formation has been extensively reviewed elsewhere.38,73
Figure 3.
Mutant p53 gains various oncogenic functions to support tumorigenesis and dedifferentiation of somatic cells into cancer stem cells (CSCs). Created with BioRender.com.
ESCs and iPSCs hold great promise in cell therapy. However, even although these cells are considered non-transformed, they can form teratomas when transplanted into non-orthotopic tissues.74 It remains unclear how p53 is involved in tumorigenesis of these pluripotent stem cells. After sequencing 140 human iPSC clones, Merkle et al found that 5 of these cell lines carried dominant negative TP53 mutations.75 Prolonged passages of these cell lines increased the fractions of TP53 mutated alleles, suggesting that p53 plays essential functions in cultured iPSCs. Given the similarities between hESCs and hiPSCs, high passage hESCs might also gain TP53 mutations, therefore increasing the tumorigenic abilities and dampening the clinical applications of these cells. However, similar sequencing studies have not been performed in hESCs.
Concluding Remarks and Perspectives
In the past 2 decades, much has been learned about the functions of p53 in ESCs. The emerging view is that p53 regulates the differentiation and self-renewal of mESCs and hESCs by altering the expression of ESC-enriched factors, such as OCT4, NANOG, and SOX2. In addition to its ESC-specific functions, p53 also has non-ESC-specific functions in ESCs, for example, apoptosis and cell cycle regulation. As discussed above, these non-ESC-specific functions are tightly connected to ESC-specific functions. These functions of p53 collectively may contribute to the low mutation burden of ESCs.
Of note, controversies regarding the pro-apoptotic function of p53 in ESCs still exist. Recent studies have highlighted experimental variables, such as in vitro culture conditions, that may underlie the seemingly contradictory results reported by different groups. In addition, in vitro culture conditions for ESCs are still evolving, compounding the difficulty in comparing past and present data. We propose that the design and interpretation of results from in vitro experiments need to be complemented by in vivo experiments. Moreover, when interpreting the results from studies of p53 in ESCs, it is critical to consider whether those studies were performed in the absence or presence of stresses. In the future, the combination of in vitro and in vivo studies will help reconcile some remaining controversies regarding the roles of p53 in ESCs, embryonic development, and tumorigenesis. As the culturing conditions of hESCs and hiPSCs mature, we will see more studies using these systems to model the roles of p53 in human embryonic development and developmental diseases.
Acknowledgments
This work was supported by intramural grants, 1ZIABC011158-12 and 1ZIABC011504-08, awarded by the National Cancer Institute (NCI), USA, to J.H. We also would like to thank Brandi Carofino for providing language and science-editing service during the preparation of this manuscript.
Contributor Information
Gamze Ayaz, Cancer and Stem Cell Epigenetics, Laboratory of Cancer Biology and Genetics, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Hualong Yan, Cancer and Stem Cell Epigenetics, Laboratory of Cancer Biology and Genetics, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Navdeep Malik, Cancer and Stem Cell Epigenetics, Laboratory of Cancer Biology and Genetics, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Jing Huang, Cancer and Stem Cell Epigenetics, Laboratory of Cancer Biology and Genetics, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Funding
The work was suppurted by intramural grants, 1ZIABC011158-12 and 1ZIABC011504-08, awarded by the National Cancer Institute (NCI), NIH, to J.H.
Conflict of Interest
The authors declared no potential conflicts of interest.
Author Contributions
G.A. conducted the literature review, wrote the article, drafted the figures, revised and edited the manuscript and figures. H.Y. conducted the literature review, wrote the article, revised and edited the manuscript and figures. N.M. revised and edited the article, reviewed the manuscript and figures. J.H. provided the idea, conducted the literature review, wrote the article, revised and edited the manuscript and figures. All authors read and approved the final manuscript.
Data Availability
No new data were generated or analyzed in support of this research.
References
- 1. Zakrzewski W, Dobrzynski M, Szymonowicz M, et al. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10(1):68. 10.1186/s13287-019-1165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kobayashi T, Surani MA.. On the origin of the human germline. Development. 2018;145(16). 10.1242/dev.150433 [DOI] [PubMed] [Google Scholar]
- 3. Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145-1147. [in Eng]. 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4. Stewart R, Stojkovic M, Lako M.. Mechanisms of self-renewal in human embryonic stem cells. Eur J Cancer. 2006;42(9):1257-1272. 10.1016/j.ejca.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 5. Roos WP, Christmann M, Fraser ST, et al. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O(6)-methylguanine due to high E2F1 regulated mismatch repair. Cell Death Differ. 2007;14(8):1422-1432. 10.1038/sj.cdd.4402136. [DOI] [PubMed] [Google Scholar]
- 6. Cervantes RB, Stringer JR, Shao C, et al. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci USA. 2002;99(6):3586-3590. 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Norimura T, Nomoto S, Katsuki M, et al. p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat Med. 1996;2(5):577-580. 10.1038/nm0596-577. [DOI] [PubMed] [Google Scholar]
- 8. Vousden KH, Prives C.. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413-431. [in Eng]. 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 9. Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med. 2016;6(3):a026104. 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koifman G, Aloni-Grinstein R, Rotter V.. p53 balances between tissue hierarchy and anarchy. J Mol Cell Biol. 2019;11(7):553-563. 10.1093/jmcb/mjz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linzer DI, Levine AJ.. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43-52. 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 12. Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342(6250):705-708. 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 13. Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233-1238. 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 14. Marchenko ND, Hanel W, Li D, et al. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17(2):255-267. 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ko LJ, Prives C.. p53: puzzle and paradigm. Genes Dev. 1996;10(9):1054-1072. 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 16. Freed-Pastor WA, Prives C.. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268-1286. 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sabapathy K, Lane DP.. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol. 2018;15(1):13-30. 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- 18. Honda R, Tanaka H, Yasuda H.. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420(1):25-27. 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 19. Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429(6987):86-92. 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 20. Leng RP, Lin Y, Ma W, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112(6):779-791. 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 21. Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15(19):5349-5357. 10.1002/j.1460-2075.1996.tb00919.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J. Current developments of targeting the p53 signaling pathway for cancer treatment. Pharmacol Ther. 2021;220:107720. 10.1016/j.pharmthera.2020.107720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Menendez D, Inga A, Resnick MA.. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9(10):724-737. 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 24. Liu Y, Tavana O, Gu W.. p53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 2019;11(7):564-577. 10.1093/jmcb/mjz060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saldana-Meyer R, Recillas-Targa F.. Transcriptional and epigenetic regulation of the p53 tumor suppressor gene. Epigenetics. 2011;6(9):1068-1077. 10.4161/epi.6.9.16683. [DOI] [PubMed] [Google Scholar]
- 26. Reisman D, Takahashi P, Polson A, et al. Transcriptional regulation of the p53 tumor suppressor gene in S-phase of the cell-cycle and the cellular response to DNA damage. Biochem Res Int. 2012;2012:808934. 10.1155/2012/808934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tam WL, Lim CY, Han J, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26(8):2019-2031. [in Eng]. 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431-440. [in Eng]. 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 29. Vaseva AV, Moll UM.. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787(5):414-420. 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kastenhuber ER, Lowe SW.. Putting p53 in context. Cell. 2017;170(6):1062-1078. 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hafner A, Bulyk ML, Jambhekar A, et al. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20(4):199-210. 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 32. Marion RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149-1153. [in Eng]. 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aladjem MI, Spike BT, Rodewald LW, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8(3):145-155. 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 34. Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7(2):165-171. 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 35. Shin MH, He Y, Huang J.. Embryonic stem cells shed new light on the developmental roles of p53. Cell Biosci 2013;3(1):42. 10.1186/2045-3701-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li VC, Kirschner MW.. Molecular ties between the cell cycle and differentiation in embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(26):9503-9508. 10.1073/pnas.1408638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jain AK, Barton MC.. p53: emerging roles in stem cells, development and beyond. Development. 2018;145(8):1-10. 10.1242/dev.158360 [DOI] [PubMed] [Google Scholar]
- 38. Ghatak D, Das Ghosh D, Roychoudhury S.. Cancer Stemness: p53 at the Wheel. Front Oncol. 2020;10:604124. 10.3389/fonc.2020.604124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans MJ, Kaufman MH.. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154-156. [in Eng]. 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 40. Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631-642. 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 41. Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196-199. 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 42. Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191-195. 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 43. Maimets T, Neganova I, Armstrong L, et al. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27(40):5277-5287. 10.1038/onc.2008.166. [DOI] [PubMed] [Google Scholar]
- 44. Jain AK, Xi Y, McCarthy R, et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol Cell. 2016;64(5):967-981. 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li M, He Y, Dubois W, et al. Distinct Regulatory Mechanisms and Functions for p53-Activated and p53-Repressed DNA Damage Response Genes in Embryonic Stem Cells. Mol Cell. 2012;46(1):30-42. [in Eng]. 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jain AK, Allton K, Iacovino M, et al. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10(2):e1001268. [in Eng]. 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li M, Huang J.. A new puzzling role of p53 in mouse embryonic stem cells. Cell Cycle. 2010;9(9):1669-1670. 10.4161/cc.9.9.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grandela C, Pera MF, Grimmond SM, et al. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res. 2007;1(2):116-128. 10.1016/j.scr.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 49. Li M, Gou H, Tripathi BK, et al. An Apela RNA-containing negative feedback loop regulates p53-mediated apoptosis in embryonic stem cells. Cell Stem Cell. 2015;16(6):669-683. 10.1016/j.stem.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee KH, Li M, Michalowski AM, et al. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci USA. 2010;107(1):69-74. 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Q, Zou Y, Nowotschin S, et al. The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell. 2017;20(1):70-86. 10.1016/j.stem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Montero SP, BowlingS, Pérez-Carrasco Ret al. Levels of p53 expression determine the competitive ability of embryonic stem cells during the onset of differentiation. bioRxiv 2022;2022.02.28.482311. 10.1101/2022.02.28.482311 [DOI] [Google Scholar]
- 53. Lima A, Lubatti G, Burgstaller J, et al. Cell competition acts as a purifying selection to eliminate cells with mitochondrial defects during early mouse development. Nat Metab. 2021;3(8):1091-1108. 10.1038/s42255-021-00422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barta T, Vinarsky V, Holubcova Z, et al. Human embryonic stem cells are capable of executing G1/S checkpoint activation. Stem Cells. 2010;28(7):1143-1152. 10.1002/stem.451. [DOI] [PubMed] [Google Scholar]
- 55. Dolezalova D, Mraz M, Barta T, et al. MicroRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells. 2012;30(7):1362-1372. 10.1002/stem.1108. [DOI] [PubMed] [Google Scholar]
- 56. Suvorova, II, Grigorash BB, Chuykin IA, et al. G1 checkpoint is compromised in mouse ESCs due to functional uncoupling of p53-p21Waf1 signaling. Cell Cycle. 2016;15(1):52-63. 10.1080/15384101.2015.1120927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Coronado D, Godet M, Bourillot PY, et al. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res. 2013;10(1):118-131. 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 58. Ter Huurne M, Peng T, Yi G, et al. Critical role for p53 in regulating the cell cycle of ground state embryonic stem cells. Stem Cell Rep. 2020;14(2):175-183. 10.1016/j.stemcr.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Y, Baskerville S, Shenoy A, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40(12):1478-1483. 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marks H, Kalkan T, Menafra R, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149(3):590-604. 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20(8):471-480. 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 62. Liu JC, Guan X, Ryan JA, et al. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell. 2013;13(4):483-491. 10.1016/j.stem.2013.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Facucho-Oliveira JM, St John JC.. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep. 2009;5(2):140-158. 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 64. de Vries A, Flores ER, Miranda B, et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc Natl Acad Sci USA. 2002;99(5):2948-2953. [in Eng]. 10.1073/pnas.052713099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. He H, Wang C, Dai Q, et al. p53 and p73 regulate apoptosis but not cell-cycle progression in mouse embryonic stem cells upon DNA damage and differentiation. Stem Cell Rep. 2016;7(6):1087-1098. 10.1016/j.stemcr.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jaiswal SK, Oh JJ, DePamphilis ML.. Cell cycle arrest and apoptosis are not dependent on p53 prior to p53-dependent embryonic stem cell differentiation. Stem Cells. 2020;38(9):1091-1106. 10.1002/stem.3199. [DOI] [PubMed] [Google Scholar]
- 67. Goh AM, Lim CY, Chiam PC, et al. Using targeted transgenic reporter mice to study promoter-specific p53 transcriptional activity. Proc Natl Acad Sci USA. 2012;109(5):1685-1690. 10.1073/pnas.1114173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu G, Ying Y, Ji K, et al. p53 coordinates glucose and choline metabolism during the mesendoderm differentiation of human embryonic stem cells. Stem Cell Res. 2020;49:102067. 10.1016/j.scr.2020.102067. [DOI] [PubMed] [Google Scholar]
- 69. Menendez S, Goh AM, Camus S, et al. MDM4 downregulates p53 transcriptional activity and response to stress during differentiation. Cell Cycle. 2011;10(7):1100-1108. 10.4161/cc.10.7.15090. [DOI] [PubMed] [Google Scholar]
- 70. Lee DF, Su J, Ang YS, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11(2):179-194. 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang ZN, Chung SK, Xu Z, et al. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells. 2014;32(1):157-165. 10.1002/stem.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Han MK, Song EK, Guo Y, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2(3):241-251. [in Eng]. 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aloni-Grinstein R, Shetzer Y, Kaufman T, et al. p53: the barrier to cancer stem cell formation. FEBS Lett. 2014;588(16):2580-2589. 10.1016/j.febslet.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 74. Ben-David U, Benvenisty N.. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11(4):268-277. 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 75. Merkle FT, Ghosh S, Kamitaki N, et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature. 2017;545(7653):229-233. 10.1038/nature22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research.