Abstract
Adipose-derived cells (ADCs) from white adipose tissue are promising stem cell candidates because of their large regenerative reserves and the potential for cardiac regeneration. However, given the heterogeneity of ADC and its unsolved mechanisms of cardiac acquisition, ADC-cardiac transition efficiency remains low. In this study, we explored the heterogeneity of ADCs and the cellular kinetics of 39,432 single-cell transcriptomes along the leukemia inhibitory factor (LIF)-induced ADC-cardiac transition. We identified distinct ADC subpopulations that reacted differentially to LIF when entering the cardiomyogenic program, further demonstrating that ADC-myogenesis is time-dependent and initiates from transient changes in nuclear factor erythroid 2-related factor 2 (Nrf2) signaling. At later stages, pseudotime analysis of ADCs navigated a trajectory with 2 branches corresponding to activated myofibroblast or cardiomyocyte-like cells. Our findings offer a high-resolution dissection of ADC heterogeneity and cell fate during ADC-cardiac transition, thus providing new insights into potential cardiac stem cells.
Keywords: adipose, derived cells, single, cell sequencing, leukemia inhibitory factor (LIF), ADC, cardiac transition
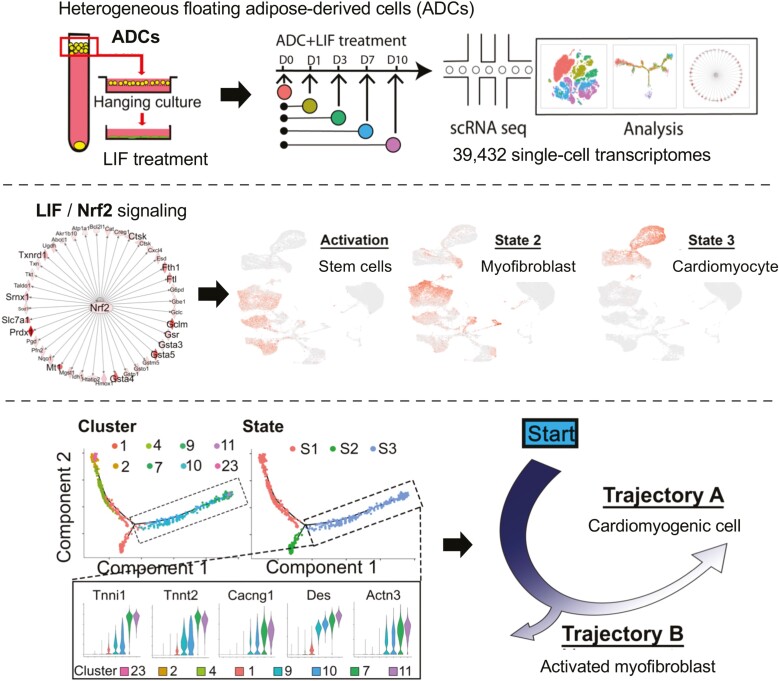
Graphical Abstract
Heterogeneous floating adipose-derived adipose cells (ADCs) undergo ADC-cardiac transition in response to leukemia inhibitory factor. Dissection of the ADC heterogeneity showed differential response to LIF when entering the cardiomyogenic program, as initiated by transient changes in nuclear factor erythroid 2-related factor 2 signaling. Pseudotime analysis navigated a trajectory with 2 branches corresponding to activated myofibroblast or cardiomyocyte-like cells.
Significance Statement.
Cells derived from fat are promising stem cell candidates due to large reserves and potential of generating organ-specific cells. However, such stem cells are heterogenous and their transitions unclear. We used single-cell transcriptomes and the response to leukemia inhibitory factor to explore the heterogeneity and the transition to cardiac cells. We identified distinct cell populations and signaling pathways used to navigate a trajectory with 2 branches corresponding to fibrotic versus cardiac cells. Our results are significant for clarification of stem cell heterogeneity and entrance into the cardiac trajectory, thereby allowing new strategies to promote cardiac health.
Introduction
Heart failure is a growing global health concern caused by irreversible myocardium damage that results from various types of injuries, such as acute myocardial infarction (MI).1-4 The past 2 decades have witnessed the development of cell-based therapies to generate new, functional cardiomyocytes through the exogenous supply of stem or progenitor cells, including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs),5-7 bone marrow-derived cells,8 peripheral blood stem cells,9 endothelial progenitors,10 mesoangioblasts,11 and adventitia resident stem cells.12,13 However, the availability, safety, and transition efficiency of noncardiac cells to functional cardiomyocytes remain a significant challenge.
Adipose tissue has drawn research attention as a safe source of multipotent stem cells for autologous cell therapy in the infarcted myocardium.14 The potential of the therapeutic use of adipose-derived cells (ADCs) is an attractive prospect because of their straightforward availability, scalability, practical conservation of bioactivity after cryopreservation, and strong paracrine effects.15-17 Our previous research and those of others indicate that ADCs can act as sources of spontaneously contracting cardiomyocyte-like cells in vitro.18-21 However, the optimal therapeutic efficacy of ADCs for ischemic heart diseases has been hindered by their poor survival and low retention rates in vivo,22 mainly because of their heterogeneity, which reduces the number of functional progenitors delivered to the injured heart. Distinct ADC populations residing in the same adipose tissue depot appear to be programmed toward alternate fates and differential responses to paracrine signals.23 Studies have attempted to characterize ADCs using cell surface markers via flow cytometry analysis but have not yet identified unique markers for each cluster.24-26 Recent studies applying single-cell RNA sequencing (scRNA-seq) to mouse white adipose tissue (WAT) identified several major subsets of ADCs that differ in their trajectories toward adipogenic differentiation,27,28 suggesting unique responsive potentials to metabolic and endocrine signals. Thus, heterogeneous ADCs in infarcted myocardium may follow several cell lineages instead of transitioning to functional cardiomyocytes in a coordinated fashion. Identifying the optimal ADC subpopulation for cardiac induction and triggering signals may be the key to improving cardiac regeneration.
Among the cardiac paracrine factors secreted after MI, leukemia inhibitory factor (LIF) is one that demonstrably enhances the cardioprotection, neovascularization, and recruitment of endogenous stem cells. Leukemia inhibitory factor induces dimerization of glycoprotein 130 (gp130) and its receptor (LIFR), which in turn phosphorylate and activate the STAT3, ERK, or PI3K signaling pathway,29 thereby protecting against oxidative stress and cell death.30,31 Cardiac STAT3 is essential for maintaining metabolic homeostasis, and loss of STAT3 in cardiomyocytes renders the heart more susceptible to pathological insult by disrupting glucose metabolism and protective networks via the upregulation of specific miRNAs.32,33 Leukemia inhibitory factor also contributes to the homing of bone marrow–derived cardiac progenitors to the injured myocardium and promotes resident cardiac stem cell differentiation and neovascularization.34-37
This study applied high-throughput scRNA-seq38-41 to systematically analyze the ADC heterogeneity and cellular dynamics in LIF-induced cardiac transition across different stages. With scRNA-seq, we identified 3 distinct ADC subpopulations that reacted differentially to LIF, among which platelet-derived growth factor receptor α (Pdgfrα)+CD55+ ADCs demonstrated a strong cardiomyogenic potential, as determined by their expression of cardiomyogenic lineage markers, along with their morphology, sarcomeric organization, and cardiomyocyte-like function in vitro and in vivo. Cardiac transition was initiated on day 1 from a cluster of ADCs enriched in Nrf2-mediated oxidative stress-response signaling. Nrf2 signaling activation is a transient but indispensable event that switches on the ADC-cardiac transition program. Nrf2 gene deletion significantly decreased cardiac transition efficiency. In the later stages, pseudotime analysis of scRNA-seq from LIF-stimulated ADCs depicted a finely resolved cascade of myocyte cell progression and identified a series of transcriptional events throughout the cardiac transition process. We identified 2 separate pathways in the transition routes associated with either cardiomyogenic fate or intermediately activated myofibrogenic cell fate. Taken together, our findings provide strategies to identify new autologous cell sources and cell signals in cardiac regeneration.
Material and Methods
For Key Resources, see Supplementary Table S1.
Contacts for Reagent and Resource Sharing
Requests for further information, resources, and reagents should be directed to corresponding authors Xiuju Wu (XiujuWu@mednet.ucla.edu) and Kristina I. Boström (kbostrom@mednet.ucla.edu). Sharing of primary samples is based on availability, and may be subject to Material Transfer Agreements and will require appropriate research ethics board certifications.
Animals
Adipoq Cre-Ert2 (BL/6-Tg(Adipoq-cre/ERT2)1Soff/J), RosatdTomato (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J), NKX2.5-eGFP (Tg(Nkx2-5-EGFP)3Sho/SmwuJ), and wild-type mice on a C57BL/6J background were obtained from Jackson Laboratory. AdipoqCre-Ert2RosatdTomatoNKX2.5-eGFP were generated and injected intraperitoneally with 100 µL of tamoxifen (20 mg/mL) for 5 consecutive days to trigger Cre-mediated recombination and permanent labeling of Adipoq+ cells. Genotypes were confirmed via polymerase chain reaction (PCR),42 and experiments were performed at 8-10 weeks. All mice were exposed to a standard 12:12 light/dark cycle and fed a standard chow diet (Diet 8604; HarlanTeklad Laboratory). The animal studies were reviewed and approved by the Institutional Review Board and conducted according to the animal care guidelines of the University of California, Los Angeles, USA. This investigation also conformed to the National Research Council’s, Guide for the Care and Use of Laboratory Animals.43
Myocardial infarction was induced by ligation of the left anterior descending (LAD) artery in wild-type C57BL6/J male mice at 12 weeks of age, as described previously.44,45 Either green fluorescent protein-expressing (GFP+) ADC-derived cardiomyocyte-like cells or phosphate-buffered saline (PBS) were injected into the infarct zone immediately after LAD artery ligation. Sham controls underwent the same surgery but without LAD artery ligation. Three weeks after the surgery, myocardial function was evaluated by echocardiography.
Cell Isolation and Culture
Isolation of adipocytes and ADCs was carried out by modifying the aforementioned method described previously.46 Briefly, approximately 1 g of fat tissue was minced and digested in 0.1% (w/v) collagenase II solution (Sigma C6885) at 37 °C for 1 h with gentle agitation. After filtration and centrifugation at 1000 rpm for 5 minutes, the floating top layer comprising adipocytes and ADCs was washed twice and seeded onto a 40 µm cell strainer (Corning) on a bottom well pre-coated with 0.1% gelatin for 30 minutes. Differentiation was induced using complete media containing Dulbecco’s modified eagle medium (DMEM) supplemented with 20% FBS, 1% GlutaMAX, 1% sodium pyruvate, 1% MEM nonessential amino acids solution, 0.1% 2-mercaptoethanol (Thermo Fisher Scientific), and 1000 U/mL LIF (Millipore ESG1107). The cell strainer with remains of adipocytes was removed after 5 days. The medium was changed every 4 days on the cells that were attached to the bottom until they were used for experiments. Collagenase-digested adipose tissues were centrifuged at 1000 rpm for 5 minutes to isolate adipose stromal cells (ASCs). The pellet was washed with phosphate-buffered saline (PBS) and resuspended in a complete medium. Cells (5 × 106) were incubated at 37 °C in 5% CO2 in 25-cm2 culture flasks, and the medium was changed every 4 days until they reached confluence. After splitting, the cells were used for experiments. Leukemia inhibitory factor signaling inhibition was achieved by adding 2 μg/mL LIF receptor (LIFR)-neutralizing antibodies (R&D Systems) or compounds (AG490 10 μΜ, UO126 10 μΜ, LY294002 5 µM) every other day starting on day 0. Additionally, 1 μg/mL of the isotype IgG or 5 mM dimethyl sulfoxide (DMSO) (Sigma-Aldrich) was added to the media as controls.
Fluorescence-Activated Cell Sorting
Cells were detached from the culture dish with 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA), centrifuged at low speed, and stained with fluorescence conjugated antibodies. Nonspecific fluorochrome- and isotype-matched IgGs (BD Pharmingen) served as controls. Flow cytometer gates were set using unstained cells and the isotype-matched controls. Cells were gated by forward scatter versus side scatter to eliminate debris. Hochest33342, which emits blue fluorescence when bound to dsDNA, was used as a live stain. A minimum of 10 000 events were counted for each analysis. All fluorescence-activated cell sorting (FACS) analyses were performed with a flow cytometer (LSR II; BD Biosciences). Fluorescence-activated cell sorting files were then exported and analyzed using FlowJoTMv10.7 software.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and processed as described previously.47 For immunofluorescence, the cells were permeabilized, blocked, and stained with α-actinin (Sigma, 1:500), cTnT (Thermo Fisher, 1:200), cTNI (Abcam 1:400), and Gja1 (Millipore 1:200) at 4 °C overnight. The mouse paraffine sections were dewaxed and rehydrated, and antigens were retrieved using an unmasking solution (Vector Laboratories). Sections were then incubated with primary antibodies Pdgfrα (R&D Systems, 1:400), Perilipin (Millipore, 1:500), CD31 (R&D Systems, 1:250), CD34 (Abcam, 1:250), CD44 (R&D System, 1:100), scavenger receptor class A (Scara) (R&D System, 1:100), CD142 (Sino Biological, 1:50), GFP (Abcam, 1:200), and α-actinin (Sigma, 1:200) at 4 °C overnight. Alexa Fluor–conjugated secondary antibodies (Invitrogen) were applied to cells and costained with 4ʹ,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Staining without primary antibodies served as a control. Images were taken with an inverted Nikon Eclipse Ti–S microscope (Nikon Corporation, Tokyo, Japan) and analyzed using ImageJ software.
Quantitative Reverse Transcription PCR
Total RNA was extracted from cells using the RNeasy mini kit (Qiagen). cDNA was produced with a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Relative quantitative (q)PCR was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) using TaqMan Universal PCR Master Mix (Thermo Fisher Scientific). Cycle conditions were one cycle at 50 °C for 2 minutes, one cycle at 95 °C for 10 minutes, and then 40 cycles at 95 °C for 15 seconds and 60 °C for 1 minute. Threshold cycles of specific cDNAs were normalized to the housekeeping gene Gapdh and translated to relative values. Primers are listed in Supplementary Table S1.
Contractility Assay
Contractility was assayed using the xCELLigence RTCA Cardio System with 5% CO2 at 37 °C. The Cardio System measures the impedance of Cor.At cardiomyocytes and transforms the values to cell index (CI) values, which reflect changes in beating activity. Prior to recording, cells were left to equilibrate for 15 minutes at the given setting. Video imaging of beating ADC-derived cardiomyocyte-like cell monolayers was recorded from 2 to 3 separate locations in each well. The average CI values for each recording period (20 seconds) were displayed.
siRNA Transfection
siRNA was purchased from Qiagen and transfected using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer’s instructions. For knockdown experiments, siRNAs were added on the day of plating. qRT-PCR was performed on D3 to evaluate the knockdown efficiency, using the primers listed in Supplementary Table S1.
10x Library Preparation, Sequencing, and Alignment
Single-cell RNA-sequencing libraries were generated with the Chromium Single Cell 3ʹ v3 assay (10x Genomics). Libraries were sequenced using the NovaSeq 6000 platform (Illumina) to a depth of approximately 300 million reads per library with a 2 × 50 read length. Raw reads were aligned to the mouse genome (mm10) and cells were called using cellranger count (v3.1.0). Individual samples were then aggregated to generate the merged digital expression matrix using cellranger aggr (v3.1.0).
Cell Clustering and Cell-Type Annotation
The R package Seurat (v3.1.2) was used to cluster the cells in the merged matrix. First, cells with less than 300 transcripts or 100 genes detected were first filtered out as low-quality cells. Genes expressed in at least one cell across the datasets were included to involve as many genes as possible. Afterward, gene counts for each cell were divided by the total gene counts for the cell and multiplied by a scale factor of 10 000, and natural-log transformation was then applied to the counts. The FindVariableFeatures function was used to select variable genes with default parameters, whereas the ScaleData function was used to scale and center the counts in the dataset. Principal component analysis (PCA) was performed using the RunPCA function on the variable genes, and the first 20 PCs were used for cell clustering (resolution = 0.5) with the FindNeighbors and FindClusters functions. Uniform manifold approximation and projection (UMAP) dimensional reduction was applied using the RunUMAP function. Cluster markers or time point genes were found using the FindAllMarkers function. The average expression for each gene was calculated for each cluster, upon which hierarchical clustering was then applied. Nuclear factor erythroid 2-related factor 2 (Nfr2) module scores were calculated by aggregating the genes’ expressions in the Nfr2 pathway using the AddModuleScore function with default parameters. To uncover the genes correlated with the Nfr2 pathway, a correlation coefficient was calculated between the module scores and the expression of every gene in the datasets. Genes with a correlation coefficient over 0.2 or less than −0.2 were plotted in the heatmap.
Pseudotime Trajectory Construction
Pseudotime trajectories were constructed using the R package Monocle (v2.10.1). The raw counts for cells were extracted and normalized by the estimateSizeFactors and estimateDispersions functions with the default parameters. Genes with average expression greater than 0.5 and detected in at least 10 cells were retained for further analysis. Variable genes were determined by the differentialGeneTest function with a model against the time point identities. The orders were determined by the orderCells function, and the trajectory was constructed by the reduceDimension function with default parameters. Differential expression in pseudotime was carried out using the differentialGeneTest function with a model against pseudotime, in which a likelihood ratio test was applied to get a P-value for each gene. The Benjamini-Hochberg procedure was then used to adjust for multiple testing to obtain a q value. A filter of q value of <0.05 was used to identify the genes that were significantly differentially expressed along the pseudotime, which were then plotted on the heatmap, and were grouped into 6 clusters using the function plot_pseudotime_heatmap. Differential expression analyses between states were carried out using the Seurat function FindMarkers. Differentially expressed genes (DEGs) with the fold changes were used to plot the heatmap showing the bifurcation expression patterns. The bifurcation heatmap was generated using the function plot_genes_branched_heatmap.
RNA Velocity Analysis
To estimate the RNA velocities of single cells, velocyto was used to distinguish unspliced and spliced mRNAs in each sample. The Python package scVelo was then used to recover the directed dynamic information by leveraging the splicing information. Specifically, the data were first normalized using the filter_and_the normalize function. The first- and second-order moments were computed for velocity estimation using the moments function. The velocity vectors were obtained using the velocity function and were then projected into a lower-dimensional embedding using the velocity graph function. Finally, the velocities were visualized in the pseudotime trajectory embedding using the velocity_embedding_stream function. All scVelo functions were used with default parameters.
Data Availability
Single-cell RNA sequencing datasets are accessible from GEO with accession number GSE164410 (Reviewer access token: ixonwocufzaflot).
Statistics
Statistical analyses were performed using GraphPad InStat (version 3.0; GraphPad Software Inc.). Data were analyzed by either unpaired 2-tailed Student’s t-test, 1-way, or 2-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test for statistical significance. Data are represented as mean ± SD. P values of <.05 were considered significant, and all experiments were repeated in triplicate.
Results
Establishment of a Highly Efficient LIF-Induced ADC-Cardiac Transition System
Our previous studies found that adipose cells in a hanging culture system contained cells with a cardiac transition potential,19,45 but the exact cell population contributing to the cardiac lineage was unclear. Therefore, we took advantage of the cardiomyocyte lineage transcription factor Nkx2.5 to determine whether adipocytes in the floating layer of adipose cells could generate Nkx2.5-positive cells. We developed an adiponectin (Adipoq) CreERT2-tdTomatofl/flNkx2.5-Egfp mouse model to trace the adipocytes in ADC-to-cardiac transition. Adipocytes in this transgenic mouse model were marked with tomato red fluorescent protein (tdTomato) after tamoxifen injection, whereas the Nkx2.5-positive cardiomyocytes were tagged with GFP (Fig. 1A). After ascertaining the specificity of Adipoq CreERT2-tdTomatofl/fl in adipocytes (Supplementary Fig. S1), cells floating after centrifugation contained tdTomato-positive adipocytes and other small-sized ADCs (Figs. 1A and Supplementary Fig. S2). These floating cells were collected and treated with LIF in a hanging culture system (Figs. 1A and Supplementary Fig. S1). Two weeks after LIF treatment, over 50% of the ADCs from the hanging cultures were Nkx2.5-Egfp-positive but tdTomato-negative (Fig. 1B); thus, the adipocytes were excluded as the cells of origin in this cardiac transition. Second, FACS analysis with polystyrene particles identified ADCs ranging from 2 to 16.5 µm in the floating viable adipose cells (Supplementary Fig. S2), which responded to LIF and generated beating cardiomyocyte-like cells. These results suggest that ADCs other than adipocytes respond to LIF and transition to cardiomyocyte-like cells.
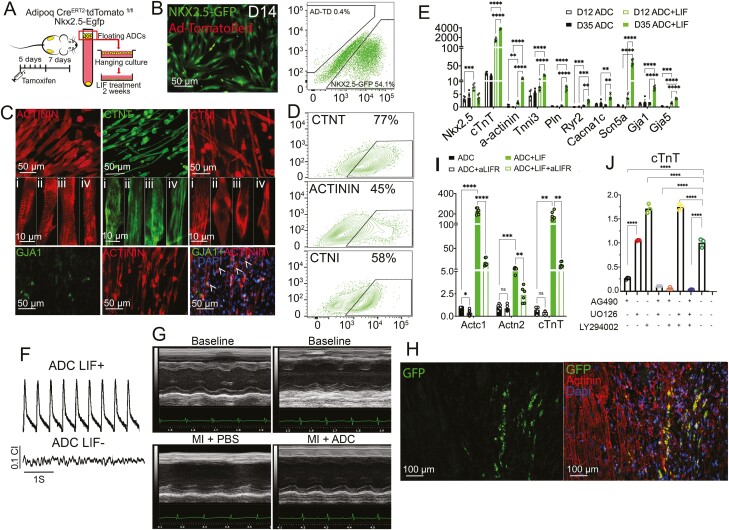
Figure 1.
LIF Induces ADC-Cardiac Transition. (A) Schematic of experimental design for collecting floating ADCs from Adipoq CreERT2-tdTomatofl/flNkx2.5-Egfp mouse (see details in Methods). (B) Immunofluorescence and flow cytometry of AD-TD (Adipoq CreERT2-tdTomato)+ and Nkx2.5-GFP+ cells 14 days (D14) and 6 days, respectively, post-LIF treatment of ADCs. (C,D) Representative immunofluorescence (C) and flow cytometry (D) of ADC-derived cardiomyocyte-like cells for α-ACTININ (sarcomere marker, red), CTNT (cardiac troponin T, green), CTNI (cardiac troponin I, red), and GJA1 (Gap junction α-1 protein, green) after LIF treatment for 21 days. Arrowheads indicate GJA1 and α-ACTININ double-positive cells. (E) Expression of cardiac markers in ADCs at day (D)12 and D35 with or without LIF treatment, as shown via real-time PCR. Gene expression is calculated as fold change compared with the ADCs on day 0 (n=6). (F) Representative traces of spontaneous beating activity of monolayer ADC-derived cardiomyocyte-like cells with or with our LIF recorded with impedance system. (G) Functional assessment of before (baseline) and 21 days after MI and injection of FBS or ADC-cardiomyocyte-like cells. See Supplementary Figure S4 for additional results. (H) Dual immunofluorescence detection of GFP+/ α-ACTININ ADC-derived cardiomyocyte-like cells at 21 days after MI at the mid-infarct level. Arrowheads show GFP and α-ACTININ double-positive cells. (I) Expression of Actc1, Actn2, and cTnT in ADCs at D10 after blocking LIFR using anti-LIFR neutralizing antibody, as shown via real-time PCR. Gene expression is calculated as fold change compared with the ADCs without treatment (n=6). (J) Expression level of cTnT in ADCs at day (D)10 after blocking of LIF signaling after exposure to vehicles AG490, UO126, and LY294002, examined using real-time PCR. Gene expression is calculated as fold change compared with the ADCs without treatment (n=3). In (E), (I), and (J), data are shown by mean ± SEM. Two way ANOVA, *P < .05, **P < .01, ***P < .001, ****P < .0001.
Leukemia inhibitory factor-treated ADCs exhibited a robust cardiac differentiation potential and, within 4 weeks, were positive for cardiac troponin T, troponin I, cardiac α-actinin, and the gap junction marker Gja1 (Fig. 1C and 1D). Moreover, LIF-treated ADCs expressed a higher level of mature cardiac markers, including cardiac sarcomeres, the myofilament markers α-actinin and Tnni3, and the gap junction proteins Gja1 and Gja5 (Fig. 1E). Furthermore, mature cardiac markers related to the calcium ion channels (Pln, Ryr2, and Cacn2) and the sodium ion channel (Scn5a) showed a higher expression in LIF-treated ADCs than in untreated ADCs (Fig. 1E). Other lineage markers, including pluripotency, adipose, endothelial, and neuronal markers, were not expressed in the LIF-treated ADCs along the differentiation course (Supplementary Fig. S3), suggesting that LIF-induced adipose-cardiac differentiation bypassed dedifferentiation and transdifferentiation. Spontaneous beating activity of LIF-treated ADCs was observed and recorded by the xCELLigence system RTCA cardio instrument (Supplementary Video 1, Fig. 1F). Leukemia inhibitory factor-treated ADCs were expanded with their subsequent incorporation into cardiomyocyte-like cells, which had a positive effect on cardiac function in hearts infarcted by permanent ligation of the left ascending coronary artery (Figs. 1G and Supplementary Fig. S4). In a limited study, significant increases were also observed after injury in the ejection fraction (from 27.7% to 41.4%) and fractional shortening (from 12.0% to 19.2%) after injection of PBS versus ADCs, respectively (Supplementary Fig. S4). Additionally, there was a significant decrease in the diastolic left ventricular (LV) volume after ADC injection (Supplementary Fig. S4). Green fluorescent protein-positive LIF-treated ADCs were detected in the host myocardium after injury, as visualized by costaining of GFP and the myocyte marker α-actinin (Figs. 1H and Supplementary Fig. S5). Approximately 30% of the GFP-positive cells showed some degree of α-actinin staining (Supplementary Fig. S5). These results suggest that the injection of LIF-treated ADCs may support cardiac function following infarction.
Next, we aimed to elucidate the pathway determining LIF-induced cardiac transition. Leukemia inhibitory factor induces dimerization of glycoprotein 130 (gp130) and its receptor (LIFR), which in turn phosphorylates and activates the STAT3, ERK, or PI3K signaling pathway.48 Leukemia inhibitory factor administration at a concentration of 10 ng/mL from days 0 to 4 triggered the cardiac transition program (Supplementary Fig. S6), whereas blocking of the LIFR using neutralizing antibodies reduced the expression of the cardiac markers (Fig. 1I). Regarding downstream pathways, LIF induced STAT3 phosphorylation within 30 minutes, which peaked at 1 h and decreased after 2 h, whereas AKT2 and MAPK phosphorylation increased after 1 h of treatment (Supplementary Fig. S7). Using chemical inhibition of the downstream effectors JAK2, MAPK, and AKT2, we found that the cardiac transition was mainly triggered by JAK2/STAT3 but not by MAPK or AKT2 (Figs. 1J and Supplementary Fig. S7). Collectively, these results demonstrated that the administration of LIF to ADCs substantially facilitates access to the cardiac transition, offering a valuable platform for further dissection.
Single-Cell RNA Sequencing Analysis of LIF-induced ADC-Cardiac Transition
To molecularly characterize the LIF-induced cardiac transition, we performed scRNA-seq navigating stage-specific ADCs along the transition using the Chromium system (10x Genomics). Floating ADCs from 8 mice were collected and treated with LIF for 0, 1, 3, 7, and 10 days. Day 0 was used as the point of comparison since attachment and early differentiation already occur within the first 24 h. We then carried out a scRNA-seq analysis on the cells (Fig. 2A). We detected 23 543 unique genes from a total of 39 432 cells, with an average of 2899 genes and 13 530 unique molecular identifiers (UMIs) per cell. Unsupervised uniform manifold approximation and projection (UMAP) algorithm clustering49,50 identified 25 cell clusters attributing to their putative identities (Fig. 2B) and hierarchical similarities based on expression patterns (Fig. 2C). We analyzed the cluster gene expression at each stage to define the ADC clusters primed toward cardiac differentiation. At the initiation stage (day 0), most cells were adipocytes with enriched adipose markers, including Apoe, Adipoq, and Fabp4 (Fig. 2D, 2F). Other cell types at this stage included adipose progenitor cells (C23: Pdgfrα+Icam1+CD34+), endothelial cells (C19: Pecam1+Cdh5+Kdr+), and contaminating immune cells (Fig. 2C). At the activation stage, fibroblast activation was marked by Pdgfrα, ly6a, and Thy1 expression at varying proportions and levels (Fig. 2C and Supplementary Fig. S8). We also found Prrx1 enrichment at this stage, which is expressed in mesenchymal cells in bone marrow with multilineage differentiation potential,51 suggesting that the ADCs were activated for differentiation (Figs. 2C, 2F and Supplementary Fig. S8). Interestingly, we found 2 EC clusters that transiently appeared at this stage, in which cluster 8 expressed the venous markers Sox17 and vWf and cluster 22 expressed the arterial marker Notch152 (Fig. 2A-2C and Supplementary Fig. S8). However, ECs were only transiently observed between days 1 and 3 and disappeared after day 7. At the myogenic stage, most cells exhibited a lower expression of stem cell markers and began expressing cardiomyogenic and myofibroblast markers, including cTnT, Tnni, Acta2, Postn, and Cthrc1 (Figs. 2C, 2F and Supplementary Fig. S8).
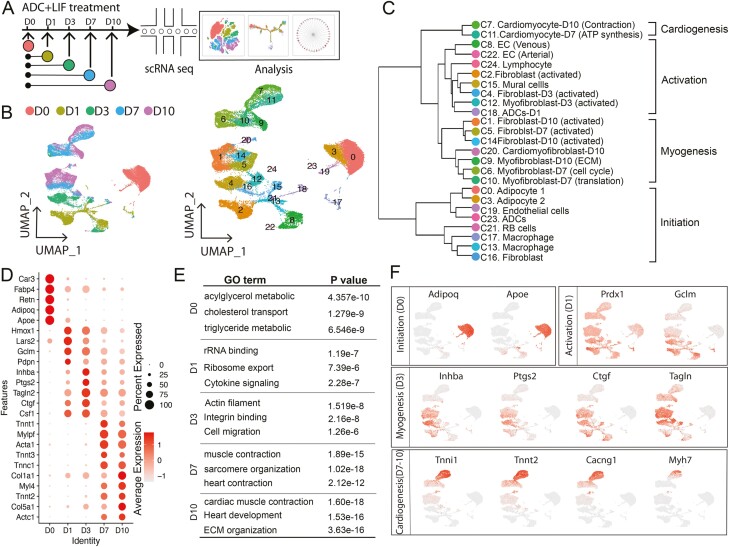
Figure 2.
High-resolution dissection of ADC-cardiac transition using scRNA-Seq. (A) Schematic diagram of scRNA-seq analysis strategy during ADC-cardiac transition. Overall, 10 545, 6460, 5087, 7673, and 9667 cells were analyzed for ADC day (D)0, ADC D1, ADC D3, ADC D7, and ADC D10 (indicated by different colors), respectively. The flow chart of the scRNA-seq analysis was adopted from 10x Genomics. (B) UMAP projection of all 39 432 individual cells during the ADC-cardiac transition process, colored by origin (left) and each subpopulation based on the transcriptomic phenotype (right). (C) Hierarchal clustering analysis of all clusters from scRNA-seq. (D) Expression of top genes at the indicated time points. (E) Gene ontology (GO) analysis of genes specifically expressed in the indicated time points. (F) UMAP projection of cells and typical gene expression at the indicated time points.
We then analyzed the scRNA-seq results from each day to dissect the cardiac transition in a time-specific manner. The LIF-induced myogenic program was activated from day 1 (Fig. 1F) with the expression of several cardiac protective genes, including Prdx1 and Gclm53-55 (Fig. 2D-2F). From day 3, the myogenic program was activated and maintained at a high level to the end of the transition stage (Fig. 2D-2F). A unique cell cluster identified on day 3 marked extracellular matrix integrin binding family and Inhba/activin, which are reportedly required for ADC differentiation56-58 and cardiac morphology.59,60 Moreover, the cardioprotective genes Hmox1, Ptgs2/Cox2, and Ctgf were activated at this stage (Fig. 2D-F), previously shown to be induced by STAT3 canonical signaling in acute ischemic stress.61 These results suggest that LIF-induced ADC-cardiac transition might have short-term effects by activating ECM and cardioprotective genes. From days 7 and 10, LIF-treated ADCs began expressing Actc1, cTnT, Tnni1, and other cardiac structural and regulatory genes. Cluster 7 from day 10 expressed the cardiomyocyte markers Myh7, calcium voltage-gated channel Atp2a2, and Cacng2, suggesting that the cells at this stage exhibit more mature cardiomyocyte characteristics. Cells at this stage were also enriched in genes involved in extracellular matrix deposition and organization, a finding supported by the identification of the myofibroblast markers col3a1 and Des (Fig. 2D-2F). The coexistence of different populations is common in cardiac differentiation, supporting the heterogeneity of cell fate transitions.40,62,63 Taken together, our data depicted a model timeline of stem cell activation and maturation in LIF-induced ADC-cardiac destiny.
Cellular Composition of Initial Floating Adipose Cells
To detect the ADC population that responds to LIF and transition to cardiomyocyte-like cells, we examined the cellular composition of the initial floating adipose cells from day 0 and identified 5 subpopulations (cluster C0, C3, C17, C19, and C23) (Fig. 3A). We excluded adipocytes, endothelial cells, and neuronal cells since cardiomyocyte-like cells are not derived from these populations (Figs. 1B, 2F, 3A, Supplementary Figs. S1, and S3). Importantly, we found that C23 expressed the canonical mesenchymal progenitor markers Pdgfrα, Icam1, and Thy-1, suggesting stem cell potential (Fig. 3A and Supplementary Fig. S9). We compared 2882 mouse cell surface markers in C23, C3, and C0 to identify specific cell surface markers for C23. We then selected 13 cell surface makers that were only expressed in C23 were selected, including the mesenchymal markers Pdgfrα, CD34, CD55, Dpp4, Pi16, and Scara5, and the fibroblast markers Col1a1, Col3a1, Col6a1, and Fbn1 (Fig. 3A and Supplementary Fig. S9). The enrichment of stem cell markers suggests that C23 may be heterogeneous and comprise molecular and functional divergent subpopulations. Additionally, we found that Pdgfra and Il6st (gp130) were robustly expressed in the ADC cluster (cluster 23) (Supplementary Fig. S10) and LIFR (Cd118) at a lower level. Immunofluorescence confirmed the presence of Pdgfra+LIFR+ cells scattered in WAT (Supplementary Fig. S10).
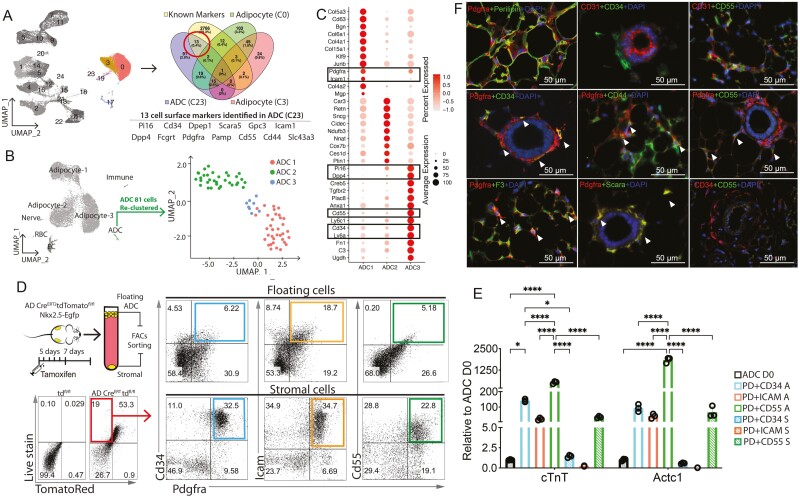
Figure 3.
Identification of LIF responsive ADCs from AD-D0. (A) UMAP (colored) highlighted 10,545 ADC cells from day (D)0 (left) were analyzed to identify cell surface markers. Venn diagram indicating the number of differentially (FDR <0.01) expressed genes across ADC cluster C23, adipocyte cluster C0, adipocyte cluster C0, and published cell surface makers and the overlap between each set of genes. A total of 13 markers were identified as ADC C23-specific expressed cell surface markers. (B) UMAP from AD-D0 highlighting the ADC cluster and reclustered at right. (C) Expression of top genes in the indicated ADC clusters. (D) AD-TD negative and Hoechst 33342-positive populations from floating ADCs and stromal cells were further analyzed in Pdgfrα+CD34+, Pdgfrα +Icam1+, and Pdgfrα+CD55+ populations. (E) Expression of cardiac markers in FACS sorted Pdgfrα+CD34+, Pdgfrα +Icam1+, and Pdgfrα+CD55+ floating ADCs (A) and stromal cells (S) 28 days post-LIF treatment, as shown via real-time PCR (n = 3). Data are shown by mean ± SEM. Two-way ANOVA, *P < .05, ***P < .001, ****P < .0001. (F) Immunostaining of identified cell surface markers in mouse subcutaneous fat tissue. Arrowheads indicate double-positive cells. Scale bars = 50 μm.
To examine the cellular composition of C23, cells were reclustered into 3 groups: ADC1, ADC2, and ADC3 (Fig. 3B). ADC1 (red) expressed fibroblast markers (Col5a3, Col6a1, and Col4a1) and progenitor cell markers (Pdgfrα and Icam1), whereas ADC3 (blue) expressed the canonical mesenchymal progenitor markers CD34, Pdgfrα, Ly6a, CD55, Pi16, and Dpp4, suggesting a stem cell population. ADC2 (green) expressed Car3, along with several adipocyte genes, including Plin1, Retn, and Nnat, suggesting that these cells could be preadipocytes (Fig. 3C). Our findings are consistent with recent scRNA-seq findings of adipose progenitor cell heterogeneity in mouse subcutaneous and visceral WAT. Progenitor subpopulations exhibit unique molecular signatures, metabolic conditions, and hierarchical differentiation potentials.27,28,64 The Dpp4+CD55 adipose stromal cells identified in WAT exhibited significant stem cell-like properties and differentiated into CD142+ adipogenesis regulate cells (Aregs) and preadipocytes.27,28,64-67 C23 contains both Dpp4+CD55+ ASCs and Icam1+ preadipocytes in the upper layer of the adipose cells, suggesting different potentials for either multipotency or adipogenesis. A reanalysis of the data set from Merrick et al64 also revealed clusters of progenitor cells that expressed Pdgfra and the LIF receptors such as Il6st (Gp130) (Supplementary Fig. S11).
Besides adipose tissue, Pdgfrα+Dpp4+CD55+CD34+ cells were recently identified in muscles, where they are referred to as interstitial stromal cells (ISCs) and exhibit multipotency in line with those reported for subcutaneous ASCs.67 The ISC subpopulations showed distinct differentiation potential depending on the CD55 expression level,67 pointing to the possibility that multipotent CD55+ may possess cardiomyogenic potential. We isolated Pdgfrα+CD34+, Pdgfrα +Icam1+, and Pdgfrα+CD55+ cells from both floating ADCs and WAT stromal cells to assess the cardiomyogenic potential of each cluster (Fig. 3D). The sorted cells were cultured with LIF for 28 days, and the expression of cardiac markers was monitored. Compared with unsorted ADCs (ADC D0), all the Pdgfrα+CD34+, Pgfra+Icam1+, and Pdgfrα+CD55+ ADCs from the floating layer expressed cTnT and Actc1, from which the Pdgfrα+CD55+ ADCs exhibited the highest expression level (Fig. 3E). Interestingly, although sorted with the same markers, the adipose stromal subpopulations exhibited a much lower expression of cardiac markers than their floating cell counterparts. Again, the Pdgfrα+CD55+ stromal population had the highest expression of cardiac markers compared with the other 2 stromal populations (Fig. 3E), suggesting that this population may react to LIF and possess cardiomyogenic potential.
Given the divergent stemness in Pdgfrα+ populations, it is important to determine the distribution of subclusters in vivo. Perivascular Pdgfrα+ ASCs have been shown to house multipotent stem cells with a tendency to participate in osteogenesis,68 fibrosis,69,70 and adipogenesis.71 We checked the anatomic relation of the stromal cells in inguinal WAT with immunofluorescence. Pdgfrα+ cells were located between mature perilipin+ adipocytes or around CD31+ small vessels and distributed throughout the inguinal WAT (Fig. 3F). Pdgfrα+CD34+ double-positive cells were also found around the vessels, mainly in the adventitia, in which Pdgfrα+CD44+ and Pdgfrα+Scara+ cell populations also resided (Fig. 3F). This finding is consistent with a previous report on multipotent ASCs.68 However, Pdgfrα+CD55+-positive cells were not among the stromal cells surrounding the vessels, suggesting that myogenic ADCs are not from the multipotent adventitial populations. CD55, a 70-kDa membrane-bound C regulator that accelerates the decay of the C3 convertase,72 was expressed on red blood cells and ADCs spread around the adipocytes (Fig. 3F). Recent findings from a scRNA-seq indicate that reticular interstitium ASCs (Dpp4+CD55+) give rise to both preadipocytes (Icam1+) and adventitial adipose regulatory cells (CD142+).64 Our results indicated that this population surrounds the adipocytes in the WAT and reacts to LIF to exhibit myogenic potential.
Pseudotime Analysis Reveals ADC-Cardiac Transition Trajectories
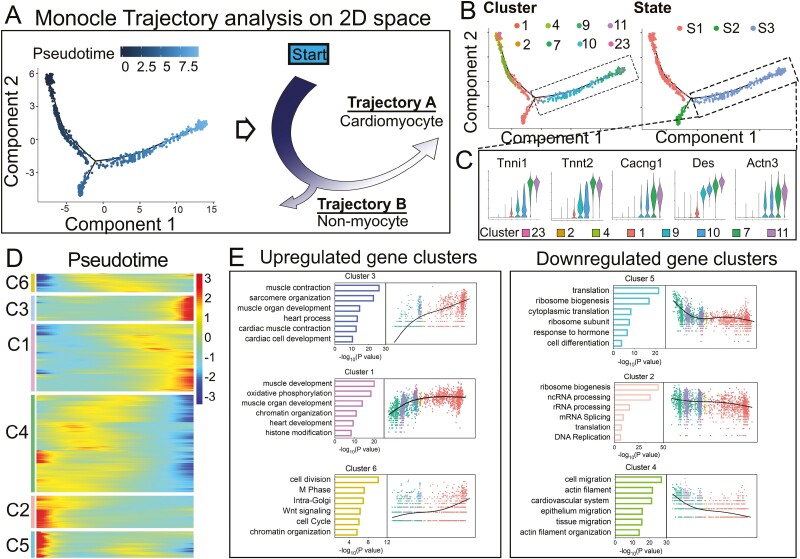
To elucidate the progression from ADCs to cardiomyocyte-like cells, we performed pseudotime analysis using R Package Monocle 2. We excluded clusters with a high cell cycle score (C4, C5, C6, C14, C20, and C24) (Supplementary Fig. S12), which is known to strongly affect the transcriptional profiles.73,74 Adipocyte clusters and contaminating cell clusters (endothelial cells, neuronal cells, and immune cell clusters) were also excluded. Monocle 2 was used to construct a trajectory containing a starting point and 2 branches (Fig. 4A-C). The starting point of the trajectory consisted of C23 from day 0 and C2 from day 1 (Figs. 2B and 4B), suggesting C2 as an initiation point in ADC-cardiac transition(state 1, S1). Along the transition course, a progression from the starting points (C2 and C23) toward each cluster was observed, with the first branch giving rise to nonmyocytes (state 2, S2), and the second branch giving rise to the cardiomyocyte-like cells with an expression of the cardiomyogenic markers, including Tnni1, cTnT, Cacng1, Des and Actn3 (state 3, S3, Fig. 4A-C). Thus, the cell cluster pseudotime reflected the cardiac transition course in the LIF-responsive populations.
Figure 4.
Reconstruction of ADC-cardiac transition trajectory in a pseudotime manner. (A) The arrangement of cells on the tree (left) shows that cells on the left side of the tree (dark blue) are less differentiated than those on the right side (light blue). Based on the pseudotime, the schematic diagram (right) indicates the transition direction. (B) Cells on the trajectory tree are colored by cluster assignment and state. (C) Violin plot displaying the expression of representative cardiac markers in state 3. (D) Heatmap to display differentially expressed gene (DEG) clusters along the pseudotime trajectory. (E) GO analysis and signature gene expression dynamics in each cell cluster.
We analyzed 7615 DEGs and observed 6 gene clusters with variations across the trend to obtain an insight into the gene regulation dynamics along the trajectory. Among the upregulated gene clusters, cluster 3 was sharply upregulated in cardiomyogenic processes, whereas cluster 1 showed a gradual upregulation of muscle gene expression, suggesting temporal waves of cell identity transitions. Interestingly, cluster 6 with a smoothly increasing trend suggested a stem cell gene expression mode in cell division, cell cycle, and chromatin organization (Fig. 4D, 4E). For the downregulated gene clusters, clusters 2 and 5 were enriched in rRNA processing, mRNA splicing, ribosome biogenesis, and translation complex formation, suggesting that translational control regulates the ADC transition. These proved to be crucial for accommodating proteome remodeling in response to changes in ADC fate (Fig. 4D, 4E). Collectively, these data depicted the trajectory of the ADC-cardiac transition and revealed the ordered activation of transcriptional waves throughout the process.
Nrf2 Signaling Is Required to Initiate ADC-Cardiac Transition
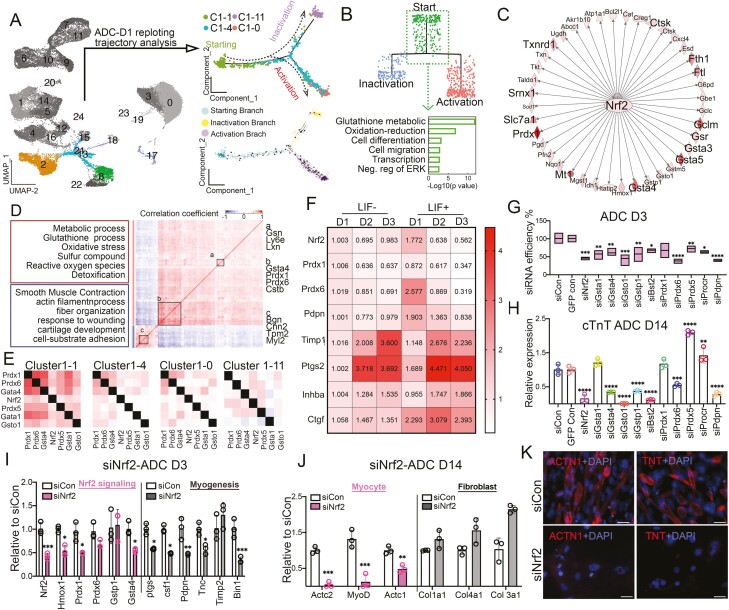
To elucidate the crucial events of the initiation stage (S1), we combined Monocle 2 and RNA velocity to analyze the day 1 ADC trajectory75,76 (Fig. 5A). The combined trajectory analysis identified cluster 1 from day 1 (C1-1) as the starting point of the LIF-induced cardiac transition, which later transitioned to either myofibroblasts (C1-11) or cardiomyocyte-like cells (C1-0) (Fig. 5A and Supplementary Fig. S12). Surprisingly, the predominant gene expression in starting cluster C1-1 was enriched in genes involved in glutathione metabostasis and oxidation-reduction (Figs. 4A and 5A, 5B). Traditionally, glutathione metabolism-dependent generation of reactive oxygen species (ROS) and its associated oxidative damage have been linked to impaired homeostasis and cellular death.77 Beyond the adverse effects of ROS accumulation, increasing evidence implicates redox status as a regulator of vital cellular processes in stem cells.78 However, how ROS regulates myogenesis from adipose progenitor cells is largely unknown.
Figure 5.
Identification of Nrf2 signaling pathway in initiation of ADC-cardiac transition fate triggered by LIF. (A) UMAP (colored) highlighted 6460 ADC cells from day (D)1 (left) were replotted and further analyzed ADC populations by Monocle 2 and RNA velocity. Arrows in RNA velocity (lower right) indicated the direction of the average velocity at a grid of points along the trajectory. (B) GO analysis (lower) of starting branch identified by dendrogram plots from the hierarchical clustering (upper). (C) Gene correlation network in cardiac transition stage identified Nrf2 signaling predicted by ingenuity pathway analysis (IPA). (D) Heatmaps showing Pearson correlation coefficient and representative GO terms enriched in positively or negatively correlated target genes, whose expression significantly correlated (P < .05) to Nrf2. (E) Coexpression measured by redundant Jaccard index clustering of Nrf2 signaling genes in clusters C1-1, C1-4, C1-0, and C1-11. (F) Heatmap of Nrf2 signaling expression level along LIF treatment time course via qPCR. (G) ADCs were transfected with siRNAs (5 nM) at D0. Cells were lysed on day 3 and real-time RT-PCR was performed using the indicated TaqMan gene expression assays. Knockdown data are expressed relative to data from cells transfected with control siRNA (n = 3). (H) Expression of cTnT in ADCs at D14 with siRNAs, as shown by real-time PCR (n = 3). Gene expression is calculated as fold change compared with the siCON ADCs at D14. (I) Expression of Nrf2 signaling and myogenic markers in ADCs at D3 with siNrf2, as shown via real-time PCR (n = 3). Gene expression is calculated as fold change compared with the siCON ADCs at D3. (J) Expression of myocyte and fibroblast markers in ADCs at D14 with siNrf2, as shown by real-time PCR (n = 3). Gene expression is calculated as fold change compared with the siCON ADCs at D14. (K) Representative ICC staining of ADC-derived cardiomyocyte-like cells for ACTN1(red) and TRIT (red) in ADCs with or without siNrf2. Scale bar = 50 μm. In (G), (H), (I), and (J), data are shown as mean ± SEM. One-way ANOVA, *P < .05, **P < .01, ***P < .001, ****P < .0001.
To understand how ROS and oxidative stress regulate the ADC-cardiac transition, we analyzed day 1 scRNA-seq results through top-scoring ingenuity pathway analysis (IPA). The IPA analysis suggested that Nrf2 was the core factor of the canonical pathway (Fig. 5C), which was recently shown to influence stem cell integrity by regulating multiple downstream antioxidant and metabolic pathways.78 Furthermore, Spearman correlation analysis between Nrf2 expression and its downstream targets79,80 in C1-1 cells suggested that Nrf2 in C1-1 regulated the expression of phase II detoxifying enzymes including NADPH, NAD(P)H quinone oxidoreductase 1, glutathione peroxidase, ferritin, heme oxygenase-1 (HO-1), and antioxidant genes79,80 (Fig. 5D). Moreover, the Gene Ontology (GO) results and coexpression patterns indicated that Nrf2 initiated oxidative stress signaling and was negatively correlated to the muscle’s filament structure (Fig. 5D), suggesting an event before myofibroblast activation. When comparing Nrf2 targeted genes coexpression heatmap in the day 1-ADC clusters, Nrf2 was found to correlate with Prdx1, Prdx6, Gsta4, Gsta1, and Gsto1 in all 4 clusters of D1, whereas Prdx1 was associated with Prdx6, Gsta4, Prdx5, Gsta1, and Gsto1 in C1-1, but undetectable in C1-4, C1-0 and C1-11, supporting a transient expression mode (Fig. 5E).
More importantly, STAT3-activated Nrf2 signaling protects against stem cell depletion in response to stress and aging,81 indicating that activated Nrf2 signaling in ADCs may be related to LIF induction. We first examined the oxidative gene expression level along the LIF treatment to investigate this potential relationship. LIF induced a sudden increase in the expression of the known oxidative genes (Nrf2, Prdx1, Prdx6, and Pdpn), which gradually decreased during the transition. Conversely, the expression of myogenic genes (Timp1, Ptgs, Inhba, and Ctgf) was relatively low initially but increased in the later transition course (Fig. 5F). These data suggested a reverse association between LIF-STAT3-Nrf2 signaling and the myogenic process. Second, we screened a set of siRNA in ADCs that targeted 14 genes in the Nrf2 pathways. The silencing efficiency on day 3 remained high, allowing us to investigate Nrf2 signaling in ADC-cardiac transition due to its transient expression within 48 h of LIF treatment (Fig. 5F). We identified Nrf2, Gsta4, Gsto1, Bst2, and Pdpn as the top candidates for initiating the cardiac transition program (Fig. 5G and 5H). Notably, knockdown of Nrf2 in ADCs consistently and significantly decreased cardiac transition efficiency, suggesting Nrf2 is a pivotal factor in cardiac induction (Fig. 5H). Third, Nrf2 silencing decreased the expression of both downstream target genes (Hmox1, Prdx1, and Gsta4) and myogenic genes (Ptgs, Csf1, Pdpn, Tnc, and Bin1), as examined on day 3 (Fig. 5I). Fourteen days after Nrf2 silencing, myocyte markers including the cardiac markers cTnT and Actc1, the smooth muscle marker Actc2, and the skeletal muscle marker MyoD were significantly reduced when compared with those of the siRNA controls (Fig. 5J). Conversely, Nrf2 silencing did not affect fibroblast gene expression, supporting the involvement of Nrf2 in myogenesis (Fig. 5J). IHC staining of Actn1 and TnT on siNrf2-treated ADCs further confirmed that losing Nrf2 jeopardized the ADC-cardiac transition potential (Fig. 5K). Thus, these observations collectively suggested that Nrf2 is required to initiate the ADC-derived cardiac transition program.
Single-Cell Trajectory Branch Points Identify Key Molecular Determinants of ADC-Cardiac Transition
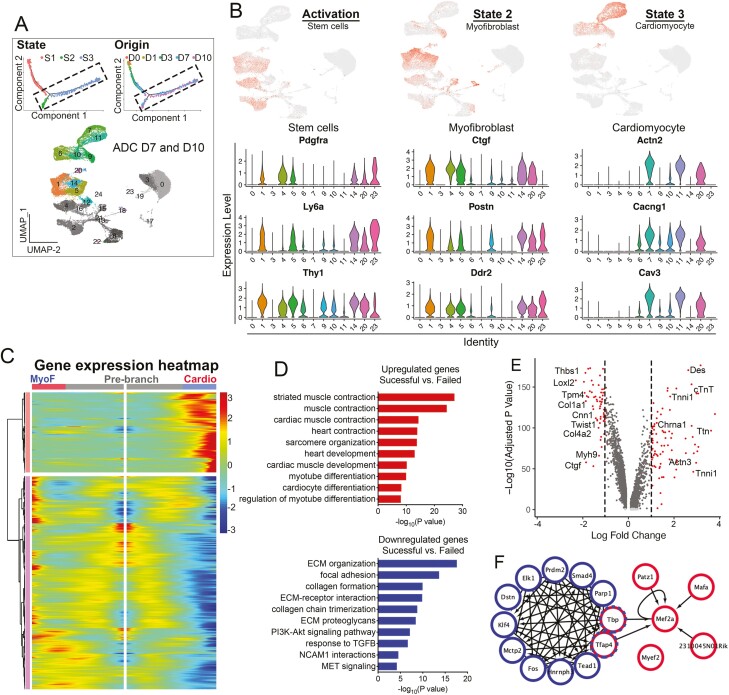
The ADC-cardiac trajectory contained 2 termini corresponding to 2 distinct outcomes: cardiomyocyte-like cells (S3) and myofibroblasts (S2), mainly from AD-day 7 and AD-day 10 (Figs. 4A-4C and 6A). S2 and S3 cells were more mature than cells at the activation stage (S1), as indicated by their low expression of stem cell markers Pdgfrα, Ly6a, and Thy1 (Fig. 6B). After 7 days, S2 cells began expressing activated myofibroblast markers Ctgf, Postn, and Ddr2 in C1, C5, C14, and C20, suggesting myofibroblast differentiation. For S3 cells, cardiac-specific genes Actn2, Cacng1, and Cav3 were expressed in clusters C6, C7, C9, C10, C11, and C20, forming the cardiac trajectory state (Fig. 6A, 6B).
Figure 6.
Molecular census of myofibroblast and cardiomyocyte stages. (A) Cells on the Monocle trajectory tree colored by state and time point origin (upper). UMAP (colored) highlighted 7673 ADC cells with D7 color, and 9667 ADC cells with D10 color (lower). (B) Expression of select genes in populations from different stages as visualized in UMAP (upper) and vln (lower) plots. (C) Gene expression heatmap of DEGs in a pseudotime-temporal order. Myofibroblast (MyoF) and cardiomyogenic (cardio) transition trajectories (including pre-branch) are shown on the left and right, respectively. (D) GO analysis of upregulated and downregulated genes comparing the successful (cardio) with failed (myofibroblast) branches. (E) Volcano plot displaying DEGs comparing stages 2 and 3. Red dots represent genes that are differentially expressed by >2 fold. (H) Transcriptional gene correlation network in ADC-cardiac transition from stages 2 to 3.
We performed branched expression analysis modeling (Beam) to identify genes determining the 2 different trajectory directions (Fig. 6C). Compared with the myofibroblast branch, cells in the cardiac branch had a higher expression of genes enriched in GO terms “muscle system process,” “muscle contraction,” “heart contraction,” and “cardiac muscle development,” indicating that LIF-treated ADCs transdifferentiated to cardiomyocyte-like cells highly expressed cardiac-specific genes such as Actc1, Tnni, and cTnT (Fig. 6A-6E). Downregulated genes exhibited myofibroblast GO terms such as “extracellular matrix organization,” “cross-linking of collagen fibrils,” and “collagen chain trimerization,” suggesting myofibroblast activation (Fig. 6A-6D). The activated myofibroblastic cells were enriched in myofibroblast genes, such as col6a1 and lox, and distributed in the days 3, 7, and 10 plots, whereas cardiomyocyte-like cells labeled with cTnT were scattered on the days 7 and 10 clusters (Fig. 6E).
We next investigated the transcriptional network in the decision of the bifurcated cell fates (Fig. 6F). Two distinct interacting subnetworks revealed critical transcriptional regulation in each as well as their potential connections. A total of 12 key transcription factors were identified at the myofibroblast activation state, including Klf4, Fos, and Smad4, which are pivotal in regulating TGF-β-induced fibrosis.82,83 Interestingly, we found that Prdm2, an H3K9 methyltransferase, was a key transcription factor in regulating myofibroblast activation (Fig. 6F). Prdm2 is considered a tumor-suppressor gene and a central player in regulating developmental transitions by directing a series of chromatin modifications at the target loci.84 More recently, Prdm2 was found to be enriched in quiescent muscle stem cells in vivo and controlled reversible quiescent states in cultured myoblasts,85 suggesting its potential role in ADC-myofibroblast activation. In the cardiomyocyte state, Mef2a was identified as the core transcription factor in the modulation of the ADC-derived cardiomyocyte gene network, which is also crucial for regulating atrial and ventricular chamber-restricted genes. Importantly, Tbp and Tfap4, genes involved in the formation of transcription initiation complexes, connected the activated myofibroblast state with the cardiomyocyte state, suggesting that cells expressing both genes may remain in an intermediate state with the potential to transfer from myofibroblasts to cardiomyocytes. Taken together, these studies depicted the sequential molecular dynamics facilitating the transition of ADCs to myofibroblasts and cardiomyocytes.
Discussion
In this study, we identified a cell population from WAT that can generate cardiomyocyte-like cells in response to LIF signaling. Key factors we sought to uncover in the ADC-cardiac transition were the cell identity of the LIF-responsive cells and the molecular dynamics leading to the establishment of the cardiac fate. To this end, we applied high-throughput single-cell RNA-seq to identify the cell heterogeneity, the cell differentiation trajectory, and the molecular events activated during the transition. We carried out in-depth analyses to demonstrate several salient properties of this process. First, our single-cell analysis identified and functionally defined the Pdgfrα+CD55+ ADC population in the WAT with cardiac potential. Second, with the R package Monocle, RNA velocity, and follow-up functional studies, we demonstrated that Nrf2 signaling is required to initiate the ADC-cardiac program in response to LIF. Third, we identified 2 separate pathways in the transition route associated with either distinct cardiomyocyte fate or intermediately activated myofibroblast cell fate. Our results raise the possibility that a new autologous cell source exists in the adipose tissue that may be a strong candidate for heart tissue regeneration.
Adipose progenitor cells have long been considered promising mesenchymal stem cell candidates that express cell surface markers such as Pdgfrα, CD29, CD34, Sca1, Dpp4, and CD24.86 However, it remains unclear which population possibly possesses the myogenic potential. Our hanging culture model found a population of ADCs that reacted to LIF and transitioned to a myogenic fate. The size of these cells ranged from 2 to 16.5 µm, making it possible for them to coexist with adipocytes in the floating layer of centrifuge tubes after repeated washing. Single-cell sequencing analysis identified 3 progenitor populations that were further analyzed by FACS sorting. Although Pdgfrα is expressed in both ADC1 and ADC3, the 2 ADCs had distinct characteristics. Apart from Pdgfrα and Icam1, ADC1 also expressed various collagens, coinciding with the previously identified Icam1+/Col4a2 ASCs27 and group 2 Icam1+preadipocytes,64 suggesting its adipogenic potential. However, instead of generating adipocytes, the sorted Pdgfrα+/Icam1+ ADCs expressed cardiac genes, albeit at a lower level than the 2 other ADC clusters. ADC3 expressed several canonical mesenchymal progenitor markers, CD34, Ly6a, CD55, Pi16, and Dpp4, suggesting a canonical stem cell population, as previously identified to comprise fibro-inflammatory progenitors (FIPs),65 interstitial progenitors,64 and Dpp4+/Pi16+ ASCs.27 The CD142+ population was considered antiadipogenic and inhibited the differentiation of adipose progenitor cells in vitro through secreted factors, suggesting that resident stem cells regulate adipocyte accumulation.28,87 We also isolated corresponding ADCs from ASCs and induced cardiac differentiation, but only Pdgfrα+/CD55+ ADCs showed a relatively robust cardiac transition. Interestingly, Pdgfrα+/CD55+ stromal cells did not colocalize with CD34+ cells in the adventitia but were spread among the adipocytes, suggesting that other Pdgfrα+/CD55+/CD34+ cells exhibit a stronger myogenic potential than the traditionally recognized multipotent mural cells around the vessels.67 Moreover, although we also identified Pdgfrα+/CD55+ ADCs in the stromal fraction in WAT, its cardiogenic potential was significantly lower than that of the floating ADCs (Fig. 3E,F). We speculate that cell size may be a differentiating factor between floating ADCs and stromal ADCs. Although the existence of small pluripotent stem cells in adult tissue remains controversial,88 our results indicate that otherwise similar cell populations where the cell size is reduced (10 µm) exhibit a stronger cardiomyogenic potential. Nevertheless, a more detailed analysis will be required to elucidate the molecular differences related to cell size and differentiation potential.
Our finding that activation of Nrf2 signaling is an indispensable event in the initiation of ADC-cardiac transition, together with previous reports of its critical role in redox and metabolic regulation in ESC and adult stem cells,78,89-91 highlights the critical role of Nrf2 signaling in cell fate determination. Nrf2 activation occurs within 24 h of LIF administration (Fig. 5A-5F), which is consistent with previous reports on adipose and osteogenic differentiation, suggesting that Nrf2 activation is a stage-specific process.92,93 Our results established a connection between the LIF-STAT3-Nrf2 signaling and the myogenic process (Fig. 5D, 5E). The addition of LIF promoted p-STAT3 signaling in the presence of Nrf2 activation, whereas blocking Nrf2 transcription by siRNA led to a reduction of p-STAT3/JAK-induced ADC-cardiac transition (Fig. 5F). It is possible that LIF/STAT3 signaling triggers alterations in the intracellular redox homeostasis of adult stem cells in response to cytokines and growth factors, thereby regulating lineage differentiation.81,94,95 This finding suggests that LIF/STAT3/NRF2 is indispensable for triggering ADC-cardiac transition in our system. Interestingly, Nrf2 also affects basic processes related to differentiation including cell cycle regulation, energy metabolism, and stem cell maintenance,78,96 which could indirectly help activate or suppress specific differentiation in multipotent cells such as the ADCs. Cardiovascular diseases such as hypertension and ischemia, and anticancer drugs induce the generation of ROS and/or reactive nitrogen species, leading to the release of Nrf2 from Keap1 translocation to the nucleus to initiate a wide range of cell defense processes against this pathological oxidative stress.97-100 At the same time, LIF is among the cardiac paracrine factors secreted after myocardial injury and has been shown to enhance cardioprotection and recruit endogenous stem cells.34-37 However, it remains largely unknown whether LIF induces the activation of endogenous stem cells via the Nrf2 pathway, which could support cardioprotection. Further research on the LIF-STAT-NRF2 signaling during activation of local stem cells may aid our understanding of cardiac regeneration.
Moreover, our trajectory results revealed that starting from day 3, ADCs navigated a trajectory with 2 branches that corresponded to cardiomyogenic fate or intermediately activated myofibrogenic cell fate (Fig. 6), which is in line with the previous findings from a study that directly reprogrammed human or mouse fibroblasts to induce cardiomyocyte-like cells.101,102 Fu et al102 took advantage of a chemical cocktail that included LIF to generate beating cardiomyocyte-like cells with a fibroblast origin. Nevertheless, they did not specify whether LIF was responsible for the induction of cardiomyogenic differentiation or other aspects such as cell maintenance. It has also been reported that LIF effectively increases the number of beating clusters.102 We speculate that there is a strong expansion of a small cell population highly responsive to LIF, functioning either as an inducing agent or as a stepping stone for other factors to induce cardiomyogenesis. The low number of cells in the scRNA-seq analysis may also result from the isolation methodology. Fibroblast is a crucial noncardiomyocyte cell type in the heart and is recognized to play complex roles in heart development, homeostasis, and disease.103 Although activated cardiac fibroblasts and resulting pathological fibrosis have long been considered to lead to impaired mechanical function associated with heart failure, arrhythmias, and sudden cardiac death,104 more recent studies have posited that cardiac fibroblasts may protect heart function by facilitating cardiomyocyte maturation in both neonatal mice and human ESC-derived cardiomyocytes.105 Deletion of fibronectin, which is mainly generated by cardiac fibroblasts, disrupted FN1-integrin interaction and consequently impaired human IPSC-derived cardiomyocyte growth.106 In line with these findings, the existence of myofibroblasts in the ADC-cardiac transition may be necessary to facilitate cardiomyocyte maturation. These results call for further identification of secreted factors and key transcriptional signals in the activated myofibroblasts and their derivatives.
Summary
Using scRNA-seq analysis to dissect the cell fate trajectories during the LIF-induced ADC-cardiac transition, we defined the cell heterogeneity, the cell differentiation trajectory, and the molecular events activated during the transition. Single-cell resolution mapping in a differentiation/developmental background will likely lead to the identification of novel cell populations, their lineage trajectory, and the molecular signatures, thereby paving the way to leveraging signaling to manipulate resident stem cells in development and disease.
Supplementary Material
Contributor Information
Jiayi Yao, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Feiyang Ma, Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA, USA; Chongqing International Institute for Immunology, Chongqing, People’s Republic of China.
Li Zhang, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Ching Zhu, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Medet Jumabay, Division of Allergy, Immunology Center for Immunity, Infection, and Inflammation Pediatrics, Dept of Medicine, University of California, San Diego, San Diego, CA, USA.
Zehao Yao, Peking Union Medical College, Chinese Academy of Medical Science & Peking Union Medical College, Beijing, People’s Republic of China.
Lumin Wang, Institute of Precision Medicine, the Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, People’s Republic of China.
Xinjiang Cai, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Daoqin Zhang, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Xiaojing Qiao, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Kalyanam Shivkumar, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Matteo Pellegrini, Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA, USA; Dept of Molecular, Cell and Developmental Biology, University of California, Los Angeles, Los Angeles, CA, USA.
Yucheng Yao, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Xiuju Wu, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA.
Kristina I Boström, Division of Cardiology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA; Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA, USA.
Funding
This study was funded by National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI): grant number HL30568 (K.I.B.), NIH/NHLBI: grant number HL81397 (K.I.B.), NIH/NHLBI: grant number HL154548 (K.I.B.), NIH/National Institute on Neurological Disorders and Stroke (NINDS): grant number NS79353 (Y.Y.), NIH/NHLBI: grant number HL139675 (Y.Y.). X.C. was supported in part by the UCLA Specialty Training and Advanced Research (STAR) fellowship program and by the NIH training grant T32HL007895.
Conflict of Interest
The authors declared no potential conflicts of interest.
Author Contributions
J.Y.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; F.M.: collection and assembly of data, data analysis, and interpretation; L.Z.: collection and assembly of data, data analysis and interpretation, manuscript writing; C.Z.: collection and assembly of data, data analysis, and interpretation; M.J.: collection and assembly of data; Z.Y., L.W., X.C., D.Z., X.Q.: collection and assembly of data; K.S.: provision of study material, data analysis, and interpretation; M.P.: data analysis and interpretation; Y.Y.: financial support, data analysis, and interpretation; X.W.: administrative support, data analysis and interpretation, and manuscript writing; K.I.B.: conception and design, financial support, administrative support, data analysis and interpretation, and manuscript writing.
Data Availability
The single-cell RNA sequencing datasets are accessible from GEO with accession number GSE164410.
References
- 1. van Berlo JH, Molkentin JD.. An emerging consensus on cardiac regeneration. Nat Med. 2014;20(12):1386-1393. 10.1038/nm.3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crea F, Libby P.. Acute coronary syndromes: the way forward from mechanisms to precision treatment. Circulation. 2017;136(12):1155-1166. 10.1161/CIRCULATIONAHA.117.029870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cahill TJ, Choudhury RP, Riley PR.. Heart regeneration and repair after myocardial infarction: translational opportunities for novel therapeutics. Nat Rev Drug Discov. 2017;16(10):699-717. 10.1038/nrd.2017.106 [DOI] [PubMed] [Google Scholar]
- 4. Bassat E, Mutlak YE, Genzelinakh A, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547(7662):179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laflamme MA, Murry CE.. Heart regeneration. Nature. 2011;473(7347):326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes [in Eng]. Nature. 2012;485(7400):593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ebrahimi B. In vivo reprogramming for heart regeneration: a glance at efficiency, environmental impacts, challenges and future directions. J Mol Cell Cardiol. 2017;108:61-72. [DOI] [PubMed] [Google Scholar]
- 8. Lunde K, Solheim S, Aakhus S, et al. Intracoronary injection of mononuclear bone marrow cells in acute myocardial infarction [in eng]. N Engl J Med. 2006;355(12):1199-1209. [DOI] [PubMed] [Google Scholar]
- 9. Zimmet H, Porapakkham P, Sata Y, et al. Short- and long-term outcomes of intracoronary and endogenously mobilized bone marrow stem cells in the treatment of ST-segment elevation myocardial infarction: a meta-analysis of randomized control trials [in Eng]. Eur J Heart Fail. 2012;14(1):91-105. [DOI] [PubMed] [Google Scholar]
- 10. Badorff C, Brandes RP, Popp R, et al. Transdifferentiation of blood-derived human adult endothelial progenitor cells into functionally active cardiomyocytes [Research Support, Non-U.S. Gov’t] [in Eng]. Circulation. 2003;107(7):1024-1032. [DOI] [PubMed] [Google Scholar]
- 11. Minasi MG, Riminucci M, De Angelis L, et al. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development. 2002;129(11):2773-2783. [DOI] [PubMed] [Google Scholar]
- 12. Fioret BA, Heimfeld JD, Paik DT, et al. Endothelial cells contribute to generation of adult ventricular myocytes during cardiac homeostasis. Cell Rep. 2014;8(1):229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mekala SR, Worsdorfer P, Bauer J, et al. Generation of cardiomyocytes from vascular adventitia-resident stem cells. Circ Res. 2018;123(6):686-699. [DOI] [PubMed] [Google Scholar]
- 14. Telukuntla KS, Suncion VY, Schulman IH, et al. The advancing field of cell-based therapy: insights and lessons from clinical trials. J Am Heart Assoc. 2013;2(5):e000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le T, Chong J.. Cardiac progenitor cells for heart repair [in Eng]. Cell Death Discov. 2016;2:16052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Litwinowicz R, Kapelak B, Sadowski J, et al. The use of stem cells in ischemic heart disease treatment. Kardiochir Torakochirurgia Pol. 2018;15(3):196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsuji W, Rubin JP, Marra KG.. Adipose-derived stem cells: implications in tissue regeneration. World J Stem Cells. 2014;6(3):312-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jumabay M, Zhumabai J, Mansurov N, et al. Combined effects of bone morphogenetic protein 10 and crossveinless-2 on cardiomyocyte differentiation in mouse adipocyte-derived stem cells. J Cell Physiol. 2017;233(3):1812-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jumabay M, Zhang R, Yao Y, et al. Spontaneously beating cardiomyocytes derived from white mature adipocytes. Cardiovasc Res. 2010;85(1):17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang G, Xiao Z, Ren X, et al. Obtaining spontaneously beating cardiomyocyte-like cells from adipose-derived stromal vascular fractions cultured on enzyme-crosslinked gelatin hydrogels. Sci Rep. 2017;7:41781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takashima S, Usui S, Inoue O, et al. Myocyte-specific enhancer factor 2c triggers transdifferentiation of adipose tissue-derived stromal cells into spontaneously beating cardiomyocyte-like cells. Sci Rep. 2021;11(1):1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ii M, Horii M, Yokoyama A, et al. Synergistic effect of adipose-derived stem cell therapy and bone marrow progenitor recruitment in ischemic heart. Lab Invest. 2011;91(4):539-552. [DOI] [PubMed] [Google Scholar]
- 23. Luong Q, Huang J, Lee KY.. Deciphering White adipose tissue heterogeneity. Biology (Basel). 2019;8(2):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pandzic Jaksic V, Grizelj D.. Under the surface of subcutaneous adipose tissue biology. Acta Dermatovenerol Croat. 2016;24(4):250-260. [PubMed] [Google Scholar]
- 25. Lynes MD, Tseng YH.. Deciphering adipose tissue heterogeneity. Ann N Y Acad Sci. 2018;1411(1):5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee KY, Luong Q, Sharma R, et al. Developmental and functional heterogeneity of white adipocytes within a single fat depot. EMBO J. 2019;38(3):e99291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burl RB, Ramseyer VD, Rondini EA, et al. Deconstructing adipogenesis induced by β3-adrenergic receptor activation with single-cell expression profiling [in Eng]. Cell Metab. 2018;28(2):300-309.e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwalie PC, Dong H, Zachara M, et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559(7712):103-108. [DOI] [PubMed] [Google Scholar]
- 29. Zgheib C, Zouein FA, Kurdi M, et al. Differential STAT3 signaling in the heart: impact of concurrent signals and oxidative stress. JAKSTAT. 2012;1(2):101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zouein FA, Kurdi M, Booz GW.. LIF and the heart: just another brick in the wall? Eur Cytokine Netw. 2013;24(1):11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirota H, Chen J, Betz UA, et al. Loss of a gp130 cardiac muscle cell survival pathway is a critical event in the onset of heart failure during biomechanical stress. Cell. 1999;97(2):189-198. [DOI] [PubMed] [Google Scholar]
- 32. Ricke-Hoch M, Bultmann I, Stapel B, et al. Opposing roles of Akt and STAT3 in the protection of the maternal heart from peripartum stress. Cardiovasc Res. 2014;101(4):587-596. [DOI] [PubMed] [Google Scholar]
- 33. Halkein J, Tabruyn SP, Ricke-Hoch M, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest. 2013;123(5):2143-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mohri T, Fujio Y, Maeda M, et al. Leukemia inhibitory factor induces endothelial differentiation in cardiac stem cells. J Biol Chem. 2006;281(10):6442-6447. [DOI] [PubMed] [Google Scholar]
- 35. Mohri T, Fujio Y, Obana M, et al. Signals through glycoprotein 130 regulate the endothelial differentiation of cardiac stem cells. Arterioscler Thromb Vasc Biol. 2009;29(5):754-760. [DOI] [PubMed] [Google Scholar]
- 36. Iwakura T, Mohri T, Hamatani T, et al. STAT3/Pim-1 signaling pathway plays a crucial role in endothelial differentiation of cardiac resident Sca-1+ cells both in vitro and in vivo. J Mol Cell Cardiol. 2011;51(2):207-214. [DOI] [PubMed] [Google Scholar]
- 37. Kanda M, Nagai T, Takahashi T, et al. Leukemia inhibitory factor enhances endogenous cardiomyocyte regeneration after myocardial infarction. PLoS One. 2016;11(5):e0156562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shalek AK, Satija R, Adiconis X, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675-685. [DOI] [PubMed] [Google Scholar]
- 40. Liu Z, Wang L, Welch JD, et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551(7678):100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakanishi M, Mitchell RR, Benoit YD, et al. Human pluripotency is initiated and preserved by a unique subset of founder cells. Cell. 2019;177(4):910-924 e922. [DOI] [PubMed] [Google Scholar]
- 42. Bostrom KI, Guihard P, Blazquez Medela AM, et al. Matrix Gla protein limits pulmonary arteriovenous malformations in ALK1 deficiency. Eur Respir J. 2015;45(3):849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. National Research Council. Guide for the Care and Use of Laboratory Animals. 8th ed. The National Academies Press; 2011. [Google Scholar]
- 44. Kolk MV, Meyberg D, Deuse T, et al. LAD-ligation: a murine model of myocardial infarction [Video-Audio Media] [in Eng]. J Vis Exp. 2009;32:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jumabay M, Abdmaulen R, Urs S, et al. Endothelial differentiation in multipotent cells derived from mouse and human white mature adipocytes [in Eng]. J Mol Cell Cardiol. 2012;53(6):790-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sugihara H, Yonemitsu N, Miyabara S, et al. Primary cultures of unilocular fat cells: characteristics of growth in vitro and changes in differentiation properties [in Eng]. Differentiation. 1986;31(1):42-49. [DOI] [PubMed] [Google Scholar]
- 47. Yao J, Guihard PJ, Wu X, et al. Vascular endothelium plays a key role in directing pulmonary epithelial cell differentiation. J Cell Biol. 2017;216(10):3369-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Levy DE, Darnell JE.. Stats: transcriptional control and biological impact [in Eng]. Nat Rev Mol Cell Biol. 2002;3(9):651-662. [DOI] [PubMed] [Google Scholar]
- 49. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38-44. [DOI] [PubMed] [Google Scholar]
- 50. Karimzadeh M, Ernst C, Kundaje A, et al. Umap and Bismap: quantifying genome and methylome mappability. Nucleic Acids Res. 2018;46(20):e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kfoury Y, Scadden DT.. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16(3):239-253. [DOI] [PubMed] [Google Scholar]
- 52. Kalucka J, de Rooij LPMH, Goveia J, et al. Single-cell transcriptome atlas of murine endothelial cells [in eng]. Cell. 2020;180(4):764-779.e720. [DOI] [PubMed] [Google Scholar]
- 53. Ahmed MS, Gravning J, Martinov VN, et al. Mechanisms of novel cardioprotective functions of CCN2/CTGF in myocardial ischemia-reperfusion injury [in Eng]. Am J Physiol Heart Circ Physiol. 2011;300(4):H1291-H1302. [DOI] [PubMed] [Google Scholar]
- 54. Kobayashi T, Watanabe Y, Saito Y, et al. Mice lacking the glutamate-cysteine ligase modifier subunit are susceptible to myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2010;85(4):785-795. [DOI] [PubMed] [Google Scholar]
- 55. Aeby E, Ahmed W, Redon S, et al. Peroxiredoxin 1 protects telomeres from oxidative damage and preserves telomeric DNA for extension by telomerase. Cell Rep. 2016;17(12):3107-3114. [DOI] [PubMed] [Google Scholar]
- 56. Ravaud C, Pare M, Azoulay S, et al. Impairment of the activin A autocrine loop by lopinavir reduces self-renewal of distinct human adipose progenitors. Sci Rep. 2017;7(1):2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zaragosi LE, Wdziekonski B, Villageois P, et al. Activin plays a critical role in proliferation and differentiation of human adipose progenitors. Diabetes. 2010;59(10):2513-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morandi EM, Verstappen R, Zwierzina ME, et al. ITGAV and ITGA5 diversely regulate proliferation and adipogenic differentiation of human adipose derived stem cells. Sci Rep. 2016;6:28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mittal A, Pulina M, Hou SY, et al. Fibronectin and integrin alpha 5 play requisite roles in cardiac morphogenesis. Dev Biol. 2013;381(1):73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mittal A, Pulina M, Hou SY, et al. Fibronectin and integrin alpha 5 play essential roles in the development of the cardiac neural crest. Mech Dev. 2010;127(9-12):472-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bolli R, Stein AB, Guo Y, et al. A murine model of inducible, cardiac-specific deletion of STAT3: its use to determine the role of STAT3 in the upregulation of cardioprotective proteins by ischemic preconditioning. J Mol Cell Cardiol. 2011;50(4):589-597. [DOI] [PubMed] [Google Scholar]
- 62. Friedman CE, Nguyen Q, Lukowski SW, et al. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell. 2018;23(4):586-598 e588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. DeLaughter DM, Bick AG, Wakimoto H, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39(4):480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Merrick D, Sakers A, Irgebay Z, et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364(6438):eaav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hepler C, Shan B, Zhang Q, et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cho DS, Lee B, Doles JD.. Refining the adipose progenitor cell landscape in healthy and obese visceral adipose tissue using single-cell gene expression profiling. Life Sci Alliance. 2019;2(6):e201900561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ferrero R, Rainer P, Deplancke B.. Toward a consensus view of mammalian adipocyte stem and progenitor cell heterogeneity. Trends Cell Biol. 2020;30(12):937-950. [DOI] [PubMed] [Google Scholar]
- 68. Wang Y, Xu J, Meyers CA, et al. PDGFRα marks distinct perivascular populations with different osteogenic potential within adipose tissue [in Eng]. Stem Cells. 2020;38(2):276-290. [DOI] [PubMed] [Google Scholar]
- 69. Iwayama T, Steele C, Yao L, et al. PDGFRα signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity [in Eng]. Genes Dev. 2015;29(11):1106-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Marcelin G, Ferreira A, Liu Y, et al. A PDGFRα-mediated Switch toward CD9 [in Eng]. Cell Metab. 2017;25(3):673-685. [DOI] [PubMed] [Google Scholar]
- 71. Berry R, Rodeheffer MS.. Characterization of the adipocyte cellular lineage in vivo [in Eng]. Nat Cell Biol. 2013;15(3):302-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ruiz-Arguelles A, Llorente L.. The role of complement regulatory proteins (CD55 and CD59) in the pathogenesis of autoimmune hemocytopenias. Autoimmun Rev. 2007;6(3):155-161. [DOI] [PubMed] [Google Scholar]
- 73. Tung PY, Blischak JD, Hsiao CJ, et al. Batch effects and the effective design of single-cell gene expression studies. Sci Rep. 2017;7:39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155-160. [DOI] [PubMed] [Google Scholar]
- 75. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018;560(7719):494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Svensson V, Pachter L.. RNA velocity: molecular kinetics from single-cell RNA-seq. Mol Cell. 2018;72(1):7-9. [DOI] [PubMed] [Google Scholar]
- 77. Trachootham D, Lu W, Ogasawara MA, et al. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10(8):1343-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dai X, Yan X, Wintergerst KA, et al. Nrf2: redox and metabolic regulator of stem cell state and function. Trends Mol Med. 2020;26(2):185-200. [DOI] [PubMed] [Google Scholar]
- 79. Malhotra D, Portales-Casamar E, Singh A, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38(17):5718-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chorley BN, Campbell MR, Wang X, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40(15):7416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Murakami S, Suzuki T, Harigae H, et al. NRF2 activation impairs quiescence and bone marrow reconstitution capacity of hematopoietic stem cells. Mol Cell Biol. 2017;37(19):e00086-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lin L, Han Q, Xiong Y, et al. Krupple-like-factor 4 attenuates lung fibrosis via inhibiting epithelial-mesenchymal transition. Sci Rep. 2017;7(1):15847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang Y, Feng XH, Derynck R.. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394(6696):909-913. [DOI] [PubMed] [Google Scholar]
- 84. Hohenauer T, Moore AW.. The Prdm family: expanding roles in stem cells and development. Development. 2012;139(13):2267-2282. [DOI] [PubMed] [Google Scholar]
- 85. Cheedipudi S, Puri D, Saleh A, et al. A fine balance: epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene [in eng]. Nucleic Acids Res. 2015;43(13):6236-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hepler C, Vishvanath L, Gupta RK.. Sorting out adipocyte precursors and their role in physiology and disease. Genes Dev. 2017;31(2):127-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vishvanath L, Gupta RK.. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J Clin Invest. 2019;129(10):4022-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ratajczak MZ, Ratajczak J, Kucia M.. Very Small Embryonic-Like Stem Cells (VSELs). Circ Res. 2019;124(2):208-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang J, Nuebel E, Daley GQ, et al. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11(5):589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ito K, Suda T.. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev. 2014;15(4):243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mathieu J, Zhou W, Xing Y, et al. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14(5):592-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chartoumpekis DV, Ziros PG, Sykiotis GP, et al. Nrf2 activation diminishes during adipocyte differentiation of ST2 cells. Int J Mol Med. 2011;28(5):823-828. [DOI] [PubMed] [Google Scholar]
- 93. Tao J, Wang H, Zhai Y, et al. Downregulation of Nrf2 promotes autophagy-dependent osteoblastic differentiation of adipose-derived mesenchymal stem cells. Exp Cell Res. 2016;349(2):221-229. [DOI] [PubMed] [Google Scholar]
- 94. Lu C, Xu W, Shao J, et al. Nrf2 induces lipocyte phenotype via a SOCS3-dependent negative feedback loop on JAK2/STAT3 signaling in hepatic stellate cells. Int Immunopharmacol. 2017;49:203-211. [DOI] [PubMed] [Google Scholar]
- 95. Si ZP, Wang G, Han SS, et al. CNTF and Nrf2 are coordinately involved in regulating self-renewal and differentiation of neural stem cell during embryonic development. iScience. 2019;19:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Murakami S, Motohashi H.. Roles of Nrf2 in cell proliferation and differentiation. Free Radic Biol Med. 2015;88(Pt B):168-178. [DOI] [PubMed] [Google Scholar]
- 97. Muthusamy VR, Kannan S, Sadhaasivam K, et al. Acute exercise stress activates Nrf2/ARE signaling and promotes antioxidant mechanisms in the myocardium. Free Radic Biol Med. 2012;52(2):366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yu J, Wang L, Akinyi M, et al. Danshensu protects isolated heart against ischemia reperfusion injury through activation of Akt/ERK1/2/Nrf2 signaling. Int J Clin Exp Med. 2015;8(9):14793-14804. [PMC free article] [PubMed] [Google Scholar]
- 99. Shen Y, Liu X, Shi J, et al. Involvement of Nrf2 in myocardial ischemia and reperfusion injury. Int J Biol Macromol. 2019;125:496-502. [DOI] [PubMed] [Google Scholar]
- 100. Dodson M, de la Vega MR, Cholanians AB, et al. Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol. 2019;59:555-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhou Y, Liu Z, Welch JD, et al. Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell. 2019;25(1):149-164 e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fu Y, Huang C, Xu X, et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails [Original article]. Cell Res. 2015;25:1013-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ieda M, Tsuchihashi T, Ivey KN, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16(2):233-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kohl P, Gourdie RG.. Fibroblast-myocyte electrotonic coupling: does it occur in native cardiac tissue?. J Mol Cell Cardiol. 2014;70:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang Y, Yao F, Wang L, et al. Single-cell analysis of murine fibroblasts identifies neonatal to adult switching that regulates cardiomyocyte maturation. Nat Commun. 2020;11(1):2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Miao Y, Tian L, Martin M, et al. Intrinsic endocardial defects contribute to hypoplastic left heart syndrome. Cell Stem Cell. 2020;27(4):574-589 e578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA sequencing datasets are accessible from GEO with accession number GSE164410 (Reviewer access token: ixonwocufzaflot).
The single-cell RNA sequencing datasets are accessible from GEO with accession number GSE164410.