Significance
Acetate-producing microorganisms (acetogens) have long been hypothesized as among the earliest evolving organisms. Concomitantly, serpentinite formations have been widely considered the most likely habitat to have supported early life. Yet, little is known of acetogens in contemporary serpentinites. Here, evolutionary and genome-guided metabolic reconstructions provide insights into the characteristics of two deeply branching, uncultured acetogens that are found in serpentinite-hosted waters. The two types are differentially distributed among serpentinite fluids and exhibit physiological traits unique to their respective environments, with the “type II” acetogens specialized for growth on substrates generated by water–rock interaction. Collectively, these data help bridge a knowledge gap between hypotheses of early acetogenic life on Earth and contemporary acetogen characteristics in early Earth analog environments.
Keywords: serpentinization, acetogenesis, Acetothermia, origin of life, metagenomics
Abstract
Little is known of acetogens in contemporary serpentinizing systems, despite widely supported theories that serpentinite-hosted environments supported the first life on Earth via acetogenesis. To address this knowledge gap, genome-resolved metagenomics was applied to subsurface fracture water communities from an area of active serpentinization in the Samail Ophiolite, Sultanate of Oman. Two deeply branching putative bacterial acetogen types were identified in the communities belonging to the Acetothermia (hereafter, types I and II) that exhibited distinct distributions among waters with lower and higher water–rock reaction (i.e., serpentinization influence), respectively. Metabolic reconstructions revealed contrasting core metabolic pathways of type I and II Acetothermia, including in acetogenic pathway components (e.g., bacterial- vs. archaeal-like carbon monoxide dehydrogenases [CODH], respectively), hydrogen use to drive acetogenesis, and chemiosmotic potential generation via respiratory (type I) or canonical acetogen ferredoxin-based complexes (type II). Notably, type II Acetothermia metabolic pathways allow for use of serpentinization-derived substrates and implicate them as key primary producers in contemporary hyperalkaline serpentinite environments. Phylogenomic analyses indicate that 1) archaeal-like CODH of the type II genomes and those of other serpentinite-associated Bacteria derive from a deeply rooted horizontal transfer or origin among archaeal methanogens and 2) Acetothermia are among the earliest evolving bacterial lineages. The discovery of dominant and early-branching acetogens in subsurface waters of the largest near-surface serpentinite formation provides insight into the physiological traits that likely facilitated rock-supported life to flourish on a primitive Earth and possibly on other rocky planets undergoing serpentinization.
The geological process of serpentinization has been argued to have created conditions conducive to the origin of life (1, 2). Key to this process is the presence of ultramafic rock that is common in Earth’s mantle, is less common on contemporary Earth’s surface, and contains abundant iron-bearing silicate minerals that can react with water to produce biologically important electron donors such as hydrogen (H2) (3). When H2 is accumulated to high enough concentrations, it can abiotically reduce dissolved inorganic carbon (DIC; ΣCO2/HCO3−/CO32−) to generate reduced carbon compounds including methane (CH4), formate (HCOO−), and carbon monoxide (CO) (3, 4), and perhaps even longer chain carbon compounds like acetate and pyruvate (5). Microbial metabolisms that are dependent on serpentinization-derived substrates, including methanogenesis (methane producing) and acetogenesis (acetate producing), have been argued as the likely first microbial metabolisms on Earth, based on several lines of physiologic and phylogenetic evidence. This includes their use of a highly similar linear pathway (i.e., the reductive acetyl-CoA or Wood–Ljundgahl [WL] pathway) for both energy conservation and biosynthesis (2, 6–10). Indeed, recent experimental evidence has confirmed the close resemblance of the reductive acetyl-CoA pathways of acetogens and methanogens via the disruption of methane-producing genes downstream of the reductive acetyl-CoA pathway in the methanogen Methanosarcina acetivorans, leading to its conversion to an acetogen (11). Consequently, serpentinization-influenced environments are commonly invoked as one of the most likely habitats to have hosted the earliest life on Earth and those that could potentially support life on other planetary systems where ultramafic rock is present and available to undergo serpentinization, such as on Enceladus or Mars (12, 13).
Recent studies of serpentinization-influenced ecosystems have begun to provide insight into the diversity and physiology of microorganisms inhabiting terrestrial (14–20) and marine (16, 21–24) serpentinites. Despite the prevalence of low-potential electron donors such as H2, HCOO−, CO, and CH4, fracture waters from serpentinites typically host low abundance communities (15) with low diversity (14, 25, 26). This has been attributed to the polyextreme conditions imposed by serpentinization reactions that include hyperalkaline pH, DIC limitation, and oxidant limitation. This combination of conditions has also been shown to correspond to streamlining of genomes, more reduced inferred proteomes, and physiological characteristics that enable more efficient utilization of lithogenic substrates (15, 18, 27). Furthermore, many of the taxa identified in serpentinites by cultivation-independent approaches correspond to uncultured bacterial taxa, with many belonging to little-characterized groups, including those possibly involved in acetogenesis (14, 18). For example, heterotrophic acetogenesis (i.e., acetate production from organic sources) activity has been implicated in the hyperalkaline, serpentinite springs in The Cedars of California (28, 29), has been inferred from a metagenome-assembled genome (MAG) also recovered from The Cedars (29), and has been suggested to be used by actinobacterial populations in the serpentinization-influenced Hakuba Happo hot springs of Japan (30). Further, genes encoding key proteins involved in acetogenesis, including acetyl-CoA synthase (Acs) and other components of the WL pathway, have been detected in metagenomes from serpentinite waters (15–18, 31). Consistently, acetate is one of the most abundant small-molecular-weight organic compounds detected in serpentinite fluids (14, 32). Despite possible detection of putative acetogens in serpentinites and possible products of their activities, little is known of how acetogenesis occurs under the polyextremophilic conditions created by serpentinization, the metabolic and physiological traits that enable robust growth in these environments, how they influence carbon and energy flow in such ecosystems, and the potential for similar conditions to have supported acetogenic life on early Earth or on other planetary bodies.
Ophiolites are sections of upper mantle and oceanic crust bedrock that have been emplaced onto continents, thereby offering opportunities to investigate microbial environments within settings undergoing modern water–rock interaction that are otherwise typically inaccessible. The Samail Ophiolite (SO), Sultanate of Oman, is the largest ophiolite in the world and is actively undergoing serpentinization (33). Importantly, fracture waters from the SO display a gradient of influence by serpentinization reactions that ranges from slightly alkaline meteoric-like waters to hyperalkaline, highly serpentinized waters (14, 34), allowing for the identification of metabolic and physiological traits that enable habitation of those environments. Acs-encoding genes were recently detected across a range of alkaline and hyperalkaline fluids from wells within the SO, suggestive of populations dependent on the WL pathway (15). Moreover, 16S ribosomal RNA (rRNA) gene-based analyses of microbial communities in fracture waters from the SO indicated the dominance of populations associated with the uncultured Acetothermia group, in particular, within the most hyperalkaline waters (14). Members of the candidate division Acetothermia (formerly the OP1 clade and also referred to as the “Bipolaricaulota”) have been suggested to be capable of autotrophic and/or heterotrophic acetogenesis (35, 36). Here, Acetothermia genomes were reconstructed from metagenomic sequences generated from SO subsurface fracture fluids that spanned geochemical gradients collected across multiple years. The physiological potential for acetogenesis and autotrophy was inferred based on metabolic reconstructions of MAGs. Further, the evolutionary histories of key proteins that underpin the WL pathway and acetogenesis were examined. The results provide insight into physiological characteristics that permit deeply rooted acetogenic Bacteria to dominate communities inhabiting serpentinized waters and shed light on how serpentinization could have supported the first acetogenic cells on Earth.
Materials and Methods
Sample Collection.
Biomass was collected from waters pumped from wells in the SO, Sultanate of Oman, in 2015 and 2017 using a submersible pump, as previously described (14, 15). Briefly, after flushing the pump system, water samples were collected at least 20 m below the water table in each well, with samples collected at two depth intervals in well NSHQ14 in 2017 (NSHQ14B at 50 m and NSHQ14C at 85 m). Biomass was filtered from ∼100 L using 4-mm 0.2-µm Millipore polycarbonate filters. Filtered biomass was also collected from waters in well BA1A from discrete depths of 108 m to 132 m and >250 m in 2020 (BA1A108 and BA1A250, respectively). These samples were collected using a down-borehole packer system and submersible pump using methods outlined elsewhere (37). Lastly, two wells sampled in 2015 and 2017 (WAB188 and NSHQ14) were sampled again in 2020, with the water from NSHQ14 collected at 9 m to 30 m. Filters were aseptically transferred to sterile cryovials and were frozen on site with liquid nitrogen for transport back to the laboratory.
Metagenomic Sequencing and Analyses.
DNA was extracted from filters and subjected to metagenomic sequencing as components of previous studies (14, 15, 20). Briefly, genomic DNA was extracted from the 2015 filtered biomass samples using a MoBio PowerSoil kit (MoBio), while genomic DNA was extracted from the 2017 samples using a Zymo Research Xpedition Soil/Fecal DNA MiniPrep Extraction kit, both according to manufacturer instructions. The 2015 libraries were sequenced on the Illumina MiSeq platform (2 × 150 bp). Deeper sequencing was conducted on the 2017 libraries that were prepared from triplicate DNA extracts using the Nextera XT kit (Illumina Inc.) and sequenced on the Illumina HiSeq 2500 Rapid Run platform (2 × 250 bp). The raw sequence reads from the eight 2015 and eight 2017 metagenomes were quality filtered, trimmed of adapters, and assembled, as previously described (15, 20). The four 2020 metagenomes subjected to analysis here were sequenced on the Illumina NovaSeq platform (2 × 150 bp) and processed as part of the Joint Genome Institute pipeline, as described elsewhere (38). The 2015 metagenomes comprised between 5.9 × 105 and 3.6 × 106 paired-end reads (average of 1.6 × 106), while the eight 2017 metagenomes comprised between 1.9 × 107 and 3.4 × 108 paired-end reads (average of 4.2 × 107), and the four 2020 metagenomes comprised between 3.4 × 108 and 4.6 × 108 paired-end reads (average of 3.5 × 108). Contigs from the 2015 and 2017 metagenomes were binned into draft MAGs based on tetranucleotide sequences and coverage profiles using MetaBAT (v.0.26.3) (39), as previously described (40), or via the MetaWRAP version 1.3.2 pipeline (2020 metagenomes), as described elsewhere (41). Housekeeping genes were identified within draft MAGs using Amphora2 (42), and these were used to preliminarily identify bins associated with Acetothermia, based on average amino acid identities (AAIs). MAG quality was assessed with CheckM v.1.0.7 (43), as previously described (40). Draft Acetothermia bins exhibited an estimated 0% contamination (Table 1) and were thus not modified further.
Table 1.
Assembly statistics for Acetothermia MAGs recovered from SO subsurface fracture waters
| ID (Year, Well, BinID) | Type | pH | Est. com. | Est. cont. | Est. rel. abund.* | Rank† | GC (%) | Size (Mbp) | N50 (kbp) | Contig (no.) | CDS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2015.WAB188.Bin1 | I | 8.7 | 86.2 | 0 | 26.9 | 2 | 56.1 | 1.8 | 23.6 | 127 | 1,958 |
| 2017.NSHQ14B.Bin.6‡ | I | 11.1 | 84.5 | 0 | 7.0 | 6 | 56.3 | 1.8 | 31.7 | 86 | 1,891 |
| 2017.WAB188.Bin.3 | I | 7.6 | 84.5 | 0 | 2.4 | 3 | 56.3 | 1.9 | 39.5 | 71 | 1,889 |
| 2020.BA1A108.Bin.1 | I | 9.0 | 89.7 | 0 | 0.7 | 9 | 56.1 | 2.0 | 197.7 | 16 | 1,987 |
| 2020.BA1A250.Bin.10 | I | 10.0 | 89.7 | 0 | 20.3 | 1 | 56.0 | 2.1 | 159.7 | 18 | 2,062 |
| 2020.NSHQ14.Bin.31 | I | 11.2 | 89.7 | 0 | 0.4 | 21 | 55.9 | 2.0 | 163.1 | 23 | 2,028 |
| 2020.NSHQ14.Bin.54 | I | 11.2 | 89.7 | 0 | < 0.1 | 42 | 56.0 | 2.0 | 163.1 | 19 | 1,992 |
| 2020.WAB188.Bin.124 | I | 7.5 | 89.7 | 0 | 1.7 | 5 | 55.9 | 2.0 | 165.0 | 23 | 2,032 |
| 2015.NSHQ14.Bin.1 | II | 11.3 | 89.7 | 0 | 43.6 | 1 | 47.9 | 1.6 | 30.1 | 81 | 1,526 |
| 2017.NSHQ14B.Bin.2‡ | II | 11.1 | 87.9 | 0 | 12.1 | 2 | 47.9 | 1.2 | 16.1 | 98 | 1,150 |
| 2017.NSHQ14C.Bin.1‡ | II | 11.3 | 87.9 | 0 | 32.6 | 1 | 47.8 | 1.2 | 14.9 | 112 | 1,147 |
| 2017.WAB71.Bin.4 | II | 10.6 | 75.2 | 0 | 12.2 | 1 | 47.8 | 1.19 | 11.7 | 132 | 1,143 |
| 2020.NSHQ14.Bin.16 | II | 11.2 | 89.7 | 0 | 5.4 | 2 | 48.0 | 1.44 | 19.4 | 98 | 1,356 |
| 2020.NSHQ14.Bin.82 | II | 11.2 | 82.8 | 0 | < 0.1 | 41 | 48.1 | 1.28 | 23.6 | 72 | 1,196 |
Est., estimated; com., completeness; cont., contamination; rel. abund., relative abundance; CDS., coding sequences.
*Relative abundance was estimated by percentage of overall reads mapped to type I or type II MAGs in each corresponding metagenome.
†Relative abundance rank was estimated based on estimated relative abundances among all MAGs generated for a given community.
‡Biomass from fracture waters was filtered from two depths in NSHQ14 in 2017, 50 m (NSHQ14B) and 85 m (NSHQ14C).
Phylogenetic Analyses and Abundance Estimation.
Phylogenomic reconstruction of the Acetothermia MAGs was conducted by identifying the presence of 30 universal single-copy housekeeping phylogenetic marker genes with the MarkerFinder program (v.1.1) (44) along with all publicly available Acetothermia/Bipolaricaulota within the Genome Tree Database (GTDB) (45) and within a recent division-wide analysis (36). Proteins encoded by marker genes were used for phylogenetic analysis and comprised 27 ribosomal proteins and three RNA polymerase subunits shared among all Bacteria. These proteins correspond to clusters of orthologous groups (COG) comprising COG0012, COG0096, COG0048, COG0097, COG0049, COG0098, COG0052, COG0099, COG0080, COG0100, COG0081, COG0102, COG0085, COG0103, COG0086, COG0184, COG0087, COG0185, COG0088, COG0186, COG0090, COG0197, COG0091, COG0200, COG0092, COG0202, COG0093, COG0256, COG0094, and COG0522. Individual marker gene proteins were aligned with Clustal Omega (v.1.2.4) (46), trimmed with TrimAl (v.1.4) (47) specifying a gap threshold (gt) value of 0.1 and default parameters otherwise, and then concatenated into a super alignment matrix. Only previously published Acetothermia MAGs that were estimated to be >50% complete were included in the analyses. The concatenated alignment was then subjected to maximum likelihood (ML) phylogenetic analysis using IQ-TREE (v.1.6.11) (48) after identifying the optimal amino acid substitution model (LG+F+I+R4) among 168 possible models via the Bayesian information criterion, as implemented in the model testing “TEST” function of IQ-TREE. Ten separate independent phylogenetic analysis runs were conducted and compared to maximize the final ML reconstruction that was then chosen as the final phylogenetic tree. Branch support was evaluated with 1,000 ultrafast bootstraps (49).
The phylogenetic analysis identified two distinct phylotypes among the recovered MAGs. Representative MAGs from each of the two groups were used to recruit contigs from each of the metagenome assemblies. The number of reads mapped to these contigs [as inferred by read mapping using Bowtie2 (50)] was then used to estimate population relative abundances within each of the metagenomes. Average AAIs (51) were calculated within and between the two phylotype groups in order to estimate within- and between-group genome relatedness using the AAI workflow within the CompareM software package (52).
Proteins encoded by key genes involved in acetogenesis or core energy conservation pathways were also subjected to phylogenetic analysis. Specifically, carbon monoxide/acetyl-CoA synthase complexes (Cdh/Acs) that are integral for the WL pathway and acetogenesis were subjected to phylogenetic analyses as described above. A recently published comprehensive database of Cdh/Acs complexes in Archaea and Bacteria was used for the analyses (53), along with reference homologs identified within the GenBank nr database based on protein basic local alignment search tool (BLASTp) searches of CdhA homologs encoded by the MAGs generated in this study. Given the different evolutionary histories of archaeal- and bacterial-like Cdh/Acs, separate phylogenetic analyses were conducted for each type, using concatenations of the CdhA, CdhB, and CdhC oxidoreductase subunits of the archaeal-like complexes, and the CdhA oxidoreductase subunits of bacterial-like complexes (53). The amino acid substitution model for the archaeal-like CdhABC analysis was LG+F+I+G4, while that for the bacterial AcsA analysis was LG+I+G4. In addition, a separate phylogenetic analysis was conducted using the CdhA subunits from both Acetothermia types along with the 500 most closely related CdhA homologs present in the National Center for Biotechnology Information (NCBI) nr database (excluding homologs from unpublished studies deposited within the last year; amino acid substitution model of LG+I+G4 was used for the analysis). Likewise, the homologous beta subunits of F-type ATP synthases and alpha subunits of V-type ATP synthases were subjected to phylogenetic analysis, as described above (amino acid substitution model LG+R8), and with reference against a previously published dataset of these subunits (54).
To assess the placement of types I and II Acetothermia among the bacterial phylogeny, a whole-domain phylogenomic analysis was conducted. Representative genomes (n = 722) from all bacterial phylum-level divisions (128 phyla total) from the GTDB were included in the analysis. Up to 10 representatives from every phylum were chosen for analysis (evenly chosen across different classes/orders), or all records within the phylum were chosen if < 10 genomes were present in the database (Dataset S1). The workflow for the phylogenetic analysis was the same as described above for the Acetothermia domain phylogeny (i.e., the same 30 universal bacterial markers and alignment generation strategy were used). Only alignments containing >50% of the maximum possible alignment residue positions were used in the analyses. The concatenated amino acid alignments were subjected to the same ML analysis as described above for the Acetothermia domain phylogeny, but using the LG+I+G4 amino acid substitution model, as chosen based on model selection results.
Metabolic Reconstructions.
Gene prediction and annotation of MAGs was performed with Prodigal v.2.6.3 (55), as implemented in Prokka v.1.14.5 (56), using default parameters. Preliminary functional annotations from the Prokka pipeline were then used to reconstruct metabolic pathways for each phylotype based on comparison against the Kyoto Encyclopedia of Genes and Genomes (KEGG) function database (57) with the KEGG Automatic Annotation Server (58). The MAGs were also queried using BLASTp for specific gene functions based on reference genomes. Positive identifications of protein homologs in the MAGs were those with BLASTp matches with >30% amino acid homology and with coverage >60% of the length of the query sequence. Putative homologs were also further screened based on comparisons against the NCBI conserved domain database (59). Because the assemblies exhibited very high within-group phylotype AAIs (∼98 to 100%), indicating nearly identical genomes, metabolic pathways were primarily conducted with the two most complete MAGs for either phylotype, but supplemented with the other MAGs, when necessary. Validation of key proteins was performed, where possible, by subjecting homologs to alignment using Clustal Omega (v.1.2.4), followed by identification of key conserved residues, including conserved cysteinyl motifs for [NiFe]- and [FeFe]-hydrogenases (60). [FeFe]- and [NiFe]-hydrogenase catalytic subunits were also subjected to gene neighborhood analysis and phylogenetic comparison against recently published comprehensive hydrogenase databases (61) to assess their functions.
Autotrophic capacity was evaluated in other populations in the SO water communities by surveying abundant genomes (>5% relative abundance among binned genomes in any of the eight metagenomes from 2017) for key genes associated with autotrophy. Only communities from the 2017 sampling efforts were used, because they were subjected to a higher level of metagenome sequencing effort when compared to those from the 2015 sampling efforts. Further, unlike communities from the 2020 sampling effort that targeted only three wells, those from the 2017 sampling effort spanned a broad range of water types present in the SO. MAG binning was conducted using the automated binning and refinement pipeline of MetaWRAP version 1.3.2 (62), as described previously (41). Only those MAGs exhibiting >50% estimated completion and <10% estimated contamination (estimated with CheckM) were used to survey putative autotrophic capacities. Specifically, the presence of the WL (reductive acetyl-CoA) pathway among MAGs was evaluated based on the presence of 1) formate-tetrahydrofolate ligase (Fhs), 2) methylenetetrahydrofolate dehydrogenase (FolD), 3) methylenetetrahydrofolate reductase (MetVF), and 4) a nearly complete Cdh/Acs complex (including at least the key catalytic subunits). The presence of the Calvin cycle was evaluated based on the presence of both ribulose-1,5-bisphosphate carboxylase-oxygenase large and small subunits. Lastly, homologs of ATP citrate lyase subunits A and B (AclA, AclB) was used to identify the presence of a putative reverse tricarboxylic acid (rTCA) cycle pathway. The citryl-CoA synthase route for the rTCA pathway was not identified in any of the MAGs based on the absence of citryl-CoA synthetase (CcsA and CcsB) and citryl-CoA lyase (ccl) proteins. Likewise, the hydroxypropionate–hydroxybutyrate cycle was not encoded by any of the MAGs based on the absence of the key gene 4-hydroxybutyrl-CoA dehydratase (AbfD), nor was the 3-hydroxypropionate cycle encoded by any of the MAGs based on the absence of malonyl-CoA reductase (mcr) and malyl-CoA/(S)-citramalyl-CoA lyase (mcl).
Results and Discussion
Description of Fracture Fluids Sampled from the SO.
The eight fracture waters sampled from the SO in 2015 ranged, in pH, from 8.2 to 11.4, while the eight sampled in 2017 ranged, in pH, from 8.3 to 11.3 (SI Appendix, Tables S1 and S2). The four waters sampled from 2020 ranged, in pH, from 7.5 to 11.4. The lithological setting of each well varied and included gabbro, peridotite, and “contact” areas between peridotite and gabbro settings that represented mixtures of alkaline and hyperalkaline fluid types (14). Fracture waters were previously classified based on their geochemical properties and extent of transformation into hyperalkaline fluids via serpentinization reactions (14, 34, 63). Briefly, less-reacted waters are termed “type I” and tend to have slightly alkaline pH, lower conductivity, higher oxidation reduction potentials (ORPs), and generally exhibit higher oxidant availability along with lower concentrations of reduced aqueous-phase gases like CH4 and H2 (14, 15, 34, 63). Conversely, waters that have been subjected to increased serpentinization are termed “type II” and tend to have hyperalkaline pH, higher conductivity, lower ORPs, minimal oxidant availability, low DIC availability, and much higher reduced aqueous-phase gas concentrations compared to type I waters (14, 15, 34, 63). Fracture water communities that were analyzed herein spanned a range of type I and type II waters obtained from multiple subsurface drilled water wells.
Recovery of Acetothermia MAGs from the SO.
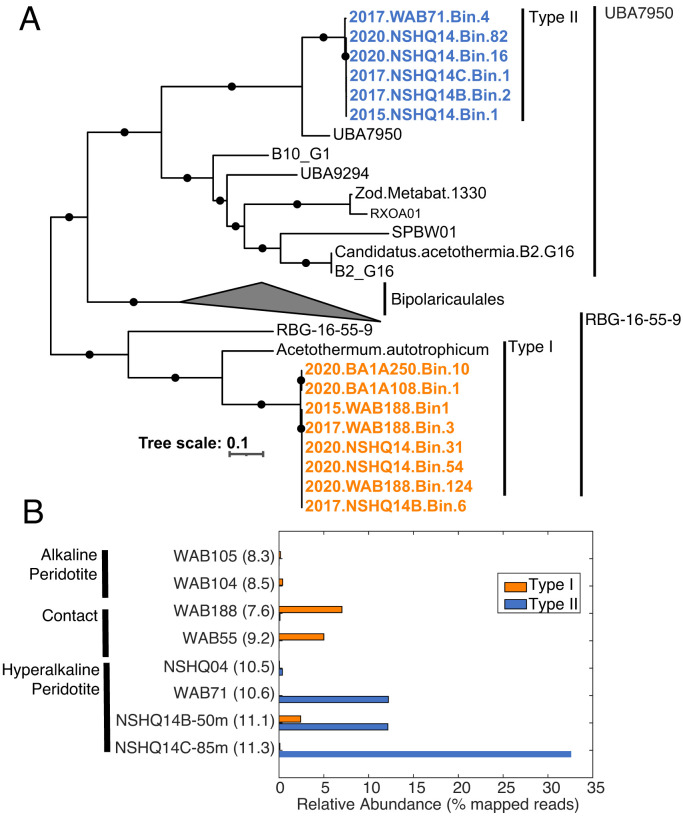
Metagenomic characterization of DNA recovered from filtered biomass collected from eight fracture waters in eight wells in 2015 (SI Appendix, Table S1), another eight fracture waters from seven wells in 2017 (SI Appendix, Table S2), and four fracture waters from three wells in 2020 yielded 14 moderate- to high-quality MAGs affiliated with Acetothermia (two from 2015 samples, five from 2017 samples, and seven from the 2020 samples; estimated completeness: 75 to 90%; Table 1). The metagenomes from the 2015 samples were sequenced at a lower read depth than those from 2017 and 2020, likely resulting in recovery of fewer MAGs. Phylogenomic analysis indicated the presence of two phylogenetically distinct populations within the Acetothermia phylum (hereafter referred to as type I and type II Acetothermia), with high within-group genomic identity in the two groups (∼98 to 100% AAI within type I and type II groups; Fig. 1A). In addition, the MAGs were nearly identical (∼100% AAI) to samples collected from the same wells across years. Such AAI values are well above commonly used thresholds to delineate species (e.g., 95% AAI) (51), and indicate essentially homogenous population-level genomic diversity across fracture waters and years for the two phylotypes.
Fig. 1.
Phylogenomic analysis and ecological distribution of Acetothermia MAGs recovered from SO subsurface fracture waters. (A) ML phylogenetic reconstruction of MAGs recovered in this study in comparison with those recovered from previous studies, with type I MAGs from the SO highlighted in orange and type II MAGs highlighted in blue. The year of the sample is shown, followed by the well designation from which the MAGs were recovered. Biomass from fracture waters was collected from 50-m (NSHQ14B) and 85-m (NSHQ14C) depths from NSHQ14 in 2017. The three proposed orders for the Acetothermia/Bipolaricualaeota are shown on the far right. Branch length is scaled to that shown in the lower left for expected substitutions per site. Bootstrap values >90% (out of 1,000 bootstraps) are indicated by black circles. Thermotogae representatives were used as the outgroup including Petrotoga mobilis SJ95, Thermotoga petrophila RKU-1, and Mesotoga inferna. (B) The abundances of types I and II populations are shown as the percentage of total metagenome reads mapped to either MAG type for each 2017 metagenome. The 2015 and 2020 metagenomes were not used for abundance calculations, given their relatively low sequencing depth and limited sampling scope, respectively. Metagenomes are arranged by the fluid type designation of their well waters and then ordered by ascending pH values for well waters, as indicated in parentheses next to each metagenome.
Comparison of type I and type II MAGs revealed overall genomic differences (∼48% AAI ± ∼10%) consistent with class- or order-level taxonomic differences (51). The type I SO clade formed a group with the previously described Candidatus Acetothermum autotrophicum MAG (35) and others recovered from subsurface environments (Fig. 1A). The type II clade formed a lineage along with a MAG generated from a Lost City hydrothermal vent fluid metagenome (UBA7950). The Lost City MAG was generated from genome mining of publicly available metagenome data (64) and was not characterized in that study. The Lost City hydrothermal system is a well-characterized marine serpentinite system (21, 22, 65) and features characteristics similar to those of highly reacted type II SO waters, including hyperalkaline pH, high concentrations of H2, and limited DIC (65). Thus, the close phylogenetic similarity between the MAGs recovered from hyperalkaline waters in the SO and the Lost City (and their phylogenetic distinction from other Acetothermia) suggests that the clade comprises serpentinite-adapted Acetothermia from globally distributed serpentinite-hosted ecosystems. The type II MAGs exhibited smaller estimated genome sizes, lower guanine and cytosine (GC) content, and fewer protein coding genes relative to the type I MAGs (Table 1), consistent with genome streamlining to minimize energy demands associated with inhabiting hyperalkaline fluids, as previously observed among genomes in serpentinization-impacted environments (15, 18).
The two populations exhibited nearly mutually exclusive distributions across fracture fluids, with the type I population more prevalent in mixed waters from “contact” wells (i.e., at gabbro/peridotite interfaces), while the type II populations were abundant in waters with hyperalkaline pH (Fig. 1B and Table 1). A notable exception was observed in waters from well NSHQ14, where the distribution of the two populations overlapped in samples taken from nearer to the surface in 2017 (50 m; NSHQ14B) and 2020, which was packed off at a depth of ∼20 m and thus only encompassed shallow waters. The type II population dominated the community in deeper waters from 2017 (85 m; NSHQ14C) and was also abundant in waters nearer to the surface in both 2017 and 2020 (Table 1), while the type I populations comprised smaller fractions of the nearer to the surface communities of 2017 and 2020 waters (Fig. 1B and Table 1). Near-surface mixing of water types closer to the surface in NSHQ14 is likely, as suggested by down-borehole geochemical data from the immediately adjacent BA3A well suggesting that the deeper waters are considerably more reducing than the surface waters (66). Thus, the near-surface waters of NSHQ14 may represent a mixture of communities typically associated with type I (or mixed) and type II waters.
Adaptations to Acetogenic Metabolism in Type I and II Acetothermia Populations.
The metabolic potentials of types I and II MAGs were investigated to evaluate whether they exhibited different adaptations to serpentinization-influenced geochemical gradients that characterize type I and II waters, respectively (SI Appendix, Tables S1 and S2). Metabolic reconstructions indicated that both type I and II populations are likely capable of autotrophic acetogenesis via the WL pathway, albeit through slight modifications that were unique to their respective environments (Fig. 2).
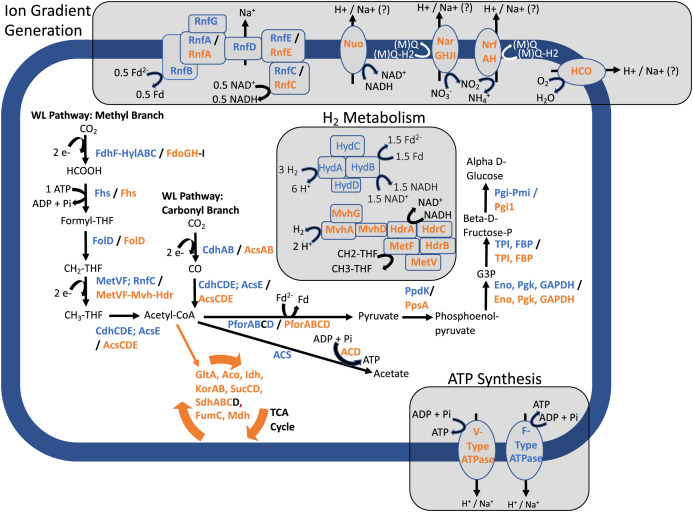
Fig. 2.
Metabolic reconstructions for Acetothermia type I and II MAGs recovered from the SO subsurface fracture waters. A composite metabolic reconstruction map is shown for type I and II MAGs, with encoded proteins/pathways in orange denoting type I MAGs and those in blue denoting the type II MAGs. Energy conservation pathways highlighted in the text are grouped in gray boxes, with the rest of the pathways representing central carbon metabolism. Question marks show areas of uncertainty in reconstructing aspects of metabolism. Subunits or proteins highlighted in black were not observed in the corresponding MAGs. Abbreviations are as follows: anaerobic carbon monoxide dehydrogenase (Coo), pyruvate Fd oxidoreductase (Pfor), ADP-forming acetyl-CoA synthetase (ACD), AMP-forming acetyl-CoA synthase (ACS), citrate synthase (GltA), aconitate hydratase (Aco), isocitrate dehydrogenase (Idh), 2-oxoglutarate Fd oxidoreductase (Kor), succinyl-CoA synthetase (Suc), succinate dehydrogenase (Sdh), fumarate hydratase (FumC), malate dehydrogenase (Mdh), pyruvate orthophosphate dikanase (PpdK), pyruvate-water dikinase (PpsA), enolase (Eno), phosphoglycerate kinase (PgK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), triosephosphate isomerase (TPI), fructose-1,6-bisphosphatase I (FBP), glucose/mannose-6-phosphate isomerase (Pgi-Pmi), glucose-6-phosphate isomerase (Pgil), Fd, reduced Fd (Fd2−), nitrate reductase (Nar), cytochrome c nitrite reductase (Nrf), heme-Cu oxidase (HCO), methyl-viologen reducing [NiFe]-hydrogenase (Mvh), and heterodisulfide reductase (Hdr).
The first step in autotrophic acetogenesis is the reduction of CO2 to CO in the carbonyl branch of the WL pathway. The type I MAGs encoded bacterial-like carbon monoxide dehydrogenase/acetyl-CoA synthase protein complex (CdhACDE/AcsE; nomenclature following ref. 53), typical of other Acetothermia (53). In contrast, type II MAGs encoded an archaeal-like carbon monoxide dehydrogenase/acetyl-CoA synthase protein complex (CdhABCDE), with an additional bacterial-like AcsE subunit (SI Appendix, Figs. S1 and S2 and Dataset S2). The two complexes are assumed to be functionally equivalent (53), although their distinct distributions between type I and II populations suggest advantages of each system in their respective environmental regimes. Moreover, the presence of both an archaeal-like CdhA colocalized with other Cdh subunits in the type II genomes, along with the existence of a separate bacterial-like (but non-Acetothermia-like) CdhA within these genomes, suggests the potential for plasticity in their use (Dataset S2). Genes encoding an ABC-type membrane transport system with components homologous to the bicarbonate transport system of Cyanobacteria were colocalized with the bacterial-like CdhA subunit in type II genomes, along with sodium bicarbonate transport protein encoding genes (sbtA) encoded by the MAGs (Dataset S2). This transport system is critical for cyanobacterial Na+-dependent bicarbonate (HCO3−) uptake under low CO2 conditions (67). Thus, it is plausible that the additional bacterial-like CdhA of type II populations is associated with DIC (HCO3−) uptake, potentially pointing to plasticity in their use of the archaeal- or bacterial-like CdhA, depending on DIC availability. Notably, no components of this transport system were identified in the type I MAGs (Dataset S2), consistent with greater availability of DIC in type I waters.
An equally important step in acetogenesis is the reduction of CO2 to HCOO− in the methyl branch of the WL pathway. Type I MAGs encoded formate dehydrogenase complexes (Fdo) that can reduce CO2 to HCOO−, whereas type II MAGs encoded homologs of the reversible NADH- and ferredoxin (Fd)-dependent, electron-bifurcating formate dehydrogenase (HylCBA-FdhF2) identified in Clostridium acidurici (68). In type II MAGs, the hylCBA-fdhF2 genes were colocalized together with genes coding for Fhs (SI Appendix, Fig. S3 and Dataset S2), a key enzyme for carbon fixation in the WL pathway. This suggests that the electron bifurcating FdhF2-Hyl complex may provide formate for the WL pathway in the type II population, and/or also contribute to the production of CO2 from formate for use in the carbonyl branch of the WL pathway to generate CO.
A critical challenge faced by the type II Acetothermia population is the availability of CO2 for reduction to CO and HCOO− in the initial steps of the WL pathway. CO2 is not readily available in SO fluids with hyperalkaline pH (pH >10) (69) due to conversion to HCO3− (pKa = 6.4) and, ultimately, CO32− (pKa = 10.2). Moreover, carbonate mineral solubility is inversely correlated to pH, resulting in substantial carbonate precipitation and DIC removal from hyperalkaline waters containing abundant Ca+ (70). However, serpentinization reactions resulting in abundant H2 can drive the generation of CO and HCOO− through the abiotic reduction of DIC (3, 4, 71). Notably, acetogens can use CO and HCOO− as initial, or sole, compounds for entry into the WL pathway (72, 73). Thus, it is plausible, if not likely, that the type II Acetothermia preferentially use CO and HCOO−, when available, rather than expending reducing equivalents from NAD(P)H or reduced Fd to reduce CO2. Similar adaptations were observed in Methanobacterium genomes and transcriptomes recovered from hyperalkaline waters in the SO (20, 27), wherein Methanobacterium were inferred to be dependent on HCOO− to overcome DIC limitation via cytoplasmic HCOO− oxidation, rather than H2, as is otherwise conserved in this methanogenic genus (27). HCOO− concentrations in the well waters hosting both Acetothermia types range from ∼1 μM to 2 μM (14, 27), suggesting that HCOO− is bioavailable in these environments. Moreover, laboratory experiments of serpentinization reactions in SO waters have demonstrated considerable HCOO− production (up to 98 μM) coinciding with H2 and CO2 drawdown (71), suggesting that abiotic HCOO− generation could support HCOO−-dependent populations in these waters.
Nevertheless, the potential use of two different enzyme systems to possibly catalyze the reduction of CO2 to HCOO− represents an intriguing difference between the two Acetothermia types. Reduction of CO2 to HCOO− (E°’ = −420 mV) by Fdo in acetogens involves NAD(P)H (E°’ = −320 mV) as an electron donor, and this reaction proceeds in vivo (despite standard state energetics) if the ratio of reduced to oxidized NAD(P)H is high. However, the reduction potential of CO2 (HCO3− or CO32−) to HCOO− becomes increasingly negative at increasingly alkaline pH (75) and becomes even less thermodynamically favorable to reduce with NAD(P)H. This reaction barrier is due to the −60 mV/per pH unit shift from standard state for a 2 e−/2 H+ charge transfer during the reduction of HCO3− to HCOO− when compared to only a −30 mV per pH unit shift from standard state due to a 2 e−/1 H+ charge transfer for NAD(P)H (75). It is possible, then, that electron bifurcation/confurcation enables type II Acetothermia cells to overcome the thermodynamics associated with CO2 (HCO3− or CO32−) reduction (E ≈ −630 mV at pH 11) by coupling the reaction with the exergonic oxidation of Fd (E ≈ −650 mV at pH 11) (8) and the endergonic oxidation of NAD(P)H (E ≈ −470 mV at pH 11), so long as the ratios of reduced to oxidized Fd and NAD(P)H in cells are high.
The other requisite proteins involved in the WL pathway (Fhs, FolD, and MetVF) were encoded in both type I and II MAGs. Given the minimal energy conserved during acetogenesis, phosphorylation of acetyl-CoA produced from the WL pathway by phosphotransacetylase (Pta) and its subsequent dephosphorylation to produce one ATP and acetate by acetate kinase (AckA) has been traditionally considered necessary for acetogens to balance the consumption of one ATP molecule from HCOO− fixation in the methyl branch of the WL pathway, thereby rendering the WL pathway ATP consumption neutral (76). However, as observed elsewhere for other Acetothermia (36), genes encoding AckA or Pta were not identified in the genomes of either type I or type II Acetothermia. Rather, a homolog of an archaeal-like ADP-forming acetyl-CoA synthetase (ACD) (77) was identified in the type I MAGs but not in the type II MAGs (Fig. 2). Further, both the type I and type II MAGs encoded homologs of the non-ATP generating acetyl-CoA synthase [Acs, also known as the AMP-forming Acs; abbreviated here as ACS to differentiate from the Acs discussed above (78)] that is generally considered an assimilatory, but potentially reversible, acetate utilization mechanism (78). Consequently, the potential to leverage acetate production from acetyl-CoA for ATP production in the type II MAGs is unclear, raising questions of how the cellular bioenergetics of the type II population might operate.
The minimal energy conserved during acetogenesis leads to acetogens operating at the near limit of possible bioenergetics when growing autotrophically (76). Importantly, the overall energetics of autotrophic, hydrogenotrophic acetogenesis in the model acetogens Clostridium ljungdahlii and Acetobacterium woodii grown under CO2-replete conditions are directly associated with H2 partial pressures, the ratio of reduced/oxidized cofactors (e.g., Fd) within cells, and small but consequential alterations to acetogenesis pathways (76). In particular, hydrogenotrophic acetogenesis is most favorable at elevated H2 partial pressures and high ratios of reduced to oxidized Fd in cells (76). Further, acetogenesis in C. ljungdahlii and A. woodii is principally energy conserving due to the activity of the multisubunit Fd-NAD+ oxidoreductase (Rnf) complex that couples the oxidation of Fd to the reduction of NAD+, with the free energy of the reaction used to generate a transmembrane ion (sodium or proton) gradient that can drive ATP synthesis (76). Rnf complexes were encoded in type II MAGs, but not type I MAGs (Fig. 2). Low-potential reduced Fd needed to drive Rnf in type II cells could be generated by tetrameric (group 3) electron bifurcating [FeFe]-hydrogenase homologs (79, 80), of which two loci were identified in the type II MAGs (Fig. 2 and Dataset S2), also as present in A. woodii and C. ljungdahlii (76). The functionality of the [FeFe]-hydrogenase was supported by colocalized genes encoding the four subunits of tetrameric bifurcating hydrogenases (HydABCD) (79) (Dataset S2) in addition to phylogenetic comparison of the catalytic subunit (HydA) to classified [FeFe]-hydrogenases in the HydDB database (group A classification; E value < 2 × 10−129) (81). These enzyme complexes are predicted to reversibly bifurcate two electrons from H2 and couple the endergonic single-electron reduction of Fd to the exergonic single-electron reduction of NAD+ (80). Notably, the well waters contain substantial concentrations of H2 [e.g., ∼2.9 mM in NSHQ14 (14)] that would increase the overall bioenergetics of hydrogenotrophic acetogenesis. Sodium or hydrogen ions pumped from Rnf could then be harnessed via F-type (bacterial-like) ATP synthases (SI Appendix, Fig. S4) that were also detected in type II, but not type I MAGs, with the latter encoding V-type (archaeal-like) ATPases (Fig. 2 and SI Appendix, Fig. S4 and Supporting Text).
In addition to the critical WL steps described above, the reduction of methylenetetrahydrofolate (CH2-THF) to methyl tetrahydrofolate (CH3-THF) is catalyzed by MetVF, thereby generating the methyl group of the methyl branch of the WL pathway (Fig. 2) (82, 83). The type I Acetothermia encode MetVF, and these genes are colocalized in the genome with those encoding HdrA and MvhD (SI Appendix, Fig. S5 and Dataset S2), while genes encoding HdrBC and MvhAG were, together, colocalized but encoded elsewhere in the genomes. HdrABC are commonly involved in bifurcating electrons in methanogens (84, 85) and were recently proposed to be involved in bifurcating electrons from NADH (E°’ = −280 mV) for the exergonic reduction of CH2-THF (E°’ = −200 mV) and the endergonic reduction of a yet to be identified second electron donor in the model acetogen Moorella thermoacetica (83). Genes encoding MetVF are colocalized in the genome in M. thermoacetica, and these proteins have been copurified as a complex with HdrABC and the [NiFe]-hydrogenase protein MvhD, suggesting that the second electron acceptor may be a proton that is reduced to H2 (E°’ = −414 mV). This is further supported by data indicating that CH2-THF reduction activity is localized to the membrane (86), where an energy-converting hydrogenase (Ech) could function in the reduction of protons to H2. The colocalization of metVF, hdrA, and mvhD in type I Acetothermia MAGs (SI Appendix, Fig. S5) suggests the presence of a similar bifurcating mechanism in these cells, albeit through a cytoplasmic MvhAGD complex (Group 3c complex, confirmed by phylogenetic comparison of the MvhA to the HydDB server; E value = 0) (Fig. 2) since these cells do not encode a separate Ech, rendering the fate of intracellular H2 produced by this reaction unclear. In contrast, neither HdrABC nor MvhD are encoded by type II Acetothermia MAGs. Rather, several homologs of RnfC, which codes for NADH dehydrogenase functionality, are found in these genomes. As such, CH2-THF reduction in type II Acetothermia may be directly coupled with NADH oxidation, as in the acetogen A. woodii (87). It is possible that reduction of CH2-THF in type II Acetothermia occurs via NADH oxidation instead of the putative bifurcating, H2-dependent reaction of type I Acetothermia due to unfavorable thermodynamics of the bifurcating reaction due to product (H2) inhibition in the H2-rich environments inhabited by type II Acetothermia (14).
Type I and II populations both exhibited the capacity to generate chemiosmotic potentials to drive ATP synthesis, albeit through distinct pathways and ATPases that may reflect the geochemical characteristics of the environments that they differentially inhabit (SI Appendix, Fig. S4 and Supporting Text). In contrast to the Rnf complexes encoded by type II MAGs, the type I MAGs encoded a nearly complete NADH dehydrogenase (Nuo) complex that allows entry of electrons in the form of NADH into canonical electron transport respiration chains of Archaea, Bacteria, and Eukarya (88). Moreover, the type I population encoded proteins allowing for the respiration of NO3− and NO2− to N2 (or NH4+) via Nar and Nrf complexes, respectively (Fig. 2), and encoded cytochrome c oxidases necessary for aerobic respiration (Fig. 2 and Dataset S2). This expanded respiratory capacity is also coincident with expected higher oxidant concentrations (e.g., NO3− and O2) in type I waters (14). The capacity for NO3− and O2 reduction was also observed in the first described Acetothermia MAG, Candidatus Acetothermum autotrophicum (35), as well as in other related MAGs (36).
The Type II Acetothermia Are Dominant Autotrophs in Serpentinizing and Hyperalkaline Subsurface Fluids.
Both type I and II Acetothermia MAGs encoded glycolytic/gluconeogenic pathways that would allow for the conversion of acetyl-CoA to six-carbon sugars that can be used in biosynthesis, albeit with partially different complements of enzymes to complete the pathways (SI Appendix, Supporting Text) (89). Substrates capable of supporting autotrophic metabolism of type I and II Acetothermia appear to be limited to HCOO−, CO, H2, and, possibly, CO2/HCO3−. In addition, the two Acetothermia types exhibited varying capacities to incorporate exogenous organic carbon compounds, with the type I MAGs encoding a greater capacity for facultatively heterotrophic metabolism (SI Appendix, Supporting Text).
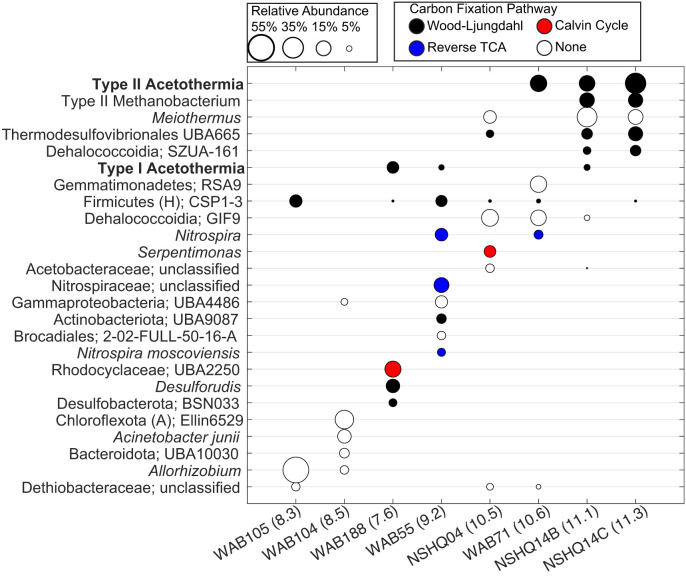
The distributions of type I and II Acetothermia were mutually exclusive among the well waters sampled in 2017 (Figs. 1B and 3), with the exception of the NSHQ14B sample taken nearer to the surface (50 m), while only the type II population was present in the deeper (85 m) waters (NSHQ14C). Type II Acetothermia were the dominant population (22%, 20%, and 35% relative abundance, respectively) in the most hyperalkaline waters analyzed in 2017 (WAB71, NSHQ14B, and NSHQ14C) and were also the dominant putative autotroph in these waters. The other putatively autotrophic populations present in high relative abundance (i.e., >5%) in the three most hyperalkaline samples collected in 2017 (Fig. 3) included those related to Methanobacterium (20, 27) (0%, 18%, and 17%, respectively), Thermodesulfovibrionales (5%, 10%, and 17%, respectively) (66), and uncharacterized Dehalococcoidia (0%, 5%, and 9%, respectively). The relative abundances of dominant taxa based on metagenomic analyses were broadly consistent with inferences from previous 16S rRNA gene analysis of these well communities from previous years (14), suggesting that they were representative of the native well communities. Consequently, the type II Acetothermia are inferred to represent key primary producers in communities inhabiting hyperalkaline SO waters and likely substantially contribute to ecosystem productivity. Consistently, these populations were also among the most abundant in the 2015 and 2020 metagenomes where they were identified (Table 1). The only autotrophic pathway identified among community members inhabiting highly serpentinized waters (pH >11.0) was the WL pathway, unlike communities inhabiting more circumneutral waters where multiple autotrophic pathways (WL, Calvin Cycle, and rTCA cycle) were identified (Fig. 3). In contrast, only minor populations of type I Acetothermia were present in lower pH wells (or the near-surface waters of NSHQ14B or NSHQ14 in 2020) (Fig. 3). In summary, both the type I and type II Acetothermia populations exhibit evidence for autotrophy that may be supplemented by organic carbon sources. However, the type II populations appear to be more dependent on substrates produced by water–rock interaction such as HCOO−, CO, or H2 for autotrophic metabolism (Fig. 3). This observation is consistent with their general dominance in environments where these substrates are more readily available (14, 69).
Fig. 3.
Putative autotrophs in subsurface fracture water communities from the SO. Each column shows the relative abundances of reconstructed MAGs (>5% relative abundance) within each well water community, with the corresponding taxonomic classification of MAGs given on the left. The circles represent individual MAGs and are scaled based on the relative abundance of that MAG among others within that community (based on the legend at the top left of the plot). The circles are colored according to the autotrophic carbon fixation pathway that is inferred to be encoded within the MAG, as based on the legend at the top right of the plot. Well names are followed by the pH of the sampled fracture waters in parentheses. Taxonomic classifications are given at the lowest characterized taxonomic designation, followed by either the specific uncultured group it belongs to (alphanumeric designations) or are otherwise followed by “unclassified” if the MAG is not related to previously characterized genomes within the Genome Toolkit Database. The type II Methanobacterium is the same as described in Fones et al. (27), and, while it encodes the WL pathway, it is dependent on formate as an electron donor and carbon source. The 2015 and 2020 metagenomes were not used for abundance calculations, given their relatively low sequencing depth and limited sampling scope, respectively.
The Type II Acetothermia Are Analogs of Early-Evolving Acetogens.
The prevalence of type II Acetothermia as putative autotrophs operating the WL pathway in hyperalkaline SO waters is consistent with previous suggestions of an ancient ancestry of the WL pathway that has its roots in primitive methanogens or acetogens that inhabited environments rich in iron sulfide minerals and that were actively undergoing serpentinization (2, 90). Arguments favoring the origin of the WL pathway in serpentinizing environments include that 1) it is a simple linear pathway that can be exergonic (91); 2) it involves reactants, intermediates, and products also involved in serpentinization (92); and 3) many of the key reactions of the WL pathway are catalyzed at iron–sulfur or nickel–iron–sulfide centers that are reminiscent of mineral catalysts that perform similar chemistry (5). These observations motivated further phylogenetic analysis of type II Acetothermia and components of their WL pathways.
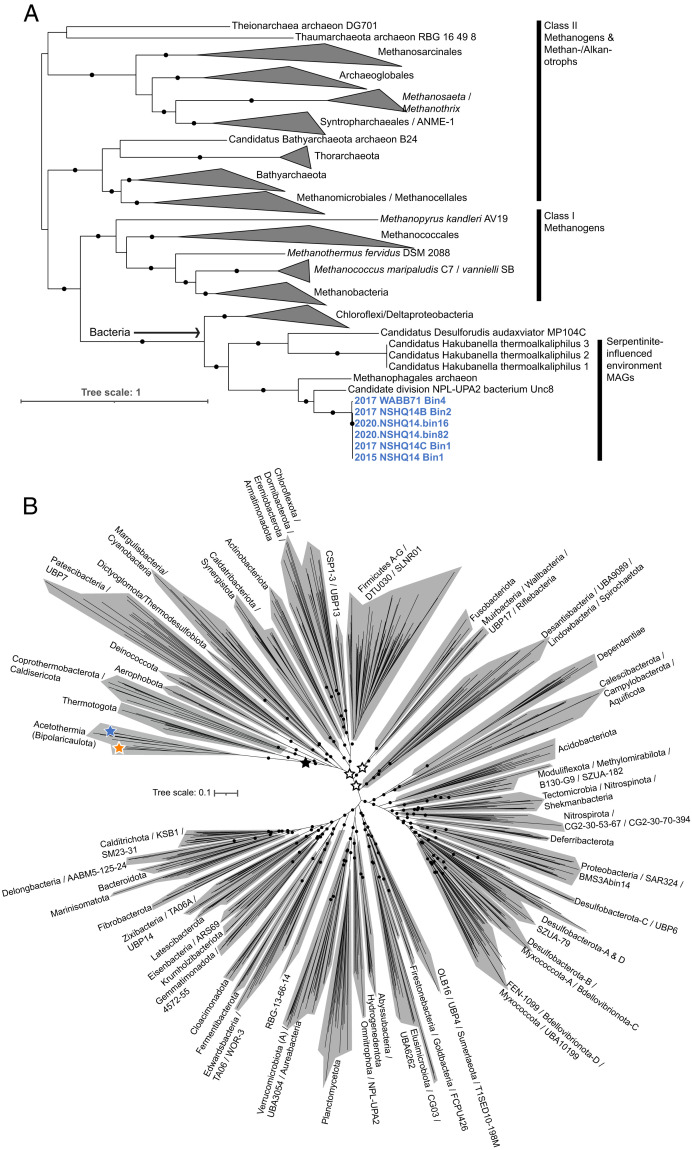
CdhABC house the nickel–iron–sulfide active site center of carbon monoxide dehydrogenase that catalyzes the initial reduction of CO2 to CO. Phylogenetic analysis of the type II archaeal-like CdhABC homologs indicated that they comprised a monophyletic group with CdhABC from other bacterial and archaeal MAGs largely from serpentinite-hosted environments (Fig. 4A). These included the putatively acetogenic Actinobacteria-related Candidatus Hakubanella thermoalkaliphilus (30), the putatively acetogenic NPL-UPA2 organism (29), an unclassified anaerobic methanotrophic euryarchaeota (ANME) MAG from the Lost City system, and a deltaproteobacterial Candidatus Desulforudis audaxviator MAG, which are commonly observed in SO fluid communities (Fig. 3) and other serpentinization-influenced communities (93). In addition, the type II CdhABC homologs were monophyletic with one another, closely related, and followed the same branching pattern as the whole genome Acetothermia phylogeny (Fig. 1A). Additional phylogenetic analysis of CdhA from the type II Acetothermia along with 500 of the most closely related homologs in the NCBI database further supported the branching of Acetothermia CdhA and those from organisms identified in serpentinite environments in association with the CdhA of Archaea that formed a group distinct to the homologous CdhA/AcsA of Bacteria (SI Appendix, Fig. S2). Together, these observations suggest that 1) Cdh were vertically inherited among the type II SO Acetothermia and have differentiated with minor taxonomic divergence of type II populations; 2) type II Acetothermia from the SO and putative acetogens from other serpentinite-hosted environments harbor Cdh that share a common evolutionary ancestor; and 3) the Cdh arose from a possible transfer (or shared evolutionary origin) between Archaea and Bacteria primarily from serpentinite-hosted environments (SI Appendix, Fig. S2), and this shared evolutionary history appears to be deeply rooted within Archaea. The latter assertion is evinced by the well-supported monophyly of bacterial Cdh from serpentinite environments (in addition to those from other bacteria including Chloroflexi/Deltaproteobacteria) with early-evolving methanogens (i.e., class I methanogens: Methanopyrales, Methanobacteriales, and Methanococcales), to the exclusion of other Archaea, including more recently evolved methanogens (i.e., Methanosarcinales and Methanomicrobiales) and recently identified methanotrophs/alkanotrophs like the Bathyarchaeota and Syntropharchaeales (Fig. 4A). This distinction is particularly striking given that Cdh complexes have been largely vertically inherited among Archaea (53), and is consistent with previous analyses hypothesizing a transfer of archaeal Cdh to Bacteria via an unidentified Euryarchaeota (29, 53). The type II MAGs also encoded bacterial-like CdhA homologs not related to the bacterial-like CdhA in the type I MAGs (SI Appendix, Fig. S6). Rather, the bacterial-like CdhA of type II MAGs were similar to those in a deep-branching group primarily comprising ANME and Firmicutes (SI Appendix, Fig. S6). As suggested above, the presence of two evolutionarily distinct bacterial- and archaeal-like CdhA within the type II MAGs may indicate physiological plasticity in the WL pathway, perhaps related to the availability of specific carbon substrates. Importantly, while it is not currently possible to determine whether the catalytic Cdh subunits share an origin with deeply diverging Archaea or whether they were laterally transferred from or into the type II lineage, the deep node connecting the primarily serpentinite-hosted bacterial Cdh lineage and that of all Archaea nevertheless suggests an early origin of the Cdh within the type II Acetothermia lineage.
Fig. 4.
Phylogenetic placement of key proteins involved in acetogenesis and Acetothermia lineages recovered from the SO subsurface waters among other bacterial lineages. (A) ML phylogeny of the oxidoreductase subunits of the carbon monoxide/acetyl-CoA synthase (CODH/ACS) complex (CdhABC) encoded by the type II Acetothermia in context of other archaeal-like CdhABC (alignment length of 1,908 amino acid positions). Each subunit was individually aligned, and a concatenation of the three was subjected to ML analysis. The type II Acetothermia CdhABC are highlighted in bolded blue text. CdhABC from MAGs recovered from serpentinite-influenced environments are indicated on the right. Black circles show >90% bootstrap support (out of 1,000 bootstraps). Branch length is scaled based on the expected number of substitutions per site legend at the bottom left. CdhABC clades are collapsed as triangles, with the taxonomic groups they correspond to indicated next to the triangles. The monophyletic bacterial CdhABC clade is shown by an arrow. The tree is shown with a midpoint-rooted visualization. (B) Unrooted ML phylogeny of concatenated protein alignments from 30 housekeeping single-copy genes within 722 bacterial genomes representative of 128 phyla in the Genome Taxonomy Database (filtered alignment length of 9,162 amino acid positions; see Materials and Methods for additional analytical details). Phylum-level groupings are provided next to the shaded regions, with multiple phyla delineated with slashes. Genome information for the 722 entries is provided in Dataset S1. Black circles show >90% bootstrap support (out of 1,000 bootstraps). Bootstraps are only shown for phylum or higher-level groupings for clarity in interpreting the tree. Branch length is scaled based on the expected number of substitutions per site, as indicated on the left. Type I and II Acetothermia lineages are indicated with orange and blue stars, respectively. The placement of recently hypothesized root positions for the bacterial domain is shown with black (44) or white (94) stars.
To further assess the phylogenetic placement of the SO Acetothermia among other taxonomic divisions, a comprehensive phylogenomic analysis was conducted with 722 genomes representing 128 bacterial phyla in the Genome Taxonomy Database (Fig. 4B). Phylogenomic analysis of 30 universally conserved phylogenetic marker genes including RNA polymerase subunits and ribosomal proteins (see Materials and Methods for additional details) suggested that the Acetothermia/Bipolaricaulota lineage comprised a highly supported outgroup to the Thermotogota that, together, were related to the Deinococcota and others (Fig. 4B), consistent with previous phylogenomic analyses (35). Recent attempts to identify the root of the bacterial domain using comprehensive phylogenomic analyses have placed the bacterial root between the Thermotogota and all remaining bacterial phyla (44) or with the Thermotogota, Deinococcota, and Synergistota as a group closest to the optimal rooting position within several possible rooting scenarios (94) (Fig. 4B). These results are consistent with an early phylogenomic analysis placing the first recovered Acetothermia/Bipolaricaulota genome from Candidatus Acetothermum autotrophicum near the root of the bacterial tree along with the Thermotogae (now Thermotogota) and the Deinococcus-Thermus (now Deinococcota) (35). The precise position of the root of the bacterial domain remains controversial and is dependent on the taxa included, genes considered in the analysis, and phylogenetic methods that are used. Nevertheless, the placement of the Acetothermia/Bipolaricaulota as a highly supported sister clade to the Thermotogota and recent analyses placing the Thermotogota near the root of the bacterial domain (44, 94) suggest that the Acetothermia/Bipolaricaulota are also among the earliest evolving bacterial lineages. Together, these observations are consistent with the hypothesis that ancestors of type II Acetothermia inhabited environments undergoing active serpentinization and add further support to the supposition that serpentinization played a key role in supporting the earliest acetogens.
Conclusions
Type II Acetothermia from the SO exhibit physiological/metabolic traits that allow them to conserve energy in contemporary serpentinization-influenced environments. Phylogenetic data indicate that many of the traits, including modifications to the WL pathway, originated early during the evolution of this lineage, suggesting selective pressures on this cell type have been fairly constant since the metabolism first originated in the lineage. Type II Acetothermia are characteristic inhabitants of highly serpentinized waters in the SO and are likely key contributors to ecosystem productivity in these environments. Further, these organisms encode proteins involved in key energy conservation pathways that are early branching within the Bacteria or derive from deep within the archaeal domain (e.g., ATP synthases and carbon monoxide dehydrogenases). The phylogenetic and physiological characterization of contemporary type II Acetothermia helps bridge theoretical inferences regarding the role of serpentinization in supporting the earliest forms of life on Earth and reveals the key physiological characteristics that could allow this metabolism to operate at the thermodynamic limits of life (76) by persisting with substrates derived from water–rock interaction such as H2, HCOO−, and CO. Key questions remain regarding type II Acetothermia metabolism, including how their cellular energetics are balanced under high H2 partial pressures and low DIC conditions relative to more-DIC-replete conditions used to cultivate canonical acetogens. Further, the form of carbon (e.g., CO2/HCO3−/CO32−, HCOO−, or CO) that supports acetogenesis and carbon fixation by the WL pathway remains to be elucidated and will require cultures to definitively prove.
Supplementary Material
Acknowledgments
This work was supported by NASA Grants NNA15BB02A, 80NSSC21K0489, and 80NSSC19M0150 to D.R.C., A.S.T., J.R.S., and E.S.B. We thank the Department of Energy's Joint Genome Institute for generating metagenomic sequence from the 2020 samples. We are grateful to Prof. Juerg Matter for help with equipment acquisition, sampling, permitting, and sample export, in addition to the Ministry of Regional Municipalities and Water Resources in the Sultanate of Oman for allowing sampling and export of well waters. We thank Eric Ellison, Hannah Miller, Daniel Nothaft, and Laura Bueter for their help during 2015 and 2017 field work sampling trips; Tristan Caro, Tori Hoehler, Elizabeth Fones, and Mike Kubo during 2020 field work sampling trips; and Mason Munro-Ehrlich for help with 2020 data generation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206845119/-/DCSupplemental.
Data, Materials, and Software Availability
The Acetothermia MAG assemblies generated in this study are available under the NCBI Bioproject accession PRJNA738530 (95). Specific accessions within the 2020 metagenomes are referenced in Supplementary Table 2. Additional metagenomic data for the 2020 metagenomes is available in the JGI IMG database under accessions 3300045453 (96), 3300045950 (97), 3300045482 (98), and 3300045452 (99).
References
- 1.Sleep N. H., Meibom A., Fridriksson T., Coleman R. G., Bird D. K., H2-rich fluids from serpentinization: Geochemical and biotic implications. Proc. Natl. Acad. Sci. U.S.A. 101, 12818–12823 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell M. J., Martin W., The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 29, 358–363 (2004). [DOI] [PubMed] [Google Scholar]
- 3.McCollom T. M., Seewald J. S., Abiotic synthesis of organic compounds in deep-sea hydrothermal environments. Chem. Rev. 107, 382–401 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Seewald J. S., Zolotov M. Y., McCollom T. M., Experimental investigation of single carbon compounds under hydrothermal compounds. Geochim. Cosmochim. Acta 70, 446–460 (2006). [Google Scholar]
- 5.Preiner M., et al. , A hydrogen-dependent geochemical analogue of primordial carbon and energy metabolism. Nat. Ecol. Evol. 4, 534–542 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Weiss M. C., et al. , The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Russell M. J., Hall A. J., Martin W., Serpentinization as a source of energy at the origin of life. Geobiology 8, 355–371 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Boyd E. S., Amenabar M. J., Poudel S., Templeton A. S., Bioenergetic constraints on the origin of autotrophic metabolism. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 378, 20190151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs G., CO2 fixation in acetogenic bacteria: Variations on a theme. FEMS Microbiol. Rev. 2, 181–213 (1986). [Google Scholar]
- 10.Fuchs G., Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Schöne C., et al. , Deconstructing Methanosarcina acetivorans into an acetogenic archaeon. Proc. Natl. Acad. Sci. U.S.A. 119, e2113853119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm N. G., Oze C., Mousis O., Waite J. H., Guilbert-Lepoutre A., Serpentinization and the formation of H2 and CH4 on celestial bodies (Planets, Moons, Comets). Astrobiology 15, 587–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waite J. H., et al. , Cassini finds molecular hydrogen in the Enceladus plume: Evidence for hydrothermal processes. Science 356, 155–159 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Rempfert K. R., et al. , Geological and geochemical controls on subsurface microbial life in the Samail Ophiolite, Oman. Front. Microbiol. 8, 56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fones E. M., et al. , Physiological adaptations to serpentinization in the Samail Ophiolite, Oman. ISME J. 13, 1750–1762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brazelton W. J., Nelson B., Schrenk M. O., Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front. Microbiol. 2, 268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seyler L. M., et al. , Carbon assimilation strategies in ultrabasic groundwater: Clues from the integrated study of a serpentinization-influenced aquifer. mSystems 5, e00607-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki S., et al. , Unusual metabolic diversity of hyperalkaliphilic microbial communities associated with subterranean serpentinization at The Cedars. ISME J. 11, 2584–2598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woycheese K. M., Meyer-Dombard D. R., Cardace D., Argayosa A. M., Arcilla C. A., Out of the dark: Transitional subsurface-to-surface microbial diversity in a terrestrial serpentinizing seep (Manleluag, Pangasinan, the Philippines). Front. Microbiol. 6, 44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus E. A., et al. , Molecular evidence for an active microbial methane cycle in subsurface serpentinite-hosted groundwaters in the Samail Ophiolite, Oman. Appl. Environ. Microbiol. 87, e02068-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrenk M. O., Kelley D. S., Bolton S. A., Baross J. A., Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ. Microbiol. 6, 1086–1095 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Brazelton W. J., Schrenk M. O., Kelley D. S., Baross J. A., Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl. Environ. Microbiol. 72, 6257–6270 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postec A., et al. , Microbial diversity in a submarine carbonate edifice from the serpentinizing hydrothermal system of the Prony Bay (New Caledonia) over a 6-year period. Front. Microbiol. 6, 857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frouin E., et al. , Diversity of rare and abundant prokaryotic phylotypes in the Prony Hydrothermal Field and comparison with other serpentinite-hosted ecosystems. Front. Microbiol. 9, 102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brazelton W. J., Morrill P. L., Szponar N., Schrenk M. O., Bacterial communities associated with subsurface geochemical processes in continental serpentinite springs. Appl. Environ. Microbiol. 79, 3906–3916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki S., et al. , Microbial diversity in The Cedars, an ultrabasic, ultrareducing, and low salinity serpentinizing ecosystem. Proc. Natl. Acad. Sci. U.S.A. 110, 15336–15341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fones E. M., et al. , Diversification of methanogens into hyperalkaline serpentinizing environments through adaptations to minimize oxidant limitation. ISME J. 15, 1121–1135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohl L., et al. , Exploring the metabolic potential of microbial communities in ultra-basic, reducing springs at The Cedars, CA, USA: Experimental evidence of microbial methanogenesis and heterotrophic acetogenesis. J. Geophys. Res. Biogeosci. 121, 1203–1220 (2016). [Google Scholar]
- 29.Suzuki S., Nealson K. H., Ishii S., Genomic and in-situ transcriptomic characterization of the candidate phylum NPL-UPL2 from highly alkaline highly reducing serpentinized groundwater. Front. Microbiol. 9, 3141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merino N., et al. , Single-cell genomics of novel actinobacteria with the Wood-Ljungdahl pathway discovered in a serpentinizing system. Front. Microbiol. 11, 1031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Twing K. I., et al. , Serpentinization-influenced groundwater harbors extremely low diversity microbial communities adapted to high pH. Front. Microbiol. 8, 308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang S. Q., Butterfield D. A., Schulte M., Kelley D. S., Lilley M. D., Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim. Cosmochim. Acta 74, 941–952 (2010). [Google Scholar]
- 33.Miller H. M., et al. , Modern water/rock reactions in Oman hyperalkaline peridotite aquifers and implications for microbial habitability. Geochim. Cosmochim. Acta 179, 217–241 (2016). [Google Scholar]
- 34.Leong J. A. M., Shock E. L., Thermodynamic constraints on the geochemistry of low-temperature, continental, serpentinization-generated fluids. Am. J. Sci. 320, 185–235 (2020). [Google Scholar]
- 35.Takami H., et al. , A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One 7, e30559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youssef N. H., et al. , The Wood-Ljungdahl pathway as a key component of metabolic versatility in candidate phylum Bipolaricaulota (Acetothermia, OP1). Environ. Microbiol. Rep. 11, 538–547 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Nothaft D. B., et al. , Aqueous geochemical and microbial variation across discrete depth intervals in a peridotite aquifer assessed using a packer system in the Samail Ophiolite, Oman. J. Geophys. Res. Biogeo. 126, e06319 (2021). [Google Scholar]
- 38.Clum A., et al. , DOE JGI metagenome workflow. mSystems 6, e00804-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang D. D., Froula J., Egan R., Wang Z., MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colman D. R., Lindsay M. R., Boyd E. S., Mixing of meteoric and geothermal fluids supports hyperdiverse chemosynthetic hydrothermal communities. Nat. Commun. 10, 681 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandes-Martins M. C., et al. , Ecological dichotomies arise in microbial communities due to mixing of deep hydrothermal waters and atmospheric gas in a circumneutral hot spring. Appl. Environ. Microb. 87 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu M., Scott A. J., Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28, 1033–1034 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Gutierrez C. A., Aylward F. O., Phylogenetic signal, congruence, and uncertainty across Bacteria and Archaea. Mol. Biol. Evol. 38, 5514–5527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parks D. H., et al. , A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1079–1086 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konstantinidis K. T., Tiedje J. M., Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parks D., CompareM, Vers. 0.0.23. https://github.com/dparks1134/CompareM. Accessed 14 November 2020.
- 53.Adam P. S., Borrel G., Gribaldo S., Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc. Natl. Acad. Sci. U.S.A. 115, E1166–E1173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulkidjanian A. Y., Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V., Evolutionary primacy of sodium bioenergetics. Biol. Direct 3, 13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M., KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182-5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu S., et al. , CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters J. W., et al. , [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim. Biophys. Acta 1853, 1350–1369 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Søndergaard D., Pedersen C. N., Greening C., Hyd D. B., HydDB: A web tool for hydrogenase classification and analysis. Sci. Rep. 6, 34212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uritskiy G. V., DiRuggiero J., Taylor J., MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dewandel B., et al. , A conceptual hydrogeological model of ophiolite hard-rock aquifers in Oman based on a multiscale and a multidisciplinary approach. Hydrogeol. J. 13, 708–726 (2005). [Google Scholar]
- 64.Parks D. H., et al. , Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Kelley D. S., et al. , A serpentinite-hosted ecosystem: The Lost City hydrothermal field. Science 307, 1428–1434 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Templeton A. S., et al. , Accessing the subsurface biosphere within rocks undergoing active low-temperature serpentinization in the Samail Ophiolite (Oman Drilling Project). J. Geophys. Res. Biogeo. 126, e06315 (2021). [Google Scholar]
- 67.Shibata M., et al. , Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: Function and phylogenetic analysis. J. Biol. Chem. 277, 18658–18664 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Wang S., Huang H., Kahnt J., Thauer R. K., Clostridium acidurici electron-bifurcating formate dehydrogenase. Appl. Environ. Microbiol. 79, 6176–6179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nothaft D. B., et al. , Geochemical, biological, and clumped isotopologue evidence for substantial microbial methane production under carbon limitation in serpentinites of the Samail Ophiolite, Oman. J. Geophys. Res. Biogeo. 126, e06025 (2021). [Google Scholar]
- 70.Kelemen P. B., et al. , Initial results from the Oman Drilling Project Multi-Borehole Observatory: Petrogenesis and ongoing alteration of mantle peridotite in the weathering horizon. J. Geophys. Res. Sol. Earth 126, e22729 (2021). [Google Scholar]
- 71.Miller H. M., et al. , Low temperature hydrogen production during experimental hydration of partially-serpentinized dunite. Geochim. Cosmochim. Acta 209, 161–183 (2017). [Google Scholar]
- 72.Moon J., et al. , Formate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 23, 4214–4227 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Bertsch J., Müller V., CO metabolism in the acetogen Acetobacterium woodii. Appl. Environ. Microbiol. 81, 5949–5956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thauer R. K., CO2-reduction to formate by NADPH. The initial step in the total synthesis of acetate from CO2 in Clostridium thermoaceticum. FEBS Lett. 27, 111–115 (1972). [DOI] [PubMed] [Google Scholar]
- 75.Chenault H. K., Whitesides G. M., Regeneration of nicotinamide cofactors for use in organic synthesis. Appl. Biochem. Biotechnol. 14, 147–197 (1987). [DOI] [PubMed] [Google Scholar]
- 76.Schuchmann K., Müller V., Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Musfeldt M., Schönheit P., Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: Heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184, 636–644 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolfe A. J., The acetate switch. Microbiol. Mol. Biol. Rev. 69, 12–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poudel S., et al. , Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim. Biophys. Acta 1860, 1910–1921 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Schut G. J., Adams M. W., The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: A new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Greening C., et al. , Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10, 761–777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thauer R. K., Jungermann K., Decker K., Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mock J., Wang S., Huang H., Kahnt J., Thauer R. K., Evidence for a hexaheteromeric methylenetetrahydrofolate reductase in Moorella thermoacetica. J. Bacteriol. 196, 3303–3314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R., Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Kaster A. K., Moll J., Parey K., Thauer R. K., Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 2981–2986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hugenholtz J., Ivey D. M., Ljungdahl L. G., Carbon monoxide-driven electron transport in Clostridium thermoautotrophicum membranes. J. Bacteriol. 169, 5845–5847 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertsch J., Öppinger C., Hess V., Langer J. D., Müller V., Heterotrimeric NADH-oxidizing methylenetetrahydrofolate reductase from the acetogenic bacterium Acetobacterium woodii. J. Bacteriol. 197, 1681–1689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schut G. J., et al. , The role of geochemistry and energetics in the evolution of modern respiratory complexes from a proton-reducing ancestor. Biochim. Biophys. Acta 1857, 958–970 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Shin J., Song Y., Jeong Y., Cho B. K., Analysis of the core genome and pan-genome of autotrophic acetogenic bacteria. Front. Microbiol. 7, 1531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin W. F., Hydrogen, metals, bifurcating electrons, and proton gradients: The early evolution of biological energy conservation. FEBS Lett. 586, 485–493 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Shock E. L., McCollom T., Schulte M. D., “The emergence of metabolism from within hydrothermal systems” in Thermophiles, Wiegel J., Michael A. W. W., Eds. (CRC, 1998), pp. 79–96. [Google Scholar]
- 92.Shock E. L., Boyd E. S., Principles of geobiochemistry. Elements 11, 395–401 (2015). [Google Scholar]
- 93.Tiago I., Veríssimo A., Microbial and functional diversity of a subterrestrial high pH groundwater associated to serpentinization. Environ. Microbiol. 15, 1687–1706 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Coleman G. A., et al. , A rooted phylogeny resolves early bacterial evolution. Science 372, eabe0511 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Colman D. R., et al. , Genome-resolved metagenomic analysis of Acetothermia-division genomes from Samail Ophiolite subsurface waters. BioProject. https://www.ncbi.nlm.nih.gov/bioproject?term=PRJNA738530. Deposited 16 June 2021.
- 96.Colman D. R., et al. , Groundwater microbial communities from wells in Ash Sharqiyah North Governorate, Oman. JGI. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300045453. Deposited 10 May 2021.
- 97.Colman D. R., et al. , Groundwater microbial communities from wells in Ash Sharqiyah North Governorate, Oman. JGI. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300045950. Deposited 16 June 2021.
- 98.Colman D. R., et al. , Groundwater microbial communities from wells in Ash Sharqiyah North Governorate, Oman. JGI. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300045482. Deposited 11 May 2021.
- 99.Colman D. R., et al. , Groundwater microbial communities from wells in Ash Sharqiyah North Governorate, Oman. JGI. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300045452. Deposited 10 May 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Acetothermia MAG assemblies generated in this study are available under the NCBI Bioproject accession PRJNA738530 (95). Specific accessions within the 2020 metagenomes are referenced in Supplementary Table 2. Additional metagenomic data for the 2020 metagenomes is available in the JGI IMG database under accessions 3300045453 (96), 3300045950 (97), 3300045482 (98), and 3300045452 (99).