Significance
Therapies and preclinical probes designed to drug and better understand the specific functions of intracellular protein–protein interactions (PPIs) remain an area of unmet need. This study describes the development of prototype therapeutics against the FOXP3 homodimer, a PPI essential for regulatory T cell suppressive capacity. We demonstrate that hydrocarbon stapled peptides designed to block this interaction can dampen regulatory T cell (Treg cell) suppressive function and lead to genetic signatures of immune reactivation. This work provides strong scientific justification for continued development of FOXP3-specific peptide-based inhibitors and provides mechanistic insights into the design and delivery of specific inhibitors of the coiled-coil region of FOXP3. These studies ultimately could lead to new immunotherapeutic strategies to amplify immune responsiveness in a number of settings.
Keywords: stapled peptide, FOXP3, coiled-coil domain, inhibitor
Abstract
Despite continuing advances in the development of novel cellular-, antibody-, and chemotherapeutic-based strategies to enhance immune reactivity, the presence of regulatory T cells (Treg cells) remains a complicating factor for their clinical efficacy. To overcome dosing limitations and off-target effects from antibody-based Treg cell deletional strategies or small molecule drugging, we investigated the ability of hydrocarbon stapled alpha-helical (SAH) peptides to target FOXP3, the master transcription factor regulator of Treg cell development, maintenance, and suppressive function. Using the crystal structure of the FOXP3 homodimer as a guide, we developed SAHs in the likeness of a portion of the native FOXP3 antiparallel coiled-coil homodimerization domain (SAH-FOXP3) to block this key FOXP3 protein-protein interaction (PPI) through molecular mimicry. We describe the design, synthesis, and biochemical evaluation of single- and double-stapled SAHs covering the entire coiled-coil expanse. We show that lead SAH-FOXP3s bind FOXP3, are cell permeable and nontoxic to T cells, induce dose-dependent transcript and protein level alterations of FOXP3 target genes, impede Treg cell function, and lead to Treg cell gene expression changes in vivo consistent with FOXP3 dysfunction. These results demonstrate a proof of concept for rationally designed FOXP3-directed peptide therapeutics that could be used as approaches to amplify endogenous immune responsiveness.
Regulatory T cells (Treg cells) have been a therapeutic target of interest for decades based on early work showing that their depletion amplifies antitumor immune reactivity (1, 2). Poor clinical outcomes are predicted by increased peripheral and intratumoral Treg cells in a number of settings, including in patients who receive adoptive cellular therapies and immune checkpoint blockade (3–6). Although discovering effective approaches to modulate Treg cells remains a high priority, clinically meaningful progress has been hampered by small molecule or deletional anti-Treg cell-focused therapies lacking sufficient cellular specificity to avoid inadvertent targeting of other T cell populations (7–11). Despite clinical success in reactivating antitumor immune responses, current immune checkpoint blockades targeting PD-1, PD-L1, and CTLA-4 are not exempt from off-target effects and toxicities. In some cases, up to 60% of patients treated with checkpoint inhibitors incur severe immune-related toxicities, generally considered to result from reactivation of dormant or suppressed immune responses (12). Thus, identification and modulation of intracellular molecular targets unique to a more specific cellular immune population like Treg cells, and associated with their immunosuppressive function, should theoretically find increased success.
Efforts to identify the mechanisms responsible for Treg cell-specific immunologic function led to the discovery of disease-defining mutations in X chromosome-encoded transcription factor, FOXP3, in patients with IPEX (immunodysregulation, polyendocrinopathy, enteropathy, X-linked syndrome) and scurfy mice (13, 14). FOXP3 homodimerization, DNA binding, and its interaction with other cotranscriptional regulators are essential for Treg cell function (15). Several features of FOXP3 make it a potentially favorable drug target: (1) FOXP3 is expressed in Treg cells with little to no expression in other cells (16); (2) reduction and not absence of FOXP3 leads to dose-dependent non-Treg cell immune reactivation in mice and humans, suggesting that a fraction of FOXP3 in a cell needs to be inhibited to alter Treg cell function (17–19); (3) naturally occurring mutations within the coding region of FOXP3 in IPEX patients are primarily located in the two distinct protein-protein interaction (PPI) interfaces, highlighting critical structure/function relationships (20, 21); (4) FOXP3 sequence, structure, and overall function are highly conserved in humans and mice, reflecting shared mechanisms of action allowing for meaningful preclinical testing (13, 22–24); (5) known FOXP3 crystal structures allow for correlation between structure and drug design (25, 26); and (6) recent evidence has confirmed that the loss of FOXP3 expression in postthymic Treg cells leads to reprogramming of Treg cells into a T effector cell-like phenotype (27). Lastly, although Treg cell loss of any kind could lead to unwanted immune activation, Treg cell depletion alone in patients and adult mice does not result in autoimmunity, supporting that transient Treg cell inhibition remains a clinically viable therapeutic strategy (1, 8, 9). Given these points, we set out to develop a hydrocarbon stapled alpha-helical (SAH) peptide that would impair Treg cell activity via disruption of a key PPI domain of FOXP3 through molecular mimicry.
Transcription factor targets have traditionally been difficult to drug. Their successful pharmacological inhibition is usually achieved through targeting of upstream kinases or nuclear receptors, or through indirect or direct modulation of their expression, all of which lack specificity (28, 29). Additionally, while transcription factor modulation can also occur by altering the direct interaction with cognate DNA, this has been historically difficult to accomplish due to the highly positively charged DNA binding interfaces (30). Another option to target transcription factors is through blocking their ability to interact with specific partner proteins. Despite some successes, small molecules with drug-like properties are best at targeting PPIs with well-defined “hot spot” regions or concentrated binding foci. They often fail to target those, like transcription factor PPIs, mediated by large interfaces where binding is the summation of geographically distinct, relatively flat, or low affinity interactions and not by pocketed areas or grooves suitable for high affinity small molecule binding (31). In contrast, peptides synthesized in the likeness of their native sequence are natural choices to target such PPIs due to the fidelity of structural contact points between binding partners. However, as is often the case with alpha-helical peptides, they typically have poor secondary structure in solution and are rapidly proteolytically degraded (32). In part to overcome these hurdles, hydrocarbon-“stapled” peptides have shown great promise for disrupting alpha-helical PPIs through stabilization of a protein fragment’s natural secondary structure and imparting it with drug-like properties, including enhanced target specificity, affinity, protease resistance, and cellular uptake (33–35). Importantly, this approach has shown promise targeting other transcription factors, including NOTCH and p53 (36–38).
In light of these data, we reasoned that targeting an “undruggable” coiled-coil alpha-helical homodimerization domain (DD) of FOXP3 using hydrocarbon stapled peptides would enable pharmacologic disruption of FOXP3 activity in Treg cells. Here, we report the design, synthesis, and biochemical evaluation of single- and double-stapled SAHs of the FOXP3DD domain. Optimized SAHs bind FOXP3, inhibit FOXP3 binding to DNA, are cell permeable and nontoxic, inhibit Treg cell suppression ex vivo, and alter Treg cell expression in vivo.
Results
Design and Synthesis of SAH Peptides Targeting FOXP3.
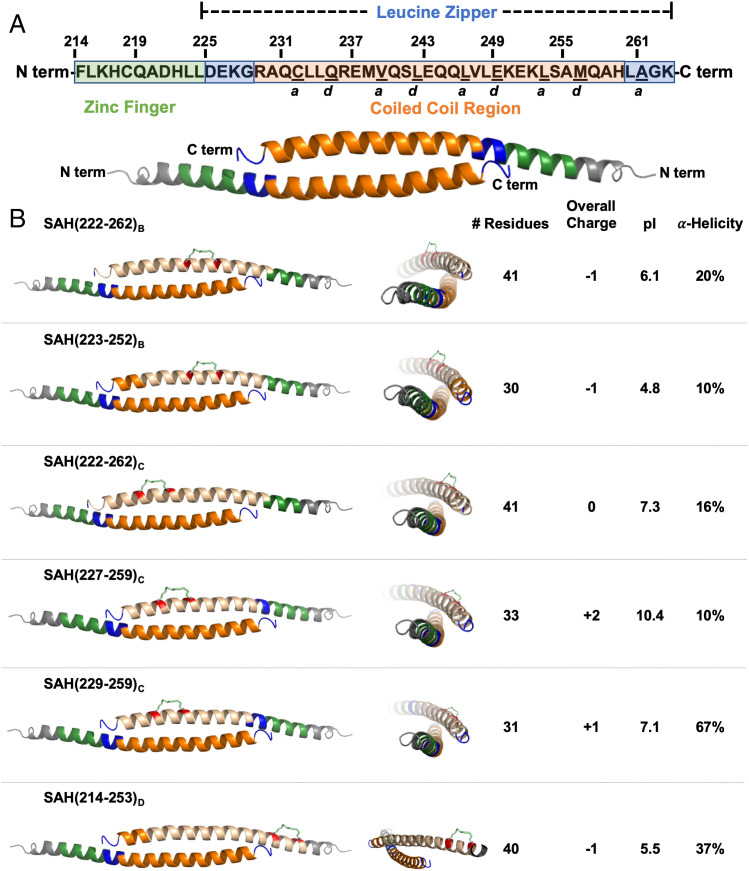
The leucine zipper coiled-coil (LZCC) of FOXP3 is necessary and sufficient for FOXP3 homodimerization and heterodimerization with FOXP1 and is critical for FOXP3’s ability to regulate transcription (39, 40). FOXP3 homodimerization is mediated by a two-stranded antiparallel alpha-helical coiled-coil interaction within this domain. Each contiguous helix is made up of the zinc finger and leucine zipper regions of FOXP3 (Fig. 1A). Within this contiguous helix, only a portion of the leucine zipper constitutes the LZCC. This antiparallel arrangement produces a generally hydrophobic coiled-coil core with flanking charged and polar residues necessary for dimer stabilization while solvent exposed polar residues enhance solubility (26). Sequence alignment of murine and human FOXP3 demonstrates that this helix is highly conserved between species (∼90%), with only five unique residues spanning the entirety of the contiguous zinc finger and leucine zipper regions (23). Most importantly, key core binding residues, Q235, E249, and K252, are 100% conserved between human and murine sequences, suggesting the ability to target both murine and human FOXP3 by drugging this region (26). Using the crystal structure of murine FOXP3 zinc finger and leucine zipper regions, additional examination of this portion of the FOXP3 sequence finds, like other coiled-coil interactions, a buried motif with amino acids (a.a.s) of a and d resonance (Fig. 1A). These a.a.s are involved in binding to an identical antiparallel helix, aligned on the hydrophobic interface of each respective helix (SI Appendix, Fig. S1). Within this helix, the terminal end residues that define the homodimer interaction are R229 and Q244 on one helix with Q244 and H259 on the corresponding antiparallel helix of another FOXP3 monomer. The interface accessible surface area of the FOXP3 coiled-coil is 966 Å2 (16% of total), compared with the average of 1,492 Å2 in other similar protein dimers, thereby classifying it as a rather flexible and dynamic interaction (26, 41).
Fig. 1.
Design and synthesis of stapled alpha-helical peptides targeting FOXP3. (A) Human FOXP3 sequence. Residues F214–K263 represent the entirety of a helix constituting the zinc finger (F214–L224; shown in green) and leucine zipper (D225–K263; shown in blue) regions. Within the leucine zipper, the coiled-coil (R229–H259; shown in orange) represents an area that is highly conserved between human and murine FOXP3. Underlined residues indicate a and d core residues, which bind each other on an identical antiparallel helix and are key contact points for homodimerization. The homodimerized FOXP3 helix is labeled according to the color codes in the sequence above. (B) Design strategy for SAHs targeting FOXP3. SAHs differing in sequence, length, staple position, overall charge, pI, and alpha-helicity are represented. Each three-dimensional structure shown is based on the crystal structure (PDB: 4I1L (26)); the upper helix represents each SAH (shown in tan) and hydrocarbon staple (insertion sites shown in red).
To determine the feasibility of blocking FOXP3 homodimerization through peptide-based molecular mimicry, a library of acetyl-capped SAHs incorporating residues within the core coiled-coil domain and N-terminal flanking region corresponding to hFOXP3(214–262) were synthesized with identical staples but with varying peptide length, charge, and staple positioning (Fig. 1B). Three differently positioned i, i + 7 hydrocarbon staples (B, C, and D) were inserted in such a way that they would extend along the peptide’s exposed face in an effort to avoid disruption of critical coiled-coil a.a. contact points (Fig. 1B and SI Appendix, Figs. S1–S3). These peptides additionally have a range of isoelectric points reflecting potential differences in solubility in various settings (e.g., in vitro versus in vivo) despite overlapping a.a. usage. Hydrocarbon staples spanning seven residues (i, i + 7) were chosen over shorter staples (e.g., i, i + 4), as longer staples have been generally shown to better stabilize alpha-helicity along long peptide fragments and can allow for increased protection against proteolysis (42). Variation of peptide length and staple positions greatly affected overall alpha-helicity as measured by circular dichroism spectroscopy (Fig. 1B). Importantly, circular dichroism was performed without the addition of organic solvent (e.g., acetonitrile), as this has been shown to enhance the estimated alpha-helical content of amphipathic helical peptides (43). Calculated alpha-helicity among these SAHs generally ranged from 10 to 40% in solution and showed only variably improved alpha-helicity compared with their sequence-matched native unstapled peptide controls (SI Appendix, Fig. S4). One notable exception was SAH(229–259)C, which was 67% alpha-helical, ∼2× greater than its native, unstapled counterpart (SI Appendix, Fig. S4). We hypothesized that greater helical stabilization would be advantageous and lead to increased mimicry of the natural FOXP3:FOXP3 PPI in addition to potential improvements in cellular permeability. To evaluate these assumptions, SAHs were next screened to measure how a.a. sequence, peptide length, and staple position affected their ability to bind FOXP3 and enter target cells.
Helicity, Peptide Length, and Staple Position Contribute to Optimized SAH-FOXP3 Target Binding.
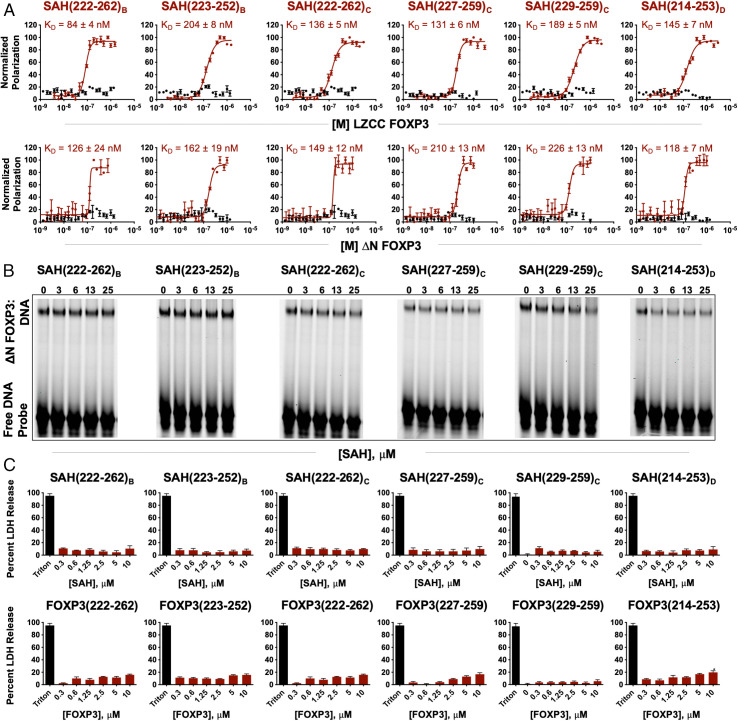
Fluorescence polarization analysis (FPA) was first used to assess binding affinities to two forms of truncated recombinant FOXP3, LZCC FOXP3 (T182-M337), and ΔN FOXP3 (T182-P431). Here, molecular binding events were measured in solution, allowing for determination of equilibrium constants reflective of specific receptor-target binding of protein and stapled peptide. While none of the native unstapled peptide controls bound LZCC FOXP3 or ΔN FOXP3, all SAHs bound LZCC FOXP3 and ΔN FOXP3 with affinities ranging from 84 nM to 226 nM (Fig. 2A). Furthermore, relative affinities were conserved for most SAHs between binding of LZCC FOXP3 and ΔN FOXP3, indicating that the measured binding activity was not restricted to the least complex target binding versions of FOXP3. SAHs and their corresponding native peptide controls were next tested for their ability to disrupt FOXP3 binding to cognate DNA using electrophoretic mobility shift assays (EMSAs). Here, a dose titration of individual SAHs was mixed with recombinant ΔN FOXP3 and a well-defined consensus FOXP3-binding oligonucleotide A’GT25 probe (25, 44). Dissociation or prevention of FOXP3 binding to DNA by SAH(229–259)C resulted in a greater dose-responsive inhibition of FOXP3:DNA binding compared with other SAHs (Fig. 2B and SI Appendix, Fig. S5). The overall charges of all SAHs ranged from −1 to +2. One possible pitfall of using stapled peptides in EMSAs is the possibility of measuring false positive DNA:target protein dissociation secondary to nonspecific binding of stapled peptides to free probe (45). This was not the case at the doses of SAH used in this assay, as there were no differences in free probe availability despite charge differences between SAHs (Fig. 2B). This is particularly interesting given that SAH(227–259)C has an overall charge of +2 and resulted in no apparent FOXP3:DNA dissociation. Thus, peptide sequence and staple position rather than overall charge appeared to drive the disruption of FOXP3:DNA binding, with potency appearing to coincide with increased alpha-helical content (SI Appendix, Fig. S4).
Fig. 2.
Helicity, peptide length, and staple position contribute to optimized SAH-FOXP3 target binding. (A) Fluorescence polarization analysis (FPA) binding curves of SAHs targeting FOXP3 (maroon) and their corresponding native unstapled peptide (black) bound to either LZCC FOXP3 or ΔN FOXP3. SAHs bind truncated FOXP3 with high affinity (KD values in the nanomolar range); native unstapled peptides are unable to bind truncated FOXP3 constructs. (B) Electrophoretic mobility shift assays (EMSAs) demonstrate that SAHs with staple position “C” are best able to disrupt FOXP3 binding to cognate DNA. Native unstapled peptides minimally bind even at high concentrations. (C) The absence of nonspecific murine CD4+ cell lysis upon SAH treatment, as assessed by LDH release at 4 h and normalized to vehicle.
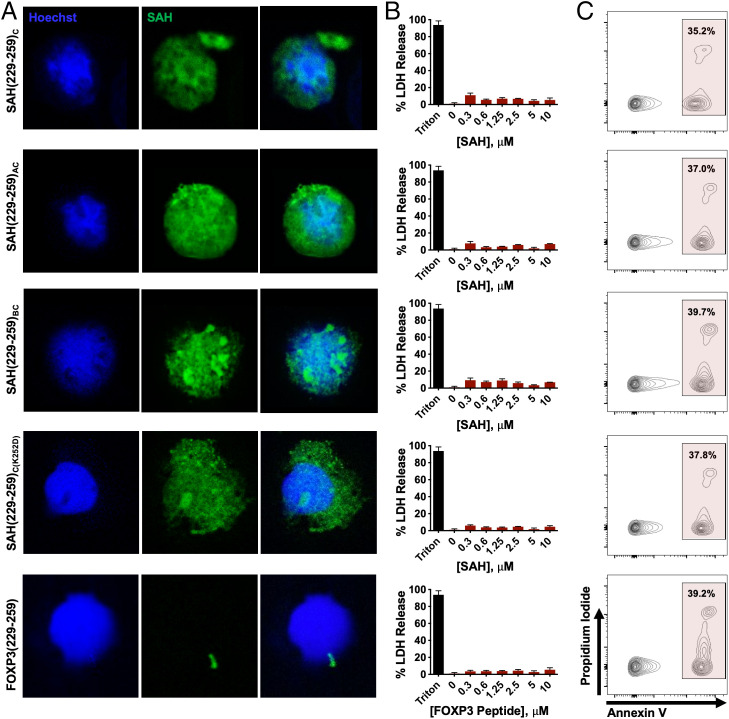
Prior to investigating further SAH optimization, all SAHs were next tested to ensure that, despite their amphipathic and helical nature, they could be incubated with cells and not cause nonspecific plasma membrane disruption or induce apoptosis. Indeed, we measured no cytoplasmic lactate dehydrogenase (LDH) release above background for any SAH tested at a broad range of doses (0–10 µM) in treated murine CD4+ T cells; nor did we detect increased apoptosis or loss of membrane integrity compared with vehicle-treated cells, as measured by annexin V/propidium iodide staining and flow cytometry (Fig. 2C and SI Appendix, Fig. S6). Cumulatively, these results indicate that a combination of sequence, staple position, and peptide length determine optimal alpha-helicity, that staple position is critical for on-target binding to FOXP3, and that SAHs are not overtly toxic to T cells.
Double Stapling of Peptides Modeled after SAH(229-259)C.
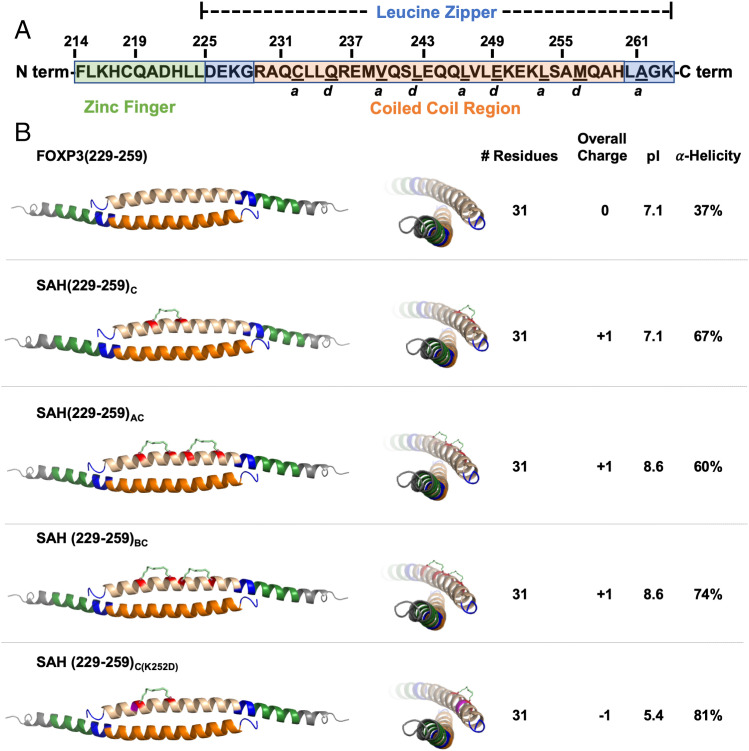
We selected SAH(229–259)C for further development based in part on its solubility profile in tissue culture media, overall charge, increased alpha-helicity compared with its unstapled analog, nanomolar binding to recombinant FOXP3 isoforms, and its ability to dose-dependently block the interaction between dimerized FOXP3 and cognate DNA. Additionally, unlike most of the other SAHs tested, SAH(229–259)C covers more core a and d resonant a.a.s and nearly the entire LZCC region of FOXP3, without encroaching on the zinc finger region. We next wondered whether insertion of additional staples N-terminally to the “C” position would increase binding to FOXP3 via induction of increased alpha-helicity or whether increased rigidity would decrease its interaction with immobilized FOXP3 (33). We therefore synthesized additional SAHs in the likeness of SAH(229–259)C, namely SAH(229–259)AC and SAH(229–259)BC with second staple positions in the “A” (a.a. 233 and 240) and “B” (a.a. 234 and 241) positions (SI Appendix, Fig. S7). We also designed a negative control point mutant that incorporated the single C staple position, SAH(229–259)C(K252D), based on a known IPEX mutation and critical contact point for FOXP3 homodimerization and function (26) (Fig. 3). Similar to our previous design strategy, staples were placed either opposite or adjacent to, but not within, the interaction interface (SI Appendix, Fig. S8A). We observed that SAH(229–259)BC had slightly improved alpha-helicity (74%) relative to the parent SAH(229–259)C (67%) as measured by circular dichroism (SI Appendix, Fig. S8B). Supporting our previous data, all SAH(229–259)C variants had significantly improved alpha-helicity relative to previously tested single-stapled SAHs, ranging from 60 to 80% compared with 10 to 37%, respectively, and continued to exhibit greatly improved alpha-helicity compared with their native, unstapled peptide controls. While the double-stapled SAHs had an identical overall charge compared with SAH(229–259)C, they had slightly different pIs based on the specific a.a.s replaced by the staples.
Fig. 3.
Double stapling of peptides modeled after SAH(229–259)C. (A) Human FOXP3 sequence. Residues F214–K263 represent the entirety of a helix constituting the zinc finger (F214–L224; shown in green) and leucine zipper (D225–K263; shown in blue) regions. Within the leucine zipper, the coiled-coil (R229–H259; shown in orange) represents an area that is highly conserved between human and murine FOXP3. Underlined residues indicate a and d core residues, which bind each other on an identical antiparallel helix and are key contact points for homodimerization. The homodimerized FOXP3 helix is labeled according to the color codes in the sequence above. (B) Design strategy for SAHs modeled after SAH(229–259)C. SAHs share sequence and length, differing in position and number of staples. These changes influence overall charge, pI, and alpha-helicity. Each three-dimensional structure shown is based on the crystal structure [PDB: 4I1L (26)]; the upper helix represents each SAH (shown in tan) and hydrocarbon staple (insertion sites shown in red).
Biochemical Characterization of SAH(229–259)C Constructs.
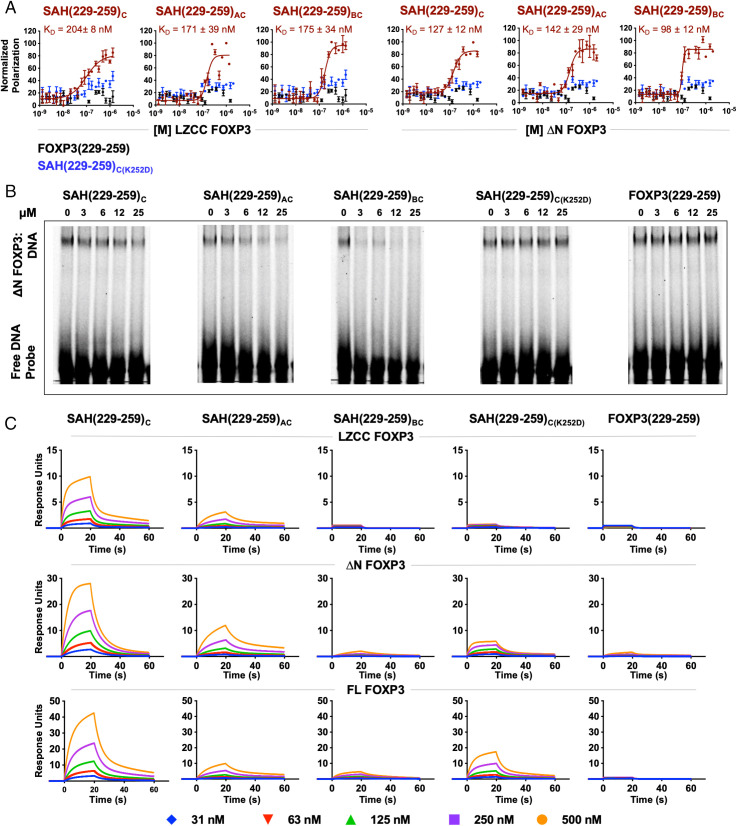
SAH(229–259)C, SAH(229–259)AC, and SAH(229–259)BC bound to LZCC FOXP3 and ΔN FOXP3 with affinities ranging from 98 nM to 204 nM, as measured by FPA (Fig. 4A). Unstapled FOXP3(229–259)Native and the point mutant control SAH(229–259)C(K252D) showed no binding to either FOXP3 isoform, reinforcing the importance of inherent helicity and sequence specificity for the SAH:FOXP3 interaction. Here, the addition of two hydrocarbon staples maintained similar high affinity, on-target binding to FOXP3 in solution. Furthermore, SAH(229–259)C and SAH(229–259)AC dose-dependently inhibited FOXP3 binding to a cognate oligonucleotide, while SAH(229–259)BC resulted in a blockade of the FOXP3:DNA interaction without a clear dose response. The point mutant and native peptide controls showed no effect (Fig. 4B and SI Appendix, Fig. S9). Again, none of the SAH(229–259)C derivatives, irrespective of overall charge, showed any nonspecific binding to the DNA probe. The natural FOXP3:FOXP3 leucine zipper interaction is thought to be rather transient and flexible. This is supported by data showing that recombinant leucine zippers of FOXP3 must be chemically crosslinked in vitro to observe durable homodimerization (26). Therefore, surface plasmon resonance (SPR) was next performed to orthogonally determine whether the binding of SAHs could be measured using a solid support approach and to determine whether double hydrocarbon stapling limited stepwise, dose-dependent binding to FOXP3 (Fig. 4C). We sought to determine whether binding changes across truncated and full-length immobilized forms of FOXP3 would identify candidate SAHs capable of drugging this dynamic PPI in a more biologically relevant cellular system. Single-stapled SAH(229–259)C dose-dependently bound to all forms of FOXP3 with the largest dynamic range, reflecting its more stable binding to FOXP3 compared with the double-stapled SAHs. The shape of the response curves to each FOXP3 construct indicated two-step, target-specific binding with fast on/off rates as measured by rapid decays in dissociations (Fig. 4C). Here, double-stapling did not improve the stability of SAH binding to FOXP3. While SAH(229–259)AC bound all forms of FOXP3, the magnitude of the interaction was approximately half that of SAH(229–259)C. The shape of the SAH(229–259)AC binding curves also indicated slower FOXP3 association at identical doses compared with SAH(229–259)C, likely reflective of its more rigid nature, as has been found in other stapled coiled-coil interactions (46). Despite promising results in EMSA testing, SAH(229–259)BC had no measurable binding to immobilized FOXP3, similar to unstapled FOXP3(229–259)Native. SAH(229–259)C(K252D) dose-dependently bound full-length FOXP3 but had a much lower dose–response compared with that of SAH(229–259)C (Fig. 4C). Thus, SAH(229–259)C appeared to strike an optimal balance of helicity and target binding in vitro and fulfilled a dual requirement for flexibility and specificity for binding as has been measured for other coiled-coil interactions (47–49).
Fig. 4.
Biochemical characterization of SAH(229–259)C constructs. (A) FPA binding curves of SAHs (maroon), their point mutant control (blue), and native unstapled peptide (black) bound to either LZCC FOXP3 or ΔN FOXP3. SAHs bind to protein (KD) in the nanomolar range. Point mutant control and native unstapled peptides do not bind either FOXP3 construct. (B) Reduction in ΔN FOXP3 binding to consensus DNA was observed following treatment with SAH(229–259)C derivatives, but not native peptide or point mutant control, as measured by EMSAs. (C) Surface plasmon resonance (SPR) of SAHs shows differences in binding kinetics between single- and double-stapled peptides against LZCC FOXP3, ΔN FOXP3, and full-length FOXP3.
Lead SAH(229–259)C Constructs Are Cell Permeable and Nontoxic to T Cells.
The ability of appropriately designed SAHs to penetrate cells is well established (35, 50, 51). Staple placement, alpha-helicity, and charge have been shown to confer differences between stapled peptides in their ability to interact with plasma membranes, directly penetrate cells, and traffic via microvesicles or endosomes (45, 52, 53). The dominant mechanism of SAH cellular uptake is thought to involve energy-dependent pinocytosis (51). Confocal microscopy found that all SAH(229–259)C compounds localized to the cytosol and nuclei of human in vitro induced Treg cells (iTreg cells) (Fig. 5A). Interestingly, staple position appeared to, at least in part, influence the intracellular patterning of individual SAHs. Treatment of cells with SAH(229–259)C, SAH(229–259)AC, and SAH(229–259)C(K252D) resulted in diffuse cellular staining, while SAH(229–259)BC resulted in more punctate patterning, perhaps reflecting aggregation or vesicular entrapment compared with SAH(229–259)C, SAH(229–259)AC, and SAH(229–259)C(K252D). In contrast, unstapled FOXP3(229–259)Native peptide showed no ability to penetrate or be retained in treated cells. FOXP3 is known to traffic from the cytosol and into the nucleus through binding additional cofactors, making these patterns biologically relevant (54). Importantly, human iTreg cells were morphologically unaltered following SAH treatment and maintained their round shape and high nuclear-to-cytoplasmic ratio. Treatment of murine thymocytes with these SAHs at a wide dose titration resulted in no LDH release or apoptotic cell death above background control-treated cells (Fig. 5 B and C).
Fig. 5.
Lead SAH(229–259)C constructs are cell permeable and nontoxic to T cells. (A) Cellular uptake by human iTreg cells incubated with FITC-labeled SAHs. Hoechst was used as a nuclear dye. SAH uptake exhibits distinct patterns, ranging from diffuse to more punctate fluorescence within the cell, including the nuclei and cytoplasm. (B and C) Absence of disruptive membrane lysis by (B) lack of nonspecific murine thymocyte membrane lysis as measured by LDH release at 4 h and normalized to vehicle and (C) apoptosis as measured by annexin V (APC) and Propidium Iodide (PI) staining and flow cytometry.
Single-Stapled SAH(229–259)C Induces Greater Gene and Protein Expression Alterations in Treg Cells Compared with Double-Stapled SAHs.
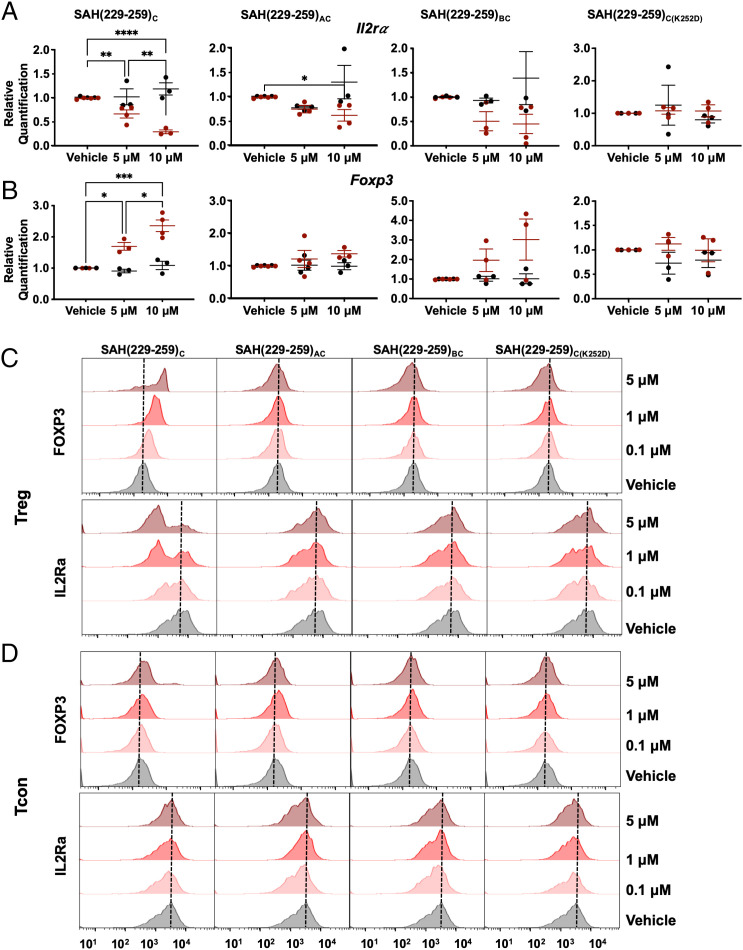
We next sought to determine whether single- and double-stapled SAH(229–259)C constructs could perturb normal FOXP3-regulated genetic changes in Treg cells through analysis of key FOXP3 transcriptional targets. Consistent with FOXP3 inhibition, SAH(229–259)C resulted in a dose-dependent decrease in Il2rα and increase in Foxp3 mRNA expression in Treg cells, but not in conventional CD4+ T cells (Tcon cells) (Fig. 6 A and B). Treatment with double-stapled SAH(229–259)AC and SAH(229–259)BC did not produce statistically significant changes in Il2rα and Foxp3 expression, consistent with their differences in binding to FOXP3 and despite their ability to block FOXP3:FOXP:DNA association extracellularly compared with SAH(229–259)C, again highlighting the importance for multimodal and orthogonal testing of developmental therapeutics (Fig. 4B). We anticipate that these differences are due in part to disparate binding kinetic and association behaviors when comparing the different SAHs. For example, SAH(229–259)BC appears to block FOXP3 binding to DNA effectively in EMSAs, but these results are not completely dose-dependent, suggesting a nonspecific interaction may complicate these results. This is supported by the lack of measurable binding in SPR and modest gene expression changes in treated Treg cells compared with SAH(229–259)C. Further, our data suggest that, despite improved binding against recombinant protein in EMSAs, the increased rigidity may not recapitulate the native PPI, making them ultimately poorer at regulating more finely tuned PPI-based regulation of target gene expression. As predicted, no changes were observed in cells treated with the single-stapled SAH(229–259)C(K252D) point mutant control, confirming on-target binding of SAH(229–259)C. These results support superior intracellular on-target efficacy of single-stapled SAH(229–259)C compared with its double-stapled counterparts. We next wanted to determine whether these genetic changes would extend to protein-level alterations in ex vivo expanded Treg cells. Reflecting our observed transcriptional changes, dose-dependent decreases in IL-2Rα and increases in FOXP3 protein expression were measured in SAH(229–259)C-treated Treg cells and not in double-stapled or point mutant control peptides (Fig. 6C). Expanded Tcon cells showed no changes in IL-2Rα or FOXP3 protein levels following treatment with any SAH at equivalent doses (Fig. 6D). The changes in FOXP3 expression in treated Treg cells is particularly intriguing, as FOXP3 is known to bind its own promoter and participate in the regulation of its own gene expression (27, 55).
Fig. 6.
Single-stapled SAH(229–259)C induces greater gene and protein expression alterations in Treg cells compared with double-stapled SAHs. (A and B) SAH(229–259)C dose dependently altered expression of (A) Il2rα and (B) Foxp3 in Treg cells (red), but not in Tcon cells (black) isolated from C57BL/6FOXP3-IRES-GFP mice measured by qRT-PCR. Double-stapled SAHs trended toward similar expression patterns, while point mutant and native peptide controls did not alter transcript expression in Treg cells or Tcon cells. (C and D) SAH(229–259)C treatment dose dependently altered IL-2Rα and FOXP3 protein in Treg cells (C), but not Tcon cells (D) as measured by flow cytometry. Asterisks indicate P < 0.05 determined by an ordinary one-way ANOVA with multiple comparisons. *P < 0.05, **P < 0.005, ***P < 0.001, ***P < 0.0001.
SAH(229-259)C Results in Greater Dampening of Treg Cell–Mediated Immune Suppression Compared with Double-Stapled SAHs.
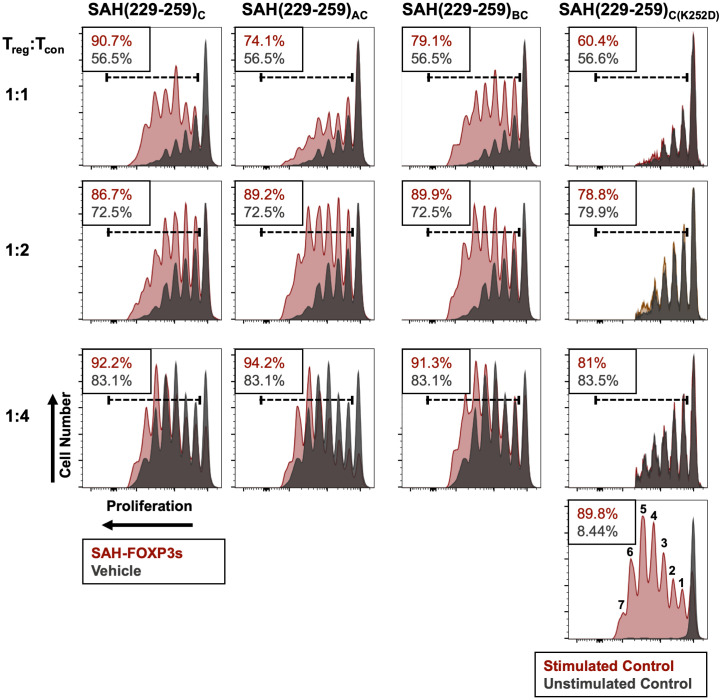
Given that SAH(229–259)C resulted in greater Treg cell transcript and protein expression compared with its double-stapled iterations, we next assessed whether similar differences would be reflected in their ability to induce dysfunctional Treg cell–mediated suppression ex vivo. To measure this, we assessed whether pretreatment of Treg cells with SAH(229–259)C constructs would inhibit proliferation of stimulated Tcon cells in culture. Pretreatment of Treg cells with single- and double-stapled SAH(229–259)C peptides dampened Treg cell-mediated ability to control target proliferation over the course of seven generations of proliferation (represented by individual peaks in each graph) compared with vehicle-treated Treg cells (Fig. 7). However, SAH(229–259)C exerted the greatest effect by completely inhibiting Treg cell-mediated suppression of target proliferation when mixed at a 1:1 ratio. Stark contrasts exist not only when comparing total proliferation relative to a vehicle-treated control but also in comparing the pace of expansion where SAH(229–259)C treatment resulted in the greatest proportion of target cells in later generations. Here, Tcon cell proliferation incubated with SAH-treated Treg cells equaled that of Tcon cells incubated in the absence of Treg cells (90.7% and 89.8%, respectively), while Tcon cells incubated with untreated Treg cells had far less proliferation (56.5%), reflecting normal Treg cell-mediated suppression. While SAH(229–259)AC and SAH(229–259)BC also inhibited Treg cell–mediated suppression (74.1% and 79.1% proliferation, respectively), they were nearly 13–20% less effective than SAH(229–259)C. Importantly, Treg cells pretreated with SAH(229–259)C(K252D) point mutant control and unstapled peptide showed no ability to prevent Treg cell–mediated suppression of Tcon cell proliferation (Fig. 7). In all cases, the SAH-induced inhibition of Treg cells was most measurable when Treg cells were cocultured with Tcon cells at equal ratios (1:1), though differences between SAH-treated and vehicle-treated Treg cells at higher titrations of Treg cells:Tcon cells indicated that there was measurable potency and durability to SAH-induced Treg cell dysfunction. While SAH(229–259)AC and SAH(229–259)BC resulted in less Treg cell transcriptional and protein expression changes compared with SAH(229–259)C in shorter-term cultures (6 h), they were able, albeit to a lesser degree, to still negatively impact Treg cell-mediated suppressive function following longer-term incubation (72 h). This supported our previous data showing that double-stapled SAHs resulted in muted trends in transcript expression compared with SAH(229–259)C. Collectively, these data indicate that specific inhibition of FOXP3 is feasible using stapled peptides, resulting in reduced acute Treg cell suppressive function in vitro.
Fig. 7.
SAH(229–259)C results in greater dampening of Treg cell–mediated immune suppression compared with double-stapled SAHs. Ex vivo suppression assays demonstrate dampened suppressive capacity of Treg cells treated with SAHs (maroon) compared with vehicle (black) in inhibiting target cell proliferation at a series of cell titrations. SAH(229–259)K252D had no effect on Treg cell suppressive function. Respective Tcon cell target proliferation is listed in the Top Corner of each plot. Stimulated and unstimulated target cell controls with numbered generations of cells are shown in the Bottom Right histogram.
SAH(229–259)C Alters Treg Cell mRNA Expression in vivo.
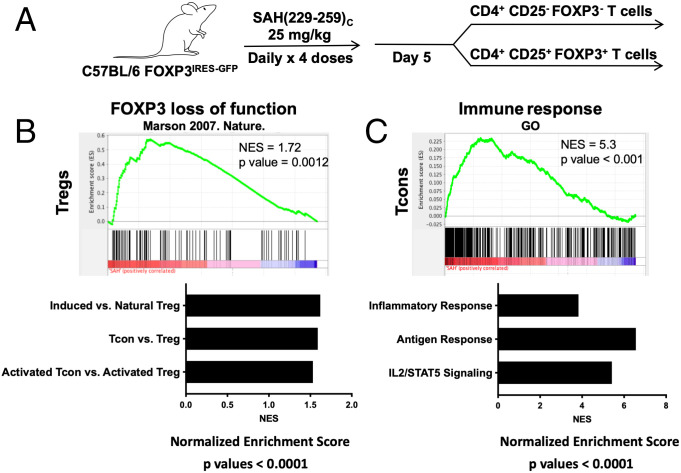
Based on correlation between target binding, genetic and protein alterations, intracellular localization, and functional Treg cell inhibition, we next tested the ability of SAH(229–259)C to alter Treg cell and Tcon cell expression patterns in vivo. Mice were treated with four daily intraperitoneal injections of SAH(229–259)C (Fig. 8A). Twenty-four hours following the final dose, Treg cells and Tcon cells were sorted and subjected to comprehensive transcriptional profiling. Gene set enrichment analysis (GSEA) found that Treg cells from SAH(229–259)C-treated animals had expression profiles that mirrored murine Foxp3-deficient hybridomas (56) (Fig. 8B). Further, GSEA also indicated that the transcriptional profiles of Treg cells from SAH(229–259)C-treated animals were more similar to induced rather than natural Treg cells (57), Tcon cells rather than Treg cells (58), and activated Tcon cells as opposed to activated Treg cells (57). Collectively, these data demonstrated that SAH(229–259)C led to transcriptional changes in Treg cells that made them inherently less like natural, thymic-derived Treg cells and more like Tcon cells. Interestingly, Tcon cells isolated from SAH(229–259)C-treated animals had enrichment of gene sets corresponding to proinflammatory responses and T cell activation (59) (Biocarta, Hallmark) (Fig. 8C). These expression patterns support that treatment with stapled peptides targeting FOXP3 are capable of inducing transcriptional reprogramming in Treg cells, at least in the short term, which may, in turn, induce a bystander T cell proinflammatory state as has been measured in patients with Treg cell dysfunction.
Fig. 8.
SAH(229–259)C alters Treg cell mRNA expression in vivo. (A) Treatment and isolation strategy of C57BL/6FOXP3-IRES-GFP mice with SAH(229–259)C in preparation for RNA sequencing. (B and C) Treatments of SAH(229–259)C altered global gene expression programs in (B) Treg cells and (C) Tcon cells with P < 0.0001. Following RNA sequencing analysis and subsequent GSEA, Treg cell gene expression mimicked gene sets corresponding to FOXP3 loss, among others. Tcon gene expression alterations mimicked gene sets corresponding to increased inflammatory responses, among others. NES, normalized enrichment score; GO, gene ontology.
Discussion
Despite great interest in the therapeutic targeting of Treg cells as a modality to improve antitumor immunity, major barriers have prevented effective clinical use of a number of compounds targeting molecules associated with Treg cell function (60). Challenges have included a combination of hurdles, such as a lack of specificity for Treg cells, compensation by other inhibitory molecules and inhibitory cytokines expressed by Treg cells, and off-target cellular toxicity in non-Treg T cells (61). Furthermore, many theoretical ideal targets include large PPIs that current small molecule approaches have been extremely difficult to target due to their more complex topographies and expansive geographies (62, 63).
Previous work targeting a separate FOXP3 PPI, that between FOXP3 and NFAT, demonstrated that nonstructured peptides spanning a loop domain and the Wing1 region of the FKH domain of FOXP3 (FOXP3 393–403) could partially inhibit FOXP3:NFAT and decrease tumor burden in mice coincidentally treated with chemotherapy or a tumor adjuvant vaccine (64). While encouraging, this peptide was not cell permeable and required plasmid transfection for cellular introduction. In another study, the same group utilized phage display to identify a random 15-mer peptide, P60, able to putatively bind somewhere within the contiguous FOXP3 ZF-LZ helix (a.a. 177–331) (65). While P60 showed some efficacy at doses of 100 µM, greater than an order of magnitude above doses used here, treated cells had indicators of apoptosis with observably higher cytoplasm:nucleus ratios and condensed chromatin. Treatment of mice with murine colon carcinoma with a cyclized version of the peptide emulsified in Freund adjuvant showed efficacy in a fraction of mice (66). We believe that the SAHs described in this study provide several promising advancements beyond this previous work, including inhibition of a new PPI, identification of specific binding location, improvement on required ex vivo dosage, and conclusive evidence of impacted FOXP3-mediated transcription.
In the present study, hydrocarbon stapling created stabilized alpha-helical peptide mimetics targeting the FOXP3 homodimer. Interestingly, while double-stapled SAHs may confer biophysical advantages in other systems, including reduced proteolytic degradation and helical stability compared with single-stapled peptides, our data suggest that increased rigidity may be detrimental depending upon the native PPI. In the case of FOXP3:FOXP3, the coiled-coil appears to be best recapitulated through mimicry with a more flexible SAH, as measured in other coiled-coil interactions. Whether such SAHs would result in ideal in vivo bioavailability is currently an area of active study.
One important area for future study also includes better understanding of details surrounding cellular localization of SAHs following uptake. We intend to confirm whether or not SAHs are getting trapped to some degree in cytoplasmic organelles, such as endosomes or lysosomes, and thus limiting their action. FOXP3 requires binding of various cofactors in order to traffic into the nucleus (54). It is important to understand whether SAHs are preferentially binding FOXP3 in the cytoplasm or nucleus and whether this has distinct effects on the ability of FOXP3 to bind nuclear factors or traffic to the nucleus. The findings of this current study also warrant further biochemical and cellular studies in order to elucidate how these SAHs may perturb other signaling pathways and identify their downstream mechanism(s) of action. For example, while not expressing FOXP3 to sufficient levels that induce a suppressive phenotype, both CD4+ Tcon cells and CD8+ cytotoxic T cells express low levels of FOXP3 that are transiently upregulated upon activation. Thus, it is conceivable that signaling pathways could also be dysregulated in these cells in some cases. Additionally, FOXP3 does not exist solely as a homodimer; thus, it is also reasonable to focus efforts on resultant transcriptional changes due to reduced or inhibited FOXP3, both as a homodimer or in interactions with other binding partners. One such PPI that could prove interesting is the heterodimer FOXP1:FOXP3, which binds in a similar location (FOXP3 LZDD) and could also be impacted by our SAHs.
Furthermore, the studies reported here involved normal mice. Additional studies exploring antitumor efficacy in preclinical tumor-bearing models would provide further justification for the continued study and exploration of SAHs targeting FOXP3, where reprogramming of Treg cells within the tumor may lead to a more proinflammatory environment. As has been discovered across a wide variety of targets and cancer types, Treg cell targeted therapy alone may not be effective against all tumors, particularly those with low neoantigen loads. This has been identified in murine models, where tumors derived from certain cell lines do not regress following Treg cell depletion (1). A critical next step will be identifying biomarkers of tumors in which Treg cells play an essential role in dampening antitumor immunity in order to better increase the chance of a successful Treg cell targeting therapy. As with many other therapeutic strategies, rational combination regimens are likely to be another reasonable approach, perhaps as in this case, the combination of FOXP3 SAHs with immune checkpoint blockade, or tumor-cell vaccination strategies.
SAHs targeting FOXP3:FOXP3 represent active preclinical probes of FOXP3 signaling pathways that have been demonstrated to reduce Treg cell suppressive function and alter global transcriptional profiles, reprogramming the immune system toward immune activation and away from suppression. We believe that these mimetics will enable future studies of FOXP3 domain-specific and protein partner-specific control of expression programs, many of which have not been fully elucidated. Current challenges that may also need to be overcome include determining optimal delivery mechanisms and application of medicinal chemistry approaches to facilitate clinical translation. To this end, we are currently developing methods of delivering stapled peptides into target cells using antibody-laden nanoparticles.
These challenges are not unique to SAHs. Indeed, prototype small molecules can often have initial low affinity (micromolar) that can be improved orders of magnitude through structure-based optimization. It is reasonable to expect that similar medicinal chemistry approaches could further improve SAH affinity. This has precedent, based on the evolution of SAHs targeting p53 (36, 67). Following subsequent iterations, there are currently a number of clinical trials utilizing this targeting strategy in adult and pediatric cancer patients (NCT02264613, NCT02909972, NCT03654716, NCT03725436, and NCT04022876). We are actively assessing additional sequences against this PPI to improve upon their activity, specifically developing peptides that are nonoverlapping and thus may be presumably less likely to dimerize in solution or within treated cells, and may improve their efficacy. One could also use in silico screening to help identify high-affinity hot-spot minimal peptide epitopes and maximize compound solubility through tools such as Druggable Protein-Protein Interaction Assessment System (68, 69) and ANCHOR (70, 71) and through the use of databases such as Search Tool for the Retrieval of Interacting Genes/Proteins (72), TIMBAL (73), and PICCOLO (74). Other approaches that can be taken to improve activity include addition of charged a.a.s or targeting domains. However, while such manipulations may improve solubility, we have also found that they may significantly dampen target affinity (75).
We have shown that designing peptide mimetics after known FOXP3 PPI crystal structures provides a promising starting point for the future development of similar transcription factor inhibitors, a class of drug targets that continues to be largely considered undruggable. Similarly designed molecular tools may also enable novel preclinical interrogation of other intracellular signaling pathways, leading to new areas of discovery.
Materials and Methods
Mice.
Ex vivo–treated Tcon cells and Treg cells were isolated from Foxp3IRES-GFP or C57BL/6 mice originally purchased from the Jackson Laboratory (C.Cg-Foxp3tm2Tch/J, C57BL/6J). All animal experiments were approved by and performed in accordance with the guidelines and regulations set forth by the Institutional Animal Care and Use Committee of the University of Chicago.
Murine T Cell Isolation and Expansion.
Foxp3IRES-GFP mice were sacrificed and their spleen, thymi, and lymph nodes removed, and single-cell suspensions were prepared via disruption of organs through a 40-μM filter. CD4+ cells were enriched through negative selection using a CD4+ T cell isolation kit following the manufacturer’s protocol (Miltenyi Biotec No. 130–104-454). CD4+ cells were stained with CD4-phycoerythrin (PE) (BD Biosciences) and CD25-allophycocyanin (APC) (BD Biosciences) flow cytometric antibodies diluted 1:100 in phosphate-buffered saline (PBS); Tcon cells (CD4+ CD25−) and Treg cells (CD4+CD25+ GFP+) were sorted on a BD FACSAria (University of Chicago Flow Cytometry Core). Sorted populations were >98% pure. Ex vivo–sorted Tcon cells and Treg cells were maintained and expanded in Advanced Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin/streptomycin, 5 μg/mL gentamicin solution, 2 mM L-glutamine, 1× nonessential a.a.s, 10 mM Hepes, human IL-2 (500 U/mL Tcon cells, 2,000 U/mL Treg cells), and CD3/CD28 Dynabeads at a ratio of 1 bead:1 cell (Tcon cells) or 3 beads:1 cell (Treg cells).
Human T Cell Isolation and iTreg Cell Induction.
Blood was drawn from healthy donors (University of Chicago institutional review board–approved protocol IRB14-0221) and was diluted 1:1 with PBS and layered over Ficoll–Paque PLUS density gradient media (GE Life Sciences). All donors signed informed consent prior to collection, and samples were deidentified prior to use. Following centrifugation for 30 min at 400g at 20 °C with brakes off, the peripheral blood mononuclear cell layer was removed and CD4+ cells were isolated using the CD4+ T Cell Isolation Kit (Miltenyi Biotec No. 130–045-101) per manufacturer’s directions. In order to induce Treg cells (iTreg cells), CD4+ cells were expanded in TEXMACS (Miltenyi Biotec No. 130–097-196) media containing 1,500 U IL-2, 100 nM all-trans retinoic acid, 2 ng/mL transforming growth factor-β, and anti-CD28 (Biolegend No. 302914) at 1:1,000 on CD3-coated (Biolegend No. 317302) 6-well plates (5 μg/mL).
Stapled Peptide Synthetic Reagents.
Fluorenylmethoxycarbonyl (Fmoc)-protected natural a.a.s, N-methyl-2-pyrrolidone (NMP), dichloromethane (DCM), and N,N-dimethylformamide (DMF) were purchased from Gyros Protein Technologies. The activating reagent (1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate [HATU]) was purchased from GenScript. The following were obtained from Sigma Aldrich: Rink amide AM resin LL (0.26 mmol/g), Grubbs Catalyst First Generation, anhydrous 1,2-dichloroethane (DCE), acetic anhydride (Ac2O), N,N-diisopropylethylamine (DIPEA), piperidine, and artificial a.a.s Fmoc-(R)-2-(7-octenyl)alanine (“R8”) paired with S5 to produce i, i+7 staples. Chemical reagents and solvents were used as received.
Stapled Peptide Synthesis and Purification.
Native and hydrocarbon stapled peptides were synthesized by Fmoc-based solid-phase peptide synthesis as described (35). Briefly, resin was swollen with DCM followed by DMF. Piperidine (20% vol/vol) in NMP was used for Fmoc deprotection with 2 × 10 min treatments. To the resin beads, an equal volume was added of a 0.3 M solution of a.a. in NMP, followed by a 0.285 M solution of HATU coupling agent in NMP and finally a 0.6 M solution of DIPEA in NMP. Typically, for standard couplings, a 10× excess of a.a.s relative to resin loading was used, and the reaction was allowed to proceed for 30 min. For addition of S5 or R8, a 5× excess of a.a.s were used for 1 h of coupling. For the a.a.s immediately following S5 or R8, the N terminus was deprotected with 4 × 10 min piperidine treatments followed by 4 × 1 h coupling reactions. After completion of synthesis, the amino terminus was either acetylated by reaction with capping solution (4:1:0.1 NMP:Ac2O:DIPEA) or left with an Fmoc-protected amine for fluorescein isothiocyanate (FITC) derivatization (see below). The olefin metathesis step was carried out by first swelling resin with anhydrous DCE, followed by exposure to 20 mol % Grubbs first-generation catalyst in DCE at 4 mg/mL for 3 × 2 h under nitrogen bubbling. Of note, native methionines were replaced with norleucine to optimize activity of the ruthenium catalyst. For FITC-functionalized peptides, resin was deprotected, and beta-alanine and FITC were added to the N terminus. The resin was then washed extensively with DCM and dried, and the peptides were then cleaved off the resin and deprotected using 95% trifluoroacetic acid (TFA), 2.5% water, and 2.5% triisopropylsilane. Double-stapled peptides were synthesized similarly, as described previously (33). Following synthesis, peptides were purified to >95% purity with high-performance liquid chromatography/mass spectrometry using a C18 column with mobile phases of water + 0.1% formic acid and acetonitrile. Acetonitrile was reduced in the pure fractions by rotary evaporation, and the peptides were then lyophilized. Aliquots were quantified by amino acid analysis.
Circular Dichroism Spectroscopy.
Dimethyl sulfoxide (DMSO) stock solutions of peptide were diluted in Milli-Q water. Each stock was aliquoted to two vials, then lyophilized for 2 d. Each sample was dissolved in 1 mL 5 mM sodium acetate buffer (pH 3.6) to bring samples to 2.5–10 μM. Each sample was put through 1 h of heated sonication to assist in dissolution. A blank sample following the same protocol was prepared to take the blank background reading for all samples. Data were acquired in five repetitions at 20 °C from 260 nm to 190 nm in a 1-mm path length quartz cell using 1-nm wavelength increments and a response time of 4 s on a Jasco J-815 Circular Dichroism Spectropolarimeter (Easton, MD, USA). The data were converted to per-residue molar ellipticity, and percent alpha-helicity was calculated as described previously (35).
Additional detailed methods can be found in (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
The authors acknowledge Megan McNerney, MD, PhD at the University of Chicago for advising on sequencing analysis pipelines. The authors thank Anjana Rao, PhD at the La Jolla Institute for Immunology for the gift of plasmids and probes. The authors thank Charles Benton for his assistance with experiments. We were supported by the University of Chicago Cytometry and Antibody Technology Core Facility (Cancer Center Support Grant P30CA014599) and the University of Chicago BioPhysics Core, where we specifically acknowledge the guidance of Elena Solomaha, PhD. We thank the University of Chicago Integrated Light Microscopy Core and Vytas Bindokas, PhD, supported by the Cancer Center Support Grant (P30CA014599). This work was supported by grants from the American Cancer Society (J.L.L.), Hyundai Hope on Wheels (J.L.L.), Lois R. and Maurice J. Beznos (J.L.L.), the University of Chicago Cancer Biology training grant/NIH T32CA009594 (K.M.H.), the University of Chicago Medical Science Training Program training grant T32GM007281 (M.R.S.), NIH F30 fellowship F30CA221250 (M.R.S.), NIH grant R35CA197583 (L.D.W.), and NIH grant R50CA211399 (G.H.B.).
Footnotes
Reviewers: K.A., University of Colorado Boulder; and P.H., Massachusetts Institute of Technology.
Competing interest statement: M.V.T. and K.A. were coauthors on an editorial in 2018 (Biomacromolecules 2018, 19, 3151−3162).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209044119/-/DCSupplemental.
Data, Materials, and Software Availability
The RNA sequencing data that support the findings in this study are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through accession No. GSE201116.
RNAseq data have been deposited in NCBI GEO (GSE201116) (76).
References
- 1.Onizuka S., et al. , Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor alpha) monoclonal antibody. Cancer Res. 59, 3128–3133 (1999). [PubMed] [Google Scholar]
- 2.Shimizu J., Yamazaki S., Sakaguchi S., Induction of tumor immunity by removing CD25+CD4+ T cells: A common basis between tumor immunity and autoimmunity. J. Immunol. 163, 5211–5218 (1999). [PubMed] [Google Scholar]
- 3.Bachanova V., et al. , Clearance of acute myeloid leukemia by haploidentical natural killer cells is improved using IL-2 diphtheria toxin fusion protein. Blood 123, 3855–3863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curiel T. J., et al. , Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Berntsen A., Brimnes M. K., thor Straten P., Svane I. M., Increase of circulating CD4+CD25highFoxp3+ regulatory T cells in patients with metastatic renal cell carcinoma during treatment with dendritic cell vaccination and low-dose interleukin-2. J. Immunother. 33, 425–434 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Shen Z., et al. , Higher intratumoral infiltrated Foxp3+ Treg numbers and Foxp3+/CD8+ ratio are associated with adverse prognosis in resectable gastric cancer. J. Cancer Res. Clin. Oncol. 136, 1585–1595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty G. L., et al. , First-in-human phase I study of the oral inhibitor of indoleamine 2,3-dioxygenase-1 epacadostat (INCB024360) in patients with advanced solid malignancies. Clin. Cancer Res. 23, 3269–3276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannull J., et al. , Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J. Clin. Invest. 115, 3623–3633 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rech A. J., et al. , CD25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci. Transl. Med. 4, 134ra62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simpson T. R., et al. , Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 210, 1695–1710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vargas F. A., et al. , Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell 33, 649–663.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins F., et al. , Adverse effects of immune-checkpoint inhibitors: Epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 16, 563–580 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Bennett C. L., et al. , The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Hori S., Nomura T., Sakaguchi S., Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Luo C. T., Li M. O., Transcriptional control of regulatory T cell development and function. Trends Immunol. 34, 531–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devaud C., Darcy P. K., Kershaw M. H., Foxp3 expression in T regulatory cells and other cell lineages. Cancer Immunol. Immunother. 63, 869–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balandina A., Lécart S., Dartevelle P., Saoudi A., Berrih-Aknin S., Functional defect of regulatory CD4(+)CD25+ T cells in the thymus of patients with autoimmune myasthenia gravis. Blood 105, 735–741 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura Y., et al. , Association of Foxp3 regulatory gene expression with graft-versus-host disease. Blood 104, 2187–2193 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Wan Y. Y., Flavell R. A., Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007). [DOI] [PubMed] [Google Scholar]
- 20.d’Hennezel E., Bin Dhuban K., Torgerson T., Piccirillo C. A., The immunogenetics of immune dysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J. Med. Genet. 49, 291–302 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Lopes J. E., et al. , Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J. Immunol. 177, 3133–3142 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Arvey A., et al. , Genetic and epigenetic variation in the lineage specification of regulatory T cells. eLife 4, e07571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolzer K., Käser T., Saalmüller A., Hammer S. E., Molecular characterisation of porcine Forkhead-box p3 (Foxp3). Vet. Immunol. Immunopathol. 132, 275–281 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Wildin R. S., et al. , X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27, 18–20 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Bandukwala H. S., et al. , Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity 34, 479–491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X., et al. , Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep. 1, 665–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charbonnier L. M., et al. , Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat. Immunol. 20, 1208–1219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caboni L., Lloyd D. G., Beyond the ligand-binding pocket: Targeting alternate sites in nuclear receptors. Med. Res. Rev. 33, 1081–1118 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Fontaine F., Overman J., François M., Pharmacological manipulation of transcription factor protein-protein interactions: Opportunities and obstacles. Cell Regen. (Lond.) 4, 2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushweller J. H., Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 19, 611–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchwald P., Small-molecule protein-protein interaction inhibitors: Therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations. IUBMB Life 62, 724–731 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Wójcik P., Berlicki Ł., Peptide-based inhibitors of protein-protein interactions. Bioorg. Med. Chem. Lett. 26, 707–713 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Bird G. H., et al. , Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc. Natl. Acad. Sci. U.S.A. 107, 14093–14098 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadji A., et al. , Preferential targeting of MCL-1 by a hydrocarbon-stapled BIM BH3 peptide. Oncotarget 10, 6219–6233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaBelle J. L., et al. , A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J. Clin. Invest. 122, 2018–2031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernal F., et al. , A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell 18, 411–422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y. S., et al. , Stapled α-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 110, E3445–E3454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moellering R. E., et al. , Direct inhibition of the NOTCH transcription factor complex. Nature 462, 182–188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chae W. J., Henegariu O., Lee S. K., Bothwell A. L., The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 9631–9636 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B., et al. , FOXP3 ensembles in T-cell regulation. Immunol. Rev. 212, 99–113 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Reynolds C., Damerell D., Jones S., ProtorP: A protein-protein interaction analysis server. Bioinformatics 25, 413–414 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Lau Y. H., de Andrade P., Wu Y., Spring D. R., Peptide stapling techniques based on different macrocyclisation chemistries. Chem. Soc. Rev. 44, 91–102 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Greenfield N. J., Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh K. P., Sundrud M. S., Rao A., Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLoS One 4, e8109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards A. L., et al. , Challenges in targeting a basic helix-loop-helix transcription factor with hydrocarbon-stapled peptides. ACS Chem. Biol. 11, 3146–3153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crone N. S. A., Kros A., Boyle A. L., Modulation of coiled-coil binding strength and fusogenicity through peptide stapling. Bioconjug. Chem. 31, 834–843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenfield N. J., et al. , The structure of the carboxyl terminus of striated alpha-tropomyosin in solution reveals an unusual parallel arrangement of interacting alpha-helices. Biochemistry 42, 614–619 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Singh A., Hitchcock-DeGregori S. E., Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin. Structure 14, 43–50 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Truebestein L., Leonard T. A., Coiled-coils: The long and short of it. BioEssays 38, 903–916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards A. L., et al. , Cellular uptake and ultrastructural localization underlie the pro-apoptotic activity of a hydrocarbon-stapled BIM BH3 peptide. ACS Chem. Biol. 10, 2149–2157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walensky L. D., et al. , Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bird G. H., et al. , Biophysical determinants for cellular uptake of hydrocarbon-stapled peptide helices. Nat. Chem. Biol. 12, 845–852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walensky L. D., Bird G. H., Hydrocarbon-stapled peptides: Principles, practice, and progress. J. Med. Chem. 57, 6275–6288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hancock W. W., Ozkaynak E., Three distinct domains contribute to nuclear transport of murine Foxp3. PLoS One 4, e7890 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birzele F., et al. , Next-generation insights into regulatory T cells: Expression profiling and FoxP3 occupancy in Human. Nucleic Acids Res. 39, 7946–7960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marson A., et al. , Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445, 931–935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haribhai D., et al. , A central role for induced regulatory T cells in tolerance induction in experimental colitis. J. Immunol. 182, 3461–3468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill J. A., et al. , Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity 27, 786–800 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Goldrath A. W., Luckey C. J., Park R., Benoist C., Mathis D., The molecular program induced in T cells undergoing homeostatic proliferation. Proc. Natl. Acad. Sci. U.S.A. 101, 16885–16890 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen B. J., Zhao J. W., Zhang D. H., Zheng A. H., Wu G. Q., Immunotherapy of cancer by targeting regulatory T cells. Int. Immunopharmacol. 104, 108469 (2022). [DOI] [PubMed] [Google Scholar]
- 61.Avarbock A. B., et al. , Lethal vascular leak syndrome after denileukin diftitox administration to a patient with cutaneous gamma/delta T-cell lymphoma and occult cirrhosis. Am. J. Hematol. 83, 593–595 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Xiao Y., et al. , Histone acetyltransferase mediated regulation of FOXP3 acetylation and Treg function. Curr. Opin. Immunol. 22, 583–591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudra D., et al. , Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 13, 1010–1019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozano T., et al. , Inhibition of FOXP3/NFAT interaction enhances T cell function after TCR stimulation. J. Immunol. 195, 3180–3189 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Casares N., et al. , A peptide inhibitor of FOXP3 impairs regulatory T cell activity and improves vaccine efficacy in mice. J. Immunol. 185, 5150–5159 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Lozano T., et al. , Blockage of FOXP3 transcription factor dimerization and FOXP3/AML1 interaction inhibits T regulatory cell activity: Sequence optimization of a peptide inhibitor. Oncotarget 8, 71709–71724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernal F., Tyler A. F., Korsmeyer S. J., Walensky L. D., Verdine G. L., Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J. Am. Chem. Soc. 129, 2456–2457 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehner B., Fraser A. G., A first-draft human protein-interaction map. Genome Biol. 5, R63 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sugaya N., Furuya T., Dr. PIAS: An integrative system for assessing the druggability of protein-protein interactions. BMC Bioinformatics 12, 50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuller J. C., Burgoyne N. J., Jackson R. M., Predicting druggable binding sites at the protein-protein interface. Drug Discov. Today 14, 155–161 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Meireles L. M., Dömling A. S., Camacho C. J., ANCHOR: A web server and database for analysis of protein-protein interaction binding pockets for drug discovery. Nucleic Acids Res. 38, W407–W411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szklarczyk D., et al. , The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higueruelo A. P., et al. , Atomic interactions and profile of small molecules disrupting protein-protein interfaces: The TIMBAL database. Chem. Biol. Drug Des. 74, 457–467 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Bickerton G. R., Higueruelo A. P., Blundell T. L., Comprehensive, atomic-level characterization of structurally characterized protein-protein interactions: The PICCOLO database. BMC Bioinformatics 12, 313 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schnorenberg M. R., et al. , Activating the intrinsic pathway of apoptosis using BIM BH3 peptides delivered by peptide amphiphiles with endosomal release. Materials (Basel) 12, 2567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hawley K. M., et al. , Inhibition of FOXP3 by Stapled Alpha-Helical Peptides Dampens Regulatory T Cell Function. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201116. Deposited 20 April 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data that support the findings in this study are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through accession No. GSE201116.
RNAseq data have been deposited in NCBI GEO (GSE201116) (76).