Significance
Z-ring formation by the protein FtsZ controls cell division in rod-shaped bacteria. The C-terminus of FtsZ encompasses a disordered C-terminal linker (CTL) and conserved CT17 motif. Both modules are essential for Z-ring formation and proper localization of FtsZ in cells. Previous studies suggested that generic intrinsically disordered regions (IDRs) might be suitable functional replacements for naturally occurring CTLs. Contrary to this suggestion, we find that the sequence-encoded conformational properties of CTLs help mediate autoregulatory interactions between covarying regions within FtsZ. Functional properties of the CTL are encoded via evolutionarily conserved, nonrandom sequence patterns. Disruption of these patterns impairs molecular functions and cellular phenotypes. Our findings have broad implications for discovering functionally consequential sequence features within IDRs of other proteins.
Keywords: intrinsically disordered proteins, autoregulation, autoinhibition, polymerization, covariation
Abstract
Intrinsically disordered regions (IDRs) can function as autoregulators of folded enzymes to which they are tethered. One example is the bacterial cell division protein FtsZ. This includes a folded core and a C-terminal tail (CTT) that encompasses a poorly conserved, disordered C-terminal linker (CTL) and a well-conserved 17-residue C-terminal peptide (CT17). Sites for GTPase activity of FtsZs are formed at the interface between GTP binding sites and T7 loops on cores of adjacent subunits within dimers. Here, we explore the basis of autoregulatory functions of the CTT in Bacillus subtilis FtsZ (Bs-FtsZ). Molecular simulations show that the CT17 of Bs-FtsZ makes statistically significant CTL-mediated contacts with the T7 loop. Statistical coupling analysis of more than 1,000 sequences from FtsZ orthologs reveals clear covariation of the T7 loop and the CT17 with most of the core domain, whereas the CTL is under independent selection. Despite this, we discover the conservation of nonrandom sequence patterns within CTLs across orthologs. To test how the nonrandom patterns of CTLs mediate CTT–core interactions and modulate FtsZ functionalities, we designed Bs-FtsZ variants by altering the patterning of oppositely charged residues within the CTL. Such alterations disrupt the core–CTT interactions, lead to anomalous assembly and inefficient GTP hydrolysis in vitro and protein degradation, aberrant assembly, and disruption of cell division in vivo. Our findings suggest that viable CTLs in FtsZs are likely to be IDRs that encompass nonrandom, functionally relevant sequence patterns that also preserve three-way covariation of the CT17, the T7 loop, and core domain.
Intrinsically disordered regions (IDRs) contribute to a multitude of protein functions (1–4). IDRs often have autoregulatory roles when tethered to folded domains either as tails or as linkers between folded domains (5–12). Of particular interest are IDRs tethered to folded domains that are enzymes (7, 13, 14). Several studies demonstrate that IDRs tethered to folded domains can function as autoregulators (12), specifically as autoinhibitors of enzymatic activities (13, 15, 16). One such example is the C-terminal tail (CTT) of the essential GTPase that controls and regulates bacterial cell division (17). The CTT encompasses a disordered C-terminal linker (CTL) and an alpha-helix-forming C-terminal peptide.
Cell division in bacteria is initiated by assembly of the cytokinetic ring at the nascent division site (18–26). Polymers formed by the essential GTPase filamenting temperature-sensitive mutant Z (FtsZ) are the foundation of this ring, which is also known as the Z-ring (27–32). FtsZ is a prokaryotic homolog of tubulin. It forms single-stranded protofilaments upon binding GTP in vitro (33). Linear polymers of FtsZ, which also undergo bundling via lateral associations, serve as a platform for the cell division machinery composed of at least 30 different proteins (19, 32, 34–39). FtsZ polymers also undergo treadmilling in vivo, driven by the turnover of subunits that occurs on the order of seconds (40).
Previous in vitro experiments showed that FtsZ polymerization belongs to a class of phase transitions known as reversible polymerization (41). A defining hallmark of reversible phase transitions, with subunit concentration as the conserved order parameter, is the presence of at least one threshold concentration for the occurrence of a specific phase transition. Cohan et al. recently identified two distinct threshold concentrations for Bacillus subtilis FtsZ (Bs-FtsZ) phase transitions occurring in the presence of GTP (17). In agreement with previous work on Escherichia coli FtsZ, Bs-FtsZ forms single-stranded protofilaments when the first threshold concentration, denoted as cA, is crossed (42–47). The second threshold concentration, denoted as cB where cB > cA, characterizes the threshold for bundling of protofilaments.
Bs-FtsZ encompasses two domains: a folded N-terminal core and a CTT (Fig. 1A). The core domain forms a complete GTPase upon dimerization whereby the T7 loop of one protomer is inserted into the nucleotide binding site of the complementary protomer. The interface between the T7 loop and the nucleotide binding site is the active site for GTP hydrolysis (48). The CTT is further composed of an intrinsically disordered CTL and a 17-residue C-terminal peptide (CT17). The CT17 was previously termed CTP (17), and it includes a conserved “constant region” and a “variable region” (CTV) (30, 49, 50). The CT17 can form alpha-helical conformations (51–53) and is thus an alpha-molecular recognition element (54) that enables a precise network of homotypic and heterotypic protein–protein interactions. Whereas the CT17 includes a conserved region (33), the CTL is hypervariable across orthologs, varying in length, amino acid composition, and sequence (33, 49, 55, 56). Mutations in the CTL and the CTV of Bs-FtsZ disrupt lateral interactions between protofilaments (49, 56).
Fig. 1.
Modular architecture of B. subtilis FtsZ includes a disordered CTT. (A) The electrostatic potential (103) is mapped onto the core domain in red and blue for regions of negative and positive potential, respectively. The T7 loop is highlighted in green. The CTT includes a CTL that connects the 17-residue C-terminal peptide (CT17) to the core domain. (B) The CTT is predicted to be disordered using IUPRED (60). The CTT sequence is shown with negatively and positively charged residues of the CTL in red and blue, respectively. The CT17 sequence is shown in gray. (C) Ensemble-averaged secondary structure contents of the CTT obtained from atomistic simulations. (D) UV-CD spectra of the four Bs-FtsZ constructs. See Materials and Methods for details on scaling of the [θ*].
Consistent with previous work (57–59), Cohan et al. showed, through systematic deletions of each module, that the core domain of Bs-FtsZ is the main driver of GTP binding-induced polymerization (17). Deletion of the CT17 (ΔCT17), previously referred to as ΔCTP, increases cA while also shifting cB upward by at least threefold. Internal deletion of the CTL (ΔCTL) decreases cA and this construct forms mini rings stabilized by cohesive interactions of the CT17. Overall, the CTL weakens the driving forces for linear polymerization and bundling, whereas the CT17 appears to be the primary driver of lateral associations. Deletion of the CTT (ΔCTT) lowers cA by over an order of magnitude and forms long, single-stranded polymers. Cohan et al. also showed that ΔCTT is the most efficient GTPase, whereas the wild-type Bs-FtsZ is the least efficient enzyme of the four constructs studied (17).
The picture that emerges is of the CTT as an autoregulator of Bs-FtsZ assembly and an autoinhibitor of enzymatic activity (17). Here, we uncover a molecular-level, mechanistic understanding of how the distinctive functions of CTTs are achieved.
Results
The CTL and CT17 Interact with the Core Domain at Mutually Exclusive Sites.
The CTT is predicted to be disordered with IUPRED scores being above 0.5 for the entirety of the CTT (Fig. 1B) (30, 60). We performed atomistic simulations to map conformational preferences of the isolated CTT peptide from Bs-FtsZ. These simulations show that the CTT of Bs-FtsZ samples a heterogenous ensemble of conformations with alpha-helical and random-coil character (Fig. 1C). A large fraction of the alpha-helical signal arises from the CT17, suggesting that it is an example of an alpha-helical molecular recognition element/feature (α-MoRE or α-ΜοRF) (61, 62). Consistent with the predictions from simulations, ultraviolet circular dichroism (UV-CD) measurements for the isolated CTT peptide indicate that it is conformationally heterogeneous (Fig. 1D and SI Appendix, Fig. S1).
The CD spectrum for the ΔCTT construct, which refers to the core domain without the CTT, is consistent with a mixture of structures with minima at 208 and 222 nm, indicating the presence of alpha-helical and beta-sheet structures. The spectrum of full-length FtsZ closely resembles that of the ΔCTT construct with a mixture of structures. The sum of the spectra for the core domain and the CTT does not reproduce the spectrum of a full-length protein. This suggests that the CTT likely undergoes conformational changes when it is tethered to the core domain by forming intermodule interactions with the core.
We also performed atomistic simulations of the full-length wild-type (WT) Bs-FtsZ protomer to investigate the presence and types of interactions among the core domain, the CTL, and the CT17. The core–CTL and core–CT17 interactions were defined by the presence of residues within the CTL or CT17 being within 10 Å of a core residue. The frequencies of contacts were calculated from simulations. These are shown as a heat map that maps the contacts being made onto the structure of the core domain. The CT17 fluctuates into and out of making contacts with the T7 loop (Fig. 2A), whereas contacts between the CTL and the core exclude the T7 loop and mainly involve residues that are spatially proximal to the region on the core where the CTT is N-terminally tethered (Fig. 2C). Synergies between the core–CTL and core–CT17 interactions were definitively unmasked in simulations of the different deletion constructs, viz., the ΔCTL and ΔCT17 that were studied by Cohan et al. (17). The CT17 does not interact with the T7 loop when the CTL is deleted. Instead, alternate contacts form with the residues that are spatially proximal to the site where the CT17 is N-terminally tethered to the core domain in the ΔCTL construct (Fig. 2B). Conversely, when the CT17 is deleted, the interactions between the CTL and the core that form in the full-length WT Bs-FtsZ protomer are preserved. Additionally, there are statistically significant contacts between the CTL and the T7 loop in the absence of CT17.
Fig. 2.
The CT17 and the CTL interact with mutually exclusive sites on the surface of the core domain. Color bars show the contact frequency between the core and (A and B) CT17 (blue) or (C and D) CTL (red) in WT, ΔCTL, and ΔCT17 constructs. The T7 loop is shown in space-filling model. The C terminus of the core domain where the CTT emanates is noted.
Overall, the simulations show that the CTL and the CT17 can compete for interactions with the T7 loop of the core domain. However, in the full-length protomer, interactions between the CT17 and the T7 loop are favored over those with the CTL. Thus, the CTL becomes the facilitator of distal contacts between the CT17 and the T7 loop of the core domain. These results suggest the presence of “on” and “off” states whereby in the “on” state the CT17 occupies the T7 loop, and in the “off” state the CT17 is dissociated from the T7 loop. Importantly, as discussed in SI Appendix, Fig. S2, the CTL-mediated contacts cannot be explained purely based on the intrinsic flexibility of the disordered CTL. Instead, these contacts appear to be sequence-specific. Accordingly, we investigated the evolutionary basis for sequence-specific, CTL-mediated contacts.
The CTL Is under Independent Selection whereas the T7 Loop and CTP Belong to the Same Covarying Sector.
While the core domain and the CT17 are conserved, the CTL is hypervariable in length and amino acid composition across FtsZ orthologs (55). We investigated covariations of sequence regions within FtsZ to ask if the interaction between the T7 loop and the CT17 modulated by the CTL is manifest as a statistically significant covariation. For this, we applied the statistical coupling analysis (SCA) first developed by Lockless and Ranganathan. This approach identifies groups of covarying residues, termed sectors, and helps determine the contribution of correlation to positional sequence conservation or covariation (63–65). First, the full sequences of 1,208 FtsZ orthologs were collected and aligned using multiple sequence alignment (MSA) (66, 67). The results from the MSA were used for the SCA along with the structure of Bs-FtsZ (48) as a reference for residue positions. Then, a covariance matrix between all pairs of sequence positions, weighted by the extent of conservation, was computed (SI Appendix, Fig. S3). Five sectors were identified from the SCA matrix that show correlations across groups. The sectors are arbitrarily named Sector 1 through 5 (Fig. 3B). As expected, the CTL is not represented in any of the sectors. This implies that it might be under independent selection, distinct from other parts of FtsZ. On the other hand, both the core domain and the CT17 are represented by one of the five sectors.
Fig. 3.
Protein sectors identified within FtsZ show covariation of the CT17 and most of the core domain surface, including the T7 loop. (A) Structure of Bs-FtsZ is shown (Protein Data Bank ID: 2VAM) (48) with cartoon-like schematic of CTT emanating from the C terminus of the core domain as a reference for B and C. The T7 loop is highlighted in cyan and the CT17 in magenta. (B) Five protein sectors are identified from SCA of 1,208 FtsZ orthologs. Each of Sectors 1 through 5 is colored in red, orange, yellow, green, and blue, respectively. (C) Only Sector 1 is shown in red, which includes both the T7 loop and the CT17.
Sector 1 is represented by 43% of the core domain residues that are prominently represented on the surface of FtsZ, including a part of the T7 loop (residues 205 and 206), in addition to an eight-residue region in the CT17 (Fig. 3C). Of particular interest is whether the representation of both the T7 loop and the CT17 in the same sector indicates covariation of the two regions. To examine this, we performed SCA on the sequences of only the core domains that were extracted from the same set of FtsZ orthologs. Interestingly, in the core-only analysis, the T7 loop does not belong to any of the sectors, thus appearing as a nonsector module in terms of the core alone (SI Appendix, Fig. S4). However, the nucleotide binding pocket within the core domain, which forms one-half of the dimerization site, remains part of Sector 1 (SI Appendix, Fig. S4). The presence of both the T7 loop residues and the nucleotide binding pocket in Sector 1 or any of the sectors is only realized in the presence of CT17, indicating covariation of the T7 loop and the CT17 regions. These results suggest that tethering enables covariation of regions that engage in autoinhibitory interactions. The next question is if the CTL, which enables the tethering, is a random, flexible IDR or if there are distinct sequence patterns that facilitate optimal autoinhibitory and other functions.
Identifying Nonrandom Sequence Patterns within the Bs-FtsZ CTL.
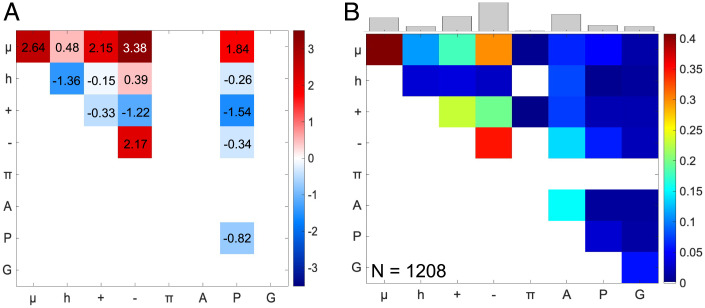
We asked if there are nonrandom sequence patterns within the CTL of Bs-FtsZ. Our goal was to identify sequence patterns that are likely facilitators of CTT interactions with the core across FtsZ orthologs. For this, we utilized the recently described NARDINI algorithm to analyze the CTL of Bs-FtsZ and extract nonrandom binary patterns using this alignment-free approach (68). NARDINI quantifies the extents of mixing vs. segregation of different pairs of amino acid types and ascribes a z-score to each of the binary patterns. A pattern is deemed to be nonrandom if the associated z-score meets the criteria of −1.5 > zxy or zxy > 1.5, where xy refers to a specific pair of residue types. This analysis shows that the CTL of Bs-FtsZ is characterized by nonrandom linear segregation of polar residues x ≡ (Gln, Ser, Thr, Asn, His) with respect to negatively charged residues y ≡ (Asp, Glu), positively charged residues y ≡ (Arg, Lys), and proline residues (Fig. 4A). Additionally, negatively charged residues also show nonrandom linear segregation with respect to all other residue types. The nonrandom linear segregation of polar and negatively charged residues appears to explain the observed statistically significant interactions with the core (Fig. 2). Clusters of negatively charged residues are likely to repel the acid-rich surface of the core (Fig. 1A), whereas clusters of polar residues are likely to enable weak interactions with the core. These features of the CTT, specifically the CTL, appear to be facilitators of the observed interactions between the T7 loop and CT17. Interestingly, positively charged residues are randomly distributed along the sequence of the CTL (−1.5 < z++ < 1.5), indicating a selection against clusters of positively charged residues that can interact favorably with the negatively charged surface of the core (Fig. 1A).
Fig. 4.
Uncovering nonrandom binary patterns in the Bs-FtsZ CTL and from CTLs across orthologs. (A) Z-score matrix for the CTL of Bs-FtsZ shows statistically significant nonrandom sequence patterns. Color bar indicates the z-scores. The axes describe the residue or residue types for which the z-scores were calculated: μ = polar, h = hydrophobic, + = positively charged, − = negatively charged, π = aromatic, A = alanine, P = proline, G = glycine. (B) Frequency of observing nonrandom segregation of different types of residue pairs (zxy > 1.5) in CTLs from 1,208 FtsZ orthologs. The histogram on top shows relative cumulative frequencies of nonrandom features for segregation of each residue/residue type.
Despite the demonstrated hypervariability of CTL sequences (55), we find that several binary sequence patterns are conserved across CTLs derived from 1,208 distinct FtsZ orthologs (SI Appendix, Fig. S5). In accord with findings for Bs-FtsZ, we find that polar and negatively charged residues form linear clusters that are segregated from one another and all other residue types (Fig. 4B) within CTL sequences. The NARDINI analysis highlights the conservation of linear clusters of negatively charged residues and clusters of polar residues. We propose that these features enable the preservation of statistically significant CTL–core interactions that enable inhibitory interactions through contacts between the CT17 peptide and the T7 loop. The corollary of our proposal is that disruption of the conserved sequence patterns, achieved by mixing or segregating oppositely charged residues along the sequence of the CTL, will weaken native interactions, or enhance nonnative interactions, thus having phenotypic consequences in vitro and in vivo. We tested this hypothesis using sequence design whereby we generated variants, each featuring CTLs with the same amino acid composition of the WT Bs-FtsZ while titrating the extent of mixing vs. segregation of oppositely charged residues within the CTL.
Design of CTL Variants.
It is well known that IDRs where the oppositely charged residues are well-mixed in the linear sequence will prefer expanded, well-solvated conformations (69–71). Additionally, CTLs with well-mixed oppositely charged residues in the linear sequence are expected to weaken native CTT interactions with the negatively charged core. Conversely, IDRs where the oppositely charged residues are segregated with respect to one another have the potential to engage in strong nonnative intra-CTL/intra-CTT interactions, nonnative interactions with the FtsZ core, and nonnative inter-CTL interactions between different FtsZ molecules.
The linear mixing/segregation of oppositely charged residues can be quantified using different parameters (69, 72, 73) including κ+−, where 0 ≤ κ+− ≤ 1 (69). Oppositely charged residues that are uniformly mixed within a linear sequence will yield values of κ+− ≈ 0, whereas a separation of oppositely charged residues into blocks will yield values of κ+− ≈ 1. To test the impact of the linear segregation vs. mixing of oppositely charged residues on FtsZ functionalities, we designed a set of CTL variants using the amino acid composition of the CTL from Bs-FtsZ. The designed sequences span κ+− values between 0.14 and 0.72 (SI Appendix, Table S1). The calculation of CTT κ+− values includes both the CTL and CT17 modules. Each of the designed Bs-FtsZ variants is denoted as Kx, where x is hundred times the κ+− value of the CTT in question. The sequences of the core domain and the CT17 were not altered.
Increasing κ+− of the CTT Introduces Intra-CTT Interactions and Enhances CTT–Core Interactions.
The working hypothesis is that increased charge segregation enhances nonnative intra-CTT, thereby weakening CTT–core interactions and reducing the potential for autoinhibition. To test this hypothesis, we performed two types of simulations of the Kappa variants of Bs-FtsZ. In the first mode, we performed simulations of CTT variants as autonomous units. In the second mode, we interrogated the conformations accessible to full-length FtsZ molecules with the CTT tethered N-terminally to the core. Results from simulations in mode 1 show a decrease of ensemble-averaged radii of gyration (Rg) as κ+− is increased (Fig. 5A). This derives from the increased favorability of intra-CTT interactions between blocks of oppositely charged residues within the CTL as the segregation of oppositely charged residues is increased. We tested predictions from simulations by performing fluorescence correlation spectroscopy (FCS) measurements to quantify the ensemble-averaged hydrodynamic radii (Rh) of CTT peptides. We observe a clear one-to-one correspondence between the calculated Rg and measured Rh values. It follows, in accord with previous observations (69, 70, 74), that increasing the linear segregation of oppositely charged residues drives the compaction of IDRs through cohesive interactions within the IDRs.
Fig. 5.
Increased linear segregation of oppositely charged residues leads to compaction. (A) Computed Rg values from simulations and measured Rh values from FCS experiments as a function of κ+− values of the Kappa variants. (B) Calculated Shannon entropy (s′) values of the Kappa variants from computed bivariate distributions of Rg and asphericity (δ*) values. (C) Normalized Rg of the Kappa variants from simulations performed in two modes, isolated (open symbols) and tethered to the GTPase core (solid symbols). CTT sequences with lower κ+− values undergo compaction upon being tethered to the folded core. (D) DLS data, shown as a checkerboard plot, quantifies the percent likelihood of observing a scatterer of a specific size for each of the Kappa variants.
Two-dimensional histograms of normalized Rg and asphericity values of the CTT variants show evidence of conformational heterogeneity as manifest in the broad distribution of the sizes and shapes (55) (SI Appendix, Fig. S6). We used these distributions to compute information theoretic measures of conformational heterogeneity such as relative Shannon entropies (s′) (55) (Fig. 5B). Despite a doubling of κ+− from 0.20 and 0.41, the values of s′ for the relevant CTT variants are bounded between 0.65 and 0.7. This implies that despite the decrease in Rg with increasing κ+−, the overall conformational heterogeneity is not greatly impacted because enhancements in shape fluctuations offset any diminution in size fluctuations (55). However, we observe a clear reduction in s′ values as κ+− increases beyond 0.41.
Simulations of full-length proteins with the CTTs attached N-terminally to the core domain (Fig. 5C) show that the CTT undergoes additional compaction for κ+− < 0.47. This derives from contacts formed between the CTT and the core domain. However, this additional compaction is not observed in variants where the CTT κ+− is greater than 0.41. This is because the interactions between oppositely charged blocky residues within the CTT are stronger than CTT–core interactions as κ+− increases beyond 0.41. The simulations support the hypothesis that segregation of oppositely charged residues into distinct blocks strengthens intra-CTT interactions thereby minimizing CTT–core interactions. This should weaken the autoinhibitory functions of the CTT, leading instead to alternative interactions in cis and in trans via the CTT.
CTT–CTT Interactions Can Drive Nonnative FtsZ Assemblies In Vitro.
The preceding discussion shows that linear segregation of oppositely charged residues within the CTL introduces nonnative interactions within the CTL. We reasoned that increased segregation of oppositely charged residues within the CTL also has the potential of introducing nonnative, CTT-mediated interdomain interactions in trans. These are likely to involve a combination of CTT–core and CTT–CTT interactions in trans that are nonexistent for the WT FtsZ. To probe for these nonnative interactions, we used dynamic light scattering (DLS) measurements of the different Bs-FtsZ variants to assess the contributions of nonnative intermolecular interactions between FtsZ protomers. These measurements were performed in the absence of GTP to detect the presence of nonnative interactions in the designed variants. We observed two groups of apparent hydrodynamic diameters (Dh) (Fig. 5D). Diameters smaller than 102 nm correspond to variants whose CTT κ+− values are lower than 0.3. In contrast, Dh values larger than 102 nm were observed for Bs-FtsZ variants with CTT κ+− values that are greater than 0.4. This increase in assembly size indicates a gain of nonnative interactions arising from altered sequence patterns within the CTL.
The contributions of nonnative CTT–CTT and CTT–core interactions to the overall FtsZ assembly in the absence of GTP can be inferred by quantifying correlations between Dh and the effective interaction strengths of CTT–CTT and CTT–core interactions. The weighted mean of the apparent Dh from DLS measurements was used as a quantitative proxy for the strengths of driving forces for Bs-FtsZ assembly in the absence of GTP. The effective strengths of CTT–CTT and the CTT–core interactions were extracted from statistical analysis of simulation results. First, the effective strengths εinter-CTT of CTT–CTT interactions were determined by extracting Rg values using snapshots where the CTT is not in contact with the core. This helps us examine how strongly the CTT interacts with itself. The quantity of interest was defined as , where Rg is the construct specific mean radius of gyration and Rg,max and Rg,min are the maximum and minimum values of the radii of gyration across κ+− variants, respectively (Fig. 6A). The derived values of εinter-CTT are positively correlated with Dh (linear regression Pearson R2 = 0.62), indicating that inter-CTT interactions contribute to the FtsZ assembly in the absence of GTP. Second, the effective strengths of CTT–core interactions were calculated over snapshots where the CTT is in contact with the core using pc, the mean per residue probability of finding any CTT residue in contact (5 Å) with any residue on the core in the simulated ensemble. Thus, pc is large if many CTT residues interact with the core and the parameter of interest is defined as . Here, pc,max and pc,min are the maximum and minimum values of pc across Kappa variants. The correlation between εcore-CTT and Dh is poor (linear regression Pearson R2 = 0.21). The inference is that CTT–core interactions on their own make negligible contributions to the driving forces for FtsZ assembly in the absence of GTP (Fig. 6B).
Fig. 6.
CTT–CTT and CTT–core interactions contribute to FtsZ assembly in the absence of GTP. Correlation between the mean apparent Dh measured using DLS and (A) εintra-CTT, (B) εcore-CTT, (C) εtot, and (D) κ+−. Here, the gray area is the 95% confidence region of the linear regression. (E) Fractional contributions of εinter-CTT (white bars) and εcore-CTT (blue bars) to εtot.
Next, we calculated the coupled contributions of the two parameters to GTP-independent assembly of FtsZ variants. For this, we used a mixture model, , where 0 ≤ f ≤ 1. Here, f is the fraction of conformations in which the CTT is not in contact with the core. Joint consideration of the contributions of both types of interactions yields a strong positive correlation between εtot and Dh (linear regression, R2 = 0.81) (Fig. 6C) providing we extract a unique, Kappa-variant specific value of f (Fig. 6E). The correlation between Dh and κ+− is weaker than that between Dh and the total effective drive to assemble (linear regression Pearson R2 = 0.68; Fig. 6D). This is because κ+− only describes the CTT–CTT interaction strength according to the positioning of oppositely charged residues, whereas the mixture model accounts for the contributions of CTT–core interactions. Taken together, our results point to a combination of variant-specific CTT–CTT and CTT–core interactions as contributors to nonnative FtsZ assembly in the absence of GTP. CTT–core interactions within the Kappa variants are not identical even if the values of f are similar. For example, f ∼ 0.5 for K14, WT, K26, and K47, and yet the interaction between the CT17 and the T7 loop is abolished in K47 (SI Appendix, Fig. S7 C and D). Instead, we observe a gain of nonnative CTL interactions with the T7 loop in this variant (SI Appendix, Fig. S7 A and B). Overall, the pattern of nonnative interactions will be influenced by the totality of nonrandom sequence patterns within the designed CTLs (Fig. 4 and SI Appendix, Fig. S7).
Nonnative Interactions Driven by Redesigned CTLs Promote Large and Diverse Assemblies That Are Less Active in GTP Hydrolysis than WT.
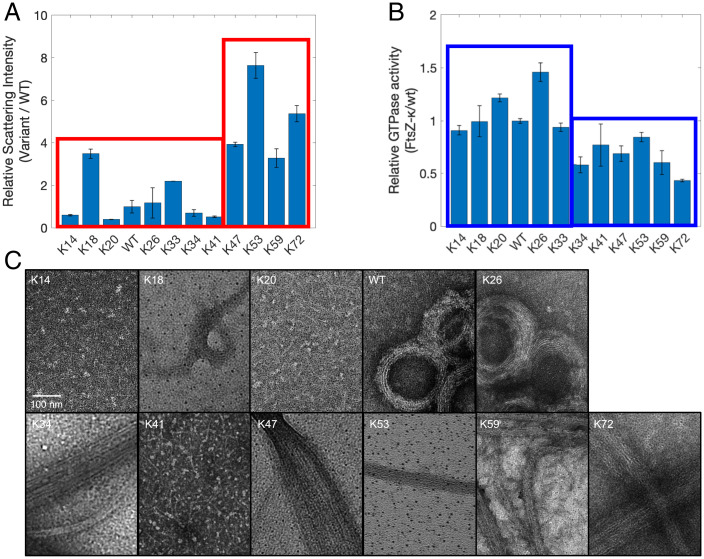
We investigated the effects of altered sequence patterns in the Kappa variants on GTP-dependent assembly and GTP hydrolysis. In vitro, Bs-FtsZ forms single-stranded protofilaments that associate laterally to form bundles (17, 49, 75–78). Right-angle light scattering is a sensitive method for studying FtsZ polymerization and bundling of FtsZ polymers in the presence of GTP (56). We measured scattering intensities of the Kappa variants and normalized these to measured values of WT for comparison. We observed two broad categories of variants using a threefold increase in scattering intensities when compared to WT as a threshold (Fig. 7A). In general, the variants where the CTT κ+− is ≤0.41 show lower scattering intensities and those with the CTT κ+− values greater than 0.41 show higher scattering intensities. However, K18 is an exception. Despite having a CTT κ+− value that is close to that of WT, the K18 variant exhibits high scattering intensities, indicating the formation of large assemblies.
Fig. 7.
Changes to the linear mixing vs. segregation of oppositely charged residues within the CTL affects GTP-dependent FtsZ assembly and GTPase activity. (A) Scattering intensities of 5 µM of WT FtsZ and different variants were measured in the presence of 1 mM GTP. Values reported here were normalized to those of WT. The red outlines indicate two broad categories of the Kappa variants that show less than or more than threefold increase in scattering intensities compared to WT. (B) Rates of GTP hydrolysis of the Kappa variants are normalized to that of WT. Blue outlines indicate two broad categories of the Kappa variants that show faster or slower rates compared to the WT. (C) Negative-stain TEM micrographs show the diverse morphologies formed by the Kappa variants.
Negative-stain transmission electron microscopy (TEM) imaging captures the diverse morphologies of higher-order assemblies formed by the Kappa variants (Fig. 7C). Those that show lower scattering intensities correspond to protofilaments—see data for K14, K20, K33 (56), and K41—or rings composed of laterally associating bundled protofilaments (WT, K26). On the other hand, stronger nonnative interactions driven by segregation of oppositely charged residues within the CTL lead to increased self-association of FtsZ variants via inter-CTT interactions among filaments. These interactions generate profoundly different assemblies when compared to morphologies observed for WT Bs-FtsZ (17, 30, 31, 79–81). Increase in the CTT κ+− leads to the formation of long, linear filamentous tracts characterized by interfilament interactions that appear to involve the CTTs—see data for K47, K53, K59, and K72 in Fig. 7C and SI Appendix, Fig. S8. These long FtsZ polymers are likely to compromise treadmilling, thereby suppressing the exchange of FtsZ subunits in polymers upon GTP hydrolysis.
Next, we measured the rates of GTP hydrolysis of Kappa variants using a series of independent continuous assays for GTPase activity. Each measurement was performed with 5 µM FtsZ in buffer MES with 1 mM GTP. As with the right-angle scattering results, the Kappa variants can be grouped into two broad categories based on their GTPase activity relative to WT (Fig. 7B). The concentration-specific GTPase activities are likely to be a convolution of many variables, including multiple assembly states of FtsZ and CTL-dependent variations to Michaelis–Menten parameters. However, at identical concentrations of FtsZ subunits, the sequence-encoded features within different CTLs directly influence the functions of the GTPase core domain with extreme segregation of oppositely charged residues reducing the GTPase activity.
Taken together, our results suggest that the sequence patterning, and the loss of emergent native contacts and / or de novo nonnative contacts that are formed in cis (intra-CTL and CTT-core) and trans (CTT–CTT and CTT–core) can alter the functions of Bs-FtsZ variants. We find that extreme segregation or mixing of oppositely charged residues in the CTL cause significant deviations from WT-like behavior in FtsZ assembly and enzymatic activity in vitro. Interestingly, we also observe robustness of both functions for CTT κ+− values between 0.19 and 0.34.
Extremes of Mixing or Segregation of Oppositely Charged Residues Show Pronounced In Vivo Phenotypes.
In contrast to the assayed in vitro functionalities, in vivo functions of FtsZ are reliant on factors such as protein stability in the cell and interactions with other components of the division machinery. To evaluate in vivo functionalities of the Kappa variants, we used a strain that would allow us to examine variants as the sole copy of FtsZ regardless of their ability to support cell growth and division. This strain was made by cloning variant alleles into a B. subtilis strain where the sole WT ftsZ gene is under the control of a xylose-inducible promoter and the variant allele is under the control of an independent isopropyl β-d-1-thiogalactopyranoside (IPTG)–inducible promoter (42). We can then deplete WT FtsZ and induce the expression of the variants in a controlled manner.
We first evaluated the stability of FtsZ in cells using quantitative immunoblots since we observed notable changes in conformational ensembles with changes in CTT κ+− values (Fig. 5C and SI Appendix, Fig. S6), and it is reasonable to expect that changes to protein structure can lead to degradation in vivo. Indeed, the ClpX chaperone is reported to modulate assembly of FtsZ in B. subtilis, and the altered conformations of CTT may lead to increased susceptibility for ClpX-mediated proteolysis by ClpP (82, 83). Furthermore, the ClpXP complex is reported to interact with the disordered CTT of FtsZ to enhance degradation in E. coli (84). Samples were prepared from midexponential phase cultures (optical density at 600 nm [OD600] of ∼0.5) that were back-diluted into IPTG then grown to and harvested at midexponential phase. Immunoblots were normalized to total protein levels. All but three variants accumulated near or above WT levels. The unstable variants are those with CTTs featuring extreme κ+− values (Fig. 8A), viz., K14, K53, and K72. These variants appear to be substantially degraded (<20% WT FtsZ levels), indicating that they are more susceptible to proteolysis.
Fig. 8.
Changes to the extent of segregation vs. mixing of oppositely charged residues within the CTL affects protein stability, cell growth, and division. (A) Quantitative immunoblots of the Kappa variants. Intensities from the gel (Above) are quantified in the histogram (Below) and normalized to the WT. WT (−) and WT (+) indicate uninduced and induced conditions, respectively. (B) Cell growth profiles of the Kappa variants. (C) In the IFM images, FtsZ is artificially colored in green, and the cell wall is in red. (Scale bar: 2 µm.) (D) The extent of condensation of FtsZ Kappa variants in cells is quantified using cumulative distribution functions. (E) We quantified cell lengths, L/R ratios, and percentage of cells with spirals from the IFM images. Data for at least 200 cells were used for each of the Kappa variants. The L/R data for WT(−) and K72 were plotted at 0 since neither rings nor spirals were observed.
Consistent with poor stability, strains expressing K72 were unable to support growth of B. subtilis (Fig. 8B). In a clear contrast, the control strain (PAL3171) expressing only WT ftsZ under the IPTG-inducible promotor exhibited a standard growth curve with exponential and stationary phases. As expected, the same control strain without ftsZ expression grew for approximately 3 h before plateauing and slightly decreasing in optical density (85). Interestingly, all Kappa variants except for K47 and K72 exhibited robust WT-like growth profiles despite degradation of K14 and K53 in cells. Strains expressing K47 were able to support growth but grew more slowly than the WT despite having FtsZ levels equivalent to WT. This suggests that K47 may interfere with both cross-wall synthesis and synthesis of the longitudinal cell wall that leads to defects in growth.
To gain insight into FtsZ assembly in vivo, we used immunofluorescence microscopy (IFM) to quantify cell length and image FtsZ ring formation. Cultures were prepared as described for immunoblots but harvested after roughly four doublings (OD600 of 1.5 to 0.3) for fixation and imaging (49). Longer cell lengths indicate impaired cell division. Cells expressing the three unstable variants, K14, K53, and K72, were 30%, 56%, and 376% longer than cells expressing WT (Fig. 8E and SI Appendix, Fig. S9). Despite the high κ+− value of its CTT, the average length of cells expressing K59 was akin to that of WT, while cells expressing the K18 allele were ∼13% longer than WT cells. The variants with extreme segregation or mixing of the oppositely charged residues in their CTTs that exhibited longer cell lengths also exhibited poor Z-ring formation. The length-to-ring ratio (L/R) metric was used to evaluate the ability of FtsZ to form Z-rings and is defined as the ratio between the cumulative length of all measured cells and the cumulative numbers of rings in those cells (86). K14 and K47 showed higher L/R ratios, K72 was unable to form any rings, whereas K59, despite the higher κ+− value of the CTT, showed robust WT-like protein stability, cell growth profile, cell length, and L/R.
Images of cells expressing Kappa variants revealed the presence of unusual FtsZ localization patterns in many of the strains (Fig. 8C). WT FtsZ forms well-defined medial rings with residual localization at cell poles. In contrast, most variants displayed loose, spiral-like structures in addition to distinct rings. Previous work identified spirals as being indicative of defects in lateral interactions between single-stranded polymers which interferes with condensation into precisely localized Z-rings (87–90). Accordingly, we quantified the spiral phenotype by quantifying the percentage of cells with spirals. This quantity, which we termed Spiral %, was higher in all variants when compared to cells expressing only WT Bs-FtsZ. We also computed, the Gaussian width (Fig. 8D), which is a measure of spiral width relative to cell length. Spiral formation was independent of FtsZ levels, cell length, and L/R ratios. Only K72 lacked spirals, possibly due to having insufficient levels of FtsZ for assembly (Fig. 8A). This suggests that the Kappa variants are unable to condense efficiently. Instead, deviation from WT-like sequence patterns leads to a mixture of rings and spiral structures (SI Appendix, Fig. S10). The latter are likely to be intermediate states that represent CTL-driven defects in condensation and/or localization.
Discussion
Considerable attention has focused on the lengths of CTLs and their impacts on FtsZ functions (81, 91–93). Conclusions from such studies can be confounding because the CTLs are not simple homopolymers. As a result, titrating lengths in arbitrary ways creates a multivariable problem where the composition and sequence patterning of residues are also varied, thereby confounding the inferred relationships between length and function. To avoid such problems, we focused on a single sequence parameter κ+− whereby we fixed the length and amino acid composition and directly modulated the interplay between intra- and intermolecular interactions. Our results show that changes to sequence patterns within the CTL have clear biophysical, biochemical, and biological consequences. We generally observe deleterious consequences in FtsZ functionalities when the oppositely charged residues of the CTL are segregated with CTT κ+− values greater than 0.34, leading to aberrant assemblies. High segregation of oppositely charged residues often leads to a loss of native and gain of nonnative CTT–core and CTT–CTT interactions between FtsZ molecules. Our results suggest that the loss of native interactions between the T7 loop and the CT17 can be detrimental, as seen for K47. In contrast, the preservation of native contacts in K59 helps maintain WT-like functions in vivo because the cognate autoinhibitory interactions are maintained for this variant. Thus, any sequence solution that mediates the native interaction between the T7 loop and the CT17 is a viable CTL. Here, we find at least one sequence solution that does so with nonrandom segregation of negatively charged residues. Other solutions may exist for sequences that accommodate the conserved nonrandom sequence patterns.
Taken together, our results suggest that the evolutionary drive to limit interactions between the CTL and the core is conserved in FtsZ orthologs. This is achieved via nonrandom segregation of negatively charged residues and dispersion of positively charged residues along the CTL sequences. These features contribute to the minimization of associations that lead to aberrant multivalent interactions among segregated blocks of charge within CTTs and between the CTT and core. Although we focused on titrating the linear segregation/mixing of oppositely charged residues, one can imagine a generalization of our approach that uses sequence libraries or directed evolution to query how large- and small-scale changes to the assortment of conserved sequence patterns alter the functionalities of IDRs. Such an approach was deployed in recent studies to identify rules underlying the functions of disordered transactivation domains of transcription factors (71, 94, 95). Investigations of additional sequence variants that impact the conformational ensemble via alternative CTL-mediated interactions such as those mediated by polar or polyglutamine tracts (96, 97) will be helpful to assess the impact of different types of polar interactions.
Our findings indicate that along with composition and length an optimal CTL requires the balancing of strengths of several parameters including intermodule interactions in cis and in trans, the nonrandom/random sequence patterns, and conserved sequence-ensemble relationships (55). Indeed, the CTT/CTL encoded effects may need to be just right—a Goldilocks effect—given that the CTT and its two distinct modules play multiple regulatory roles. Similar results have been uncovered in a recent study of hypervariable linkers within viruses (98).
Materials and Methods
All B. subtilis strains were derived from the strain JH642 (99). Cloning and genetic manipulation were performed using standard techniques (100, 101). All cloning was done using the E. coli strain AG1111 derivative PL930 (102). PL930 contains the low-copy plasmid pBS58 expressing E. coli ftsQAZ, which facilitates cloning of B. subtilis FtsZ. Vent DNA polymerase was used for PCR (New England Biolabs). All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. All genomic DNA extractions were performed using the Wizard Genomic DNA Purification Kit (Promega). Plasmid preparations were made using the NucleoSpin Plasmid Kit (Macherey-Negel). Gel/PCR purifications were performed using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Negel). Cells were grown in Luria–Bertani medium at 37 °C unless otherwise noted. Antibiotics were used at the following concentrations: ampicillin = 100 µg/mL, spectinomycin = 100 µg/mL, and chloramphenicol = 5 µg/mL.
Additional details regarding cloning, strain construction, growth conditions, immunoblotting, measurements of growth curves, IFM, sequence analysis, and details of the simulation setup are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Stephen Vadia and Catherine Kornacki for cloning and strain constructions. TEM imaging was performed in part using the Washington University Center for Cellular Imaging (WUCCI) supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), and the Foundation for Barnes-Jewish Hospital. M.K.S. was a CSELS postdoctoral fellow of the erstwhile Center for Science & Engineering of Living Systems (CSELS) in the James McKelvey School of Engineering at Washington University in St. Louis. This work was supported by grants FA9550-20-1-0241 from the Air Force Office of Scientific Research and MCB1614766 from the NSF to R.V.P. and by grant 5R35GM127331 from the NIH to P.A.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2211178119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Brodsky S., Jana T., Barkai N., Order through disorder: The role of intrinsically disordered regions in transcription factor binding specificity. Curr. Opin. Struct. Biol. 71, 110–115 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Morffy N., Strader L. C., Structural aspects of auxin signaling. Cold Spring Harb. Perspect. Biol. 14, a039883 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyson H. J., Wright P. E., NMR illuminates intrinsic disorder. Curr. Opin. Struct. Biol. 70, 44–52 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondos S. E., Dunker A. K., Uversky V. N., Intrinsically disordered proteins play diverse roles in cell signaling. Cell Commun. Signal. 20, 20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desjardins G., et al. , Synergy of aromatic residues and phosphoserines within the intrinsically disordered DNA-binding inhibitory elements of the Ets-1 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 111, 11019–11024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Currie S. L., et al. , Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5. Nucleic Acids Res. 45, 2223–2241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugge K., et al. , Interactions by disorder—A matter of context. Front. Mol. Biosci. 7, 110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittal A., Holehouse A. S., Cohan M. C., Pappu R. V., Sequence-to-conformation relationships of disordered regions tethered to folded domains of proteins. J. Mol. Biol. 430, 2403–2421 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Chen K., Li J., Wang C., Wei Z., Zhang M., Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions. eLife 6, e29150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Borrajero C., et al. , The biophysical basis for phosphorylation-enhanced DNA-binding autoinhibition of the ETS1 transcription factor. J. Mol. Biol. 431, 593–614 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Kang H.-S., et al. , An autoinhibitory intramolecular interaction proof-reads RNA recognition by the essential splicing factor U2AF2. Proc. Natl. Acad. Sci. U.S.A. 117, 7140–7149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillén-Boixet J., et al. , RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeon J. H., Heinkel F., Sung M., Na D., Gsponer J., Systems-wide identification of cis-regulatory elements in proteins. Cell Syst. 2, 89–100 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Paquette D. R., Tibble R. W., Daifuku T. S., Gross J. D., Control of mRNA decapping by autoinhibition. Nucleic Acids Res. 46, 6318–6329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bista M., Petrovich M., Fersht A. R., MDMX contains an autoinhibitory sequence element. Proc. Natl. Acad. Sci. U.S.A. 110, 17814–17819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor S. S., et al. , The tails of protein kinase A. Mol. Pharmacol. 101, 219–225 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohan M. C., Eddelbuettel A. M. P., Levin P. A., Pappu R. V., Dissecting the functional contributions of the intrinsically disordered C-terminal tail of bacillus subtilis FtsZ. J. Mol. Biol. 432, 3205–3221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straight A. F., Field C. M., Microtubules, membranes and cytokinesis. Curr. Biol. 10, R760–R770 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Haeusser D. P., Margolin W., Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 14, 305–319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolliday N., Bouquin N., Li R., Assembly and regulation of the cytokinetic apparatus in budding yeast. Curr. Opin. Microbiol. 4, 690–695 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Huckaba T. M., Pon L. A., Cytokinesis: Rho and formins are the ringleaders. Curr. Biol. 12, R813–R814 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Baluska F., Menzel D., Barlow P. W., Cytokinesis in plant and animal cells: Endosomes ‘shut the door’. Dev. Biol. 294, 1–10 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Guizetti J., Gerlich D. W., Cytokinetic abscission in animal cells. Semin. Cell Dev. Biol. 21, 909–916 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Rincon S. A., Paoletti A., Molecular control of fission yeast cytokinesis. Semin. Cell Dev. Biol. 53, 28–38 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Thieleke-Matos C., Osório D. S., Carvalho A. X., Morais-de-Sá E., Emerging mechanisms and roles for asymmetric cytokinesis. Int. Rev. Cell Mol. Biol. 332, 297–345 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Addi C., Bai J., Echard A., Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr. Opin. Cell Biol. 50, 27–34 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Lutkenhaus J., FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 9, 403–409 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Rothfield L., Justice S., García-Lara J., Bacterial cell division. Annu. Rev. Genet. 33, 423–448 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Janakiraman A., Goldberg M. B., Recent advances on the development of bacterial poles. Trends Microbiol. 12, 518–525 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Erickson H. P., Anderson D. E., Osawa M., FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang K. H., Durand-Heredia J., Janakiraman A., FtsZ ring stability: Of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859–1868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier E. L., Goley E. D., Form and function of the bacterial cytokinetic ring. Curr. Opin. Cell Biol. 26, 19–27 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Buske P. J., Mittal A., Pappu R. V., Levin P. A., An intrinsically disordered linker plays a critical role in bacterial cell division. Semin. Cell Dev. Biol. 37, 3–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bisson-Filho A. W., et al. , Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monteiro J. M., et al. , Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams D. W., Errington J., Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Egan A. J. F., Vollmer W., The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 1277, 8–28 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Lutkenhaus J., The ParA/MinD family puts things in their place. Trends Microbiol. 20, 411–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Söderström B., Daley D. O., The bacterial divisome: More than a ring? Curr. Genet. 63, 161–164 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Anderson D. E., Gueiros-Filho F. J., Erickson H. P., Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186, 5775–5781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer S. C., Reversible polymerizations and aggregations. Annu. Rev. Phys. Chem. 53, 173–200 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Romberg L., Simon M., Erickson H. P., Polymerization of Ftsz, a bacterial homolog of tubulin. is assembly cooperative? J. Biol. Chem. 276, 11743–11753 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Caplan M. R., Erickson H. P., Apparent cooperative assembly of the bacterial cell division protein FtsZ demonstrated by isothermal titration calorimetry. J. Biol. Chem. 278, 13784–13788 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Matsui T., Han X., Yu J., Yao M., Tanaka I., Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem. 289, 3501–3509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagstaff J. M., et al. , A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. mBio 8, e00254-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miraldi E. R., Thomas P. J., Romberg L., Allosteric models for cooperative polymerization of linear polymers. Biophys. J. 95, 2470–2486 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corbin L. C., Erickson H. P., A unified model for treadmilling and nucleation of single-stranded FtsZ protofilaments. Biophys. J. 119, 792–805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliva M. A., Trambaiolo D., Löwe J., Structural insights into the conformational variability of FtsZ. J. Mol. Biol. 373, 1229–1242 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Buske P. J., Levin P. A., Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945–10957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaughan S., Wickstead B., Gull K., Addinall S. G., Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58, 19–29 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Schumacher M. A., Huang K. H., Zeng W., Janakiraman A., Structure of the Z ring-associated protein, ZapD, bound to the C-terminal domain of the tubulin-like protein, FtsZ, suggests mechanism of Z ring stabilization through FtsZ cross-linking. J. Biol. Chem. 292, 3740–3750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schumacher M. A., Zeng W., Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. U.S.A. 113, 4988–4993 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosyak L., et al. , The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19, 3179–3191 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohan A., et al. , Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Cohan M. C., Ruff K. M., Pappu R. V., Information theoretic measures for quantifying sequence-ensemble relationships of intrinsically disordered proteins. Protein Eng. Des. Sel. 32, 191–202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buske P. J., Levin P. A., A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249–263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Löwe J., Amos L. A., Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Schumacher M. A., Ohashi T., Corbin L., Erickson H. P., High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP. Acta Crystallogr. F Struct. Biol. Commun. 76, 94–102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jindal B., Panda D., Understanding FtsZ assembly: Cues from the behavior of its N- and C-terminal domains. Biochemistry 52, 7071–7081 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Erdős G., Pajkos M., Dosztányi Z., IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 49 (W1), W297–W303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng Y., et al. , Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 46, 13468–13477 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oldfield C. J., et al. , Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 44, 12454–12470 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Halabi N., Rivoire O., Leibler S., Ranganathan R., Protein sectors: Evolutionary units of three-dimensional structure. Cell 138, 774–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lockless S. W., Ranganathan R., Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Rivoire O., Reynolds K. A., Ranganathan R., Evolution-based functional decomposition of proteins. PLOS Comput. Biol. 12, e1004817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goujon M., et al. , A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohan M. C., Shinn M. K., Lalmansingh J. M., Pappu R. V., Uncovering non-random binary patterns within sequences of intrinsically disordered proteins. J. Mol. Biol. 434, 167373 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Das R. K., Pappu R. V., Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 110, 13392–13397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherry K. P., Das R. K., Pappu R. V., Barrick D., Control of transcriptional activity by design of charge patterning in the intrinsically disordered RAM region of the Notch receptor. Proc. Natl. Acad. Sci. U.S.A. 114, E9243–E9252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Staller M. V., et al. , A high-throughput mutational scan of an intrinsically disordered acidic transcriptional activation domain. Cell Syst. 6, 444–455.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sawle L., Huihui J., Ghosh K., All-atom simulations reveal protein charge decoration in the folded and unfolded ensemble is key in thermophilic adaptation. J. Chem. Theory Comput. 13, 5065–5075 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Sawle L., Ghosh K., A theoretical method to compute sequence dependent configurational properties in charged polymers and proteins. J. Chem. Phys. 143, 085101 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Das R. K., Huang Y., Phillips A. H., Kriwacki R. W., Pappu R. V., Cryptic sequence features within the disordered protein p27Kip1 regulate cell cycle signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 5616–5621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gündoğdu M. E., et al. , Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 30, 617–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Popp D., Iwasa M., Narita A., Erickson H. P., Maéda Y., FtsZ condensates: An in vitro electron microscopy study. Biopolymers 91, 340–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gueiros-Filho F. J., Losick R., A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mukherjee A., Lutkenhaus J., Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181, 823–832 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu G., et al. , In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One 5, e12682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guan F., et al. , Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division. eLife 7, e35578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huecas S., et al. , Self-organization of FtsZ polymers in solution reveals spacer role of the disordered C-terminal tail. Biophys. J. 113, 1831–1844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haeusser D. P., Lee A. H., Weart R. B., Levin P. A., ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J. Bacteriol. 191, 1986–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weart R. B., Nakano S., Lane B. E., Zuber P., Levin P. A., The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol. Microbiol. 57, 238–249 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Viola M. G., et al. , An enhancer sequence in the intrinsically disordered region of FtsZ promotes polymer-guided substrate processing by ClpXP protease. Protein Sci. 31, e4306 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arjes H. A., et al. , Failsafe mechanisms couple division and DNA replication in bacteria. Curr. Biol. 24, 2149–2155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weart R. B., et al. , A metabolic sensor governing cell size in bacteria. Cell 130, 335–347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Squyres G. R., et al. , Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol. 6, 553–562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Michie K. A., Monahan L. G., Beech P. L., Harry E. J., Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J. Bacteriol. 188, 1680–1690 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ben-Yehuda S., Losick R., Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109, 257–266 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Ma X., Margolin W., Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181, 7531–7544 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sundararajan K., et al. , The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 6, 7281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gardner K. A., Moore D. A., Erickson H. P., The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol. 89, 264–275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barrows J. M., Sundararajan K., Bhargava A., Goley E. D., FtsA regulates Z-ring morphology and cell wall metabolism in an FtsZ C-terminal linker-dependent manner in Caulobacter crescentus. J. Bacteriol. 202, e00693-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Staller M. V., et al. , Directed mutational scanning reveals a balance between acidic and hydrophobic residues in strong human activation domains. Cell Syst. 13, 334–345.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ravarani C. N., et al. , High-throughput discovery of functional disordered regions: Investigation of transactivation domains. Mol. Syst. Biol. 14, e8190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Das R. K., Ruff K. M., Pappu R. V., Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 32, 102–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williamson T. E., Vitalis A., Crick S. L., Pappu R. V., Modulation of polyglutamine conformations and dimer formation by the N-terminus of huntingtin. J. Mol. Biol. 396, 1295–1309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.González-Foutel N. S., et al. , Conformational buffering underlies functional selection in intrinsically disordered protein regions. Nat. Struct. Mol. Biol. 29, 781–790 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Perego M., Spiegelman G. B., Hoch J. A., Structure of the gene for the transition state regulator, abrB: Regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2, 689–699 (1988). [DOI] [PubMed] [Google Scholar]
- 100.Harwood C. R., Cutting S. M., Molecular Biological Methods for Bacillus (Modern Microbiological Methods, Wiley, New York, 1990), p. 581. [Google Scholar]
- 101.Sambrook J., Russell D. W., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 3, 2001). [Google Scholar]
- 102.Wang X., Lutkenhaus J., The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9, 435–442 (1993). [DOI] [PubMed] [Google Scholar]
- 103.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.