Significance
Cells display on their surface peptides, which are derived from their expressed genes, bound to MHC I molecules. This process allows CD8 T lymphocytes to detect and eliminate virally infected and cancer cells that are making antigenic proteins. Here, we describe that tetraspanin-5 (Tspan5) is a component of this pathway that functions to organize MHC I molecules into clusters that are more stimulatory for CD8 T cells. This finding reveals a mechanism that increases the effectiveness of antigen presentation and provides insight into why MHC I molecules can be clustered on cells.
Keywords: tetraspanin, Tspan5, MHC I, antigen presentation, cluster
Abstract
MHC molecules are not randomly distributed on the plasma membrane but instead are present in discrete nanoclusters. The mechanisms that control formation of MHC I nanoclusters and the importance of such structures are incompletely understood. Here, we report a molecular association between tetraspanin-5 (Tspan5) and MHC I molecules that started in the endoplasmic reticulum and was maintained on the plasma membrane. This association was observed both in mouse dendritic cells and in human cancer cell lines. Loss of Tspan5 reduced the size of MHC I clusters without affecting MHC I peptide loading, delivery of complexes to the plasma membrane, or overall surface MHC I levels. Functionally, CD8 T cell responses to antigen presented by Tspan5-deficient dendritic cells were impaired but were restored by antibody-induced reclustering of MHC I molecules. In contrast, Tspan5 did not associate with two other plasma membrane proteins, Flotillin1 and CD55, with or the endoplasmic reticulum proteins Tapasin and TAP. Thus, our findings identify a mechanism underlying the clustering of MHC I molecules that is important for optimal T cell responses.
Antigen presentation is necessary for T cell immune surveillance of infected and transformed cells. CD8 T cell activation is primarily driven by the presentation of peptides from endogenously expressed proteins on MHC class I molecules (1, 2). Exogenous antigens can also be presented on MHC class I molecules by professional antigen presenting cells (APC) using a process termed cross-presentation (3). T cells can be stimulated by low numbers of their specific MHC I-presented peptides. How small numbers of antigenic complexes present in the context of much more abundant nonactivating (self) peptides can efficiently stimulate CD8 T cells has been a subject of much interest (4–6).
Antigen-specific T cell stimulation is initiated by direct contact of the T cell receptor (TCR) with their cognate peptide-bound MHC complexes (pMHC) on APCs. Multimerization of the TCR has been proposed to be a requirement for signaling through the TCR–CD3 complex (7, 8). TCR signaling occurs primarily in dense clusters of these receptors on the surface of T cells (9). TCRs may preexist as aggregates (10–12), although this has been questioned (13), and may be clustered as a result of interaction with pMHC complexes (9). The mechanisms that underlie the clustering of TCR are incompletely understood.
The observation that in the absence of TCR, MHC molecules reside in preformed nanoclusters on APCs has suggested that these antigen complexes are poised to aggregate cognate TCRs (14). Formation of MHC nanoclusters on APCs has been demonstrated in both resting and activated APCs (14–20), and such clusters are thought to play a role in T cell activation by increasing the sensitivity of the T cell response (21, 22). The mechanisms that underlie the clustering of MHC I molecules on APCs are incompletely understood.
Tetraspanins (Tspans) are a large family of proteins containing four transmembrane domains that have been shown to regulate protein trafficking and compartmentalization (23, 24). Tspans can help organize the plasma membrane (PM) into Tspan-enriched microdomains (TEM), in which certain membrane proteins reside. A number of Tspan proteins (CD9, CD37, CD53, CD81, and CD82) have been shown to associate with MHC II molecules in various professional APCs (25–27). Where examined, the loss of these Tspans does not reduce MHC II clustering (28). Therefore, whether Tspans are needed to form and maintain MHC II clusters isn’t clear. Even less is known about Tspans and MHC I molecules. In two previous reports, Tspan proteins (CD53, CD81, and CD82) have been observed to weakly associate with MHC I molecules (25, 29), although the functional significance of this interaction was not resolved.
Previously, we had performed a genome-wide forward genetic screen in dendritic cells (DC) to identify novel genes regulating MHC I cross-presentation (BioStudies S-BSST306) (30). One of the gene candidates that was identified was the Tspan family member, Tspan5. A role for Tspan5 in antigen presentation was not previously recognized. Here we validate Tspan5’s role in antigen presentation. Our data show that Tspan5 directly associates with MHC I molecules and that this association is needed for the formation of large MHC I clusters on DCs, and that these clusters are needed for optimal stimulation of CD8 T cells.
Results
Tspan5 in DCs Is Necessary for Optimal Stimulation of CD8 T Cells by pMHC I Complexes.
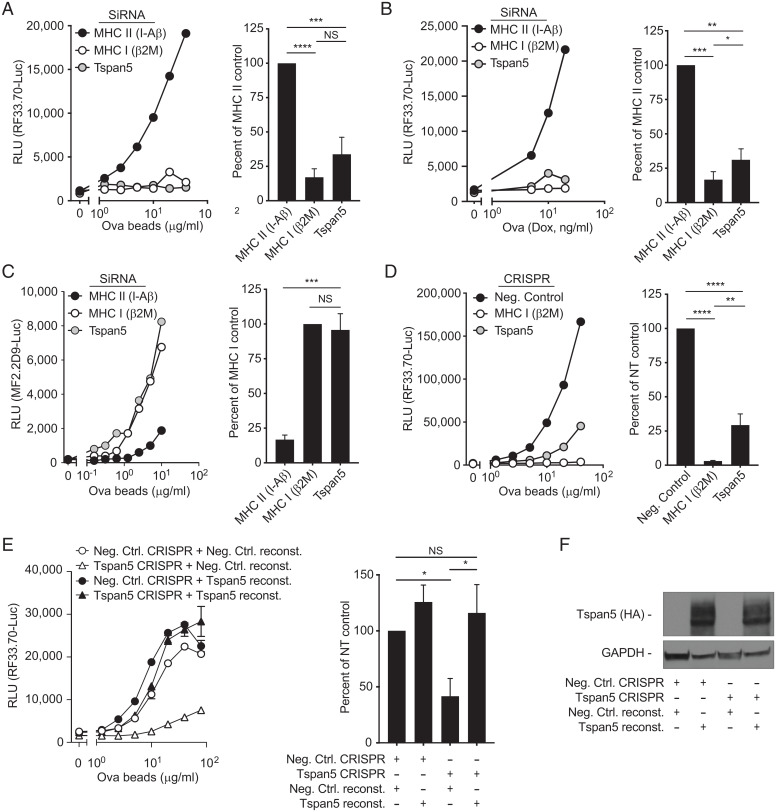
In order to determine the role of Tspan5 in antigen presentation, Tspan5 small-interfering RNA (siRNA) were transfected in a mouse DC clone (DC3.2), which resulted in a marked (98 ± 2.3%) reduction in Tspan5 mRNA and surface expression (SI Appendix, Fig. S1A). Alternatively, these DCs were transfected with a negative control siRNA targeting the MHC II protein H-2 I-A β-chain (this protein is not involved in MHC I presentation) or positive control siRNA targeting the light chain of the MHC I molecule, β-2-microglobulin (β2M). siRNA-treated DCs were incubated with ovalbumin (Ova)-conjugated beads, as a source of particulate antigen for cross-presentation, and cultured with a CD8 T cell hybridoma (specific for H-2Kb bound to Ova257–264). This hybridoma (hereafter referred to as RF33.70-Luc) contains luciferase under the control of the interleukin (IL)-2 promotor element NFAT and produces luciferase in proportion to the amount of TCR-stimulation (30). Stimulation of the Ova-specific, MHC I-restricted CD8 hybridoma by the Tspan5-deficient and β2M-deficient (positive control) DCs was significantly reduced compared to the negative control DCs (MHC II I-A β-chain) (Fig. 1A).
Fig. 1.
Loss of Tspan5 expression selectively inhibits MHC class I presentation. (A) DC3.2 cells were treated with siRNA for the MHC class II gene H-2 I-A β-chain (black), the MHC class I light chain β2M (white), or Tspan5 (gray). A titration of Ova-conjugated beads was cultured with these DCs and the CD8+ T cell hybridoma RF33.70-Luc and after 16 h, cross-presentation was assayed by measuring luciferase activity. The line graph shows an antigen titration for a representative experiment and the bar graph shows averages of greater than or equal to three experiments taken at a single antigen dose. (B) Similar to A, except instead of Ova-beads, endogenous cytosolic Ova (NS-Ova) expression was induced in DC2.4 cells using doxycycline for 4 h, at which time BFA (to block further egress of pMHC I complexes) and CD8 T cells were added for the remaining 16 h. (C) Similar to A, except instead of measuring MHC I presentation, MHC II presentation was measured with the CD4+ hybridoma MF2.2D9-Luc and siRNA for β2M (MHC I, white) was used as the negative control. (D) Cross-presentation was assessed in DC3.2 cells gene-edited with CRISPR/Cas9 transduction without a guide (Neg Control, black), with guides targeting β2M (MHC I (white), or with guides targeting Tspan5 (gray). Ova-beads were used as a source of antigen and assayed as described in A. (E) DCs were transfected with Cas9, without guide RNA (Neg Ctrl. CRISPR, circle) or with Tspan5 targeted guides (Tspan5 CRISPR, triangle) as shown. Neg Control or Tspan5 knockout DCs were then transfected with an empty plasmid (Neg. Ctrl. reconst.) or with a mutant form of Tspan5 that cannot be targeted by Tspan5 guide RNAs (Tspan5 reconst.) and presentation of Ova was assayed as in D. Left graph shows representative data over a dose range of Ova-conjugated beads, while the Right bar graph shows luciferase activity normalized to the nontargeting control with the mean + SD from three independent assays. (F) Western blot of lysates from DCs shown in D were probed for the Tspan5 expression (HA-tag). (GAPDH serves as the loading control). T cell activation was assessed by the production of luciferase under control of an NFAT promoter in all assays. RLU refers to luciferase activity expressed as relative light units. Data in A–D are representative of greater than or equal to three independent experiments. Statistics were calculated from independent experiments using one-way ANOVA. P values were based on Tukey’s multiple comparison test; NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, we tested whether silencing of Tspan5 also blocked presentation of endogenously expressed Ova. For this purpose, we used another DC (DC2.4) that had been engineered to express a nonsecreted form of Ova (NS-Ova) under the control of a doxycycline (Dox)-inducible promoter. Stimulation of the CD8 T cell by DC2.4 expressing the endogenous Ova antigen was also inhibited by the loss of Tspan5 and β2M (Fig. 1B). These results indicate that Tspan5 influences CD8 T cell stimulation by both endogenous antigen, antigen presented by the classic or direct pathway, and exogenous antigen, which is cross-presented.
In contrast, opposite results were obtained when silenced DCs were incubated with the same exogenous antigen but then incubated with a CD4 T cell hybridoma (MF2.2D9-Luc, specific for I-Ab bound to Ova258–276) to assay for MHC II antigen presentation. Stimulation of the Ova-specific, MHC II-restricted CD4 hybridoma by the I-A–silenced DCs (MHC II) was inhibited compared to the Tspan5-deficient and β2M-deficient DCs (Fig. 1C). Note that for MHC II presentation, the β2M-deficient DCs serve as a negative control (since β2M is not needed for MHC II presentation), while the MHC II-deficient DCs now serve as a positive control. These results indicate that loss of Tspan5 did not affect MHC class II-restricted presentation, and therefore Tspan5 was selectively required for responses to MHC I-presented antigen.
We also knocked out Tspan5 using CRISPR/Cas9 to determine if the loss of MHC I antigen presentation was replicated by an independent technique. In these cells, TIDE analysis revealed that the Tspan5 gene was disrupted in greater than 90% of cells, resulting in a significant loss of Tspan5 (SI Appendix, Fig. S1B). Antigen presentation with Tspan5-deficient DCs incubated with exogenous particulate antigen and the CD8 T cell hybridoma was significantly reduced as compared to control CRISPR/Cas9 DC cells (negative control [Ctrl]) (Fig. 1D), which is a similar result to that observed with siRNA-silencing of Tspan5 (Fig. 1A).
To prove that these effects were due to loss of Tspan5 expression, we reconstituted Tspan5 expression through transduction of Tspan5 with synonymous mutations that prevented it from being targeted by our guide RNAs. Reconstitution of Tspan5 expression (Fig. 1 E and F), but not a control protein (Negative Ctrl reconst.), completely restored CD8 T cell activation over a wide dose of antigen (Fig. 1E). Together, these data confirm that the loss of MHC class I presentation is specifically due to the loss of Tspan5 expression. These data together with our knockdown data (Fig. 1 A and B) demonstrated that blocking Tspan5 expression resulted in an inhibition of MHC I presentation.
The Defect in Antigen Presentation Is Not Due to a Loss of Peptide-Loading or Trafficking of MHC I Complexes.
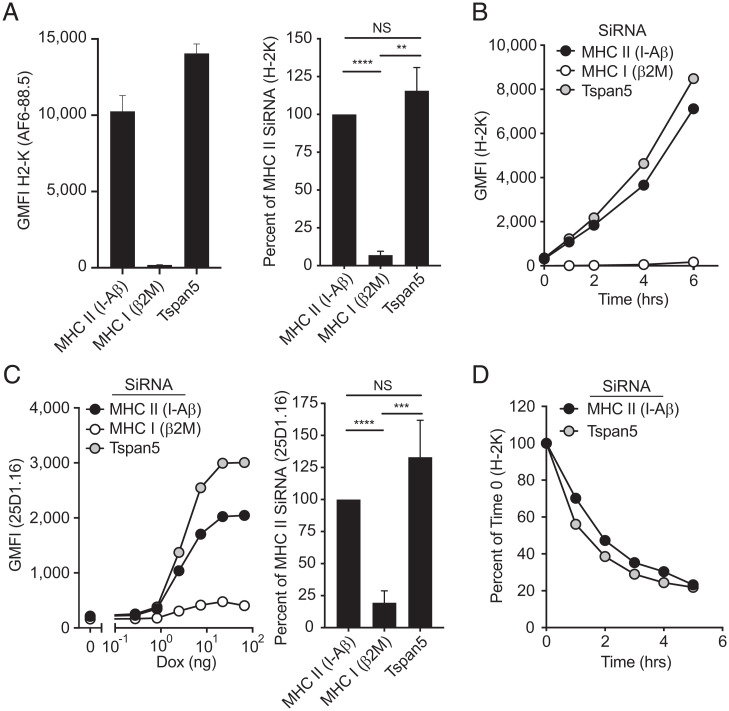
Tspan5 was previously shown to regulate ADAM-10 membrane expression, by controlling endoplasmic reticulum (ER) to membrane trafficking and membrane retention of this metalloprotease (31, 32). The inhibition in MHC I presentation we observed in Fig. 1 could have been a result of a defect in MHC trafficking or limited peptide supply/loading (which are required for release of MHC I from the ER) leading to suppressed MHC class I at the PM. However, in the absence of Tspan5, MHC I expression in steady-state conditions was not altered (Fig. 2A and SI Appendix, Fig. S2A). Next, we exposed DCs to low pH to denature the pMHC I complexes on the PM and then quantified by flow cytometry the reappearance of new MHC I complexes on the cell surface over time. This assay essentially measures the production and transport kinetics of new pMHC I complexes. Loss of Tspan5 did not reduce the reexpression of surface MHC I complexes (Fig. 2B). Since this regeneration of MHC I complexes requires nascent MHC I molecules to assemble, load with peptide, and then to be transported to the PM, these results indicated that none of these processes require Tspan5 (Fig. 2B).
Fig. 2.
Expression and peptide-loading of MHC class I molecules are not affected by the loss of Tspan5 expression. (A) DC2.4 cells were treated with siRNA for the MHC class II gene H-2 I-A β-chain (MHC II), the MHC class I light chain, β2M (MHC I), or Tspan5. H-2K expression at the PM was assessed by flow cytometry 48 h after silencing. Data shown are from a representative experiment (Left) or normalized data expressed as the percent MHC I expression compared to MHC II (I-Aβ) siRNA treated control with means + SD of six independent experiments (Right). (B) Similar to A, except surface MHC class I molecules were removed from silenced DCs by acid stripping and then allowed to recover over time (MHC II, black), the MHC class I light chain (MHC I, white) or Tspan5 (gray). Data are representative of n > 3 independent experiments. (C) DCs were silenced for 48 h and then antigen synthesis was induced with increasing concentrations of Dox. Production and egress of Ova-MHC I complexes was measured by analyzing surface Ova peptide-bound H-2K molecules by flow cytometry (25D1.16 antibody). The line graph is a representative experiment, and the bar graph shows means ± SD of greater than or equal to three experiments taken at a single dose of Dox (Right). (D) DCs were treated with siRNA targeting H-2 I-A β-chain (MHC II [I-Aβ], black) or Tspan5 (gray). Cells were treated with BFA to block egress of new MHC I molecules from the ER. Remaining surface MHC I molecules were analyzed over time by flow cytometry. Data are representative of three independent experiments. Statistics were calculated using one-way ANOVA. P values were based on Tukey’s multiple comparison test; NS = not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, we further tested whether the loss of Tspan5 affected the levels of surface peptide-loaded MHC I complexes that are generated from endogenously expressed Ova. We induced expression of endogenous Ova with Dox and subsequently stained control or Tspan5-deficient cells with an antibody (25D1.16) that detects S8L peptide-bound H-2Kb molecules. Similar to total class I levels (Fig. 2 A and B), the levels of these specific pMHC I complexes were also not altered by the lack of Tspan5 expression (Fig. 2C). Finally, we explored whether Tspan5 plays a role in the membrane retention or internalization of class I molecules by following class I expression after blocking transport of new complexes from the ER with Brefeldin A (BFA). Our data show no change in the rate of internalization of MHC class I in the presence or absence of Tspan5 (Fig. 2D and SI Appendix, Fig. S2B). Taken together these data indicate that the loss of Tspan5 does not affect the assembly, peptide-loading, ER-to-membrane transport, or membrane retention of MHC I molecules.
Tspan5 Selectively Affects pMHC I-Stimulated Responses.
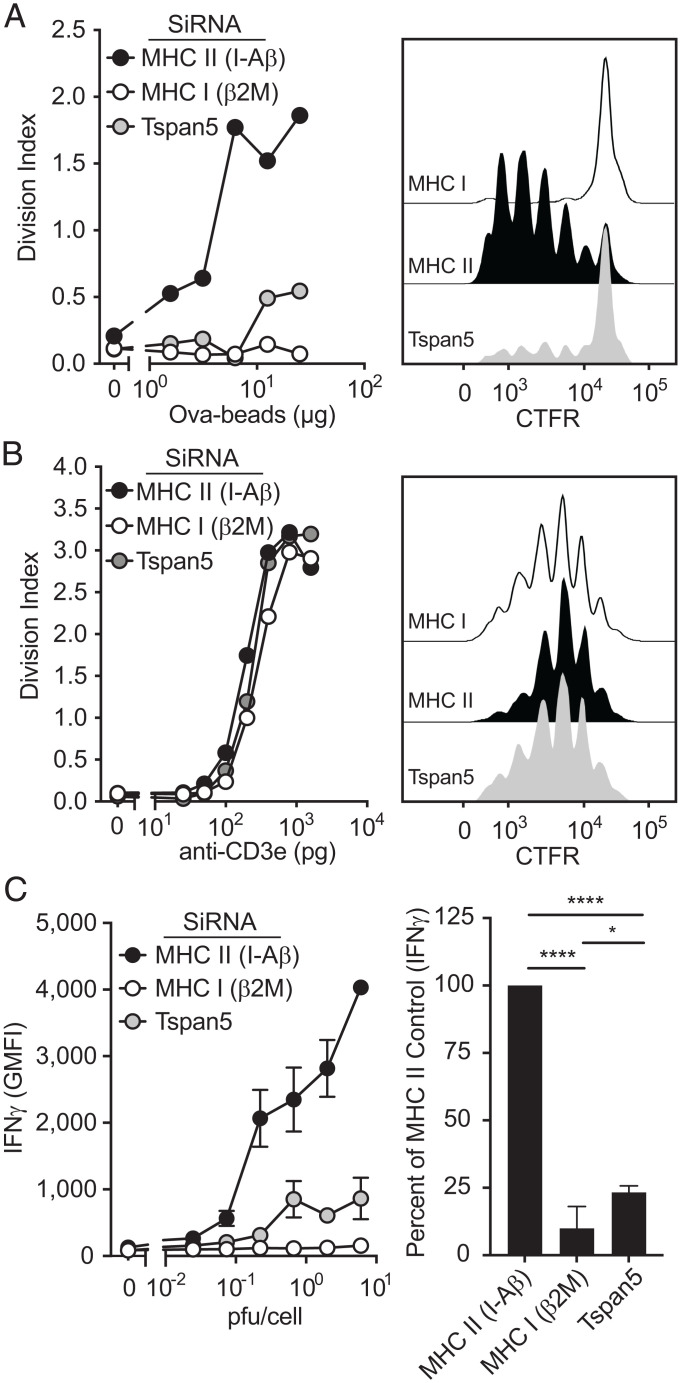
Our findings that the overall levels of pMHC I complexes were not reduced in Tspan5-deficient cells, raised the possibility that Tspan5 was contributing to T cell stimulation in ways other than through antigen presentation (e.g., affecting costimulation or adhesion molecules). We first examined the effect of Tspan5-loss on the expression of major costimulatory and adhesion molecules and found that expression was not altered by the loss of Tspan5 (SI Appendix, Fig. S3A). Since T cell hybridomas generally don’t require costimulation, a Tspan5-effect on costimulatory molecules was unlikely to be the basis for reduced T cell stimulation. To further determine whether the defect in T cell responses with Tspan5-deficient DCs was due to effects on MHC I molecules directly versus an alteration in other receptor-ligand interactions, we activated T cells in a way that bypassed the requirement for MHC I molecules, but still required costimulatory molecules and adhesion molecules (33). We used naïve TCR transgenic CD8 T cells (OT-1) for this analysis because these cells are dependent on adhesion and costimulation for activation (34). First, we confirmed that activation of these primary CD8 T cells through pMHC (Ova-conjugated beads) was reduced in the absence of Tspan5 (Fig. 3A). In contrast, when the requirement for MHC class I was bypassed using anti-CD3 bound to Fc-receptors expressed on DCs, OT-1 T cell activation was unaffected by the loss of Tspan5 over the entire anti-CD3 dose–response curve (Fig. 3B). Furthermore, we included a group with an anti–LFA-1 blocking antibody to confirm that LFA-1 was required for DC stimulation of naïve T cells through anti-CD3, and it inhibited the T cell response, as expected (SI Appendix, Fig. S3B).
Fig. 3.
Loss of Tspan5 selectively reduces MHC class I antigen presentation. (A) DC3.2 cells were silenced for the indicated genes. After 48 h, DCs were γ-irradiated and cultured with increasing concentrations of Ova-conjugated beads as a source of antigen and CTFR-labeled naïve OT-1 T cells. Proliferation was assessed 72 h later by measuring dilution of CTFR in CD8 T cells by flow cytometry. (B) Similar to A, except a titration of anti-CD3 was used instead of Ova-conjugated beads. (A and B, Left) Average number of cell division (division index) over the full titration of Ova (A) or anti-CD3e (B). (Right) Histograms of OT.1 proliferation by dilution of CTFR label at 10 μg/mL Ova beads (A) and 1 ng/mL anti-CD3e (B). (C) DCs were silenced as indicated in A. Cells were then infected with a titration of LCMV, washed, and coincubated with effector p14 CD8 T cells from p14 TCR transgenic mice. T cell activation was assessed by intracellular staining for IFN-γ in cells gated for CD8 expression. The line graph shows an antigen titration for a representative experiment and the bar graph shows averages of greater than or equal to three experiments taken at a single antigen dose (∼0.6 to 2 pfu per cell) (Right). Statistics were calculated from independent experiments using one-way ANOVA. P values were based on Tukey’s multiple comparison test; *P ≤ 0.05, ****P < 0.0001.
These results, together with our findings that loss of Tspan5 does not affect peptide-MHC II-stimulated CD4 T cell responses (that are also dependent on the same accessory receptor–ligand interactions), argue that Tspan5 is contributing to T cell stimulation through effects on MHC I molecules directly. To determine whether Tspan5 regulates antigen presentation of a different antigen (LCMV) and a different MHC I class I molecule (H-2D), we generated effector T cells from p14 TCR transgenic mice. DCs silenced for Tspan5 or for control proteins were then infected with LCMV, which generates GP33–41 peptides presented on H-2D molecules. Similar to the defects observed with Ova presentation (SI Appendix, Fig. S3C), LCMV-infected DCs were also defective in presenting antigen in the absence of Tspan5 (Fig. 3C). These data generalize the requirement of Tspan5 for optimal activation of multiple primary T cells being stimulated by different MHC I molecules.
Tspan5 Association with MHC Class I Molecules.
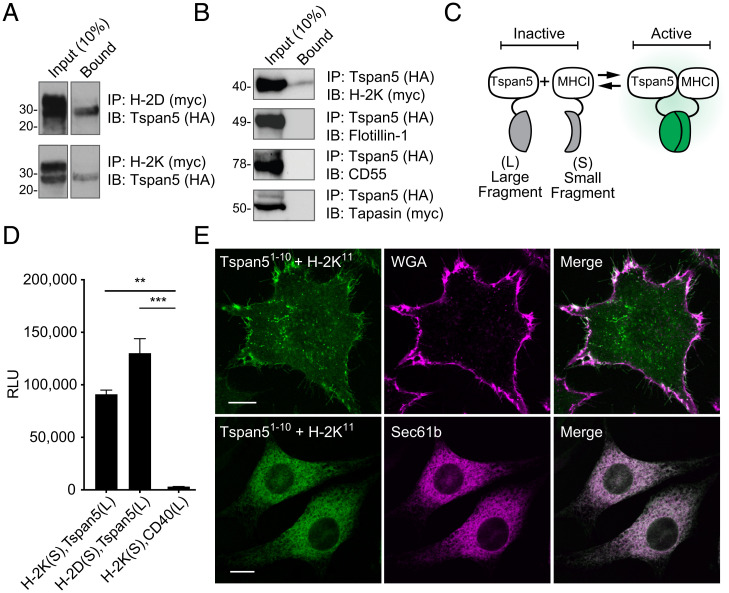
Given the selective effects of Tspan5-deficiency on MHC I-stimulated responses, we investigated whether Tspan5 molecules associated with MHC I complexes using several different techniques. First, we analyzed whether Tspan5 coimmunoprecipitated with MHC I molecules. In these experiments, we expressed HA-tagged Tspan5 and Myc-tagged MHC I in 293T cells and then immunoprecipitated MHC I molecules from clarified detergent cell lysates. The immunoprecipitates were then analyzed for the presence of Tspan5 by Western blot. In these experiments, Tspan5 coimmunoprecipitated with both H-2K and H-2D MHC I molecules (Fig. 4A). Alternatively, we immunoprecipitated Tspan5 and confirmed its association with H-2K by Western blot (Fig. 4B). In contrast, two other PM-associated proteins, Flotillin1 and CD55, did not comimmunoprecipitate with Tspan5 (Fig. 4B) and similar negative controls with ER proteins will be described below. Therefore, Tspan5 is selectively associating with the MHC molecules. These experiments suggest that MHC I molecules are directly associated with Tspan5 and are present in Tspan5-enriched membrane domains. These findings led us to perform additional experiments to probe the Tspan5–MHC I interaction.
Fig. 4.
Tspan5 associates with MHC class I molecules. (A) Myc epitope-tagged MHC I molecules, H-2D (Upper) or H-2K (Lower), were coexpressed with HA-tagged Tspan5 in 293T cells. Cells were lysed and association with Tspan5 was determined by immunoprecipitation (IP) and Western blotting (IB) as indicated. Input = 10% of lysate and Bound = eluted from myc antibody. (B) Same as A except that HA-tagged Tspan5 was immunoprecipitated and coprecipitated proteins were detected from the same lysate and eluate fractions by immunoblotting for myc-tagged H-2K, endogenous Flotillin-1, endogenous CD55 (DAF), or myc-tagged Tapasin. (C) Diagram of the complementation assay with either split luciferase or split GFP. (D) Constructs fused with the short (S) or long (L) NanoBit luciferase fragments were cotransfected in 3T3 cells as indicated. Protein–protein interaction was assessed by measuring luciferase activity. **P < 0.01, ***P < 0.001. (E) Split complementation was adapted for GFP to assess where protein–protein interaction occurs within L929 cells. GFP fluorescence indicates Tspan5 fused with GFP-exons 1 to 10 (Tspan51-10) complementing with the MHC class I fused with GFP-exon 11 (H-2K11, Left). Subcellular distribution was determined by staining the cell membrane with Alexa647-conjugated WGA (Upper) or by cotransfection with ER resident protein Sec61b-mCherry (Lower). The confocal images shown in the Upper panels were taken in an optical plane near the coverslip in order to visualize complementation in the plasma membrane and the ones in the Lower panels were taken through the center of the cell to visualize the ER. (Scale bar, 5 µm.) Data shown are representative of at least three independent experiments.
In a second approach, we used a split luciferase complementation assay to test for molecular associations in situ in living DCs (35). In this assay, fragments of luciferase, which are not themselves enzymatically active (Fig. 4C), are fused to the intracellular domains of test proteins. If the test proteins are in close proximity (35), then the two different luciferase fragments can interact and become catalytically active (Fig. 4C). In our experiments, MHC class I molecules, H-2K or H-2D, were fused to an inactive small fragment of the NanoLuc luciferase molecule (S), while the larger inactive fragment of luciferase (L) was fused to Tspan5 (Fig. 4C). When these constructs were expressed in 3T3 cells, both H-2K and H-2D interacted with Tspan5 to reconstitute luciferase activity (measured by generation of luminescence from furimazine) (Fig. 4D). In contrast, luciferase activity was not generated between a control membrane protein (CD40) and Tspan5 split luciferase fusions (Fig. 4D). These findings further demonstrate that Tspan5 and MHC I molecules interact in living cells.
In a third set of experiments, we used a different molecular complementation approach to further confirm the above results and address in what subcellular compartments class I associated with Tspan5. This system used a split GFP complementation assay, which exhibited rapid folding properties (36); one part of GFP was fused to MHC I molecules and the other portion to Tspan5. When GFP fragments (exons 1 to 10) fused to Tspan5 or to MHC I (GFP exon 11) are coexpressed with control proteins in L929 cells, no fluorescence was detected, as expected (SI Appendix, Fig. S4 A, Left and Center). However, a strong fluorescence signal was generated when the MHC I and Tspan5 GFP fusion constructs were coexpressed (SI Appendix, Fig. S4 A, Right). These results further confirmed that MHC I and Tspan5 were in very close proximity in living cells. When analyzed by confocal microscopy, a portion of the GFP fluorescent signal was present on the cell membrane and colocalized with WGA, a marker for the PM (Fig. 4 E, Upper).
In addition, in optical sections through the ER, which was marked by mCherry-Sec61b, there was substantial colocalization of the Tspan5–MHC I complementing GFP fluorescence with Sec61b (Fig. 4 E, Lower). When we quantified the intracellular pool of MHC I by flow cytometry, it was similar whether the MHC I and Tspan5 molecules were fused with GFP-complementation fragments (SI Appendix, Fig. S4C), or small epitope tags (SI Appendix, Fig. S4D), or cells were just transfected with MHC I without Tspan5 (SI Appendix, Fig. S4E); therefore, the intracellular pool of MHC I was not an artifact of the GFP fusion or overexpression of Tspan5. Also, as described above, the transport of newly synthesized MHC I molecules (Fig. 2B) to, and steady-state levels (Fig. 2A) on, the cell surface were not affected by loss of Tspan5, indicating that Tspan5 does not arrest MHC I molecules in the ER. In independent experiments, we evaluated whether Tspan5 associated with MHC I in the ER, by analyzing the maturation of its glycans. The Tspan5 that coimmunoprecipitated with MHC I molecules had both endoglycosidase H-sensitive (a property of proteins that are in the ER or early Golgi) and resistant (a property of proteins in that have passed through the late Golgi) species (SI Appendix, Fig. S4F), which confirmed an association between these molecules in the ER and thereafter.
After assembling in the ER, MHC I molecules incorporate into a peptide-loading complex (PLC), wherein they bind Tapasin and TAP. To gain further insight into when Tspan5 was associating with MHC I in the ER, we investigated whether it was associating with the PLC. We first assayed for molecular proximity between these molecules using the split-luciferase complementation system. We detected no luciferase complementation between Tspan5 and either Tapasin or TAP (SI Appendix, Fig. S4 B, Right). In contrast, and as expected, complementation was detected between MHC I and Tspan5, Tapasin and TAP, which served as positive controls (SI Appendix, Fig. S4 B, Left). Consistent with these results, in a further experiment we could not detect coimmunoprecipitation of Tspan5 with Tapasin (Fig. 4B). Together, these data suggest that Tspan5 interacts with MHC I molecules after they are released from the PLC.
These data show that Tspan5 associated with MHC I molecules in the ER, where newly synthesized class I molecules first form and bind peptide, and that this association was maintained on the PM, where pMHC complexes are available to interact with cognate TCRs on CD8 T cells.
Loss of Tspan5 Disrupts MHC I Nanoclusters.
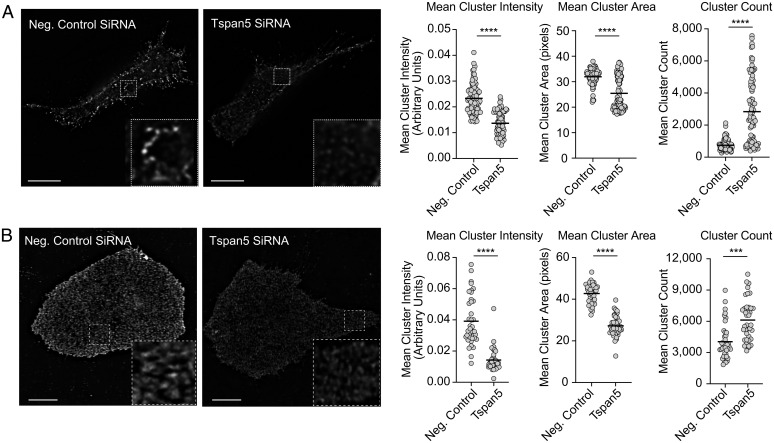
A number of other Tspans have been shown to affect the clustering of membrane proteins (37–39). These findings, together with our data showing that the loss of Tspan5 affects MHC I-dependent responses without reducing overall surface levels of MHC I, led us to examine by immunofluorescence microscopy how Tspan5 influenced the distribution of MHC I on the surface of DCs. To eliminate potential artifacts of bivalent antibody-induced movement of receptor proteins even after fixation (40–42), we used a direct-labeled Fab-fragment specific for the class I molecule, H-2K (Y3-Fab). When analyzed by confocal microscopy, MHC I was found to be distributed in clusters on the cell surface, as has been reported by others (15, 21, 43, 44) (Fig. 5A). In Tspan5-deficient DCs, MHC I clusters were smaller, less intense, and more numerous (Fig. 5). Although control and Tspan5-deficient cells have the same amount of MHC I on their cell surface (Fig. 2A and SI Appendix, Figs. S2A and S5C), fluorescent confocal micrographs of Tspan5-deficient cells appear less bright to the eye (Fig. 5A). Presumably, this optical appearance is due to their clusters being smaller and less intense and also because these cells may have more MHC I-distributed diffusely outside of clusters. Using image analysis software, we quantified changes in the intensity, number and mean area of MHC I clusters. In the absence of Tspan5, the intensity of molecules per cluster (which is proportional to the number of MHC I molecules within a cluster) decreased by ∼50% (Fig. 5 A, Left graph). Fig. 5A also shows that in the absence of Tspan5, the area of clusters decreased as compared to control-treated cells (Fig. 5 A, Right graph), while the total number of MHC I clusters was increased. Therefore, Tspan5 is needed to form and maintain large MHC I nanoclusters and in its absence MHC I is organized into more smaller clusters with fewer MHC I molecules per cluster. To exclude the possibility that fixation of cells was influencing MHC I clustering, we performed the same analysis on live cells and observed similar results (SI Appendix, Fig. S5A).
Fig. 5.
Tspan5 regulates MHC class I receptor organization. (A) DC2.4 cells were transfected with siRNA targeting Tspan5 or the MHC II IA beta chain (Neg. Control SiRNA). The distribution of H-2K molecules on the cell surface was assessed using fluorophore conjugated Fab-fragment (Y3-Fab). Fluorescent images were acquired from multiple 0.125-μm z-sections followed by deconvolution using the DV OMX V4 microscope, as described. Images shown are representative of membrane staining at the coverslip. (Scale bar, 10 µm.) Inset images (4.2x magnification) show representative differences in cluster intensity and size. Graphs show the mean intensity of MHC I clusters per cell from representative experiments (arbitrary units, Left graph), mean area of MHC I clusters (pixels, Center graph), or cluster count (Right graph). Data in the graphs are from >45 independent images and are representative of ≥3 independent assays. (B) Similar to A except using human U2OS cells instead of DC2.4 cells and a fluorophore conjugated Fab-fragment (W6/32-Fab) instead of Y3-Fab. Magnification shown in B is 3.3x. (Scale bar, 10 µm.) P values determined by two-tailed, unpaired t test; ***P < 0.001, ****P < 0.0001.
We extended our studies to the human osteosarcoma cells, U2OS. Tspan5 knockdowns were equally as efficient in human U2OS cells as in mouse DCs (SI Appendix, Fig. S5 B and C). We visualized the distribution of MHC I molecules by staining with fluorophore-labeled Fab fragments of the HLA-specific antibody, W6/32. Our data demonstrated that in the absence of Tspan5, MHC I cluster intensity and area were also significantly reduced and cluster number was significantly increased (Fig. 5B). Our findings are consistent with other studies showing that changes in Tspan expression or mutations affecting function of Tspans can affect cluster size/area and the density of associated proteins in clusters (37). Taken together, our data indicate that Tspan5 is influencing the distribution of MHC I molecules on the cell surface.
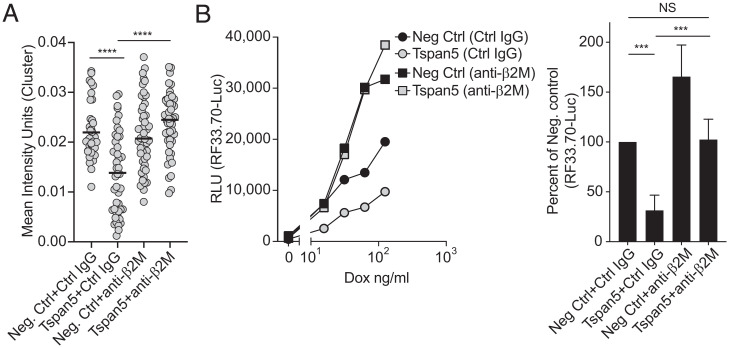
Since clustering of TCRs and their MHC ligands is needed for optimal T cell activation, the reduction in MHC I cluster size in Tspan5-deficient cells could explain why the loss of Tspan5 inhibits the ability of DCs to stimulate CD8 T cell responses. To test this hypothesis, we examined whether increasing the clustering of MHC I molecule through antibody-mediated cross-linking could restore the ability of Tspan5-deficient cells to stimulate T cells. For this purpose, we used antibodies to the MHC I light chain (β2M) because this protein is not part of an interaction surface with either the TCR or CD8 molecules. We found that when antibodies to β2M were added to Tspan5-deficient DCs, the size of MHC I clusters was restored to levels at or above those observed in the presence of Tspan5 (negative Ctrl siRNA-treated cells) (Fig. 6A). Importantly, addition of the anti-β2M antibody restored the ability of Tspan5-deficient cells to stimulate the CD8 T cells to the level of WT DCs (negative Ctrl) (Fig. 6B). This effect was specific for MHC I-stimulated responses because anti-β2M did not alter stimulation of the CD8 T cells by anti-CD3 on DCs (SI Appendix, Fig. S5D). Together, these data demonstrate that Tspan5-mediated clustering of peptide loaded MHC I molecules is necessary for optimal activation of CD8 T cells.
Fig. 6.
Antibody cross-linking of MHC I molecules restores MHC I clustering and T cell stimulation. (A) Tspan5 or negative control siRNA (Neg. Ctrl) were transfected into DC3.2 cells. After 48 h, cells were treated with a control antibody (Ctrl IgG) or with an antibody to the class I light chain (anti-β2M) for 4 h. Cells were fixed and stained with H-2K reactive Y3-Fab. Total internal reflection microscopy was used to image MHC I clusters at the PM. Graph shows the mean intensity of stained clusters/cell (gray circle) as assessed by Cell Profiler for >40 images per condition. Mean values per treatment are shown with a black horizontal line. P values were determined by an unpaired, two-tailed t test; *** P < 0.001, ****P < 0.0001. (B, Left) Ova expression was induced in DC2.4 cells by the addition of Dox for 2 h followed by the addition of BFA and RF33.70-Luc T cells plus control antibody (Control IgG) or MHC I-crosslinking antibody (anti-β2M). After 16 h of incubation, T cell activation was assessed by measuring luciferase activity. Graph shown is representative for antigen (Dox) titration. (Right) Same as the Left panel, except the percent of T cell activation was compared to the negative control siRNA + negative control antibody treated samples at 30 ng/mL Dox. Data are the means + SD from greater than three independent experiments. P values were determined by two-tailed, paired t test; NS = not significant, ***P < 0.001.
Discussion
MHC I molecules can be found in small nanoclusters on the surface of APCs and these clusters are present prior to any interaction with T cells. However, what caused these clusters to form and persist, and their role in antigen presentation was largely unknown. Herein, we report a mechanism that promotes MHC I-clustering and is needed for optimal stimulation of CD8 T cells. Our major findings are that Tspan5 associates with MHC I in the ER and these molecules remain associated at the PM. Loss of Tspan5 did not affect peptide processing or MHC I peptide-loading, nor did it affect the kinetics of pMHC delivery to, or overall levels on, the surface of APCs. Instead, Tspan5 influenced the organization of MHC I into nanoclusters on the PM. Moreover, in the absence of Tspan5, pMHC I stimulation of CD8 T cells was significantly reduced but could be restored by the reexpression of Tspan5 or by reclustering MHC I with antibodies. These results elucidate a mechanism that underlies the generation of MHC I clusters and demonstrate its importance for optimal T cell stimulation.
We could not detect direct binding and close association of Tspan5 with Tapasin or TAP, although under these same conditions, we detected interactions of MHC I with Tapasin and TAP. Therefore, Tspan5 does not appear to bind MHC I molecules in, or be a component of the PLC. This is not surprising because the cryoelectron microscopic structure of the PLC revealed that MHC I is surrounded almost completely by Tapasin, TAP, calreticulin, and ERP57 (45), and these molecular interactions would likely sterically interfere with Tspan5 binding to the MHC I molecule. In any case, our data suggest the interaction of Tspan5 with MHC I occurs upon or after the release of MHC I from the PLC.
Tspans are a large family of membrane multispanning proteins that have a broad tissue distribution, with individual members having some unique and other overlapping functions (28). Various Tspans have been shown to influence molecular trafficking and organization, thereby influencing a variety of biological processes (23). Tspans can function by forming intermolecular complexes with other Tspan proteins, as well as with other proteins, resulting in formation of large membrane domains termed TEMs (28). Proteins localize to TEMs through their interactions with Tspans or possibly other mechanisms (46). Our data do not resolve whether Tspan5 directly binds MHC I molecules. However, our GFP complementation data show that these two molecules are in very close proximity in living cells. The maximum intermolecular distance over which split GFP constructs can complement (i.e., reconstitute fluorescence) was reported to be the length of the GFP-attached linker sequences, in an extended conformation (47). If this is correct, then based on our use of flexible linkers of 10 and 11 amino acids, complementation would be occurring between Tspan5 and MHC I molecules that are within ≤8 nm of each other.
Tspan5 is widely expressed, including in DCs and other APCs. Tspan5 is a highly conserved protein, and in fact its amino acid sequence is identical in mouse and human. Tspan5 is a member of the C8 family of Tspans, all of which have eight cysteines in their large extracellular loop (48, 49). There have been some previous investigations of Tspan5 and its related TspanC8 family members. These studies have revealed that the TspanC8 proteins can interact with one another (50). In one well-studied system, Tspan5 and other TspanC8 family members have been shown to bind the metalloprotease ADAM-10 in the ER and regulate its trafficking to the PM (31, 50). In the absence of Tspan5, ADAM-10–mediated shedding of Notch-1 resulted in the suppression of osteogenesis (51) and of VE-Cadherin–dependent T cell migration (31, 52). In neurons, Tspan5 interaction with the adhesion molecule neuroligin-1 promoted its clustering and was shown to be necessary for maturation of neuronal dendrite spines (39). However, there have been no reports of Tspan5 interaction with MHC I or its playing a role in antigen presentation. Tspans CD53, CD81, and CD82, were reported to interact with MHC I, but the functional importance of these interactions was not explored (25, 29). It will be of interest in future studies to examine whether additional Tspans participate with Tspan5 in MHC I antigen presentation.
Some Tspans (e.g., CD37, CD53, CD81, and CD82) have also been reported to associate with MHC II molecules (25, 53). None of these MHC II-associated Tspans have been reported to affect MHC II clustering. In fact, the presence of many of these Tspans seemed to negatively regulate the ability of APCs to stimulate CD4 T cells (54, 55). Loss of Tspan5 did not reduce antigen-MHC II stimulation of CD4 T cells. It will be of interest to explore whether these or other Tspans play a positive role in antigen presentation, similar to what we have observed for Tspan5 and MHC I, or if they have redundant functions.
MHC I molecules are delivered to the cell surface as nanoclusters, which are estimated to contain 25 to 250 complexes, and may then coalesce to form larger clusters (microclusters) upon engagement with T cells expressing the appropriate TCR (15, 19, 21, 44, 56). These clusters were suggested to form in the ER or cis-Golgi compartment, where MHC I molecules assemble and bind peptides (43, 44). Consistent with this suggestion, we found that Tspan5 associated with MHC I in the ER, as evidenced in a Tspan5–MHC I GFP-complementation assay, wherein the ER became fluorescent, and also by finding endo H-sensitive Tspan5–MHC I complexes (Endo-H sensitivity is a characteristic of proteins before their transit through the Golgi apparatus).
Several lines of evidence suggest that Tspan5 augments the effectiveness of antigen presentation through its interactions with MHC I. First, Tspan5 associated with MHC I molecules. Second, Tspan5 only augmented responses stimulated by MHC I molecules. If anti-CD3 bound to APCs was substituted for specific pMHC I complexes, then loss of Tspan5 did not reduce stimulation of CD8 T cells, even under limiting conditions. Moreover, Tspan5’s augmenting effect was selective for CD8 responses, as Tspan5 did not augment CD4 T cell stimulation by MHC II-presented peptides. These findings indicated that Tspan5 was not affecting any of the APC accessory molecule interactions, such as adhesion or costimulatory molecules, that were needed in common for both CD4 and CD8 T cells or APC-dependent anti-CD3 responses. Third, and most importantly, loss of Tspan5 reduced the size of MHC I nanoclusters and T cell stimulation. Importantly, reconstituting MHC I clusters via antibody cross-linking restored both the MHC I cluster formation and T cell stimulation.
The role of Tspan5 in stimulating T cells by organizing MHC I clusters is of considerable interest because of the likelihood that these preformed clusters are promoting the aggregation of cognate receptors on CD8 T cells (TCRs and CD8). Indeed, one threshold for T cell activation is based on the size of TCR clusters (8, 9, 57, 58), which has been shown to influence signal strength and the magnitude of the subsequent T cell response (10, 59). It has been reported that dimerization is the minimal requirement for T cell stimulation, and higher-order clustering further enhances signaling and responses (7, 8, 19, 57, 60, 61). Our finding that Tspan5 is likely binding pMHC I complexes upon release from the PLC provides a potential mechanism to allow cohorting of pMHC I complexes being produced at any snapshot in time (44). Since the expression of a particular protein and subsequent production of its antigenic peptides can vary at different times (e.g., over the course of a viral infection), we speculate that Tspan5 association with complexes upon release from the PLC could help cocluster more pMHC complexes with the same peptide and thereby further enhance the clustering/stimulation of cognate TCRs. MHC I clusters may also facilitate rebinding of CD8 to cognate and noncognate complexes (62, 63) and clustering of CD8-associated Lck has been reported to enhance stimulation (64, 65). Presumably, Tspan5–MHC I clusters enhance T cell stimulation through their effects on TCR and CD8 clustering, but could also possibly affect other parameters, such as TCR mechanosensing (66–68). It is interesting that in our experiments approximately twofold reductions in larger MHC I clusters have a strong effect on CD8 T cell stimulation. Perhaps this is due to a threshold effect, as it has been observed that when T cells recognize pMHC I complexes, signaling was observed in dense TCR microclusters (size/number), but not smaller less dense ones (8, 9). It is also possible that Tspan5 is affecting T cell stimulation in additional ways, although this seems less likely, given our finding that antibody-mediated reclustering of MHC I restores the stimulatory activity of Tspan5-deficient cells.
In summary, our results have identified Tspan5 as a component in the MHC I antigen presentation pathway. Our findings also reveal a mechanistic dimension to the MHC I antigen presentation process. Specifically, while it was recognized that MHC I molecules could be in clusters, the functional significance of this was not entirely clear. Although earlier reports of cohorting of specific MHC I complexes hinted at increased stimulatory activity, there was no underlying mechanism to account for the phenomenon or to directly show its functional importance (19, 21, 43, 44). Our results provide evidence that the clustered topology of MHC I complexes is indeed important to the effectiveness of the antigen presentation process.
Materials and Methods
Mice.
All mouse strains were maintained in specific pathogen-free facilities at the University of Massachusetts Chan Medical School (UMMS) in accordance with approved guidelines set forth by the UMMS Department of Animal Medicine and Institutional Animal Care and Use Committee. C57BL/6, OT-1 TCR transgenic (recognizing Ova257–264 on H-2Kb) and p14 TCR transgenic mice (recognizing GP33–41 on H-2Db) were acquired from Jackson Laboratories and bred in our facility.
Cell Lines.
Mouse DCs DC3.2 and DC2.4 isolated from C57BL/6 mice (69) were maintained in complete growth media (RPMI-1640, 10% FCS, 10 mM Hepes, 1× MEM nonessential amino acids, 100 U/mL penicillin and 100 μg/mL streptomycin, 2 mM l-glutamine, and 54 μM 2-mercaptoethanol). Mouse L929 cells and 3T3 cells, as well as human embryonic kidney (293T) and bone osteosarcoma epithelial cells (U2OS) were cultured in RPMI-1640 supplemented with 10% FCS, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The CD8 T cell hybridoma RF33.70 recognizes the chicken Ova peptide SIINFEKL (S8L; Ova257–264) in the context of the class I molecule H-2Kb (70). The CD4 T cell hybridoma, MF2.2D9, recognizes Ova peptide (Ova258–276) in the context of the class II molecule H-2 I-Ab (71). Both T cell hybridomas were maintained in complete growth media and were engineered to express luciferase under the control of the NFAT promoter, as described elsewhere (referred to as RF33.70-Luc and MF2.2D9-Luc) (30, 72).
Flow Cytometry.
Antibodies recognizing the following molecules were used: H-2Kb (AF6-88.5, BioLegend; or Y3, Bio X Cell), H-2Db (KH95, BioLegend), ICAM-1 (YN1/1.7.4, BioLegend), interferon (IFN)-γ (XMG1.2, BioLegend), pan-HLA antibody (W6/32, BioLegend), CD80 (16-10A1, BD Biosciences), CD86 (GL1, BD Biosciences), CD40 (3/23, BD Biosciences), CD8 (53-6.7, ThermoFisher), and CD11c (N418, eBioscience). Ova257–264 bound to H-2Kb (25D1.15) (73) and Tspan5 [clone 16B8, a kind gift of Eric Rubenstein, INSERM, Paris, France (74)] were used for immunofluorescence staining. Briefly, adherent cells were trypsinized, washed and incubated with ice-cold 2.4G2 hybridoma supernatant to block nonspecific binding of staining antibodies to mouse Fc receptors. Staining was performed on ice in staining buffer (PBS with 1% [vol/vol] FCS) for at least 30 min. Cells were analyzed on the LSR2 flow cytometer (BD Bioscience). Flow cytometry data were further analyzed using FlowJo software (Tree Star). To follow the egress of MHC I molecules from the ER to the PM, DCs were washed with ice-cold PBS followed by the removal of peptide by incubating cells for 90 s in acid-stripping buffer (0.123 M citric acid, 0.234 M sodium phosphate), resulting in the denaturation of surface MHC I molecules. Cells were neutralized in complete media and incubated at 37 °C to allow repopulation of MHC I. Incubations were stopped at the time points indicated by placing cells on ice and staining for H-2Kb or for peptide presentation with 25D1.16 antibodies. MHC stabilization experiments were performed by treating cells with BFA (GolgiPlug, BD Biosciences) over time, followed by staining for MHC I molecules H-2K and H-2D, as indicated. Intracellular staining was performed using the CytoFix/CytoPerm kit (BD Biosciences), in accordance with manufacturer’s instructions. To quantitate the levels of cell-surface vs. intracellular MHC I on L929 cells, cells were first incubated in PBS or acid-stripped, as described above, and then fixed in 4% PFA. Surface MHC I was stained on fixed cells ± stripping using staining buffer. Total MHC I (surface + intracellular) was stained on fixed and permeabilized cells using staining buffer supplemented with 0.25% saponin. Intracellular MHC I was stained in acid-stripped cells that were fixed and permeabilized. Antigen presentation assays involving flow cytometry are described below.
Molecular Cloning.
A nonsecreting mutant of Ova (NS-Ova, amino acids 51 to 386) was cloned into a modified Dox-inducible lentiviral vector (pTRIPz, Open Biosystems), as previously described (30). Mouse Tspan5 was amplified from DC3.2 cells and fused with an N-terminal HA-epitope tag by encoding the tag into primer sequences followed by ligation into the lentiviral expression vector pCDH (Systems Bioscience). An myc-epitope tag was incorporated into the C terminus of H-2Kb, H-2Db, CD40, Tapasin, and ligated into the lentiviral expression vector pHIV (pHIV-Zsgreen was a gift from Bryan Welm and Zena Werb, Addgene #18121, http://n2t.net/addgene:18121; RRID:Addgene_18121) and further modified with a selectable marker encoding antibiotic resistance (hygromycin or puromycin) in place of ZsGreen, as indicated. For construction of the split GFP complementing constructs, Tspan5 was fused on the C terminus to a 7-amino acid Gly-Ser (GS) rich linker peptide to exons 1 to 10 of the superfold mutant of GFP (sfGFP, a kind gift of Bo Huang, Addgene #70219, http://n2t.net/addgene:70219; RRID:Addgene_70219). The mouse MHC I molecule H-2Db was fused on the N terminus with a 13-amino acid GS linker peptide to exon 11 of sfGFP (pEGFP-GFP11, Addgene #70217). Both constructs were then cloned into the lentiviral expression vector pCDH (Systems Bioscience) expressing either puromycin or blasticidin resistance genes. For construction of the split luciferase constructs, Tspan5 was cloned at the C-terminal end of the long fragment (L) of the NanoBit luciferase construct pBit 1.1N (N198, Promega) separated by a 15-amino acid linker. H-2K, H-2D, TAP, Tapasin, or CD40 were cloned at the C-terminal or N-terminal end of the small fragment (S) of the NanoBit construct pBiT2.1C or pBiT2.1N, respectively (Promega) separated by a 16 linker. CRISPR guide sequences targeting β2M (GTGGGTGGCGTGAGTATACT, AGTATACTCACGCCACCCAC) and Tspan5 (ATTCCTGGGTGAAGTCTATG, TGATGGACGAGATGTTGGAG) were designed using Synthego (https://design.synthego.com/#/) for optimal on-target specificity. The two guide sequences were cloned into pCRoatan dual sgRNA expression vector (kind gift of Gregory Hannon, Cancer Research UK, Cambridge, UK), according to methods described previously (75).
Transfection and Lentiviral Transduction.
Lentivirus was produced by cotransfection of lentiviral vectors with packaging constructs psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259, both gifts of Didier Trono, http://n2t.net/addgene:12260; RRID:Addgene_12260 and http://n2t.net/addgene:12259; RRID:Addgene_12259, respectively) using Lipofectamine 2000 (Invitrogen) according to manufacturer instructions. After 48 h, supernatants containing virus were collected and cultured with cells, as indicated, in the presence of 8 μg/mL Polybrene (Sigma) followed by centrifugation for 60 min at 1,100 × g. After 48 h, transduced cells were selected by culturing with antibiotic or sorted by flow cytometry. Protein expression was monitored by Western blot or by flow cytometry. Transfection of pBIT split complementation plasmids was performed in 3T3 cells using Lipofectamine 2000 (Invitrogen), according to manufacturer’s recommendations. Transfection efficiencies were confirmed after 48 h by Western blot or flow cytometry.
Gene Silencing.
siRNA targeting murine Tspan5 (GGAAUAACGUUUCUUGGAA), H-2 I-A β-chain (H-2 Ab1; GGACGCAGCGCAUACGAUA) and β2M (ACAUACGCCUGCAGAGUUA) were purchased from Thermo Scientific (siGENOME). siRNA SMARTpools targeting human Tspan5 and a nontargeting control (negative Ctrl) were purchased from Horizon Biosciences. siRNAs were transfected in DC cell lines and U2OS cells by reverse transfection using Lipofectamine RNAiMAX reagent (Invitrogen). Briefly, siRNAs (50 nM per well) were mixed with RNAiMAX (0.4 μL per well) in 1× siRNA buffer (Dharmacon) in 50 μL per well of a 96-well plate for 20 min. DCs were plated in antibiotic-free media containing RPMI-1640 and 20% FCS at a density of 1.5 × 104 cells per well for 48 h. Silencing efficiency was determined by flow cytometry or by qPCR (Taqman, Applied Biosystems). Relative expression was calculated using the ΔΔCT method including mouse HPRT1 or human β-actin as housekeeping controls. CRISPR knockout DCs were generated by lentiviral transduction of Lenti-Cas9 vector (kind gift of Feng Zhang, Addgene #52962, http://n2t.net/addgene:52962; RRID:Addgene_52962) encoding Streptococcus pyogenes Cas9 along with pCRoatan vectors expressing dual guide RNAs targeting either Tspan5 or β2M, as mentioned above. Indel frequencies were determined from heterogenous cell cultures following extraction of genomic DNA (DNeasy Blood & Tissue kit, Qiagen) and PCR amplification. PCR products were purified, and sequenced using the 5′ PCR primer, followed by TIDE analysis (76). Knockout efficiency was confirmed by flow cytometry. Guide RNA producing indel frequencies <90% were eliminated from future experiments. In some experiments, WT Tspasn5 expression was restored in Tspan5 knockout DCs by cloning Tspan5 with six silent mutations in each of the two guide target sequences.
Antigen Presentation.
Cross-presentation and MHC II presentation assays were performed with mouse DC cell lines (DC3.2 and DC2.4) using chicken Ova as a source antigen (69), as indicated. Ova (Sigma) was covalently conjugated to 1.5-μm amine-coated iron oxide beads (Ova beads) using BioMag Plus Amine Protein coupling kit (Bangs Laboratories) according to the manufacturer’s recommendations. Quantification of antigen bound to beads was determined by UV spectrophotometry and confirmed by PAGE followed by Coomassie brilliant blue (ThermoFisher). Ova beads were titrated as indicated and cultured with DCs in the presence of CD4 T cell hybridoma (MF2.2D9-Luc) or CD8 T cell hybridoma (RF33.70-Luc) for 24 h. T cell stimulation was detected by measuring the expression of luciferase under the control of an NFAT promoter, as described previously (30, 72). Luciferase activity was measured using One-Glo reagent (Promega) according to manufacturer’s recommendations. For classic class I presentation, antigen expression was induced in DC2.4 cells following transfection of a nonsecreting form of Ova (NS-Ova) under a Dox-inducible promoter. Briefly, Dox was titrated, as indicated, and cultured with DCs for 3 h followed by the addition of BFA (GolgiPlug, BD Bioscience) and RF33.70-Luc T cells. Cells were incubated for an additional 18 h, followed by the measurement of luciferase activity. In some experiments, following silencing of the indicated genes by siRNA, antigen presentation was also performed in the presence of anti-β2M antibody or control antibody (to induce the cross-linking of MHC I molecules). Briefly, 48 h after silencing, DC3.2 cells were cultured with Fc-receptor blocking antibody 2.4G2, followed by addition of 1μg/mL of an isotype control anti-mouse control antibody (Santa Cruz Biotechnology) or mouse anti-β2M antibody (S19.8, BD Pharmingen) for 1 h at 37 °C. Ova-conjugated beads and RF33.70-Luc CD8 T cells were then cultured for an additional 24 h in the continued presence of the cross-linking or control antibody.
T cell stimulation was assessed by following luciferase activity as indicated above. T cell proliferation was assessed using naïve OT-1 transgenic T cells (which recognize Ova257–264 bound to H-2Kb) (77). Briefly, naïve CD8 T cells were isolated from spleens of TCR transgenic mice by negative selection (Miltenyi Biotec). CD8 T cell purity of >90% was confirmed by flow cytometry. Cells were labeled with Cell Tracer Far Red (CTFR, Invitrogen) and washed in normal media. DCs, previously silenced for the indicated genes, were γ-irradiated to induce growth arrest. Naïve CFTR-labeled OT-1 T cells were then plated with a limiting concentration of Ova-coupled beads or anti-CD3e (clone 2C11, a kind gift from Susie Swain, UMMS, Worcester, MA) for 72 h. Stimulation of T cells with anti-CD3 was also performed in the presence of a blocking antibody for LFA-1 (M17/4.2) (78), as indicated. Evaluation of T cell proliferation was assessed on a LSR2 flow cytometer (BD Bioscience) by analyzing the dilution of CTFR in proliferating CD8 T cells. Proliferation was calculated using the division index to account for dividing and nondividing cells using FlowJo software (Tree Star). Stimulation of effector CD8 T cells was performed using OT-1 T cells or p14 transgenic T cells (which recognize LCMV GP33–41 bound to H-2Db). Briefly, splenocytes from TCR Tg mice were stimulated in vitro by incubating with 100 nM of the appropriate peptide for 3 h. Splenocytes were washed and then cultured for 7 to 14 d in RPMI containing 20 ng/mL IL-7 and 20 ng/mL IL-15. DCs were silenced as described and then incubated with antigen and effector CD8 T cells. Ova beads were used to stimulate OT-1 T cells, as described, while DC3.2 cells were infected with a titration of LCMV (Armstrong strain) (a kind gift of Raymond Welsh, UMMS, Worcester, MA) for 3 h, washed and then cultured for an additional 24 h in the presence of p14 CD8 effector T cells. Cells were treated with BFA for 4 h and then stained for IFN-γ production and analyzed by flow cytometry.
Microscopy.
L929 cells transfected with GFP complementation fragments were grown on 35-mm dishes fitted with no. 1.5 coverslips (Nunc) overnight. Cells were washed and fixed with 2% PFA and were stained with Alexa Fluor-350–conjugated WGA (ThermoFisher) before washing and resuspension in PBS. Other cells were transduced with lentivirus encoding the ER localization marker Sec61b fused to mCherry (kind gift of Jennifer Lippincott-Schwartz, Addgene #90994, http://n2t.net/addgene:90994; RRID:Addgene_90994). Live L929 cells were analyzed on a Leica SP5 confocal inverted microscope fitted with a 63×/1.4 oil objective (Leica) using 405-, 488-, and 594-nm laser excitation. Confocal images were taken sequentially through the cell starting at the coverslip, followed by 0.125-μm z-sections. For MHC I cluster analysis, mouse DC3.2 cells and human U2OS cells were silenced with the indicated siRNAs and grown on no. 1.5 coverslips (Electron Microscopy Science). Cells were fixed with 2% PFA and blocked with 5% BSA in PBS (wt/vol). Cells were stained for MHC I expression using Alexa-647–labeled Fab fragments. Briefly, Fab fragments for the Y3 and W632 antibodies were generated using immobilized papain (Thermo) according to the manufacturer’s instructions. Undigested antibody and remaining Fc fragment were removed by binding digested antibody to Protein A-conjugated agarose beads (Millipore). Purified Fab fragments were labeled with Alexa Fluor-647 antibody-labeling kit (Molecular Probes) according to the manufacturer’s recommendations. The concentration of Fab fragments and dye-labeling efficiency were calculated from absorbance using the recommended calculations provided by the kit. Stained cells were washed and mounted (Prolong Glass, Invitrogen) on no. 1.5 glass slides. Images were acquired on a DeltaVision OMX V4 inverted imaging system (GE Healthcare) with pco.edge SCMOS cameras, an Olympus PlanApoN 60×/1.42 NA oil immersion objective (Olympus) in 1.514 immersion oil, 642-nm excitation laser, and 683/40-nm emission filter. Multiple z-stacks with a spacing of 0.125 μm were acquired in conventional mode or a single z-section at the coverslip for total internal reflection fluorescence (TIRF), as indicated. Deconvolution of serial z-stacks was performed postacquisition to improve the signal-to-background ratio of the images using SoftWorX (DeltaVision, GE Healthcare) and single optical slices were identified at the coverslip using WGA counterstain for orientation.
For MHC I cluster analysis on live cells, DC2.4 cells were silenced with the indicated siRNAs and grown on 35-mm dishes with no. 1.5 poly-d-lysine–coated 14-mm diameter glass coverslips (Mattek Corporation, P35GC1.514C.S). Cells were stained for MHC I expression using Alexa-647–labeled Fab fragments. After washing stained cells, culture dishes were filled with cell culture media supplemented with Prolong Live Antifade Reagent (Thermo) and imaged on the DeltaVision OMX V4 using conventional mode with solid-state illumination at 642 nm for excitation, the Olympus PlanApoN 60×/1.42 NA oil immersion objective with 1.514 immersion oil and a 683/40-nm emission filter. The z-stacks were taken with 125-nm steps and images were deconvolved using the SoftWorx software. Serial images were acquired as described previously for fixed cells. Images were further processed using Fiji/ImageJ software (79) followed by a custom pipeline designed to analyze MHC I clusters at the PM (Cell Profiler, Broad Institute) (80). For each experimental condition, at least 10 different fields of view were randomly identified and each cell within the field was then acquired. Detection settings were kept constant for all acquired images within each experiment.
Protein–Protein Interaction.
Coimmunoprecipitation assays were performed by transfecting Hek293T cells with plasmids expressing HA-tagged Tspan5, and myc-tagged H-2K or H-2D, as indicated. Briefly, cells were lysed in cell lysis buffer (1% Brij-35 or 1% CHAPS, 150 mM NaCl, and 50 mM Tris, pH 7.0). Nuclear debris was removed by centrifugation (20,000 × g for 10 min at 4 °C). Insoluble membrane and cytoskeletal debris were further removed by centrifugation at 100,000 × g for 60 min at 4 °C. Soluble protein lysates were incubated with either 1 μg rabbit control antibody (Santa Cruz Biotechnology), rabbit anti-myc antibody (Genscript) covalently conjugated to magnetic beads (Thermo), or with anti-HA antibody-conjugated beads (Thermo, Cat# 88836) in accordance with manufacturer recommendations. Immunoprecipitated protein was washed in ice-cold wash buffer (0.1% Brij-35 or 0.1% CHAPS, 150 mM NaCl, and 50 mM Tris, pH 7.0) followed by acid elution according to the manufacturer’s recommendations. Polyacrylamide gels were loaded with protein from cell lysates prior to mixing with antibody (input 10%) and with protein associated with antibody-coated beads (bound) and were separated by SDS/PAGE, transferred to a nylon membrane, and stained with antibody specific for either HA-tagged Tspan5 (clone 6E2, Cell Signaling), myc-tagged H-2K, or Tapasin (clone 9B11, Cell Signaling), endogenous Flotillin-1 (clone W16108A, BioLegend), or endogenous CD55/Complement Decay Accelerating Factor (clone M033, BioLegend), as indicated. Glycosylation of Tspan5 was determined by treating cell lysates with PNGase (New England Biolabs) or EndoH (New England Biolabs) according to the manufacturer’s recommendation. For split luciferase assay, pBiT fusion constructs were cotransfected in 3T3 cells using Lipofectamine 2000, as indicated. After 48 h, cells were transferred to 96-well plates at 100,000 cells per well. Luciferase activity was assessed using the NanoBIT (Promega) PPI substrate in accordance with the manufacturer’s recommendations. Split-GFP complementation was assessed by confocal microscopy, as described above.
Statistical Analysis.
All statistical analysis was performed using Prism software (GraphPad). Datasets comparing two treatment groups were analyzed using two-tailed, unpaired Student’s t test, as indicated. Means from multiple experiments were compared by one-way ANOVA and Tukey’s multiple comparisons test. For TIRF and confocal microscopy, cluster analysis was generated in Cell Profiler, as indicated, and compared using two-tailed, unpaired Student’s t test. Statistical analysis was documented such that P > 0.05 was considered not significant (NS) and with significance depicted with asterisks as follows: *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank L. Stern and J. Lai (University of Massachusetts Chan Medical School, Worcester, MA) for helpful discussions and review of the manuscript; and E. Rubenstein (INSERM, France), G. Hannon (Cancer Research UK, Cambridge, United Kingdom), and Raymond Welsh (University of Massachusetts Chan Medical School, Worcester, MA) for providing reagents. This work was supported by Grant AI114495 from the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122188119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the main text and SI Appendix.
References
- 1.Blum J. S., Wearsch P. A., Cresswell P., Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rock K. L., Farfán-Arribas D. J., Colbert J. D., Goldberg A. L., Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 35, 144–152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colbert J. D., Cruz F. M., Rock K. L., Cross-presentation of exogenous antigens on MHC I molecules. Curr. Opin. Immunol. 64, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yachi P. P., Lotz C., Ampudia J., Gascoigne N. R. J., T cell activation enhancement by endogenous pMHC acts for both weak and strong agonists but varies with differentiation state. J. Exp. Med. 204, 2747–2757 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krogsgaard M., Juang J., Davis M. M., A role for “self” in T-cell activation. Semin. Immunol. 19, 236–244 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dushek O., van der Merwe P. A., An induced rebinding model of antigen discrimination. Trends Immunol. 35, 153–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogsgaard M., et al. , Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature 434, 238–243 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Manz B. N., Jackson B. L., Petit R. S., Dustin M. L., Groves J., T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. U.S.A. 108, 9089–9094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pageon S. V., et al. , Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 113, E5454–E5463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schamel W. W. A., et al. , Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 202, 493–503 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillemeier B. F., et al. , TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crites T. J., et al. , TCR Microclusters pre-exist and contain molecules necessary for TCR signal transduction. J. Immunol. 193, 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossboth B., et al. , TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat. Immunol. 19, 821–827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer P. H., Pierce S. K., Evidence for dimers of MHC class II molecules in B lymphocytes and their role in low affinity T cell responses. Immunity 1, 699–707 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Matko J., Bushkin Y., Wei T., Edidin M., Clustering of class I HLA molecules on the surfaces of activated and transformed human cells. J. Immunol. 152, 3353–3360 (1994). [PubMed] [Google Scholar]
- 16.Roucard C., Garban F., Mooney N. A., Charron D. J., Ericson M. L., Conformation of human leukocyte antigen class II molecules. Evidence for superdimers and empty molecules on human antigen presenting cells. J. Biol. Chem. 271, 13993–14000 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Cherry R. J., et al. , Detection of dimers of dimers of human leukocyte antigen (HLA)-DR on the surface of living cells by single-particle fluorescence imaging. J. Cell Biol. 140, 71–79 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropshofer H., et al. , Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 3, 61–68 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Fooksman D. R., Grönvall G. K., Tang Q., Edidin M., Clustering class I MHC modulates sensitivity of T cell recognition. J. Immunol. 176, 6673–6680 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch B., Heipertz E. L., Drake J. R., Roche P. A., Major histocompatibility complex (MHC) class II-peptide complexes arrive at the plasma membrane in cholesterol-rich microclusters. J. Biol. Chem. 288, 13236–13242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferez M., Castro M., Alarcon B., van Santen H. M., Cognate peptide–MHC complexes are expressed as tightly apposed nanoclusters in virus-infected cells to allow TCR crosslinking. J. Immunol. 192, 52–58 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Lindstedt R., Monk N., Lombardi G., Lechler R., Amino acid substitutions in the putative MHC class II “dimer of dimers” interface inhibit CD4+ T cell activation. J. Immunol. 166, 800–808 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Levy S., Shoham T., The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5, 136–148 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hemler M. E., Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Szöllósi J., Horejsí V., Bene L., Angelisová P., Damjanovich S., Supramolecular complexes of MHC class I, MHC class II, CD20, and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. J. Immunol. 157, 2939–2946 (1996). [PubMed] [Google Scholar]
- 26.Unternaehrer J. J., Chow A., Pypaert M., Inaba K., Mellman I., The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc. Natl. Acad. Sci. U.S.A. 104, 234–239 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoorn Tv., Paul P., Janssen L., Janssen H., Neefjes J., Dynamics within tetraspanin pairs affect MHC class II expression. J. Cell Sci. 125, 328–339 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Saiz M. L., Rocha-Perugini V., Sánchez-Madrid F., Tetraspanins as organizers of antigen-presenting cell function. Front. Immunol. 9, 1074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagaudrière-Gesbert C., et al. , The tetraspanin protein CD82 associates with both free HLA class I heavy chain and heterodimeric beta 2-microglobulin complexes. J. Immunol. 158, 2790–2797 (1997). [PubMed] [Google Scholar]
- 30.Cruz F. M., Colbert J. D., Rock K. L., The GTPase Rab39a promotes phagosome maturation into MHC-I antigen-presenting compartments. EMBO J. 39, e102020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haining E. J., et al. , The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J. Biol. Chem. 287, 39753–39765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eschenbrenner E., et al. , TspanC8 tetraspanins differentially regulate ADAM10 endocytosis and half-life. Life Sci. Alliance 3, e201900444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wacholtz M. C., Patel S. S., Lipsky P. E., Leukocyte function-associated antigen 1 is an activation molecule for human T cells. J. Exp. Med. 170, 431–448 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachmann M. F., et al. , Distinct roles for LFA-1 and CD28 during activation of naive T cells: Adhesion versus costimulation. Immunity 7, 549–557 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Dixon A. S., et al. , NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Cabantous S., Terwilliger T. C., Waldo G. S., Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Termini C. M., et al. , The membrane scaffold CD82 regulates cell adhesion by altering α4 integrin stability and molecular density. Mol. Biol. Cell 25, 1560–1573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattila P. K., et al. , The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 38, 461–474 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Moretto E., et al. , TSPAN5 enriched microdomains provide a platform for dendritic spine maturation through neuroligin-1 clustering. Cell Rep. 29, 1130–1146.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayor S., Rothberg K. G., Maxfield F. R., Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science 264, 1948–1951 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Tanaka K. A. K., et al. , Membrane molecules mobile even after chemical fixation. Nat. Methods 7, 865–866 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Stanly T. A., et al. , Critical importance of appropriate fixation conditions for faithful imaging of receptor microclusters. Biol. Open 5, 1343–1350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pentcheva T., Edidin M., Clustering of peptide-loaded MHC class I molecules for endoplasmic reticulum export imaged by fluorescence resonance energy transfer. J. Immunol. 166, 6625–6632 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Lu X., et al. , Endogenous viral antigen processing generates peptide-specific MHC class I cell-surface clusters. Proc. Natl. Acad. Sci. U.S.A. 109, 15407–15412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blees A., et al. , Structure of the human MHC-I peptide-loading complex. Nature 551, 525–528 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F., Tetraspanin-enriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Cieri D., et al. , SPLICS: A split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ. 25, 1131–1145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boucheix C., Rubinstein E., Tetraspanins. Cell. Mol. Life Sci. 58, 1189–1205 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S., et al. , The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86, 674–684 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Dornier E., et al. , TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 199, 481–496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J., Fujiwara T., Ye S., Li X., Zhao H., Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits osteoclastogenesis in vitro. Calcif. Tissue Int. 95, 209–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyat J. S., et al. , ADAM10-interacting tetraspanins Tspan5 and Tspan17 regulate VE-cadherin expression and promote T lymphocyte transmigration. J. Immunol. 199, 666–676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Angelisová P., Hilgert I., Horejsí V., Association of four antigens of the tetraspans family (CD37, CD53, TAPA-1, and R2/C33) with MHC class II glycoproteins. Immunogenetics 39, 249–256 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Sheng K.-C., et al. , Tetraspanins CD37 and CD151 differentially regulate Ag presentation and T-cell co-stimulation by DC. Eur. J. Immunol. 39, 50–55 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Gartlan K. H., et al. , A complementary role for the tetraspanins CD37 and Tssc6 in cellular immunity. J. Immunol. 185, 3158–3166 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Blumenthal D., Edidin M., Gheber L. A., Trafficking of MHC molecules to the cell surface creates dynamic protein patches. J. Cell Sci. 129, 3342–3350 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Boniface J. J., et al. , Initiation of signal transduction through the T cell receptor requires the multivalent engagement of peptide/MHC ligands [corrected]. Immunity 9, 459–466 (1998). Erratum in Immunity 9, 891 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Deeg J., et al. , T cell activation is determined by the number of presented antigens. Nano Lett. 13, 5619–5626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minguet S., Swamy M., Alarcón B., Luescher I. F., Schamel W. W. A., Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 26, 43–54 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Cochran J. R., Cameron T. O., Stern L. J., The relationship of MHC-peptide binding and T cell activation probed using chemically defined MHC class II oligomers. Immunity 12, 241–250 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Cebecauer M., et al. , CD8+ cytotoxic T lymphocyte activation by soluble major histocompatibility complex-peptide dimers. J. Biol. Chem. 280, 23820–23828 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Anikeeva N., et al. , Quantum dot/peptide-MHC biosensors reveal strong CD8-dependent cooperation between self and viral antigens that augment the T cell response. Proc. Natl. Acad. Sci. U.S.A. 103, 16846–16851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anikeeva N., Fischer N. O., Blanchette C. D., Sykulev Y., Extent of MHC clustering regulates selectivity and effectiveness of T cell responses. J. Immunol. 202, 591–597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benard E., Nunès J. A., Limozin L., Sengupta K., T cells on engineered substrates: The impact of TCR clustering is enhanced by LFA-1 engagement. Front. Immunol. 9, 2085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossy J., Owen D. M., Williamson D. J., Yang Z., Gaus K., Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat. Immunol. 14, 82–89 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Hartman N. C., Nye J. A., Groves J. T., Cluster size regulates protein sorting in the immunological synapse. Proc. Natl. Acad. Sci. U.S.A. 106, 12729–12734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng Y., et al. , Mechanosensing drives acuity of αβ T-cell recognition. Proc. Natl. Acad. Sci. U.S.A. 114, E8204–E8213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu C., Chen W., Lou J., Rittase W., Li K., Mechanosensing through immunoreceptors. Nat. Immunol. 20, 1269–1278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen Z., Reznikoff G., Dranoff G., Rock K. L., Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 158, 2723–2730 (1997). [PubMed] [Google Scholar]
- 70.Rock K. L., Rothstein L., Gamble S., Generation of class I MHC-restricted T-T hybridomas. J. Immunol. 145, 804–811 (1990). [PubMed] [Google Scholar]
- 71.Rock K. L., Rothstein L., Gamble S., Fleischacker C., Characterization of antigen-presenting cells that present exogenous antigens in association with class I MHC molecules. J. Immunol. 150, 438–446 (1993). [PubMed] [Google Scholar]
- 72.Kincaid E. Z., et al. , Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 13, 129–135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Porgador A., Yewdell J. W., Deng Y., Bennink J. R., Germain R. N., Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity 6, 715–726 (1997). [DOI] [PubMed] [Google Scholar]
- 74.Saint-Pol J., et al. , New insights into the tetraspanin Tspan5 using novel monoclonal antibodies. J. Biol. Chem. 292, 9551–9566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erard N., Knott S. R. V., Hannon G. J., A CRISPR resource for individual, combinatorial, or multiplexed gene knockout. Mol. Cell 67, 348–354.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hogquist K. A., et al. , T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 78.Sanchez-Madrid F., Simon P., Thompson S., Springer T. A., Mapping of antigenic and functional epitopes on the alpha- and beta-subunits of two related mouse glycoproteins involved in cell interactions, LFA-1 and Mac-1. J. Exp. Med. 158, 586–602 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carpenter A. E., et al. , CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.