Significance
Unfolded protein response (UPR) is thought to coordinate cell growth and proliferation by governing endoplasmic reticulum (ER) proteostasis; however, the mechanisms underlying this fundamental adaptation process remain poorly defined. Here, by using Drosophila, mammalian models, and a human database, we uncover a dual role for Ire1/Xbp1s in development and tumorigenesis. A central element for this regulatory mechanism appears to be the association between the UPR downstream target Bip/HSPA5 and Hippo pathway transducer Yorkie (Yki)/YAP. Bip sequesters Yki in the cytoplasm to restrict its nuclear localization to impede cell proliferation, meanwhile also increasing invasive cell migration. Our results reveal an unexpected, context-dependent crosstalk between UPR and Hippo pathway in regulating tissue homeostasis and tumor progression.
Keywords: unfolded protein response, Hippo signaling, size control, tumorigenesis, triple-negative breast cancer
Abstract
Unfolded protein response (UPR) is the mechanism by which cells control endoplasmic reticulum (ER) protein homeostasis. ER proteostasis is essential to adapt to cell proliferation and regeneration in development and tumorigenesis, but mechanisms linking UPR, growth control, and cancer progression remain unclear. Here, we report that the Ire1/Xbp1s pathway has surprisingly oncogenic and tumor-suppressive roles in a context-dependent manner. Activation of Ire1/Xbp1s up-regulates their downstream target Bip, which sequesters Yorkie (Yki), a Hippo pathway transducer, in the cytoplasm to restrict Yki transcriptional output. This regulation provides an endogenous defensive mechanism in organ size control, intestinal homeostasis, and regeneration. Unexpectedly, Xbp1 ablation promotes tumor overgrowth but suppresses invasiveness in a Drosophila cancer model. Mechanistically, hyperactivated Ire1/Xbp1s signaling in turn induces JNK-dependent developmental and oncogenic cell migration and epithelial-mesenchymal transition (EMT) via repression of Yki. In humans, a negative correlation between XBP1 and YAP (Yki ortholog) target gene expression specifically exists in triple-negative breast cancers (TNBCs), and those with high XBP1 or HSPA5 (Bip ortholog) expression have better clinical outcomes. In human TNBC cell lines and xenograft models, ectopic XBP1s or HSPA5 expression alleviates tumor growth but aggravates cell migration and invasion. These findings uncover a conserved crosstalk between the Ire1/Xbp1s and Hippo signaling pathways under physiological settings, as well as a crucial role of Bip-Yki interaction in tumorigenesis that is shared from Drosophila to humans.
Cell growth and proliferation during normal organ development and tissue regeneration depend on the synthesis of new proteins (1). This process requires high levels of protein synthesis, folding, modification, and quality control, which are events orchestrated by the endoplasmic reticulum (ER) (2). It is therefore not surprising that ER homeostasis might regulate cell growth and proliferation; however, the direct mechanisms underlying this fundamental adaptation remain poorly defined. Excessive protein synthesis in the ER beyond the protein folding capacity of this organelle can evoke a cellular state of ER stress and subsequently activate the unfolded protein response (UPR). When the capacity of the UPR to sustain proteostasis is overwhelmed, cells sometimes initiate apoptosis (2). Much of the research to date on UPR has focused on its roles in cellular adaptation to stress or death. Whether or how UPR signaling contributes to cell growth and proliferation in organ size control and regeneration remains to be determined. In the context of cancer cells, insults from physiological stress and microenvironmental stress also lead to UPR activation. In general, UPR is thought to nourish tumor growth and foster malignant transformation by hijacking UPR to provide survival signals and eventually avoid cell death (3, 4). However, certain UPR pathways remain functionally antitumor, particularly those that block malignant cell proliferation or excessive regenerative response (5, 6), implying that in certain circumstances they might serve as endogenous defense mechanisms to oppose tumorigenesis.
The UPR comprises three conserved signaling branches in mammals and Drosophila: Ire1/Xbp1s, Perk/Atf4 (PEK/crc in fly), and Atf6 (7). These signaling cascades are maintained at quiescent, basal levels by Hsc70-3/Bip, a master ER-resident chaperone. The most phylogenetically conserved UPR arm is mediated by the endonuclease Ire1, which processes Xbp1 messenger RNA (mRNA) in metazoan cells to generate a spliced form of Xbp1 protein (Xbp1s). Xbp1s acts as a highly active transcription factor to induce the expression of ER chaperones (e.g., Bip) and other target genes in a cell type–specific manner (2). Most recent studies on the Ire1/Xbp1s pathway in cancer have defined its tumor-promoting roles, including maintaining tumor cell proteostasis, stimulating cell proliferation, and supporting angiogenesis (3, 4). Critically, all these studies used transplantable tumor models, which thus leaves unaddressed the capacity of Ire1/Xbp1s to fight primary autochthonous tumorigenesis. On the other hand, the intensity and duration of UPR branches might affect tumor cell fate. Harnessing the power of these UPR pathways represents a significant therapeutic challenge, and therefore, it is an urgent need to dissect how UPR alleviates cellular stress during tumorigenesis.
The Hippo pathway prominently affects organ size and regeneration by regulating the downstream transcriptional cofactor Yorkie (Yki) to coordinate cell growth, proliferation, and differentiation in developing adult tissues (8, 9). Long-term YAP (mammalian Yki ortholog) activation has been related to cancer initiation and progression in multiple cancer types (10, 11). However, YAP has been suggested as a tumor suppressor gene, because it is located in a chromosome region with frequent loss of heterozygosity (LOH) (12, 13). More recently, it has been found that the loss of YAP in triple-negative breast cancer (TNBC) cells leads to decreased primary tumor size but causes significantly more lung metastasis (14). To elucidate the Yki/YAP contradictory roles in vivo, our laboratory previously used Drosophila wing epithelium and different human cancer cell lines to demonstrate that Yki/YAP inactivation induces JNK-dependent cell invasion and epithelial-mesenchymal transition (EMT) (15). However, the precise role of YAP in breast cancer metastasis and whether YAP can crosstalk with other critical prometastasis factors remains unclear.
In this study, we reveal a dual role of the Ire1-Xbp1s-Bip signaling pathway in controlling cell proliferation and organ size and promoting developmental and oncogenic cell migration via crosstalk with Hippo signaling. Our data indicate that the UPR branch not only has important physiological significance but also has oncogenic and tumor-suppressive roles. We also demonstrate that Bip/HSPA5 are critical factors that physically interact with Yki/YAP to induce different outcomes at the crossroads of development and tumorigenesis in Drosophila and humans.
Results
UPR Signaling Is Activated in Drosophila Malignant Tumors.
Despite accumulating evidence of sustained ER stress in many cancer types, how tumors regulate UPR branches and whether UPR signaling alleviates or aggravates tumor progression in vivo remain controversial (4). For the last decades, fly geneticists worldwide, including us, have generated multiple in vivo tumors in various epithelial tissues, and these studies have collectively demonstrated that Drosophila tumor models can faithfully recapitulate numerous hallmarks of human cancer (16, 17). In Drosophila eye-antennal imaginal discs, clonal expression of oncogenic Ras (RasV12) alone induces hyperproliferation and forms benign tumors (18), whereas simultaneous inactivation of tumor-suppressor genes (e.g., scrib, l(2)gl, baz, fmt) results in more aggressive malignant tumors (16, 18) (Fig. 1A). To examine if genetic signatures exist to reflect a tumor’s ability to activate ER stress, we examined the expression levels of the chaperone Bip, a hallmark of cellular adaptation to ER stress (4, 19), using genetically engineered Drosophila tumor models. Interestingly, we noticed that Bip levels are more elevated in malignant tumors than in benign tumors, suggesting that malignant tumors experience stronger ER stress in situ (Fig. 1A). To identify which UPR branches were mediating this adaptive response to ER stress, we used two well-established in vivo reporters to examine the activation status of the Ire1 and Perk branches. Using an Xbp1 splicing reporter (7), we observed strong Ire1 activation in the RasV12; lgl−/− cells compared with control tissues (Fig. 1B). Similarly, malignant tumors also dramatically up-regulated Xbp1 mRNA expression by using an Xbp1 transcription reporter (20) (Fig. 1C). On the contrary, Perk signaling was weakly activated through a translational activation reporter of ATF4 (7) (Fig. 1D). These results indicate that UPR signaling, especially the Ire1/Xbp1s pathway, is highly activated in Drosophila malignant tumors.
Fig. 1.
Ectopic expression of Ire1 or Xbp1s blocks tumor growth by inhibiting Yki activation. (A) Eye discs bearing green fluorescent protein (GFP)-labeled clones with indicated genotypes were immunostained with anti-Bip antibody. Compared with controls or benign tumors, Bip levels are strongly up-regulated in malignant tumors. (B–D) Fluorescent micrographs of cephalic complex bearing GFP-labeled clones in eye disc dissected from third-instar larvae are shown. Compared with controls, RasV12; lgl−/− tumors increase Xbp1 splicing (B) and Xbp1 mRNA expression (C), whereas they only slightly induce translational activation of ATF4 (D). (E–G) Fluorescent micrographs of GFP-labeled pupa/larva are shown with indicated genotypes. Compared with controls and RasV12-induced benign overgrowth, larvae bearing RasV12; lgl−/−, RasV12; scrib−/−, or RafGOF; scrib−/− tumors induce massive overgrowth, which was dramatically suppressed by coexpression of Ire1, Xbp1s, or PEK. Conversely, the tumor overgrowth phenotype was inhibited by neither overexpression of ATF4 or Atf6 nor depletion of Ire1, Xbp1, PEK, ATF4, or Atf6. (H) Quantification of pupation-related penetrance observed with indicated genotypes. (I, J) Eye discs bearing GFP-labeled clones with indicated genotypes were immunostained with anti-Diap1 or anti-dMyc antibody. Ectopic expression of Ire1 or Xbp1s inhibits malignant tumor–induced Yki target genes expression, as indicated by Diap1 (I) and dMyc (J) immunostaining. (K) Quantification of relative intensity of Diap1 and dMyc with indicated genotypes. Data are mean ± SD. ****P < 0.0001 by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test (K). Scale bars: 50 μm (A, I, J), 100 μm (B–G). DAPI, 4′,6-diamidino-2-phenylindole; EGFP, enhanced GFP.
Activation of Ire1/Xbp1s Suppresses Epithelial Tumorigenesis via Repression of Yki.
Do different UPR branches modulate tumorigenesis independently? To this end, we genetically manipulated the UPR core components to test its exact roles in epithelial tumorigenesis. As anticipated, RasV12; lgl−/− and RasV12; scrib−/− animals displayed large neoplastic tumors and died as oversized larvae (Fig. 1 E, F, and H). We found that silencing of all three UPR branches or overexpression of Atf6 did not significantly ameliorate tumor progression (Fig. 1 E, F, and H). The efficacies of these UPR-related strains had been verified (SI Appendix, Reagent or Resource), and we further validated the UAS-Atf6 strain by qRT-PCR (SI Appendix, Fig. S1A). Interestingly, although PEK overexpression significantly inhibited tumor overgrowth, activation of its downstream target ATF4 did not replicate the growth-inhibiting phenotype (Fig. 1 E and H). This might be explained by ATF4-independent functions of hyperactivated PEK that cause cell-cycle arrest in the integrated stress response (21). Strikingly, coexpression of Ire1 or Xbp1s dramatically restrained the tumor overgrowth phenotype (Fig. 1 E, F, and H). Furthermore, ectopic Ire1 or Xbp1s expression also drastically inhibited the tumor overgrowth phenotype in other tumor models (Fig. 1 G and H). The above observation of the growth inhibition of Ire1/Xbp1s-activated clones was unexpected, because Ire1/Xbp1s are generally known as protumorigenic signaling.
Next, we examined the potential downstream factor(s) that mediate the antitumorigenic roles of Ire1/Xbp1s. Because hyperactivated Ire1/Xbp1s signaling triggers apoptosis (19), we coexpressed the apoptosis inhibitor p35 and confirmed that this growth-inhibiting effect was independent of cell death (Fig. 1 F and H). Next, we asked whether the tumor-suppressive role of Ire1/Xbp1s involves the Hippo, JNK, or Jak-Stat signaling pathway, each of which is a known intrinsic cue that drives malignant transformation (17). Ectopic expression of Ire1/Xbp1s significantly inhibited Yki target genes, including Diap1 and dMyc, but not the JNK target gene Mmp1 or Stat signaling (Fig. 1 I–K and SI Appendix, Fig. S1 B–E). Collectively, these results suggest that the Ire1/Xbp1s pathway blocks tumor growth by inactivating Yki.
Ire1/Xbp1s Pathway Functions Through Yki to Regulate Growth and Restrict Organ Size.
The Hippo pathway is crucial in controlling organ size during normal development (22). To further dissect the in vivo mechanisms by which the Ire1/Xbp1 branch regulates Hippo signaling, we genetically modulated Ire1 and Xbp1s expression (overexpression or down-regulation) in various tissues, including developing eyes and wings, the sizes of which are sensitive to endogenous Yki activity, thus offering a convenient model (9). Ectopic expression of Ire1 or Xbp1s resulted in smaller adult eye sizes when driven by the GMR-Gal4 driver (Fig. 2A). Conversely, depletion of Ire1 or knockdown of Xbp1 along with p35 expression modestly increased eye size (Fig. 2A). A similar phenotype was observed using a wing-specific promoter, nub-Gal4 (SI Appendix, Fig. S2A). To avoid off-target effects, we simultaneously depleted Xbp1 or Ire1 using independent transgenic RNA inhibitor (RNAi) lines and kibra or Mer (upstream component of Hippo signaling) (23, 24) and observed a significant synergistic increase in adult eye size (Fig. 2B). Together, these data imply that Ire1/Xbp1s signaling might generally regulate organ size and growth under physiological conditions.
Fig. 2.
Ire1/Xbp1s pathway acts in parallel with Yki to positively regulate Hippo signaling. (A, B) Light micrographs of Drosophila adult eye side view and quantification of the eye size with indicated genotypes. (C) Fluorescent micrographs of eye discs bearing green fluorescent protein (GFP)-labeled clones with indicated genotypes immunostained with anti–β-galactosidase (β-Gal) antibody to label diap1 transcription. Overexpression of Ire1 or Xbp1s significantly inhibits endogenous Diap1 transcription, whereas codepletion of Xbp1 rescues Ire1-induced Diap1-lacZ down-regulation. (D) Fluorescent micrographs of wing discs immunostained with anti-Diap1, anti-dMyc, and anti–β-Gal antibody to label fj transcription. Cells are labeled by GFP expression along the anterior/posterior (A/P) boundary (dashed lines) under control of the dpp driver. Quantification of relative intensity of Diap1, dMyc, and fj-lacZ with indicated genotypes. (E) Fluorescent micrographs of wing discs expressing hh-Gal4 with posterior cells labeled by expression of GFP were immunostained with anti-Diap1 antibody and anti–β-Gal antibody to label ex and ban transcription. Dashed lines mark the A/P boundaries. Quantifications of relative immunostaining intensity are shown on the right. (F) Fluorescent micrographs of eye discs containing GFP-marked MARCM clones of control, Yki overexpression, sav mutant, and wts mutant with or without Ire1 or Xbp1s overexpression. Quantification of the GFP-positive clone size with indicated genotypes. (G) Light micrographs of Drosophila adult head dorsal view and quantification of eye width with indicated genotypes. Data are mean ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P < 0.001, ****P < 0.0001 by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test (A–G). Scale bars: 20 μm (C), 50 μm (D, E), 100 μm (F), 200 μm (A, B, G). DAPI, 4′,6-diamidino-2-phenylindole; ns, not significant.
To test whether Ire1/Xbp1s-mediated growth control depends on Yki activation, we examined the expression of Yki target genes. Ire1 or Xbp1s overexpression significantly decreased endogenous expression levels of Diap1-lacZ, Diap1, dMyc, and fj-lacZ in both the eye disc and wing disc, either via the mosaic analysis with a repressible cell marker (MARCM) system or driven by dpp-Gal4, respectively (Fig. 2 C and D). Moreover, we found that knockdown of Xbp1 reversed the suppression effect of Ire1 on the expression of Diap1-lacZ, Diap1, dMyc, and fj-lacZ, suggesting that Ire1 suppresses Yki activity in an Xbp1-dependent manner (Fig. 2 C and D). We next investigated the physiological role of Ire1 and Xbp1 in regulating Yki target genes expression. In Ire1- or Xbp1-depleted cells, the expression of Diap1, ex-lacZ, and ban-lacZ was significantly increased, suggesting that Ire1/Xbp1 signaling negatively regulates Yki target genes (Fig. 2E). Furthermore, we found that the up-regulation of Diap1, ex-lacZ, and ban-lacZ caused by Xbp1 knockdown could be reversed by depletion of yki or removal of one copy of endogenous yki; however, yki heterozygosity alone had no significant effect, suggesting that yki is indispensable for Xbp1 knockdown–induced up-regulation of Yki targets genes (Fig. 2E). Overall, these results suggest that Ire1/Xbp1s signaling acts through Yki to negatively regulate expression of Yki target genes.
To further explore the mechanisms by which Ire1/Xbp1s regulate Hippo signaling, additional genetic experiments were performed. Loss-of-function mutations in the upstream components (sav, hpo, wts) of the Hippo pathway and gain of function of Yki result in similar tissue overgrowth phenotypes (9, 25). We used the MARCM system to ectopically express Ire1 or Xbp1s in sav-deleted, wts-deleted, or Yki-overexpressed clones. Overexpression of Ire1 or Xbp1s alone significantly inhibited their own clone growth at 29 °C, but it did not at 25 °C (Fig. 2F and SI Appendix, Fig. S2B). The clonal overgrowth phenotype in the developing eye caused by sav loss, wts loss, or Yki overexpression was dramatically suppressed by overexpression of Ire1 or Xbp1s at 25 °C (Fig. 2F). Consistently, Ire1 or Xbp1s overexpression also significantly suppressed the adult eye overgrowth and Diap1 up-regulation resulting from depletion of wts or Yki activation (Fig. 2G and SI Appendix, Fig. S2D), indicating that Ire1/Xbp1s signaling acts in parallel with or downstream of Yki. Consistently, we noticed that Xbp1 depletion partially reversed the eye size reduction phenotype caused by overexpression of Hpo or Wts (SI Appendix, Fig. S2C).
Taken together, these above genetic data suggest that Ire1/Xbp1s signaling may act in parallel with Yki to regulate the Hippo pathway.
Bip Restricts Yki Nuclear Localization to Control Yki Transcriptional Output and Organ Size.
As Bip is a bona fide target of Xbp1s, several studies have reported that it can mediate UPR-Xbp1s–driven cardioprotection (26, 27). Next, we examined whether Ire1/Xbp1s repress Yki activity via Bip. We found that Bip overexpression significantly reduced the expression of ex-lacZ, dMyc, and Diap1 (Fig. 3 A and B and SI Appendix, Fig. S3 A and B). Conversely, depletion of Bip was sufficient to increase the expression of ex-lacZ, which could be reversed by yki knockdown or yki heterozygosity (Fig. 3 A and B). These data suggest that genetically Bip also acts upstream of Yki. Furthermore, we found that depletion of Bip reversed the inhibitory effect on Diap1 expression caused by Xbp1s overexpression (Fig. 3C), and conversely, ectopic expression of Bip suppressed Xbp1 deletion–induced ex-lacZ and Diap1 up-regulation (Fig. 3A and SI Appendix, Fig. S3B), suggesting that Bip acts downstream of Ire1/Xbp1s signaling to regulate the Hippo pathway. Moreover, we found that ectopic Bip expression not only decreased eye and wing size (Fig. 3 D and E) but also suppressed tumor overgrowth in a dose-dependent manner under pathological conditions (Fig. 3 F–I). Finally, our genetic experiments showed that Bip overexpression significantly suppressed the eye overgrowth and Diap1 up-regulation resulting from wts depletion or Yki activation (Fig. 3J and SI Appendix, Fig. S3E), suggesting that Bip acts in parallel with or downstream of Yki. Taking the above genetic clues together, these data collectively indicate that Bip may act in parallel with Yki to regulate Yki transcriptional output and organ size.
Fig. 3.
Bip sequesters Yki in the cytoplasm to control Yki transcriptional outputs and organ size. (A) Fluorescent micrographs of wing discs expressing hh-Gal4 with posterior cells labeled by expression of green fluorescent protein (GFP) were immunostained with anti–β-Gal antibody to label ex transcription. Dashed lines mark the anterior/posterior (A/P) boundaries. (B) Quantifications of relative immunostaining intensity with indicated genotypes. (C) Fluorescent micrographs of wing discs immunostained with anti-Diap1 antibody. Cells are labeled by GFP expression along the A/P boundary under control of the ptc driver. Quantification of relative intensity of Diap1 is shown on the right. (D) Light micrographs of Drosophila adult eye side view (left) and quantification of the size (right) with indicated genotypes. (E) Light micrographs of Drosophila adult wings (left) and quantification of indicated genotypes using nub-Gal4. (F) Left, fluorescent micrographs of GFP-labeled pupa/larva are shown with indicated genotypes. Right, quantification of phenotype penetrance with the indicated genotypes. (G, H) Eye discs bearing GFP-labeled clones with indicated genotypes were immunostained with anti-Diap1 (G) or anti-dMyc (H) antibody. (I) Quantification of relative intensity of Diap1 and dMyc with indicated genotypes. (J) Light micrographs of Drosophila adult eye side view (left) and quantification of the size (right) of the indicated genotypes. (K) Fluorescent micrographs of wing pouch region immunostained with anti-Yki antibody. Cells are labeled by GFP expression along the A/P boundary under the control of ptc driver. Quantification of relative ratio of nuclear Yki intensity to total intensity with indicated genotypes shown on the right. (L–M) PLA was performed on wing discs with indicated genotypes to test for close-proximity interactions between FLAG-Yki and HA-Bip (L) or Myc-Yki and HA-Bip (M). (N) Quantification of PLA signal intensity in L and M. (O) Physical association test between FLAG-Yki and HA-Bip in adult Drosophila eyes using GMR-Gal4S. Lysates expressing the indicated constructs underwent IP and were probed with the indicated antibodies. Data are mean ± SD. **P ≤ 0.01, ****P < 0.0001 by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test (B, C, I, J, K) and two-tailed Student’s t test (D, E). Scale bars: 10 μm (K), 50 μm (A, C, G, H, L, M), 100 μm (F), and 200 μm (D, E, J). DAPI, 4′,6-diamidino-2-phenylindole; IB, immunoblotting; IgG, immunoglobulin G; ns, not significant.
Upon Hippo signaling inactivation, Yki translocates into the nucleus and forms a complex with transcription factor Scalloped (Sd) to initiate growth-related target gene transcription (8). Xbp1s or Bip overexpression significantly suppressed the nuclear accumulation of Yki resulting from wts knockdown (Fig. 3K), consistent with the observation that they were able to inhibit Yki transcriptional output and organ growth. Bip acts as a molecular chaperone to interact with multiple partners in the cytoplasm or ER to affect different signaling pathways (28). Notably, a previous study suggested Bip as a potential interacting protein of Yki (29). Therefore, we hypothesized that Bip might bind to Yki to restrict its nuclear import. Indeed, we detected a strong colocalization of FLAG-tagged Yki and HA-tagged Bip driven by ptc-Gal4 in vivo (SI Appendix, Fig. S3F). To further validate whether Bip interacts with Yki, we performed in vivo Duolink in situ proximity ligation assay (PLA), which is widely used to monitor protein-protein interactions at subcellular levels (30). We observed strong positive PLA signals between FLAG-tagged Yki or Myc-tagged Yki and HA-tagged Bip, compared with the negative controls (Fig. 3 L–N). Next, we ectopically expressed FLAG-tagged Yki and HA-tagged Bip in the developing eye and observed robust physical interaction between Bip and Yki in both directions by coimmunoprecipitation (co-IP) assays (Fig. 3O and SI Appendix, Fig. S3G). Unfortunately, because anti-Bip antibodies are not available, we were not able to examine the physiological interaction between endogenous Bip and Yki. Together, these results suggest that Bip can physically interact with Yki in vivo.
While Bip predominantly resides in the ER, there is a cytoplasmic variant that lacks the ER localization signal but retains its cytoplasmic chaperone function (31). Next, we asked whether Bip interacts with Yki to induce translocation of Yki to the ER. To this end, we performed two-color superresolution imaging to visualize the localization of nanoparticles inside cells and probe their interactions with cellular machineries at the nanoscale (32). We coexpressed FLAG-HSPA5 (mammalian Bip ortholog), HA-YAP (mammalian Yki ortholog), and mCherry-Sec61β (an ER membrane protein) in human U2OS osteosarcoma cells and found that although HSPA5 overexpression sequestrated YAP in the cytoplasm, it did not induce YAP localization to the ER (SI Appendix, Fig. S3H).
Incoherent Regulation Between Ire1/Xbp1s Signaling and Yki.
Yki can directly regulate transcription of various Hippo pathway upstream components (e.g., Mer, Ex), forming a negative feedback loop to fine tune the pathway (8). This endogenous defensive mechanism can balance Hippo signaling to maintain tissue homeostasis. This kind of regulation, wherein a transcriptional activator can be suppressed by its own targets, is called incoherent regulation (33). Indeed, Wts overexpression and yki or sd deletion completely suppressed tumor-induced Bip up-regulation, whereas blocking JNK by expression of a dominant negative form of fos (fosDN) had no effect (SI Appendix, Fig. S4 A and B), suggesting that Bip might be a downstream target of Hippo signaling. Consistently, we found that loss of wts or overexpression of Yki is sufficient to up-regulate Bip expression in an sd-dependent manner (SI Appendix, Fig. S4C). Moreover, overexpression of an activated form of Yki (YkiS168A) significantly up-regulates the transcription of Ire1, Xbp1, and Bip in an sd-dependent manner (SI Appendix, Fig. S4D). These data suggest that Yki positively regulates Ire1/Xbp1s signaling, which in turn negatively controls Yki activity to sustain stable growth input and protein homeostasis.
Ire1/Xbp1s Pathway Modulates Intestinal Homeostasis and Regeneration by Regulating Hippo Signaling.
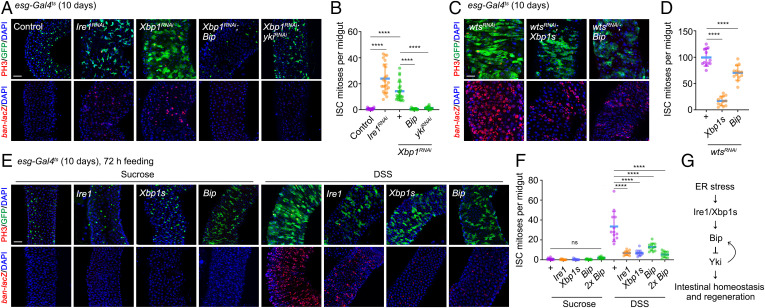
The homeostasis of self-renewal and differentiation in intestinal stem cells (ISCs) is controlled by intrinsic signals and their niche (34). The protein synthesis burden and underlying ER stress of ISCs are challenging for ISCs during this process (35). However, how UPR affects ISC proliferation under physiological and pathological conditions remains less explored. Given that Hippo signaling also plays critical roles in regulating ISC division and renewal (36), we hypothesized that the Ire1/Xbp1s pathway might modulate Yki-mediated ISC proliferation. In accordance with our hypothesis, depletion of Ire1 or Xbp1 in midgut progenitors using esg-Gal4 showed a modest up-regulation of ISC mitoses and ban-lacZ, which could be reversed by Bip overexpression or Yki depletion (Fig. 4 A and B). Additionally, ectopic expression of Xbp1s or Bip significantly suppressed the elevated ISC proliferation and ban-lacZ up-regulation induced by depletion of wts (Fig. 4 C and D). These results are consistent with those we obtained in imaginal discs and further support a model in which the Ire1-Xbp1s-Bip signaling pathway also restricts ISC proliferation by inhibiting Yki activity to maintain gut homeostasis.
Fig. 4.
Ire1-Xbp1s-Bip signaling is required for intestinal homeostasis and regeneration. (A, C, E) Fluorescent micrographs of adult midguts expressing esg-Gal4ts driver to activate/down-regulate gene expression specifically in green fluorescent protein (GFP)-labeled progenitor cells. Two- to 3-d-old adult females were shifted from 18 °C to 29 °C for 10 d before dissection. Midguts were immunostained with anti-PH3 antibody or anti–β-Gal antibody to label ban transcription. (E) Flies with indicated genotypes were shifted from 18 °C to 29 °C for 10 d and then were fed with sucrose or DSS for 72 h at 29 °C before dissection. Quantification of PH3-positive cells in adult midguts with indicated genotypes. (G) Schematic representation of Ire1/Xbp1s-Bip axis in regulating intestinal homeostasis and regeneration. Data are mean ± SD. ****P < 0.0001 by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test (D, F) or Mann-Whitney test (B). Scale bar: 50 μm (A, C, E). DAPI, 4′,6-diamidino-2-phenylindole; ns, not significant.
Tissue injury induced by dextran sodium sulfate (DSS) stimulates ISC proliferation through a cell-autonomous role of Yki in precursor cells (37). Next, we asked whether the Ire1-Xbp1-Bip pathway is activated and serves as a rheostat sufficient to antagonize tissue damage–induced, Yki-dependent ISC renewal. Interestingly, DSS treatment resulted in faster regenerative growth and up-regulation of Bip levels in the precursor cells, which was suppressed by knockdown of Yki or sd (SI Appendix, Fig. S5A). Consistently, Bip levels were also significantly increased by expressing YkiS168A in precursor cells and could be completely reverted by sd depletion (SI Appendix, Fig. S5B). These results suggest that injury-induced Bip up-regulation is Yki dependent, and this incoherent regulation also exists in the intestine. Furthermore, we found that activation of the Ire1-Xbp1s-Bip pathway significantly blocked DSS-induced ISC proliferation and decreased the levels of ban-lacZ (Fig. 4 E and F), indicating that the crosstalk between the Ire1/Xbp1s pathway and Hippo signaling is essential for intestinal homeostasis and normal regeneration (Fig. 4G). Collectively, our data suggest that Ire1/Xbp1s/Bip signaling is a widespread in vivo modulator of the Hippo pathway in multiple tissue contexts.
Ire1/Xbp1s Pathway Is Required for Loss of Cell Polarity–Induced Tumor Invasion and EMT.
Given that increased Xbp1s expression inhibits tumor overgrowth, we further deleted Xbp1 in RasV12; dlgRNAi tumors, which induce overgrowth with cell invasion into the ventral nerve cord. As expected, complete removal of Xbp1 increased tumor volume (Fig. 5A). Surprisingly, we observed a significant reduction in invasive cell migration behavior upon Xbp1 deletion (Fig. 5A). This observation of invasion promotion by Xbp1 is surprising, indicating that invasiveness is not just an indirect consequence of tumor overgrowth. To further determine the effect of Ire1/Xbp1s on cell invasion, we used a well-established cell invasion model in Drosophila wing discs (15, 38). Knockdown of scrib or dlg driven by ptc-Gal4 induced strong invasive cell migration, Mmp1 up-regulation, and Dlg and E-cadherin (EMT markers) down-regulation, all of which could be strongly reversed by codepletion of Ire1 or Xbp1 (Fig. 5B and SI Appendix, Fig. S6). These results suggest that loss of cell polarity requires Ire1/Xbp1s signaling to trigger cell invasion and EMT, which contribute to cancer invasiveness.
Fig. 5.
Ire1/Xbp1s pathway induces JNK-dependent invasive cell migration and EMT by inhibiting Yki. (A) Left, fluorescent micrographs of green fluorescent protein (GFP)-labeled tumors and ventral nerve cords (VNCs) are shown. Bottom panels, fluorescent images of dissected VNCs representing four different grades of tumor invasiveness ranging from noninvasive (score 0) to strong tumor invasion (score 4). Middle and right, quantification of tumor volume and invasion degree of RasV12; dlgRNAi or RasV12; dlgRNAi; Xbp1−/− animals. (B) Left, cells from wing discs are labeled by GFP expression along the anterior/posterior (A/P) boundary under control of the ptc driver to monitor cell migration. Right, quantification of migrating cell number with indicated genotypes. (C) Fluorescent micrographs of wing discs with indicated genotypes immunostained with anti-Mmp1, anti-dMyc, and anti–E-cadherin antibodies. Cells are labeled by GFP expression along the A/P boundary to indicate the invasion phenotype. (D) Quantification data of migrating cell number with indicated genotypes. (E) Light micrographs of adult thoraxes are shown. Compared with controls, depletion of Xbp1 in the dorsal midline driven by pnr-Gal4–induced thorax cleft phenotype was partially restored by heterozygosity of puc or yki. (F) Schematic summary of Ire1/Xbp1 axis in regulating EMT and cell migration. Loss of cell polarity induces activation of Ire1/Xbp1s signaling, which in turn up-regulates Bip to sequester Yki in the cytoplasm. Repressed Yki activation would further activate JNK signaling–dependent developmental migration, invasive cell invasion, and EMT. Data are mean ± SD. **P ≤ 0.01, ****P < 0.0001 by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test (B, D) and two-tailed Student’s t test (A). Scale bars: 50 μm (B, C) and 200 μm (A). ns, not significant.
We then asked whether hyperactivation of Ire1-Xbp1s-Bip signaling phenocopies this invasion-promoting effect. As expected, overexpression of Ire1/Xbp1s signaling induces strong cell invasion behavior and EMT (Fig. 5 C and D). In accordance with previous studies, activation of Ire1/Xbp1s triggers apoptosis, as evident by the activation of caspase-3 and Dcp1 (SI Appendix, Fig. S7A. Given that cell invasion is frequently accompanied by apoptosis (15, 38), we further blocked apoptosis by coexpressing p35 and did not observe a significant suppressive effect on Ire1/Xbp1s-induced cell invasion or Mmp1 activation, suggesting the invasion behavior is not a secondary effect of cell death (SI Appendix, Fig. S7B). To rule out the possibility that Ire1/Xbp1s may have a nonautonomous effect on Mmp1 activation, we generated Flp-out clones to coexpress Xbp1s and p35. We observed that cells within the clone displayed filopodia-like structures and exhibited molecular characteristics of EMT, including F-actin accumulation and Mmp1 activation in a cell-autonomous manner SI Appendix, Fig. S7C). Overall, these results indicate that Ire1/Xbp1signlaing is both necessary and sufficient to trigger invasive cell migration and EMT.
Ire1/Xbp1s Pathway Is Involved in Developmental and Oncogenic Cell Migration and EMT.
Next, we dissected the signaling axis downstream of Ire1/Xbp1 that is responsible for cell invasion. Mmp1 is a direct transcriptional target of JNK signaling, activation of which has a pivotal role in tumor invasion (18). Our laboratory previously identified the essential mechanism that increased Hippo signaling activity promotes JNK-dependent cell invasion (15, 38). This led us to hypothesize that hyperactive Ire1/Xbp1s signaling in turn activates JNK-dependent cell migration and invasion via repression of Yki. Indeed, we observed that blocking JNK signaling by coexpressing bskDN completely counteracted hyperactivated Ire1- or Bip-induced cell invasion behavior and EMT (Fig. 5 C and D). Interestingly, although JNK inhibition completely blocked Xbp1s activation–induced invasive cell migration (Fig. 5D), it did not fully suppress EMT phenotype (Fig. 5C). We then asked whether ectopic Yki expression could suppress hyperactivated Ire1/Xbp1s-induced, JNK-dependent cell invasion and EMT. Overexpression of wild-type Yki causes massive overgrowth in Drosophila (9), potentially masking any intrinsic difference. With this consideration, we used tubulin α1 promoter–activated Yki (tub > yki) (9, 39), thus slightly increasing Yki expression. We found that the Ire1/Xbp1s signaling–induced invasion phenotype and Mmp1 up-regulation could be significantly reversed by expressing tub > yki, indicating Yki functions upstream of JNK signaling (Fig. 5 C and D). Thorax closure serves as another in vivo model to study cell migration in Drosophila development. JNK signaling is crucial for thorax closure, because reduced JNK activity results in a dorsal cleft phenotype in the thorax (40). Remarkably, knockdown of Xbp1 using pnr-Gal4 produced a mild thorax cleft that phenocopied JNK inactivation, which was suppressed in heterozygous puc or yki mutants (Fig. 5E and SI Appendix, Fig. S8A). Therefore, Xbp1s-mediated Yki suppression is also required for physiological JNK-mediated cell migration in thorax closure.
Next, we investigated the molecular mechanism of the Yki-JNK axis in Ire1/Xbp1s-induced cell invasion and EMT. We previously found that bantam (ban) and Myc, two essential downstream targets of Yki, could both inhibit JNK-dependent tumor invasion (15, 38). Myc can form a heterodimer with Myc-associated protein X (Max) to initiate gene transcription, and this complex can directly up-regulate the transcription of puc and thus inhibit JNK-mediated cell migration (38). Our genetic epistasis analysis showed that hyperactivated Ire1/Xbp1s signaling–induced invasion phenotype and Mmp1 activation were almost completely reverted by coexpressing ban or dMyc/dMax (SI Appendix, Fig. S8B). Snail (sna) is a transcriptional repressor controlling EMT during embryogenesis and tumor progression (41). In addition, sna regulates JNK-mediated cell death in Drosophila (42). Therefore, we further proposed that sna might act downstream of JNK, which is required for Ire1/Xbp1s activation–induced cell invasion and EMT. As a positive control, blocking JNK by coexpressing bskDN totally counteracted puc loss or wild-type hep activation–induced cell migration and Mmp1 up-regulation (SI Appendix, Fig. S9A). Intriguingly, sna silencing significantly blocked JNK-mediated cell migration and produced a mild thorax cleft that was further enhanced in heterozygous bsk mutants (SI Appendix, Fig. S9 A and B). Importantly, suppressing sna dramatically blocked Ire1, Xbp1s, or Bip hyperactivation–induced cell invasion and EMT (Fig. 5 C and D). Taken together, these data suggest that hyperactivated Ire1/Xbp1 activity suppresses Yki transcriptional output, which in turn activates JNK signaling to drive developmental and oncogenic cell migration and EMT (Fig. 4F).
XBP1s/HSPA5 Pathway Regulates Growth and Invasiveness of TNBC.
As the major components of the Hippo pathway are highly conserved between Drosophila and humans, we next explored whether Xbp1s/Bip-mediated restriction of Yki activation is also conserved during human tumor progression. Firstly, we analyzed the mRNA expression levels of XBP1 and HSPA5 in various human cancers and found that they are both frequently up-regulated in most human tumor tissues and cancer cell lines, especially in breast cancer (SI Appendix, Fig. S10 A–C). Surprisingly, although total XBP1 mRNA levels were strongly increased in breast cancer, XBP1 is specifically and significantly down-regulated in basal-like/TNBC compared with the other four subtypes (Fig. 6 A–C and SI Appendix, Fig. S10D). We observed that high expression of XBP1 mRNA was significantly associated with longer distant metastasis-free or overall survival in TNBC patients (Fig. 6D and SI Appendix, Fig. S10E). Similarly, patients with high mRNA and protein expression of HSPA5 also had better outcomes, although the transcription level–related significant difference existed only until 144 mo for overall survival (Fig. 6E and SI Appendix, Fig. S10E). Interestingly, XBP1 and HSPA5 expression did not correlate with clinical outcome in HER2-enriched or luminal B breast cancer patients, while luminal A breast cancer patients with high XBP1 or HSPA5 expression had shorter overall survival time (SI Appendix, Fig. S10 F–K).
Fig. 6.
XBP1s/HSPA5 pathway suppresses TNBC cell growth through repressing YAP activation. (A–G) Patient data were obtained from RNA sequencing (RNA-seq) and microarray data from bc-GenExMiner v4.7 databases. (A–C) Violin plots show XBP1 gene expression levels in tumor tissues with different breast cancer subtypes. Analysis of XBP1 expression by RNA-seq (basal-like n = 766, HER2-enriched [HER2-E] n = 332, luminal A n = 924, luminal B n = 783, normal breast-like n = 759) and microarray (basal-like n = 2,332, HER2-E n = 916, luminal A n = 2,480, luminal B n = 1,888, normal breast-like n = 1,548) according to Hu subtypes (A). XBP1 expression analysis by RNA-seq and microarray based on PAM50 classifier (B) and immunohistochemistry (IHC) (C). (D, E) Kaplan-Meier graphs demonstrating a significant association between decreased expression of XBP1 (D) or HSPA5 (E) (purple line) and shorter distant metastasis-free (n = 1,283) or overall survival (n = 999) in TNBC patients. P values were determined by log-rank (Mantel-Cox) test. (F, G) Pearson analysis gene-expression data from TNBC patients were displayed in a correlation map (F), depicting a negative correlation between XBP1 and YAP1 target genes. Each cell corresponds to a pairwise correlation and was colored according to the correlation coefficient value, from dark blue (coefficient = −1) to dark red (coefficient = 1). Correlation analysis from RNA-seq data are shown in (G). (H–I) Nucleocytoplasmic separation (H) and immunofluorescence (I) of YAP cellular localization after overexpression of empty-vector control (Lenti-NC), XBP1s (Lenti-XBP1s), or HSPA5 (Lenti-HSPA5) using lentivirus in MDA-MB-231 cells. Immunoblot analysis of Nuc and Cyt separation with the indicated antibodies. Quantification of YAP localization in the right panel (n = C, YAP is evenly distributed; n < C, YAP is enriched in the cytoplasm; n > C, YAP is enriched in the nucleus). (J, K) Immunofluorescence (J) and Duolink PLA (K) of FLAG-HSPA5, endogenous YAP, and nuclei. Traces of fluorescence intensity spatial profiles through the dashed lines shown in (J). Quantification of endogenous YAP and FLAG-tagged HSPA5 binding (red dots) is shown (K). (L) Co-IP experiments showed that ectopically expressed FLAG-HSPA5 interacts with endogenous YAP. (M) Coomassie bright blue staining of SDS-PAGE after in vitro pull down using purified SFB-tagged YAP1 and FLAG-tagged HSPA5 proteins. Arrow indicated the protein band. Asterisk represents the nonspecific protein during purification. (N) Colony formation of Lenti-NC, Lenti-XBP1s, or Lenti-HSPA5 group. Colony number was quantified. (O, P) Left, representative image of the xenograft tumors isolated from indicated groups. Right, quantifications of tumor growth curves and tumor weight. (L, M) Representative images of immunohistochemical staining of Ki-67 (L) or immunofluorescence of YAP localization (R) in paraffin-embedded xenograft tumor tissues collected from indicated groups. Quantifications of YAP localization in the right panel. (S) Schematic model depicting the coordinate regulation of UPR and Hippo signaling in development and tumorigenesis. Data are mean ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P < 0.001, ****P < 0.0001 by Dunnett-Tukey-Kramer’s test (A), Welch’s test (B, C), two-tailed Student’s t test (I, K, N, O, P), and Mann-Whitney test (J, K). Scale bar: 10 μm (I, J, K), 20 μm (R), and 50 μm (Q). a.u., arbitrary unit; DAPI, 4′,6-diamidino-2-phenylindole; IB, immunoblotting; IgG, immunoglobulin G; GADPH, glyceraldehyde-3-phosphate dehydrogenase; HR, hazard ratio.
YAP has been implicated as an oncogene in TNBC (43). Indeed, YAP1 expression was significantly higher in TNBC compared with other subtypes, and its elevated expression correlated dramatically with poor survival in TNBC patients (SI Appendix, Fig. S11 A–D). Increased expression of XBP1 was associated with reduced expression of YAP target genes (LMNB2, BIRC5, CDX2, MYC, ANKRD1, EDN1) in human TNBC samples, and this negative correlation specifically existed only in TNBC (Fig. 6 F and G and SI Appendix, Fig. S12 A and B). The above systematic analyses imply that XBP1-HSPA5 activation might have specific prognostic value and an underlying tumor-suppressive effect by inhibiting YAP activity in TNBC.
Next, we used MDA-MB-231 cells (a well-established TNBC cell line) to dissect the function of the XBP1s/HSPA5-YAP axis. Consistent with Bip-mediated cytoplasmic sequestration of Yki in Drosophila, overexpression of XBP1s or HSPA5 restricted the translocation of YAP from cytoplasm to the nucleus (Fig. 6 H and I and SI Appendix, Fig. S13A). We detected a strong colocalization and PLA signals between endogenous YAP and FLAG-tagged HSPA5 in the cytoplasm, whereas the control group gave no obvious signal (Fig. 6 J and K). As an independent test of physical association, we performed co-IP assays and observed that FLAG-tagged HSPA5 efficiently coimmunoprecipitated with endogenous YAP in both directions (Fig. 6L). Furthermore, we purified proteins to perform in vitro pull-down assays and confirmed that HSPA5 directly interacts with YAP (Fig. 6M).
Consistent with our findings above in Drosophila, we found that XBP1s or HSPA5 overexpression markedly inhibited the proliferative capacity of MDA-MB-231 cells, as demonstrated by colony formation assay (Fig. 6N). To validate the roles of XBP1s/HSPA5 in TNBC, we established a xenograft model of MDA-MB-231 cells in nude mice. XBP1s or HSPA5 overexpression significantly blocked tumor growth (Fig. 6 O and P) and reduced tumor cell proliferation (Fig. 6Q). More importantly, XBP1s or HSPA5 overexpression almost completely sequestered YAP in the cytoplasm compared with the control (Fig. 6R). Our data suggest that HSPA5-mediated cytoplasmic retention of YAP has an important role in regulating TNBC tumorigenesis. We further tested whether activation of XBP1s/HSPA5 also promotes cell invasion in TNBC. Consistently, XBP1s or HSPA5 overexpression significantly increased the cell migration and invasion ability of MDA-MB-231 cells (SI Appendix, Fig. S13 B and C). Collectively, the above data establish previously unrecognized crosstalk between the XBP1s/HSPA5 and Hippo signaling pathways, as well as a crucial role of HSPA5-YAP interaction in tumorigenesis that is shared from Drosophila to humans.
Discussion
Despite these recent advances, the paradoxical roles of UPR signaling in both tumor-suppressive and -promoting effects as well as the molecular mechanisms associated with each remain major challenges in the field. Here, by using the Drosophila imaginal epithelium and xenograft model, we report that ectopic activation of the Ire1/Xbp1s pathway potently blocks tumor progression; we also find that deletion of Xbp1 increases tumor growth but paradoxically inhibits tumor invasion. We show that Ire1/Xbp1s signaling acts through Bip to sequester Yki in the cytoplasm, which restricts its nuclear localization to regulate transcriptional output. On one hand, Bip-Yki interaction controls organ growth and regeneration and opposes tumorigenesis. On the other hand, this Yki-inhibiting effect in turn activates JNK-dependent cell migration, invasion, and EMT. Clinically, XBP1 expression negatively correlates with YAP activation in TNBC. We further show that ectopic expression of XBP1s or HSPA5 suppresses tumor growth but promotes cell migration and invasion of TNBC cells. Our studies elucidate crosstalk between UPR and the Hippo signaling pathway in growth control under physiological settings and implicate both oncogenic and tumor-suppressive roles of such interaction in a context-dependent manner (Fig. 6S).
Crosstalk of UPR and Hippo Signaling Opens Avenues for Modulating the Yki/YAP Oncoprotein.
Genetic experiments using Drosophila imaginal discs have yielded valuable information about the exact roles of certain genes in the genesis of malignant tumors (17, 44). To determine how specific the effects of Ire1/Xbp1s signaling on tumor growth are, we examined whether Ire1/Xbp1s regulate the activity of three signaling pathways: Hippo, JNK, and Jak-Stat. Examinations in tumor clones using reporter genes for these pathways suggest that ectopic expression of Ire1, Xbp1s, and Bip represses only Yki activation. These data reveal a previously unrecognized role of the Ire1-Xbp1s-Bip pathway in inhibiting Yki activity during tumorigenesis and demonstrate that this is a pathway-specific mechanism rather than a general transcriptional effect.
Bip is thought to function primarily in the ER lumen. Under certain circumstances, Bip is also redistributed to the nucleus, cytoplasm, or plasma membrane, where it can interact with multiple partners and thus trigger different signaling pathways (28, 45). Our data suggest that the Bip-mediated Yki cytoplasm distribution may provide another important defense line to prevent aberrant nuclear accumulation and activation of Yki/Yap. In other words, this failsafe mechanism is likely to explain how UPR signaling rapidly coordinates cell growth and proliferation in organ size control and regeneration.
Feedback Between Ire1/Xbp1s Signaling and Yki Promotes Homeostatic Growth and Regeneration.
Understanding how Hippo signaling activity is regulated in normal development has been a subject of longstanding interest. While increasing studies in both Drosophila and mammals highlight that multiple upstream input integrates to modulate Yki/YAP, it remains a challenge to define the exact physiological contexts in which this biological input is specifically applied to regulate Hippo signaling (8). Does this regulatory relationship between Ire1/Xbp1s and Yki have functional consequences under physiological settings? A key finding of our study is that rapid sequestration of Yki in the cytoplasm via Bip in the Drosophila imaginal disc upon ER stress indicates that Ire1/Xbp1s are a unique class of signals capable of restricting Yki activity.
Ire1/Xbp1s signaling serves as the most phylogenetically conserved UPR branch, and their high expression alleviates ER stress and provides a guarantee for stable protein synthesis to adapt to a high proliferate rate. When concomitant ER stress reaches some threshold, this failsafe mechanism is likely to restrict cell growth and guard against excessive regeneration under conditions where Yki levels are elevated, as seen in Yki-expressing mosaic clones and DSS-induced midgut damage. We propose that positive regulation of Ire1/Xbp1s signaling by Yki and negative feedback from Ire1/Xbp1s to Yki orchestrate powerful growth input and protein homeostasis to cells and thus generate homeostatic growth and regeneration in developing tissue. It is interesting to note that the activation of the PERK branch increases ATF4 expression for the subsequent induction of YAP in hepatocellular carcinoma (46). Whether different UPR signals converge on Yki/YAP to determine cell fate and whether their dysregulation contributes to tumorigenesis must be determined in a future study.
Mechanism That Links Cell Invasion to Ire1/Xbp1s Signaling and Hippo Signaling.
Our work may shed light on how Ire1/Xbp1s signaling and Hippo signals act jointly through Bip and Yki to trigger opposing effects in tumorigenesis. Our findings may also help us to understand the seemingly contradictory observations of YAP being a good prognosis marker in breast cancer, despite its roles in promoting tumor overgrowth (14). Although the data are compelling that Ire1/Xbp1s might serve as a tumor suppresser to restrict oncogenic Yki activity during the early phases of transformation, its activity may also be critical for cell invasiveness in late stages. Future studies are necessary to evaluate whether metastatic lesions have higher Ire1 or Xbp1s expression than primary tumors in TNBC patients.
Materials and Methods
Supplemental discussion and detailed methods and materials, including genotypes for each figure, fly husbandry and fly stocks, immunofluorescence, immunohistochemistry, Duolink in situ PLA, DSS treatment, RNA isolation and RT-qPCR, virus infection and exogenous gene expression, subcutaneous xenograft experiments, soft agar colony growth, immunoblotting and immunoprecipitation, in vitro pull-down assays, two-color superresolution imaging, wound healing assay, transwell invasion assay, statistical analysis, reagents, and resources, are listed in SI Appendix.
Supplementary Material
Acknowledgments
We thank Jingnan Liu, Tian Xu, Duojia Pan, Bruce Hay, Kenneth Irvine, Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, and Developmental Studies Hybridoma Bank for providing fly stocks and reagents. We thank Shian Wu and Shang Cai for insightful suggestions. We also thank Wenhan Liu for fly stock maintenance. This work was supported by startup funds from Westlake University and grants from the National Natural Science Foundation of China (32170824) to X.M., Westlake Laboratory of Life Sciences and Biomedicine (10128A092001), and “Team for Growth Control and Size Innovative Research” (201804016).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202133119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Lee A. S., Kranzusch P. J., Cate J. H., eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M., Kaufman R. J., Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Cubillos-Ruiz J. R., Bettigole S. E., Glimcher L. H., Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell 168, 692–706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., Cubillos-Ruiz J. R., Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 21, 71–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niederreiter L., et al. , ER stress transcription factor Xbp1 suppresses intestinal tumorigenesis and directs intestinal stem cells. J. Exp. Med. 210, 2041–2056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bujisic B., et al. , Impairment of both IRE1 expression and XBP1 activation is a hallmark of GCB DLBCL and contributes to tumor growth. Blood 129, 2420–2428 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Ryoo H. D., Drosophila as a model for unfolded protein response research. BMB Rep. 48, 445–453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu F. X., Zhao B., Guan K. L., Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J., Wu S., Barrera J., Matthews K., Pan D., The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Wu J., et al. , Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature 572, 402–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C. K., et al. , Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 363, 644–649 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Carter S. L., et al. , Loss of heterozygosity at 11q22-q23 in breast cancer. Cancer Res. 54, 6270–6274 (1994). [PubMed] [Google Scholar]
- 13.Moya I. M., et al. , Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science 366, 1029–1034 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Fan H., et al. , ASB13 inhibits breast cancer metastasis through promoting SNAI2 degradation and relieving its transcriptional repression of YAP. Genes Dev. 34, 1359–1372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X., et al. , Hippo signaling promotes JNK-dependent cell migration. Proc. Natl. Acad. Sci. U.S.A. 114, 1934–1939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X., et al. , PP6 disruption synergizes with oncogenic Ras to promote JNK-dependent tumor growth and invasion. Cell Rep. 19, 2657–2664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilder D., Ong K., Hsi T. C., Adiga K., Kim J., Tumour-host interactions through the lens of Drosophila. Nat. Rev. Cancer 21, 687–700 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagliarini R. A., Xu T., A genetic screen in Drosophila for metastatic behavior. Science 302, 1227–1231 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Hetz C., Zhang K., Kaufman R. J., Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryoo H. D., Li J., Kang M. J., Drosophila XBP1 expression reporter marks cells under endoplasmic reticulum stress and with high protein secretory load. PLoS One 8, e75774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malzer E., et al. , Impaired tissue growth is mediated by checkpoint kinase 1 (CHK1) in the integrated stress response. J. Cell Sci. 123, 2892–2900 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y., Pan D., The Hippo signaling pathway in development and disease. Dev. Cell 50, 264–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamaratoglu F., et al. , The tumour-suppressor genes NF2/Merlin and expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Yu J., et al. , Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tapon N., et al. , Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Bi X., et al. , Endoplasmic reticulum chaperone GRP78 protects heart from ischemia/reperfusion injury through Akt activation. Circ. Res. 122, 1545–1554 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groenendyk J., Sreenivasaiah P. K., Kim D. H., Agellon L. B., Michalak M., Biology of endoplasmic reticulum stress in the heart. Circ. Res. 107, 1185–1197 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Casas C., GRP78 at the centre of the stage in cancer and neuroprotection. Front. Neurosci. 11, 177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon Y., et al. , The Hippo signaling pathway interactome. Science 342, 737–740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fredriksson S., et al. , Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Ni M., Zhang Y., Lee A. S., Beyond the endoplasmic reticulum: Atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J. 434, 181–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S. A., Shim S. H., He J., Zhuang X., Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 8, 499–508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P., et al. , A balance of Yki/Sd activator and E2F1/Sd repressor complexes controls cell survival and affects organ size. Dev. Cell 43, 603–617.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clevers H., Loh K. M., Nusse R., Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Zeng X., Ryoo H. D., Jasper H., Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet. 10, e1004568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregorieff A., Liu Y., Inanlou M. R., Khomchuk Y., Wrana J. L., Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Ren F., et al. , Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 21064–21069 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X., et al. , Myc suppresses tumor invasion and cell migration by inhibiting JNK signaling. Oncogene 36, 3159–3167 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Dong J., et al. , Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin-Blanco E., Pastor-Pareja J. C., Garcia-Bellido A., JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 97, 7888–7893 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su J., et al. , TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 577, 566–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C., et al. , CtBP modulates Snail-mediated tumor invasion in Drosophila. Cell Death Discov. 7, 202 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R. H., et al. , A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 31, 1088–1105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuelten C. H., Parent C. A., Montell D. J., Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 18, 296–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shim S. M., et al. , The endoplasmic reticulum-residing chaperone BiP is short-lived and metabolized through N-terminal arginylation. Sci. Signal. 11, eaan0630 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Wu H., et al. , Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat. Commun. 6, 6239 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.