Abstract
Unstructured data in the EHR contains essential patient information. Natural language processing (NLP), teaching a computer to read, allows us to tap into this data without needing the time and effort of manual chart abstraction. The core first step for all NLP algorithms is pre-processing the text to identify the core words that differentiate the text while filtering out the noise.. Traditional NLP uses a rules-based approach, applying grammatical rules to infer meaning from the text. Newer NLP approaches use machine learning/deep learning which can infer meaning without explicitly being programmed.
NLP use in nephrology research has focused on using NLP to identify distinct disease processes, such as CKD, and can be used to extract patient-oriented outcomes such as symptoms with high sensitivity. NLP can identify patient features from clinical text associated with AKI and progression of CKD. Lastly, inclusion of features extracted using NLP improved performance of risk prediction models compared to models that only use structured data. Implementation of NLP algorithms has been slow, partially hindered by the lack of external validation of NLP algorithms. However, NLP allows for extraction of key patient characteristics from free text, an infrequently used resource in nephrology.
Keywords: NLP, nephrology, machine learning
Introduction:
Given the expansion of electronic health records (EHR) in healthcare systems, clinical research utilizing EHR data has seen exponential growth in recent years. Most of this research has only used structured data such as billing codes, vital signs, and laboratory values; however, features such as billing codes were created to maximize the billing rate rather than telling the clinical history of the patient. Unstructured data such as provider notes and radiology reports, while templated to provide additional documentation for billing, often contains patient information such as symptoms and social determinants of health which is unavailable in structured data fields. As such, researchers have been trying for decades to perfect Natural Language Processing (NLP) techniques to extract this critical information from the notes.
What is NLP:
NLP is a subset of artificial intelligence encompassing the art of teaching computers to understand written text. NLP is the deep analysis of the linguistic constructs of the text in order to interpret things like sentence structure, synonyms, abbreviations, negation and inflections specifying plurality and tense. In medicine, while NLP has many potential uses, the predominant research using NLP has focused on identifying patient characteristics and diagnoses from clinical notes, generally with the goal of identifying patients matching certain criteria or identifying patient phenotypes for research or clinical decision support.1, 2
References to NLP frequently conflate the acts of identifying clinical concepts described in free text notes and analysis of these clinical concepts. This happens because to some degree the two steps can be accomplished simultaneously in modern systems, but it is important to assess the two phases independently. The first phase is feature identification of clinical concepts described in the notes, while the latter is analyses of these concepts. An overview of the steps of NLP is provided in Figure 1.
Figure 1:
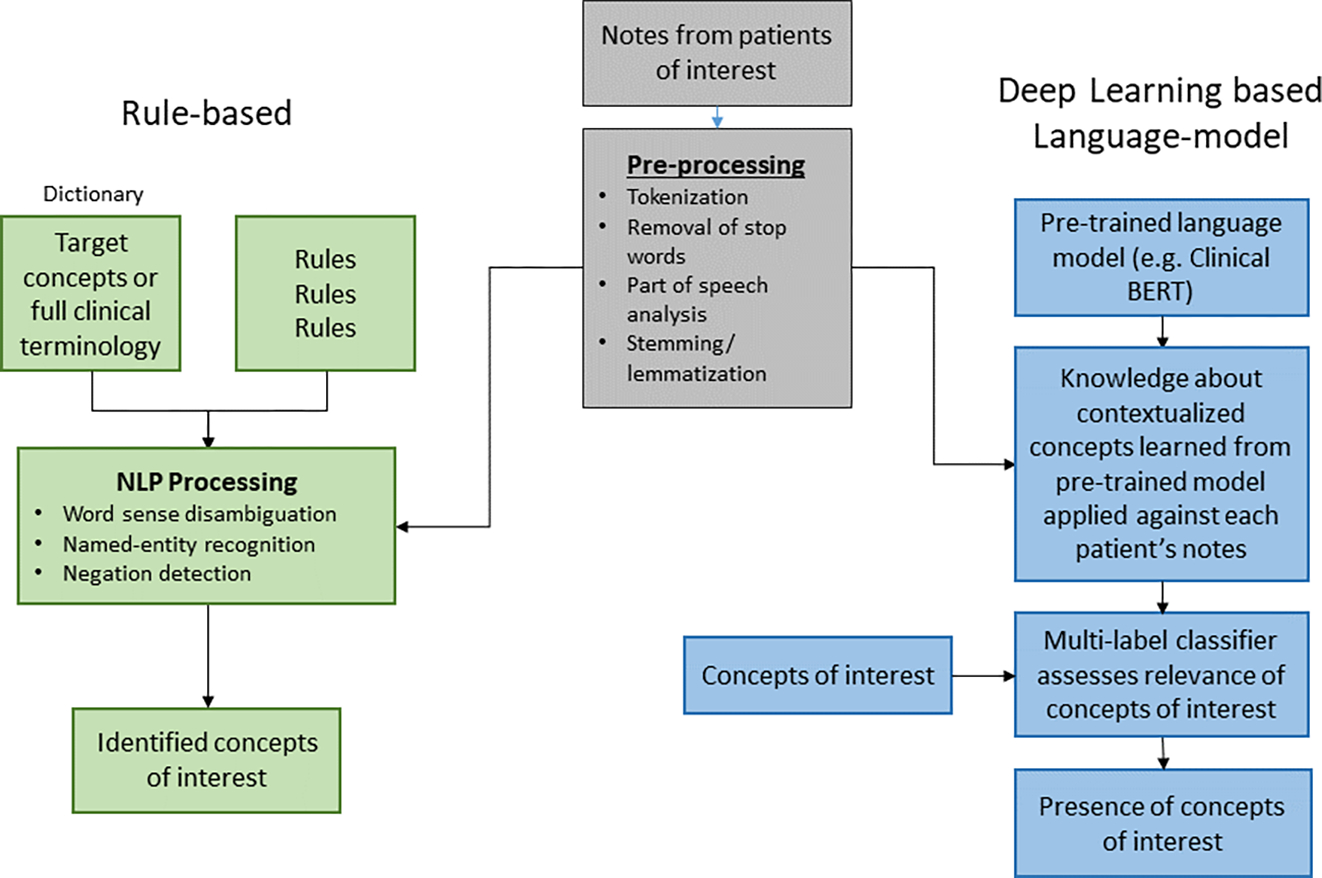
Steps that natural language processing performs to process clinical notes and extract features for analysis. Clinical notes are first pre-processed to remove noise and filter down to core words. Text is then processed by either a rules-based NLP or a Deep learning based Language-model.
Text preprocessing:
No mathematical equation, from basic statistics to deep-learning, fundamentally understands words, so the root goal of NLP is the identification of discrete concepts from a corpus that the algorithm can assess. The core first step for all methods is preprocessing the text to identify the core strings that differentiate the text while filtering the noise. This occurs in various stages, as discussed in Table 1. The most basic of these is tokenization, which is simply the process of breaking up documents into component paragraphs, sentences and words.
Table 1:
Common preprocessing stages of natural language processing with definition and examples.
| NLP Stage | Definition | Example |
|---|---|---|
| Tokenization | Breaking text down into its components | The kidney helps the body maintain homeostasis. --> The | kidney | helps | the | body | maintain | homeostasis. |
| Remove stop words | Removing common words (e.g. “the”, “a”, “and”) that do not provide information | the | kidney | helps | the | body | maintain | homeostasis --> | kidney | helps | body | maintain | homeostasis |
| Part of speech tagging | Assigning a grammatical role to a word used in a sentence. These are generally: noun, pronoun, adjective, verb, adverb, preposition, conjunction, interjection | kidney: noun helps: verb body: noun maintain: verb homeostasis: noun |
| Stemming/lem matization | Reducing inflected or derived words into their stem words or base words | kidney: kidney helps: help body: body maintain: maintain homeostasis: homeostasis |
| Named-entity recognition | Identify and locate named entities such as names, organization, and locations. | Belding Hibbard Scribner (Person) was an American (Location) physician and a pioneer in kidney dialysis. |
| Negation detection | The task of determining the presence of absence of a finding. | Mrs. Nephron did not (negation detection) require dialysis. |
Rules-based NLP:
Rules-based NLP uses hard-grammatical rules to infer meaning from text. The simplest analyses look at string presence and frequency. In these situations, the text may be simplified by eliminating extraneous words and simplifying words to their root. Free text includes many common words that do not provide information in a text document, and include words like “the”, “are”, “a”. These stop words generally confound pure text analyses and are removed from the text data. Word stemming is the process of trimming words down to their stem, e.g. “changing”, “changes” and “change” to “chang”. The most famous algorithm is likely that of M.F. Porter. 3 This throws away information that can be valuable to the interpretation of the text, however it also unifies references that would otherwise look like different things.
Lemmatization is a process designed to eliminate unnecessary variation in words as stemming attempts to do, but more intelligently. It first attempts to map to real words, for example “changing”, “changes” and “change” would map to “change”, as it understands that “changing” is a form of “change”. Second, it can understand some complex synonyms such that “cars” and “automobile” could both map to “car”. Finally, through incorporating part of speech analysis, it may differentiate similar words with legitimately different meanings. For example, the verb “accounting” should probably remain an independent concept from the noun “accountant”, despite their similarity.
There are many ambiguous words in English, such as “mean”, “plant”, “pound” and “well”. Word sense disambiguation assesses the context around homonyms to determine which meaning is appropriate. Studies show that one sense of a word is generally used within a document, so many approaches are built around this assumption.4, 5
Part of speech analysis (POS) assesses sentence structure to interpret constructs affecting the meaning of named entities referenced.6 For example, to properly understand the difference between “the physician called the patient” and “the patient called the physician”, it is critical to understand which is the subject and which is the direct object.
Negation detection is the task of determining whether a referenced finding is negated, as in “no indication of x, y or z”. The most well-known negation algorithm is NexEx, by Chapman et al., which was later revised as ConText, adding support for the identification of hypothetical references, past tense, and references to people other than the patient.7, 8
In medical NLP, where consistent identification of patient facts is the primary goal, named-entity recognition performs the critical task of identifying key entities such as names, organizations, and locations. When possible, mapping to a common dictionary strengthens the analysis by unifying references by synonyms, abbreviations and misspellings to a single identifier, and this is also easier than identifying named entities without a dictionary. This dictionary mapping is especially useful in medicine, where great effort has been gone into developing structured terminologies. One example of a dictionary is SNOMED, a comprehensive collection of medical terms that provides codes and synonyms used in clinical documentation that was developed by College of American Pathologists (CAP).9 By mapping features to concepts in these terminologies, analyses can leverage the synonyms and logical relationships between concepts captured in these efforts to normalize the features and use the hierarchy to understand things such as that a patient with Alzheimer’s Disease is by definition a patient with dementia.
Deep Learning based Language-model:
Advances in deep learning have transformed what is possible with computational learning in many applications, and clinical NLP is no exception. While rules-based NLP can be complex and require manual coding of rules for each concept of interest in the NLP algorithm, deep learning-based language models automate aspects of NLP that are very difficult to get right using traditional rule-based models by assessing word usage patterns across very large collections of sample documents and building models based on these observations. Deep learning also facilitates analyses and applications that would not be possible otherwise. For example, in medicine the order of events and the time between events is crucial for understanding patient outcomes. Much research in LSTMs (Long Short-Term Memory models) and Transformers has identified how to take these things into account in ways that would be very difficult with traditional machine learning analyses.10 These models may be applied as everything from clinical decision support tools to patient chatbots.
Pre-processing for automated deep learning approaches generally incorporates components from traditional NLP, including the removal of stop words and stemming or more robust lemmatization to simplify words. For deep learning models, patient concepts are represented in vector form, a numerical representation of the meaning of a word. In the simplest case, each vector represents a word. This is called a bag-of-words approach, creating “one-hot” vectors (where each feature is given a 0 or a 1 based on its presence for a given patient or document) representing simply the presence of a word (often stemmed and lemmatized to normalize slightly) without representing contextual information. This approach ignores grammar and word order, focusing only on if and how many times a word is present. For example, using the bag-of-words approach, the sentence “Patients on hemodialysis receive hemodialysis three times a week” would be presented as {Patient:1, hemodialysis: 2, receive:1, three:1, times:1, week:1}, and the vector would be {1,2,1,1,1,1}. This is one of the easiest models to apply but clearly fails with the earlier example using exactly the same representation for “the physician called the patient” as for “the patient called the physician”.
In more abstract models, vectors represent the meaning of each word rather than the letters making it up. These are generally known as word embeddings. Word embedding models, most popularly word2vec 11, use deep learning to infer underlying word meaning such that two lexically disjoint synonyms will be represented by a nearly identical vector, thereby reducing high dimensional vocabularies to a smaller feature space to simplify and strengthen analyses.12 Therefore, words that are related such as “queen” and “woman” would be located closer together than “queen” and “kidney”. Building on advances made with word embeddings, Transformers have been engineered to represent a finding not just as a single vector for the primary word, but they also examine surrounding words and add additional vectors representing the context of use.13 Generating these models requires vast amounts of sample narrative and computing power, but pre-trained models have been developed for general English, notably the BERT (Bidirectional Encoder Representations from Transformers) model by Devlin et al, 201814 and the GPT (Generative Pre-trained Transformer) models by OpenAI, most recently GPT-3.15 Medical variants have been trained on biomedical narratives, such as BioWordVec 16, BioBERT 17, Clinical BERT 18 and Bio+Clinical BERT 18.
Common NLP applications:
There are many applications that can be used for NLP. While an exhaustive review of the available applications is beyond the scope of this review we discuss some commonly used open-source applications. A commonly used rules-based engine is MetaMap19, which was developed by the Lister Hill National Center for Biomedical Communications at the National Library of Medicine. This application was originally developed for processing biomedical scholarly articles and can be used to map biomedical text to the Unified Medical Language System (UMLS). The UMLS is another example of a dictionary that is comprised of a set of files and software that links many health and biomedical vocabularies together, including CPT, International Classification of Diseases (ICD)-10-CM, LOINC, MeSH, RxNorm, and SNOMED CT.20
Another open-source NLP software is Apache cTAKES: clinical Text Analysis and Knowledge Extraction System.21 cTAKES, originally developed by the Mayo Clinic, combines rules-based and machine learning techniques to facilitate the extraction of information from clinical text. cTAKES is a modular system of pipelines components, where the components are executed in sequence to process the clinical text. CTAKES, like MetaMap, uses the UMLS to extract and standardize medical concepts.
Python, a popular coding language, has several Python libraries that can be used for NLP. Some commonly used ones are Natural Language Toolkit and Spacy.22, 23 While both are open-source, neither of these libraries were originally built specifically for clinical text but provide the text preprocessing steps as described previously. Subsequent Python packages such as scispaCy contain models for processing biomedical, scientific and clinical text. 24
Few studies have compared the performance of different NLP applications. One study evaluated the performance of MetaMap and cTAKES for the identification of 15 obesity comorbidities which found that cTAKES had slightly better performance.25 Wu et al. compared MetaMap, cTAKES, and Medical Language Extraction and Encoding System (MedLEE), a rules-based NLP developed at Columbia University, for handling of abbreviations from discharge summaries.26, 27 MedLEE had the best performance of the three NLP applications tested. Taggart et al. compared a rules-based NLP to machine learning NLP for identifying bleeding events from clinical notes.28 Authors found that while both methods results in similar sensitivities, the rules-based method had the over best performance with higher specificity, positive predictive values, and negative predictive value. Ultimately the choice of which NLP application to use depends on the users’ familiarity with the different the software.
Application of NLP in Nephrology:
NLP has been used in several different ways in nephrology. First, NLP can aid in identifying diseases or patient phenotypes from free text in the EHR. Second, NLP can aid in improving risk prediction models. Figure 2 provides additional potential applications of NLP in clinical care. Below we highlight several articles exemplifying uses for NLP in nephrology.
Figure 2:
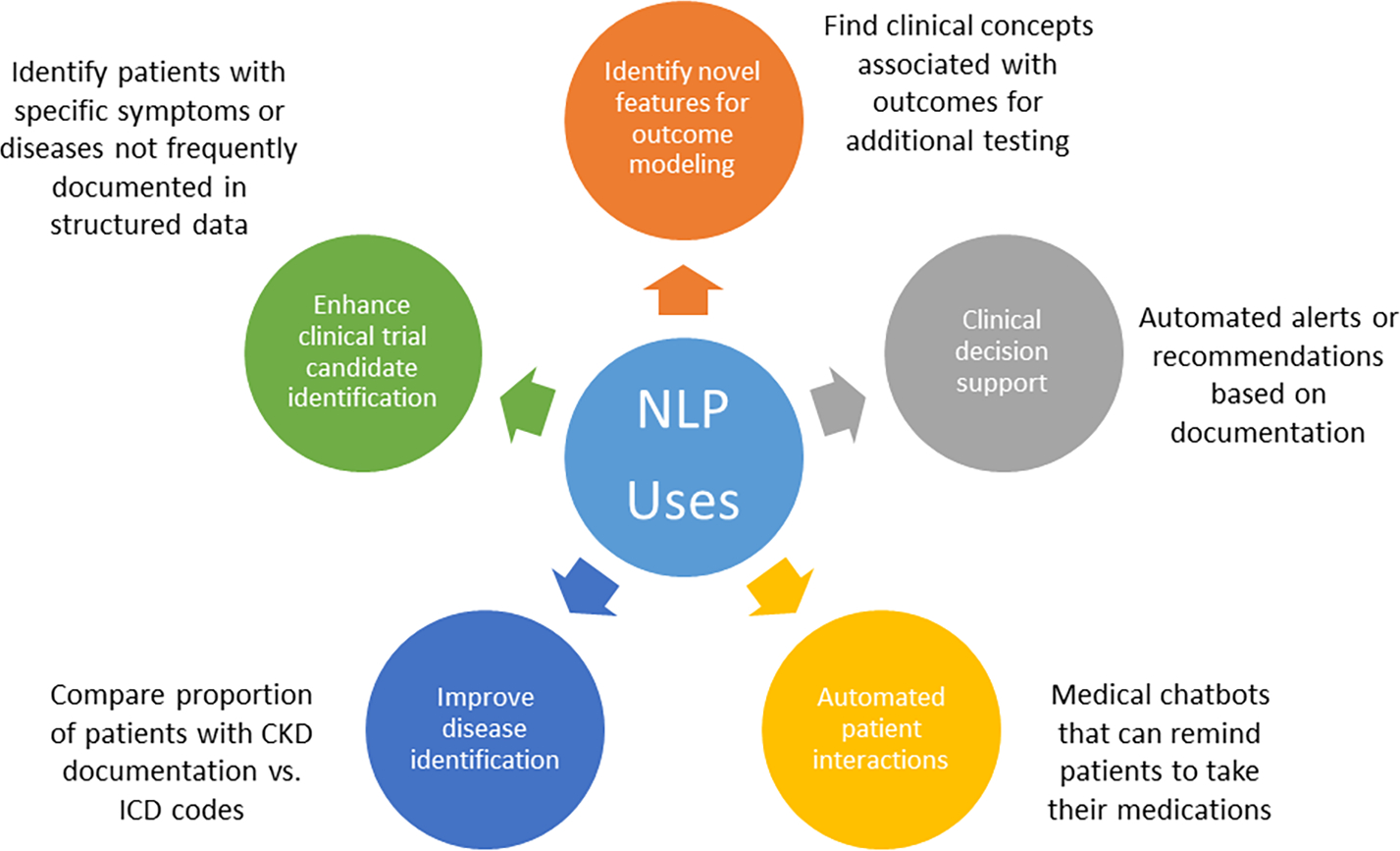
Potential applications of NLP in clinical care.
Identification
While identification of chronic kidney disease (CKD) allows for the institution of therapies to mitigate the progression of kidney disease, CKD is generally under recognized. To evaluate provider recognition of CKD, Chase et al. used NLP to identify whether CKD patients in an outpatient clinic had CKD documented by their providers.29 The authors identified a dictionary of terms associated with CKD from notes of patients with known CKD and used these terms to classify whether the patient had CKD. They applied this dictionary in two ways, using a classifier and a simpler word count method, and found that their models had a sensitivity up to 99.8% and specificity of 99.8%. Twenty-two percent of patients with an eGFR <60 ml/min/1.73m2 lacked documentation of CKD and were less likely to receive guideline-based care. The authors propose that a tool based on their NLP algorithm could be used to prompt physicians to document CKD and allow for earlier implementation of CKD care. However, as this study was done at a single center in a small number of patients and providers, and given differences in documentation by the healthcare system, external validation of these findings is necessary.
NLP can be used to identify a variety of medical conditions. Michalopoulos et al used the cTAKES, to identify dementia, diabetes, and infarction.30 They identified terms associated with these three terms from the UMLS and then used a rules-based approach to determine if the patient had a diagnosis. They were able to detect these risk factors from nephrology clinical notes with an accuracy of 99%, 84%, and 80% respectively. The study was a proof of concept for the ability of NLP to extract risk factors with the goal of developing a ML model to predict the risk of dialysis withdrawal in the future.
Hypertension management is an undisputed integral part of CKD management. While blood pressure is available as structured data in the EHR, providers will often include additional blood pressure readings (e.g. home readings or repeat in-office measurements) in the free text. Therefore, Greenberg et al. used NLP to extract blood pressure readings from the chart of patients with CKD.31 Their NLP algorithm used regular expressions, a sequence of characters which the authors used to indicate the presence of a blood pressure value to identify the blood pressure readings.32 While using only blood pressure readings available in the flowsheets identified 42.3% of patients had controlled blood pressure, when NLP was used in conjunction with flowsheet blood pressure readings, this increased to 52.6%. While their use of regular expressions allowed them to perform their study quickly, this was at the risk of relatively high false positives and false negatives. This study demonstrates that the addition of features from free text can improve the characterization of patients’ medical conditions.
NLP can aid in the identification of patient-centered outcomes such as symptoms. Dialysis patients carry a large burden of symptoms and symptoms are often discussed with providers.33 However, this information is only documented in free text. Chan et al used NLP to identify and quantify seven different symptoms experienced by two different HD cohorts.34 The NLP algorithm matched free text in progress notes to SNOMED CT, a comprehensive health care terminology resource, and compared this technique to manual review and ICD codes. The authors found that NLP was significantly more sensitive in the identification of symptoms compared to ICD codes, with similar specificity. NLP was validated by manual chart review and found to be similar with respect to the identification of symptom burden. A limitation of this study was the use of chart review for validation instead of patient survey data. However, this method provides a way to abstract patient symptoms in a high-throughput fashion.
Prediction
As demonstrated above, NLP can be used to identify patient features that are not traditionally available without substantial effort and time. This allows for the inclusion of patient features into risk prediction models, which can potentially improve model performance.
AKI is common, occurring in over 50% of ICU patients, and is associated with increased mortality.35 Performance of risk prediction models using structured features is moderate.36 Therefore, Li et al. used NLP to extract data from clinical notes within the first 24 hours of a patient’s ICU admission to predict the risk of AKI in the upcoming 72 hours.37 Their algorithm used the bag-of-words approach and extracted features from UMLS. Lastly, they also included pre-trained word embeddings into their deep learning models only. Of the combinations of NLP and classifiers, their highest performing models achieved an AUC of 0.779. On feature analysis, terms like “lasix”, “CABG”, and “labile” were some of the top predictors. In a follow-up study, the addition of laboratory data in conjunction with data from clinical notes improved model performance with an AUC of 0.835.38
While AKI is an acute process with a short time window for intervention, CKD is a more indolent process, and early identification of patients at risk for CKD progression will allow for the implementation of aggressive risk factor mitigation or referral to a nephrologist. One of the most commonly used risk prediction tools is the Tangri risk score, which incorporates 8 clinical variables to predict CKD progression to dialysis.39 Using NLP may unlock hidden data within the EHR, which may enhance our ability to predict future progression.
Singh et al. used clinical notes in the year prior to an initial nephrology consultation to find terms associated with progression to ESKD or death.40 Notes were processed using the MetaMap Software, which matches text from notes to the UMLS Concept unique identifiers. Initially, many of the concepts that were found to have highest association with progression to ESKD were intuitive, such as “chronic renal insufficiency”, “dialysis”, and “volume overload”. The authors then adjusted the data for known clinical risks of progression using the Tangri score and found new concepts that were associated with ESKD development such as “Fast food” (HR 4.34), “psoriasis” (HR 6.00), and “ascorbic acid 500 mg” (HR 5.48). After adjustments, NLP identified 885 concepts, which demonstrates that NLP allows for association testing between novel patient characteristics from the unstructured data with clinical outcomes. Recent studies have found that the addition of data from the free text can improve model performance.41 42
While the papers we have discussed so far have focused on the identification and used single clinical terms, Perotte et al. used latent Dirichlet allocation (LDA) to identify topics from clinical notes and included these topics into various prediction models to predict the risk of progression from CKD III to CKD IV.43 LDA is an unsupervised method, learned by the model and does not require manual guidance, which identifies words or phrases that share a theme.44 An example theme they identified as associated with increased risk of progression was diabetes which included terms such as insulin, Lantus, glucose, and diabetes. The authors found that a model that incorporated laboratory data over time in addition to data from clinical text extracted by NLP produced the best prediction of progression to CKD IV.
Challenges in implementing NLP in medicine
While there has been an increase in the use of NLP in research and the inclusion of data from free text has been repeatedly demonstrated to improve models, these models have not been used in clinical practice. One major limitation shared by these studies is that they were all derived in single sites. As each institution has its own note template and there are regional variations in acronym use, it is imperative that NLP algorithms be externally validated with performance assessed at different healthcare systems. However, for external validation to occur, progress notes must be shared across institutions.
Given that progress notes are rich with PHI and sharing of PHI is highly restrictive to protect patient privacy, this presents a major roadblock to the advancement of NLP. Manual deidentification can be unreliable and time-consuming.45 46 Fortunately several groups have developed deidentificaiton software to aid in this arduous task. Neamatullah et al. developed an automated deidentification software package which identified PHI using several methods including PHI look-up tables and PHI indicators such as “Dr.” or “Mrs.”. 47 Gupta et al. Also developed a deidentification software, the De-Id engine, which uses a set of rules and dictionaries to identify PHI and replace it with specific tags. 48 This engine was applied to pathology notes with good performance. Sweeney et al. developed a Scrub system which uses numerous algorithms to detect different types of PHI based on numerous lists containing common facts to remove PHI from a pediatric medical record system. 49
Conclusions:
EHR data contains a plethora of information on patients, much of which is stored as free text and not usable for research without spending massive amounts of time and energy to perform manual chart abstraction. NLP provides a way to access this data, and we have presented several ways that NLP has been using in research in nephrology. While there are challenges to the implementation of NLP into clinical care, NLP can enable the extraction of key patient characteristics from free text and allow for the inclusion of novel predictors into risk stratification models.
Clinical Summary:
Unstructured data in the EHR contains essential patient information and identifying this information has traditionally required manual chart review which is time consuming.
Natural language processing (NLP) allows us to identify patient information from free text in a high throughput manner.
In nephrology research, NLP has been used to identify different disease processes and patient-centered outcomes.
Inclusion of features extracted from clinical text by NLP has identified novel predictions of AKI and CKD progression, and inclusion of NLP extracted features improves performance of models built using structured data alone.
Acknowledgements:
LC is supported in part by a grant from the NIH/NIDDK (K23DK124645).
TTVV receives financial compensation as a consultant for Clinithink, developer of clinical natural language processing (CNLP) technology used to automate the review of electronic medical records. He also owns equity in Clinithink (a privately traded company).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1): 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5): 760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter MF. An Algorithm for Suffix Stripping. Program-Autom Libr. 1980;14(3): 130–137. [Google Scholar]
- 4.Gale WA, Church KW, Yarowsky D. One Sense Per Discourse. Speech and Natural Language. 1992: 233–237. [Google Scholar]
- 5.Yarowsky D Unsupervised Word Sense Disambiguation Rivaling Supervised Methods. 33rd Annual Meeting of the Association for Computational Linguistics: Association for Computational Linguistics; 1995:189–196. [Google Scholar]
- 6.Ferraro JP, Daume H, DuVall SL, Chapman WW, Harkema H, Haug PJ. Improving performance of natural language processing part-of-speech tagging on clinical narratives through domain adaptation. Journal of the American Medical Informatics Association. 2013;20(5): 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5): 301–310. [DOI] [PubMed] [Google Scholar]
- 8.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42(5): 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spackman KA, Campbell KE, Cote RA. SNOMED RT: a reference terminology for health care. Proc AMIA Annu Fall Symp. 1997: 640–644. [PMC free article] [PubMed] [Google Scholar]
- 10.Palangi H, Deng L, Shen YL, et al. Deep Sentence Embedding Using Long Short-Term Memory Networks: Analysis and Application to Information Retrieval. Ieee-Acm T Audio Spe. 2016;24(4): 694–707. [Google Scholar]
- 11.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems. 2013;26. [Google Scholar]
- 12.Shao Y, Taylor S, Marshall N, Morioka C, Zeng-Treitler Q. Clinical text classification with word embedding features vs. bag-of-words features. 2018 IEEE International Conference on Big Data (Big Data): IEEE; 2018:2874–2878. [Google Scholar]
- 13.Vaswani A, Shazeer N, Parmar N, et al. Attention Is All You Need. Adv Neur In. 2017;30. [Google Scholar]
- 14.Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805. 2018. [Google Scholar]
- 15.Brown TBaM, Benjamin and Ryder Nick and Subbiah Melanie and Kaplan Jared and Dhariwal Prafulla and Neelakantan Arvind and Shyam Pranav and Sastry Girish and Askell Amanda and Agarwal Sandhini and Herbert-Voss Ariel and Krueger Gretchen and Henighan Tom and Child Rewon and Ramesh Aditya and Ziegler Daniel M. and Wu Jeffrey and Winter Clemens and Hesse Christopher and Chen Mark and Sigler Eric and Litwin Mateusz and Gray Scott and Chess Benjamin and Clark Jack and Berner Christopher and McCandlish Sam and Radford Alec and Sutskever Ilya and Amodei Dario. Language Models are Few-Shot Learners. arXiv. 2020. [Google Scholar]
- 16.Zhang Y, Chen Q, Yang Z, Lin H, Lu Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Scientific data. 2019;6(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4): 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. arXiv preprint arXiv:1904.03323. 2019. [Google Scholar]
- 19.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3): 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenreider O The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue): D267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5): 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird S, Klein E, Loper E. Natural language processing with Python: analyzing text with the natural language toolkit: “ O’Reilly Media, Inc.”; 2009. [Google Scholar]
- 23.Honnibal M, Montani I. spaCy 2: Natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing. To appear. 2017;7(1): 411–420. [Google Scholar]
- 24.Neumann M, King D, Beltagy I, Ammar W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. Sigbiomed Workshop on Biomedical Natural Language Processing (Bionlp 2019). 2019: 319–327. [Google Scholar]
- 25.Reátegui R, Ratté S. Comparison of MetaMap and cTAKES for entity extraction in clinical notes. BMC medical informatics and decision making. 2018;18(3): 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Denny JC, Rosenbloom ST, Miller RA, Giuse DA, Xu H. A comparative study of current Clinical Natural Language Processing systems on handling abbreviations in discharge summaries. AMIA ... Annual Symposium proceedings. AMIA Symposium. 2012;2012: 997–1003. [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association : JAMIA. 1994;1(2): 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taggart M, Chapman WW, Steinberg BA, et al. Comparison of 2 Natural Language Processing Methods for Identification of Bleeding Among Critically Ill Patients. JAMA Network Open. 2018;1: e183451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chase HS, Radhakrishnan J, Shirazian S, Rao MK, Vawdrey DK. Under-documentation of chronic kidney disease in the electronic health record in outpatients. J Am Med Inform Assoc. 2010;17(5): 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michalopoulos G, Qazi H, Wong A, Butt Z, Chen H. Automatic Extraction of Risk Factors for Dialysis Patients from Clinical Notes Using Natural Language Processing Techniques. Stud Health Technol Inform. 2020;270: 53–57. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg JO, Vakharia N, Szent-Gyorgyi LE, et al. Meaningful measurement: developing a measurement system to improve blood pressure control in patients with chronic kidney disease. J Am Med Inform Assoc. 2013;20(e1): e97–e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. J Am Med Inform Assoc. 2006;13(6): 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisbord SD, Fried LF, Arnold RM, et al. Prevalence, severity, and importance of physical and emotional symptoms in chronic hemodialysis patients. J Am Soc Nephrol. 2005;16(8): 2487–2494. [DOI] [PubMed] [Google Scholar]
- 34.Chan L, Beers K, Yau AA, et al. Natural language processing of electronic health records is superior to billing codes to identify symptom burden in hemodialysis patients. Kidney Int. 2020;97(2): 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoste EA, Bagshaw SM, Bellomo R, et al. Epidemiology of acute kidney injury in critically ill patients: the multinational AKI-EPI study. Intensive Care Med. 2015;41(8): 1411–1423. [DOI] [PubMed] [Google Scholar]
- 36.Huang CY, Grandas FG, Flechet M, Meyfroidt G. Clinical prediction models for acute kidney injury. Rev Bras Ter Intensiva. 2020;32(1): 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Yao L, Mao C, Srivastava A, Jiang X, Luo Y. Early prediction of acute kidney injury in critical care setting using clinical notes. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM): IEEE; 2018:683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun M, Baron J, Dighe A, et al. Early Prediction of Acute Kidney Injury in Critical Care Setting Using Clinical Notes and Structured Multivariate Physiological Measurements. Stud Health Technol Inform. 2019;264: 368–372. [DOI] [PubMed] [Google Scholar]
- 39.Tangri N, Stevens LA, Griffith J, et al. A predictive model for progression of chronic kidney disease to kidney failure. JAMA. 2011;305(15): 1553–1559. [DOI] [PubMed] [Google Scholar]
- 40.Singh K, Betensky RA, Wright A, Curhan GC, Bates DW, Waikar SS. A Concept-Wide Association Study of Clinical Notes to Discover New Predictors of Kidney Failure. Clin J Am Soc Nephrol. 2016;11(12): 2150–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makino M, Yoshimoto R, Ono M, et al. Artificial intelligence predicts the progression of diabetic kidney disease using big data machine learning. Sci Rep. 2019;9(1): 11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventrella P, Delgrossi G, Ferrario G, Righetti M, Masseroli M. Supervised machine learning for the assessment of Chronic Kidney Disease advancement. Comput Meth Prog Bio. 2021;209: 106329. [DOI] [PubMed] [Google Scholar]
- 43.Perotte A, Ranganath R, Hirsch JS, Blei D, Elhadad N. Risk prediction for chronic kidney disease progression using heterogeneous electronic health record data and time series analysis. J Am Med Inform Assoc. 2015;22(4): 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. Journal of machine Learning research. 2003;3(Jan): 993–1022. [Google Scholar]
- 45.Douglass M, Clifford GD, Reisner A, Moody GB, Mark RG. Computer-assisted de-identification of free text in the MIMIC II database. Comput Cardiol. 2004;31: 341–344. [Google Scholar]
- 46.Douglass MM, Cliffford GD, Reisner A, Long WJ, Moody GB, Mark RG. De-identification algorithm for free-text nursing notes. Computers in Cardiology 2005, Vol 32. 2005;32: 331–334. [Google Scholar]
- 47.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol. 2004;121(2): 176–186. [DOI] [PubMed] [Google Scholar]
- 49.Sweeney L Replacing personally-identifying information in medical records, the Scrub system. Proc AMIA Annu Fall Symp. 1996: 333–337. [PMC free article] [PubMed] [Google Scholar]


