Abstract
The adaptor protein (AP) complexes not only form the inner layer of clathrin coats but also have clathrin-independent roles in membrane traffic whose mechanisms are unknown. HIV-1 Nef hijacks AP-1 to sequester major histocompatibility complex class I (MHC-I), evading immune detection. We found that AP-1:Arf1:Nef:MHC-I forms a coat on tubulated membranes without clathrin and determined its structure. The coat assembles via Arf1 dimer interfaces. AP-1–positive tubules are enriched in cells upon clathrin knockdown. Nef localizes preferentially to AP-1 tubules in cells, explaining how Nef sequesters MHC-I. Coat contact residues are conserved across Arf isoforms and the Arf-dependent AP complexes AP-1, AP-3, and AP-4. Thus, AP complexes can self-assemble with Arf1 into tubular coats without clathrin or other scaffolding factors. The AP-1:Arf1 coat defines the structural basis of a broader class of tubulovesicular membrane coats as an intermediate in clathrin vesicle formation from internal membranes and as an MHC-I sequestration mechanism in HIV-1 infection.
The AP-1 complex forms a tubular coat held together by Arf1 dimers and promoted by HIV-1 Nef in the absence of clathrin.
INTRODUCTION
Membrane and secretory proteins and lipids are distributed to their destinations within eukaryotic cells by the trafficking of tubular and vesicular structures. Tubular and vesicular structures are organized by proteinaceous vesicle coats, which include clathrin and its adaptor protein (AP) complexes, coat protein complex I (COPI), COPII, and retromer (1). Clathrin and COPI are vesicular coats (2), retromer is a tubular coat (3), and COPII can assemble in both tubular and vesicular geometries (2). The AP complexes AP-1 to AP-5 function in a vast array of cellular process, of which some, but not others, depend on clathrin. The AP complexes AP-1 and AP-2 form the inner layer of clathrin-coated vesicles (CCVs), connecting interior lipids and cargoes to the exterior clathrin cage. Despite that clathrin itself does not usually form tubular coats, there have been many reports of AP complexes on tubular structures, including the typically clathrin-dependent AP-1 (4–6). AP-3 is principally localized to tubular structures and, while it can associate with clathrin, AP-3 is functionally independent of it (7, 8). AP-4 does not usually interact with clathrin at all (9, 10). These observations suggest that AP-1, AP-3, and AP-4 must have some fundamental ability to assemble on membranes in the absence of clathrin. Despite extensive structural studies of the AP and related complexes (2, 11), the basis for the formation of tubular and clathrin-independent assemblies is unknown.
One example of such a coat assembly might be the AP-1–dependent sequestration of major histocompatibility complex class I (MHC-I) by the HIV-1 accessory protein Nef. Down-regulation of the antigen presentation complex MHC-I is a ubiquitous strategy used by viral pathogens to subvert host CD8+ T cell–mediated elimination of virally infected cells (12). The human and simian immunodeficiency viruses—HIV-1, HIV-2, and SIV—encode an accessory factor, Nef, that achieves this critical task as one of its various functions (13–18). Nef targets MHC-I by hijacking AP-1, along with its small guanosine triphosphatase activator, Arf1. This reroutes mature MHC-I molecules to the perinuclear region and/or endosomal sorting network instead of the plasma membrane (19–23). MHC-I is eventually degraded at the lysosome in a clathrin-dependent mechanism (20, 24). Hijacked AP-1 and MHC-I form long-lived internal membrane-associated assemblies that sequester MHC-I away from the plasma membrane (19, 25–28). The nature of the MHC-I–sequestering assemblies has remained obscure.
AP-1 and the four other human AP complexes (2 through 5) share a conserved heterotetrameric architecture consisting of a small (σ), medium (μ), and two large subunits; AP-1 consists of σ1, μ1, γ, and β1 (Fig. 1A) (29). APs are allosterically regulated and couple conformational states to cargo binding, membrane targeting, and recruitment of clathrin (30, 31). In the locked conformation, AP-1 is primarily cytosolic and has a low affinity for phosphatidylinositol 4-phosphate [PI(4)P] and cargo molecules. To become activated, myristoylated Arf1 in its guanosine triphosphate (GTP)–bound state recruits AP-1 to target membranes at the trans-Golgi network (TGN) and endosomes and stabilizes the unlocked conformation of AP-1 (32). The unlocked conformation binds cargo bearing the consensus YxxΦ motif at the μ domain and the more variable dileucine cargo motifs, (D/E)xxxL(L/I/M), at the σ domain (where “x” is any amino acid and Φ is any bulky hydrophobic residue). Once bound to and stabilized by cargo and myristoylated Arf1, AP-1 recruits clathrin through conserved clathrin binding motifs encoded within the extended C-terminal appendage of the β1 domain (33). AP-1 localizes to membrane compartments enriched in PI(4)P (34), including the TGN and endosomal system. However, Arf1 is the major determinant of subcellular localization and activation of AP-1 (35, 36). AP-3 (37) and the clathrin-independent AP-4 (38) are also Arf1-dependent. Thus, AP-1, AP-3, and AP-4 comprise a subset of Arf-dependent AP complexes that are likely to be governed by common recruitment and activation principles.
Fig. 1. AP-1, Arf1, and Nef form an ordered coat on MHC-Icyto-membranes.
(A) Cartoon overview of AP-1, Arf1, Nef, and MHC-I. Functional sites and their associated domains are labeled. (B) Representative cryo–electron tomographic slices through an AP-1βFL:Arf1myr:Nefmyr–coated vesicle incorporated with MHC-Icyto lipopeptide. Yellow arrows indicate AP-1 density. Dark blue arrows denote tracks of AP-1 γ:γ–juxtaposed molecules. Light pink arrows denote tracks of AP-1 β1:β1 –juxtaposed molecules. (C) Representative cryo–electron tomographic slices through an AP-1βFL:Arf1myr:Nefmyr–coated tube incorporated with MHC-Icyto lipopeptide. Arrows are as indicated in (B). (D) Reconstructed map and model of a continuous AP-1:Arf1:Nef coat on a section of tubular membrane. Cross-sectional view of the coat down the tube axis (left) and surface view of the coat (right). Arrows are as described in (B). Fitted AP-1, Arf1, and Nef are color-coded to match the depiction in (A). Arf1 is colored pink and purple to denote association with AP-1 β1 and γ, respectively.
In the course of reconstituting the Nef-sequestered state of AP-1 and MHC-I, we noticed that AP-1:Arf1:Nef:MHC-I could form a continuous tubular protein coat on membranes in the absence of clathrin. How AP-1, AP-3, and AP-4 might organize tubular coats is a long-standing question. Viruses are often observed to “overdrive” cellular processes in the course of hijacking them, providing windows into fundamental aspects of cell biology that would be hard to obtain otherwise. Given the potential significance for tubular coat organization across eukaryotes, we determined the structure of the coat by cryo–electron tomography and subtomogram averaging (STA). The resulting structure suggests a paradigm for clathrin-independent tubular membrane coating and stabilization by the Arf1-dependent subset of AP complexes.
RESULTS
AP-1, Arf1, and Nef self-assemble to form a tubular coat on membranes
The cytosolic tail of MHC-I subtypes A and B contains the consensus sequence Y(S/T)QA, which does not normally recruit AP-1 because of the presence of an Ala in place of the requisite larger hydrophobic residue of the YxxΦ motif (39). Nef converts this nonsubstrate into a neo-substrate by complementing the suboptimal cargo motif (40). Nef’s tandem PxxP motif and key electrostatic interactions drive recognition of the MHC-I tail onto the canonical tyrosine motif cargo binding site of μ1 (40). To reconstitute MHC-I down-regulation on membranes in vitro, liposomes containing 3 mole percent of MHC-Icyto lipopeptide in a TGN-like lipid composition were generated by extrusion through a 50-nm filter. Liposomes were incubated with AP-1, myristoylated Arf1 (Arf1myr), and myristoylated HIV-1 NL4-3 Nef (Nefmyr) (Fig. 1A and fig. S1A). Arf1 nucleotide exchange was stimulated by preincubating Arf1myr, GTP, and EDTA at 37°C for 20 min, followed by the addition of a molar excess Mg2+ over EDTA. The resulting samples were vitrified and imaged by cryo–electron microscopy (cryo-EM).
The cryo-EM images revealed a dense protein coat on the surface of membranes (fig. S1B). The protein coat was present on spherical vesicles, amorphous membranes, and membrane tubes, indicating that AP-1 binds to membranes of varying curvatures. To investigate the spatial distribution of AP-1 on the surface of membranes, we collected tilt series and reconstructed tomograms of the coated membranes (fig. S2 and table S1). The tomograms show that the protein coat forms a nearly contiguous layer on most membranes (Fig. 1, B and C, and fig. S1, C and D). The tomograms revealed a notable repeating density pattern consistent with a regular lattice, most obviously on tubular membranes (Fig. 1C and figs. S1C and S4A). The patterned array was present on a range of tube diameters and was most obvious on relatively straight tube sections and, to a lesser extent, on spherical vesicles (Fig. 1, B and C, and figs. S1, C and D, and S4A) and flat membrane sheets. We determined the structure of the assembly using STA. We focused on AP-1 assemblies on tubulated membranes. Seventy-two tilt series were collected and reconstructed into tomograms for STA (tables S1 and S2). Of the 284 annotated tubes, narrow tubes were highly enriched relative to wide tubes (fig. S1E).
Subtomograms were pooled on the basis of tube diameter and analyzed separately to maximize structural homogeneity. Aligned and averaged subtomograms from narrow tubes (20 to 30 nm in diameter) resulted in a 20 Å resolution map, which could be fit unambiguously with atomic models of the hyperunlocked AP-1core (Fig. 1D and figs. S2 and S3) (41, 42). The coat has helical symmetry with a 10-nm pitch and 10.5 AP-1 protomers per turn (table S4). Globular densities adjacent to the fitted AP-1 heterotetramers showed density extending downward toward the membrane surface, suggesting that the densities belonged to the N termini and globular domains of Arf1 and/or Nef. Placement of AP-1core models [Protein Data Bank (PDB): 6cm9] (42) within the lattice revealed two alternating tracks of AP-1 dimers propagating along the tube axis: one dimer pair created by γ:γ and the other by β1:β1 contacts (Fig. 1, B to D).
The AP-1 coat is connected by two Arf1 bridges
The dimeric interfaces connecting the protomers laterally and axially were investigated by STA. The β1:β1- and γ:γ-juxtaposed AP-1 dimers were globally resolved to 9.3 and 9.6 Å, respectively (Fig. 2, A and B, fig. S3, and table S2), with local resolution ranging from 8 to16 Å (fig. S3). At this resolution, Arf1 and Nef could be unambiguously fit to the density. Two Arf1 and one Nef molecules per AP-1 complex constitute the asymmetric unit of the lattice (Fig. 2C). The stoichiometry and interfaces between AP-1, Arf1, and Nef within the asymmetric unit match those observed in the asymmetric unit of the soluble (e.g., non-membrane–associated) closed trimeric AP-1 complex consisting of the AP-1core, N-terminally truncated Arf1, and a genetic fusion of BST2(2–20) and NL4-3 Nef (PDB: 6cm9) (42). The asymmetric unit of the closed trimer docks into the lattice density with no need for conformational adjustments and only a slight shift in the Nef core. The major interactions between the β1-bound Arf1 (hereafter, Arf1β1) and γ-bound Arf1 (hereafter, Arf1γ) with the GTP-dependent switch regions of the Arf1 molecules are identical to those seen in the BST2(2–20)-bound closed AP-1 trimer (42) and therefore not presented in detail here. The density of the lattice is fully accounted for by AP-1core, Arf1myr, and Nefmyr.
Fig. 2. Arf1 mediates AP-1 dimerization.
(A) Subnanometer resolution map and associated atomic model fit of the Arf1β1: β1 juxtaposed AP-1 dimer as viewed looking onto the tube surface. Atomic models are colored as denoted in (C). (B) Subnanometer resolution map and associated atomic model fit of the Arf1γ:γ-juxtaposed AP-1 dimer as viewed looking onto the tube surface. Atomic models are colored as denoted in (C). (C) Cartoon representation of the asymmetric unit (AP-11:Arf12:Nef1; red outline) flanked on either side by asymmetric units creating Arf1γ:γ and Arf1β1:β1 dimeric connections. (D) Zoom of the interface mediating Arf1γ: γ and γ:γ dimeric interactions. (E) Zoom of the interface mediating Arf1β1:β1 dimeric interactions. (F) Sequence alignment of μ and γ (or equivalent) domains from human and yeast APs. Yellow-highlighted residues are labeled according to their position in human AP-1 and correlate with residues shown in (D) and (E).
Placement of the asymmetric unit (AP-11:Arf12:Nef1) within each dimer revealed that the AP-1 protomers do not contact one another directly. Rather, each dimeric interface is mediated by AP-1:Arf1 and Arf1:Arf1 contacts (Fig. 2, A and B). These Arf1 dimer contacts have not been reported previously. At 9 Å resolution, side-chain densities are not visible; however, side-chain locations can be inferred from the previously determined high-resolution structures of all of the components. Arf1β1 localizes charged and polar residues (K36, E41, K59, S162, and D164) near complementary residues in the μ1 domain of an adjacent asymmetric unit (R246, K396, K400, and Q404) (Fig. 2E). Arf1γ similarly localizes E41 and T44 with γ K186 and σ1 K129/K133 (Fig. 2D). Arf1 switch 1 and 2 regions consist of residues 45 to 54 and 70 to 80, respectively. Thus, some of these contacts are at the periphery of the GTP-dependent conformational switch regions, but none are actually within the switch regions. The biochemical properties of residues at each interface, particularly the Arf1-μ1 interface, are conserved between AP-1 and its human and yeast AP homologs (Fig. 2F). This suggests that the other Arf1-dependent AP complexes could use similar contacts to form Arf1-mediated higher-order assemblies. Consistent with this, an Arf1 contact with the CTD of the μ4 subunit of AP-4 was inferred from a yeast two-hybrid binding screen (38). The only direct AP-1:AP-1 interaction found within the lattice is created by γ residues K144, K147, and S149 (Fig. 2D). AP-2, AP-3, and AP-4 also share polar and charged residues at these positions (Fig. 2F), again suggesting other APs could make similar contacts.
Next, we analyzed interactions that stabilize axial propagation of AP-1 protomers along the tube axis. The two respective axial interfaces bridging β1:β1 and γ:γ dimers were symmetry expanded and the STA maps globally refined to 9.3 and 9.4 Å, respectively (Fig. 3, A and B, fig. S3, and table S2). Each interface is primarily composed of direct Arf1:Arf1 interactions (Fig. 3, C and D), and both interfaces use a core set of residues conserved among human Arf1 homologs and yeast Arf1 (Fig. 3E), namely, E115, R117, R149, and H150 (Fig. 3, C and D). Arf1γ also makes polar contacts between its H146 and μ1 E284 of the adjacent AP-1 protomer (Fig. 3C).
Fig. 3. Arf1:Arf1 homodimerization in the narrow tube lattice.
(A) Subnanometer resolution map and associated atomic model fit of the Arf1γ: Arf1γ homodimeric interface as viewed looking onto the tube surface. Atomic models are colored as denoted in Fig. 2. Cartoon representation of the AP-1 lattice as viewed looking onto the tube surface is shown for reference. (B) Subnanometer resolution map and associated atomic model fit of the Arf1β1:Arf1β1 homodimeric interface as viewed looking onto the tube surface. Atomic models are colored as denoted in Fig. 2. Cartoon representation of the AP-1 lattice as viewed looking onto the tube surface is shown for reference. (C) Zoom of the interface mediating Arf1γ:Arf1γ homodimerization. (D) Zoom of the interface mediating Arf1β1:Arf1β1 homodimerization. (E) Sequence alignment of Arf1 (human and yeast) and human Arf1 homologs. Yellow-highlighted residues are labeled according to their position in human Arf1 and correlate with residues shown in (C) and (D). Residues linked by solid lines are proposed to interact directly.
Arf1, Nef, and AP-1 contributions to membrane binding
Each protomer is arranged on the membrane surface such that the N termini of Arf1 and Nef are proximal to and oriented toward the membrane (Fig. 4, A and B). Density for the N-terminal residues of Arf1 (2 to 17) is resolved well enough to model an amphipathic helix and unstructured chain leading to the Arf1 globular core (Fig. 4, C and D). The modeled amphipathic helix is oriented along the long axis of the membrane tube consistent with the map density (Fig. 4, C to E), although the relative orientation is more ambiguous for Arf1β1 (Fig. 4, D and E). The corresponding density for the N terminus of Nef is not resolved in any of the subnanometer resolution structures, including those with no symmetry imposed during subtomogram alignment, suggesting that the position and/or orientation of its N terminus is less constrained relative to the N terminus of Arf1.
Fig. 4. Lattice contacts with the membrane.
(A) Subnanometer resolution map and associated atomic model fit of the narrow tube lattice asymmetric unit (AP-11:Arf12:Nef1) as viewed from the side. Atomic models are colored as denoted in the legend shown in (B). The inset shows a zoom of the AP-1:membrane interface specifically highlighting μ1:membrane contacts. Nef is omitted for visual clarity. (B) A 180° rotated view of the map and model shown in (A). The inset shows a zoom of the AP-1:membrane interface specifically highlighting γ:membrane contacts. (C) Atomic model fit to the density associated with Arf1γ as viewed from the side. Density for the near-complete N terminus (2 to 17) is present, and a speculative model for the amphipathic helix is shown and highlighted (red circles). (D) Atomic model fit to the density associated with Arf1β1 as viewed from the side. As in (C), density for the near-complete N terminus (2 to 17) is present, and a speculative model for the amphipathic helix is shown and highlighted (red circles). (E) Y and Z slices through the STA structure(s) at regions of Arf1:membrane contacts. Slices emphasize the density associated with the Arf1 N terminus. The Y slice shows Arf1 density from the side, while the Z slice shows Arf1 density in-plane with the membrane. Red arrows denote density assigned to the putative Arf1 amphipathic helix. Yellow arrows denote the long axis of the tube.
The AP-1 core is oriented such that μ1 and γ are parallel with the membrane and make three points of contact. Residues R167 and K168 within the basic loop of μ1 and K346 within the basic patch of μ1 appear to make direct contact with the membrane, presumably with anionic lipid headgroups (Fig. 4A). R303 and R304 in the basic patch are also closely localized to the membrane surface (Fig. 4A). The third point of contact is observed in the γ N terminus (Fig. 4B). Residues predicted to interact with phosphoinositides, e.g., PI(4)P, including Y45, K78, and R48 are localized to membrane density near the putative PIP headgroup binding site, suggesting that AP-1 γ is engaged with PI(4)P (Fig. 4B) (43).
Nef and MHC-I engagement with the tubular coat
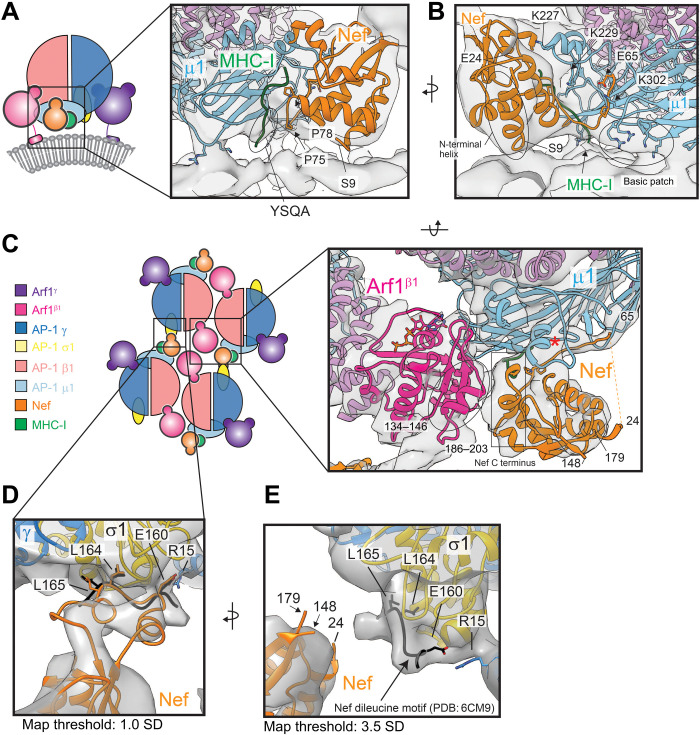
Rigid-body docking of the AP-1:Arf12:BST2-Nef structure (PDB: 6CM9) confirms that Nef is localized immediately adjacent to the AP-1 μ1 tyrosine motif binding site within the AP-1:Arf1 tubular lattice (Figs. 2A, 3B, and 5, A to C). The crystalized Nef, μ1, and MHC-I structure [PDB: 4EN2; (40)] can be docked into the STA essentially without modification (Fig. 5, A to C). The Nef 72-PxxPxxP-78 motif is positioned to complement the MHC-Ictyo tail at the μ1 cargo binding site (Fig. 5A), and the Nef N-terminal helix (9–24) is packed against the Nef core (Fig. 5, A and B). Residues upstream of the Nef N-terminal helix are proximal to the membrane and likely directly interact with the membrane in a manner consistent with nuclear magnetic resonance structures of the N-terminal membrane anchor domain of Nef (44). As seen in the Nef:MHC-I:μ1 crystal structure [PDB: 4EN2; (40)] and AP-1:Arf1:BST2-Nef complex (42), the acidic patch of Nef (62-EEEE-65) is only partially resolved in the STA density, and direct interaction can only be inferred between μ1 K302 and Nef E65 (Fig. 5B). The entire loop spanning from the end of the N-terminal helix to the acidic patch (E24-E64) is unresolved (Fig. 5C). Overall, our observations in the context of the lipid bilayer are consistent with the direct interactions between Nef, μ1, and the cytosolic tails of MHC-I and tetherin seen previously in the absence of membrane (40, 42).
Fig. 5. Nef interactions within the AP-1:Arf1 lattice.
(A) Side-view cartoon depiction of the AP-1:Arf12:Nef asymmetric unit from the narrow tube lattice with inset showing the Nef:MHC-I:μ1 interfaces. (B) A 180° rotated view of the inset shown in (A). (C) Top-view cartoon depiction of the AP-1:Arf1:Nef lattice with inset showing the Nef:μ1:MHC-I:Arf1β1 quaternary complex (map threshold, 3.5 SD). The red asterisk denotes the μ1 internal loop stabilized by Nef. (D) Inset showing the Nef ExxxLL:σ1 interface. The map density is fit with a hypothetical structural model based on the predicted and solved structures. The ExxxLL motif of Nef observed in the AP-1:Arf1:BST2-Nef closed trimer (PDB: 6CM9) is shown in black for comparison. (E) A 90° rotated view of (D). The map threshold density was set to 3.5 SD from the mean. The ExxxLL motif of Nef from PDB: 6cm9 (black) is modeled as in (D).
Nef makes two additional interactions in the lattice context that have not been previously reported. Directly adjacent to the tyrosine cargo binding site, the C-terminal residues 186 to 203 pack against a helical region of Arf1β1 consisting of residues 134 to 146 (Fig. 5C). The same C-terminal region of Nef stabilizes an internal loop on μ1 and facilitates ternary contacts between MHC-I, Nef, and μ1 (Fig. 5C) as previously observed (40)). The proximity of Arf1β1 to the cargo binding groove, at a region distinct from the YSQA binding region, hints at a potential role for Arf1 in cargo binding and/or cargo specificity, especially for cargoes with longer cytosolic tails. The other noteworthy interaction between Nef and AP-1 is mediated by the Nef dileucine motif (160-ExxxLL-165) (Fig. 5, D and E). Nef is positioned within the lattice such that its dileucine motif reaches across to an adjacent AP-1 protomer (Fig. 5, D and E). The dileucine site of σ1 is occupied (Fig. 5E) consistent with the σ1:160-ExxxLL-165 interaction observed in the AP-1:Arf1:BST-2 closed trimer (42) and its known interaction with AP-1 (26, 45, 46). Increasing the STA map threshold revealed density directly linking the σ1 site to Nef’s core (Fig. 5D). Docking of predicted and previously solved structures of Nef’s dileucine loop into the map correlate well with the density and meet the spatial requirements for a direct interaction (Fig. 5D), thus supporting Nef interactions with two distinct AP-1 molecules in the lattice.
Rearrangements stabilize the lattice on wide tubes
Ordered AP-1 coats were observed on the majority of membrane tubes, including those with much wider diameters (e.g., >60 nm; fig. S4A). To determine how the lattice adjusts to fit flatter membrane surfaces, subtomograms from wider tubes (60 to 70 nm in diameter) were aligned and averaged. The resulting maps were globally resolved to 20 Å (fig. S4B and table S3). The helical pitch on wide tubes is 70 nm (table S4), and AP-1 dimers are stacked almost coincident with the long axis of the membrane tube (Fig. 1, C and D, versus fig. S4, A and F). As on narrow tubes, the lattice is stabilized by AP-1:Arf1 and Arf1:Arf1 dimeric interactions, and asymmetric units within the lattice can be fit with the same AP-1:Arf12:Nef protomer derived from the closed, trimeric AP-1 assembly (Fig. 6A), with notable exceptions, discussed below. Interactions with the membrane are conserved between the narrow and wide tube lattices with clear and obvious density for the N termini of Arf1 extending downward from the protein coat to the outer leaflet of the bilayer (Fig. 6B and fig. S4E) and contacts between the membrane and AP-1 subunits (fig. S4, C and E).
Fig. 6. Rearrangements of the lattice on wide tubes.
(A) Composite map and atomic model of the Arf1β1-linked AP-1 dimer and Arf1γ-linked dimer on wide tubes, shown from the top view. Arf1γ’, the third Arf1 molecule within an asymmetric unit on wide tubes, is shown in cyan. (B) Side view of the STA map with atomic model fit of Arf1γ′. (C) Composite map and atomic model of the Arf1γ axial interface, shown from the top view. (D) Zoom of the Arf1γ-Arf1γ’ homodimeric interface, Arf1γ:γ interface, and Arf1γ′:γ/σ1 interface. (E) Zoom of the Arf1γ′:μ1 interface. (F) Cartoon summary of the AP-1:Arf1:Nef lattice interactions on narrow and wide tubes. The left and right cartoons depict the arrangements structurally observed on tubes. The middle cartoon illustrates the differences between the two arrangements.
Symmetry expansion and focused alignment and averaging of the asymmetric unit and individual dimeric interfaces resulted in EM density maps globally resolved to 9.2 Å for the asymmetric unit, 9.8 Å for the β1:β1-juxtaposed dimer, and 11.6 Å for the γ:γ-juxtaposed AP-1 dimer (fig. S4 and table S2). At the γ:Arf1 dimeric interface, the γ subunits are no longer close enough to directly interact, due to a ~3-nm displacement of one AP-1 protomer along the tube axis (Fig. 6, A and F, and fig. S4F). Interactions between Arf1γ and the juxtaposed γ subunit stabilize these dimeric interactions (Fig. 6D). The relative angle between asymmetric units at the γ:Arf1 dimeric interface increases from 130° on narrow tubes to 150° on wide tubes (fig. S4, D versus E) but without involving substantial changes to the asymmetric unit itself (Fig. 6A). The β1:Arf1 dimeric interface is similar to the interface on narrow tubes, with the exception that the interface is straighter by 20° (Fig. 6F and fig. S4, D versus E). The asymmetric units appear to hinge at the Arf1β1:β1 interface (fig. S4, D versus E). In this arrangement, polar and electrostatic interactions are mediated between Arf1β1 D164/Y167 and μ1 R246/K400. At the homodimeric Arf1β1 interface, the contacting residues appear to be the same as those in the narrow lattice, indicating that the interactions are conserved between narrow and wide tube lattices. Contacts between Nef, cargo, AP-1, and the membrane are also unchanged.
The most prominent deviation between wide and narrow lattices is the presence of additional protein density at the axial interface between Arf1γ:γ dimer stacks (Figs. 6C versus 3A) belonging to an additional Arf1 molecule. Because of the local resolution of the maps (10 to 12 Å; fig. S4C), rigid-body modeling of Arf1 resulted in multiple Arf1 arrangements that could satisfactorily fit the density. One solution is shown for clarity (Fig. 6, A to E). The result reveals that three Arf1 and one Nef per AP-1 constitute the asymmetric unit within the lattice on wide tubes. The auxiliary Arf1, termed Arf1γ’, is positioned to simultaneously contact μ1, γ, and Arf1γ (Fig. 6, C to E). The interfacial contacts map to the same general locations as those seen in the narrow tube lattice (Figs. 3C versus 6, D and E). The one exception is that Arf1γ’ creates a previously unknown homotypic interface with Arf1γ using residues 133 to 139, which are distinct from Arf1:Arf1 dimer contacts in the narrow lattice (Fig. 6D). On the basis of the Arf1 dimers observed on membranes in this study, we revisited our previously predicted model for a hexagonal AP-1:Arf1 coat compatible with a hexagonal clathrin lattice (47). In the new model (fig. S5), the flexibility of the Arf1 contacts observed here gives rise to a mixture of pentagons and hexagons, instead of hexagons only, and is thus fully compatible with formation of a dodecahedral clathrin cage. This mixture of pentagons and hexagons results in a model for a lower density spherical coat forming the inner layer of AP-1 clathrin vesicles, whose character thus differs markedly from the clathrin-independent tubular coat structure reported here.
AP-1 tubes are stabilized by clathrin knockdown and colocalize with Nef in cells
In cells, AP-1 and Arf1 localize to tubular and vesicular membrane compartments, such as the TGN, endosomes, and CCVs, during normal cargo trafficking (35, 48–50). Nef expression promotes AP-1 localization to tubulated membranes (22, 26). As detailed above, the AP-1 lattice is stabilized by multivalent contacts. Arf1:AP-1 interactions mediate both axial and lateral contacts within the lattice, suggesting that Nef augments but is not essential for lattice formation. Thus, we hypothesized that AP-1 and Arf1 could self-assemble into tubular coats in the absence of Nef and that Nef would localize to these structures in cells. Unlike AP-3 and AP-4, nearly all reported AP-1–dependent processes are clathrin-dependent. We thus further reasoned that the tubular clathrin-independent AP-1 coat would represent a clathrin-independent intermediate preceding the formation of CCVs, not a fully clathrin-independent pathway. This hypothesis predicts that clathrin knockdown should stabilize such an intermediate, enriching the cell with AP-1–positive tubules.
AP-1 and Nef localization was monitored in HeLa cells by Airyscan confocal microscopy. AP-1 complexes were visualized by expression of a μ1A construct C-terminally tagged with green fluorescent protein (GFP) (51), and Nef was C-terminally HALO-tagged. In the absence of Nef, AP-1 localized to spherical densities throughout the cytosol and at the perinuclear region (Fig. 7A). In the presence of Nef, the number and morphology of AP-1 densities remained virtually unchanged (Fig. 7B). Nef signals partially overlapped with AP-1 density under these conditions (Fig. 7D). The AP-1 signal more strongly overlapped with the perinuclear region in Nef-expressing cells (compare Fig. 7, A versus B).
Fig. 7. Clathrin knockdown induces formation of AP-1 tubules in cellulo.
(A) Single Z-slice images of HeLa cells transfected with AP-1–GFP construct under mock or clathrin knockdown (KD) conditions, acquired using live-cell confocal microscopy. Magnified insets of the indicated area are shown on the right of each image. Scale bars, 5 μm. (B) Single Z-slice images of HeLa cells cotransfected with AP-1 GFP and NA7-Nef-HALO construct under mock or clathrin knockdown conditions, acquired using live-cell confocal microscopy. Magnified insets of the indicated area are shown on the bottom right corner of image. Scale bars, 5 μm. (C) Western blot of mock or clathrin knockdown (si cltc) HeLa cell lysate 96 hours after siRNA treatment, probed with anti-cltc and anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies. Average % of cltc knockdown ± SD shown at the bottom. (D) Quantification of AP-1 and Nef colocalization in the presence and absence of clathrin. Line indicates mean. (E) Model. Multimodal interactions account for versatile arrangements of AP-1 and Arf1 on membranes. Lattice-like AP-1:Arf1 assemblies on membrane tubes accumulate cargo in trafficking compartments. Hexagonal and pentagonal AP-1:Arf1 assemblies interface with clathrin to mobilize cargo into vesicles. Nef hijacks the AP-1:Arf1 tubular coat to sequester cargo by promoting lattice arrangements and co-opts the clathrin-like geometries to promote trafficking of cargoes to the lysosome.
We then replicated the observation that when clathrin is depleted in cells, AP-1 is enriched on tubular endosomes (4). Clathrin was depleted from cells by small interfering RNA (siRNA), and the localization of AP-1 and Nef was monitored. Under knockdown conditions, clathrin heavy-chain levels were reduced on average by >80% as determined by Western blot (Fig. 7C). AP-1 robustly localized to tubulated membranes. Clathrin knockdown increased Nef association with both punctate and tubular AP-1 structures in a statistically significant manner (Fig. 7D). This suggests that Nef preferentially associated with the tubular form of AP-1 coat that predominates in clathrin knockdown. These observations confirm that membrane tubulation is an inherent property of AP-1 that is enhanced by the removal of clathrin. These AP-1 tubules strongly colocalize with Nef when it is expressed, yet their formation does not require Nef.
DISCUSSION
The Arf-dependent APs have long been known to have clathrin-independent functions, yet the mechanism of their clathrin-independent assembly on membranes has been unknown. It was not even clear if tubular coats were an inherent property of AP complexes or required additional factors. Here, we took advantage of the ability of the HIV-1 Nef protein to overdrive AP-1 membrane recruitment in MHC-I down-regulation. We confirmed that tubular membrane coat formation is an inherent property of AP-1, requiring only Arf1 as a cofactor. We went on the determine the structure of the coat. The coat is densely packed on membranes, similar in that respect to retromer (3), leaving few gaps on the membrane surface.
AP-1:Arf1 strongly promotes formation of high curvature tubules (fig. S1E), although it can assemble on flat membranes as well. There is extensive literature showing that Arf1 associates with tubulated membranes and can, at sufficiently high concentrations, stabilize curvature and induce tubulation by itself (52–56). Isolated Arf1 has been previously shown to dimerize (56) but not to form higher assemblies on its own. Curvature induction by Arf1 in vivo therefore requires a mechanism to cluster Arf1 at the needed high local concentration. AP-1 recruitment is intimately linked to Arf1 (35, 36). AP-1 has multiple Arf1 binding sites (32, 42), which provides a means to cross-link Arf1 into larger assemblies. On the basis of their known Arf1 dependence and structural conservation, the same can be said for AP-3 (37) and AP-4 (38). Thus, cellular Arf1-localized tubules (55) most likely consist of Arf1 dimers cross-linked by AP complexes.
The structural model of the AP-1 membrane coat was built on a body of prior crystallographic and cryo-EM analysis in the absence of membranes. Some features are as anticipated from past work, while others are not. AP-1 protomer docking on membranes is essentially as predicted (32, 41, 47). The protomer docking mode is also consistent with the docking seen in reconstructions of the AP-2 complex within CCVs (57, 58). The lattice observed here is more densely packed than that of the open hexagonal AP-1:Arf1 lattice proposed to recruit clathrin (fig. S5) (47) and is incompatible with clathrin binding. Previously, the structure of a membrane-free assembly of a closed AP-1:Arf1:Nef:BST2 trimer was determined (42), in which Arf1β1, but not Arf1γ, was disposed on the membrane-binding face. In the presence of membranes and the MHC-I tail, we find here that both Arf1β1 and Arf1γ contact membranes. The closed AP-1:Arf1:Nef:BST2 trimer (42) was also incompatible with propagation into a higher-order lattice. The fundamental biological insight here is that AP-1:Arf1 can form a dense coat and thereby tubulate membranes on its own, in the absence of clathrin. The tubular structure is denser than the clathrin-associated version of the AP-1:Arf1 assembly. The high density is made possible by previously unidentified Arf1 dimer–mediated bridging, which allows for tighter AP-1 packing than is possible with the previously known Arf1 trimers.
These new data and models put the role of Nef in MHC-I sequestration and degradation into fresh perspective. The structure shows that Arf1 cross-links AP-1 laterally and axially and is sufficient on its own to bridge the tubular coat. In cells, we observe that clathrin knockdown is sufficient to induce extensive AP-1 tubular structures, whether or not Nef is present. Once present, Nef strongly colocalizes with the tubes. Nef integration into the tubules provides a mechanism for sequestration of MHC-I (19, 25–28). We observed that the Nef dileucine motif is positioned to cross-link adjacent AP-1 protomers in the tubular lattice. Nef does not require its dileucine motif to down-regulate MHC-I (20, 59, 60). The interactions between μ1, the Nef core, and the YQSA motif of MHC-I are sufficient for down-regulation from the plasma membrane without needing the dileucine motif. However, the Nef dileucine motif does increase the time that AP-1 (and AP-3) complexes reside on endosomal membranes (26, 61). This effect is consistent with the observation that Nef colocalization with AP-1 increases upon clathrin knockdown, which favors the tubular AP-1 lattice over the open clathrin-associated lattice.
Arf1 trimers were previously observed in the context of AP-1 in the absence of membranes (42, 47) and shown to form a hexagonal lattice (47). Arf1 trimer-linked COPI coats have been directly visualized on spherical membrane vesicles (62). The different geometries and interfaces formed by Arf1 in different settings suggests that the energy barriers between the different arrangements must be small. The interfacial free energy of association of each of the molecular contacts is also likely to be small, given that they are dominated by polar residues and bury modest amounts of solvent accessible surface area. We generated a model for the AP-1:Arf1 lattice rearrangements that would match the symmetry of a clathrin coat (Fig. 7E and fig. S5). Given that clathrin depletion stabilizes AP-1 tubule formation, we infer that the dense tubular coat form of AP-1 exists normally in the presence of clathrin and absence of Nef but as a transient intermediate in CCV formation. The rearrangement to a clathrin-like geometry entails breaking one set of dimeric contacts and replacing them with trimeric contacts. The polar character of the dimer contacts helps ensure these contacts are weak and can break readily when a rearrangement is needed. The idea that a dense cylindrical AP-1:Arf1 inner coat could exist as a normal intermediate in CCV formation is consistent with new thinking about the role of the COPII inner coat at endoplasmic reticulum exit sites (63). In this model, the inner coat sets the radius of membrane curvature and initiates the subsequent recruitment of clathrin. Once clathrin begins to arrive, AP-1:Arf1 rearranges into hexagons and pentagons (Fig. 7E and fig. S5), leading to CCV formation.
Our finding that the Arf1 interaction sites are largely conserved across species and between AP-1, AP-3, and AP-4 suggests that our observations are general across biology and span disparate pathways mediated by the Arf1-dependent AP complexes. We proposed above that the AP-1:Arf1 tubular coat ordinarily functions as an intermediate on the pathway to CCV assembly. This idea is supported by the increased AP-1 tubule formation upon clathrin knockdown. We proposed that HIV-1 Nef hijacks this tubular intermediate and causes it to persist for an abnormally long time to sequester MHC-I, before its eventual packaging in CCVs and sorting to the lysosome for degradation. Because AP-3 and AP-4 activities are largely (7, 8) or completely (9, 10) independent of clathrin, we predict that these two complexes self-assemble with Arf1 into stable tubular coats. For AP-3 and AP-4, these tubular entities would presumably be the cargo carriers themselves, not merely intermediates in spherical vesicle formation. While the AP-1 tubule would be terminated by the recruitment of clathrin, the AP-3 and AP-4 tubules should persist until GTP hydrolysis, catalyzed by Arf GAPs, promotes dissociation of Arf1 from the membrane.
In summary, we found that AP-1:Arf1 can form a contiguous tubular membrane coat cross-linked by Arf1 dimers, even in the absence of clathrin, and we determined the structure of the coat. This has allowed us to structurally rationalize a large body of unexplained findings on the clathrin-independent roles of AP complexes, their residence on tubular structures, and the ability of AP-1 to sequester MHC-I during Nef-dependent down-regulation.
MATERIALS AND METHODS
DNA constructs for protein expression
The His6- and glutathione S-transferase (GST)–tagged AP-1 constructs were previously described (56). For the full-length β1 AP-1 (AP-1βFL) complex, β1 (1 to 584) was replaced with full-length β1 DNA. MHC-I (338 to 365) peptide was expressed as a Tobacco etch virus protease-cleavable C-terminal fusion to GST and included a C-terminal His6 tag and N-terminal cysteine for lipopeptide coupling. NL4-3 Nef was expressed with a TEV-cleavable C-terminal His6 tag from the pST39 expression vector. Yeast NMT or human NMT1 (81 to 496) was subcloned into the pRSFDuet-1 vector. Bovine Arf1 (no tag) was subcloned into the pHis-parallel2 vector using Nde I/Xho I sites. The bovine Arf1 protein sequence is the same as that of human.
Protein purification
The AP-1 complexes were expressed in BL21 (DE3) pLysS (Promega, Madison, WI) strains and induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 20°C overnight. The cells were lysed by sonication in 50 mM tris at pH 8.0, 300 mM NaCl, 10% glycerol, 3 mM β-mercaptoethanol (β-ME), and 0.5 mM phenylmethanesulfonyl fluoride (PMSF). The clarified lysate was first purified on a Ni–nitrilotriacetic acid (NTA) column (QIAGEN, Valencia, CA). The eluate was further purified on glutathione–Sepharose 4B resin (GE Healthcare, Piscataway, NJ). After TEV cleavage at 4°C overnight, the sample was concentrated and then loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) in 20 mM tris at pH 8.0, 200 mM NaCl, and 0.3 mM tris(2-carboxyethyl)phosphine (TCEP). The sample fractions were pooled together, adjusted to 30 mM imidazole, and passed through 1 ml of glutathione–Sepharose 4B and then onto a Ni-NTA column (QIAGEN) to capture the residual GST- and His-tag fragments. The sample was adjusted to 20 mM tris at pH 8.0, 200 mM NaCl, and 0.3 mM TCEP by buffer exchange in the concentrator, supplemented to 10% glycerol, and flash-frozen for long-term storage.
Myristoylated Nef constructs were coexpressed with human NMT in BL21 (DE3) Star cells and induced with 0.3 mM IPTG at 28°C for 8 hours. The medium was supplemented with 50 μM myristic acid, solubilized in ethanol, 30 min before IPTG induction. The cell pellet was lysed by sonication and purified on a Ni-NTA column in 50 mM tris at pH 8.0, 300 mM NaCl, 20 mM imidazole, 3 mM β-ME, and 0.5 mM PMSF. Proteins were eluted with 300 mM imidazole and further purified by ion exchange (HiTrap Q HP). Enriched fractions were pooled and loaded onto a HiLoad 16/60 Superdex 75 column (GE Healthcare) in 20 mM tris at pH 8.0, 200 mM NaCl, 5 mM MgCl2, and 0.3 mM TCEP. High-purity protein fractions were pooled, and proteins were quantified by molar absorption measurements. Myristoylation was confirmed by whole-protein mass spectrometry.
Myristoylated Arf1 constructs were coexpressed with yNMT in BL21 (DE3) Star cells and induced with 0.3 mM IPTG at 25°C overnight. The medium was supplemented with 50 μM myristic acid, solubilized in ethanol, 30 min before IPTG induction. The cell pellet was lysed by sonication, clarified by centrifugation and then filtered to 0.45 μm, and passed over Q HP Sepharose resin. Myristoylated Arf1 was enriched by ammonium sulfate precipitation (35% saturation) and then applied to HiLoad 16/60 Superdex 75 column (GE Healthcare) in 20 mM tris at pH 8.0, 200 mM NaCl, 5 mM MgCl2, and 0.3 mM TCEP. Myristoylated and nonlipidated Arf1 proteins were separated by ion exchange on a monoS 10/100 GL with a linear salt gradient in MES (pH 6.0) buffer supplemented with 1 mM MgCl2 and 1 mM dithiothreitol (DTT). Proteins were buffer exchanged into 20 mM tris (pH 7.5), 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 10% glycerol with spin concentrators before storage at −80°C. Proteins were quantified by molar absorption measurements. Myristoylation was confirmed by whole-protein mass spectrometry.
MHC-I peptide was expressed in BL21 (DE3) Star cells and induced with 0.3 mM IPTG at 20°C overnight. The cell pellet was lysed by sonication and purified on a Ni-NTA column in 50 mM tris at pH 8.0, 300 mM NaCl, 20 mM imidazole, 3 mM β-ME, and 0.5 mM PMSF. The eluate was further purified on glutathione–Sepharose 4B resin (GE Healthcare, Piscataway, NJ). After TEV cleavage at 4°C overnight, the sample was recaptured on Ni-NTA resin, eluted, and then loaded onto a HiLoad 16/60 Superdex 750 column (GE Healthcare) in 10 mM tris at pH 7.0, 10 mM NaCl, 0.1 mM EDTA, and 1 mM β-ME. Pure fractions were concentrated by evaporation on the SpeedVac, then pooled, and stored at −20°C.
Lipopeptide coupling
Peptides were treated with DTT for 1 hour at 25°C before buffer exchange on a PD-10 column (Cytiva) according to the manufacturer’s instructions. Peptide concentration was quantified by molar absorption measurements. Peptide fractions were pooled and then incubated at a molar ratio of 1:1.1 (excess lipid) with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl)butyramide] (Avanti Polar Lipids) for 3 to 12 hours at 25°C with end-over-end mixing. MPB-PE was vacuum desiccated in an amber glass vial before coupling. Lipopeptides were purified from unreacted peptides by reverse-phase liquid chromatography using C18 columns as previously described (64). Lipopeptide-containing fractions were dried under vacuum, then resuspended in 2:1 chloroform/methanol, and stored at −80°C. Lipopeptide concentrations were estimated by Coomassie stain on SDS–polyacrylamide gel electrophoresis (PAGE) gels using peptide standards.
Liposome preparation
Lipids were mixed at a ratio of 52% dioleoylphosphatidylcholine, 26% palmitoyloleoylphosphatidylethanolamine, 7% 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine, 2% PI(4)P, 10% cholesterol, and 3% lipopeptide for liposome experiments. Liposomes were prepared by resuspending the lipid mix to lipid concentrations (1 mg/ml) in HKM buffer [20 mM Hepes (pH 7.4), 125 mM potassium acetate, 1 mM DTT, and 2 mM MgCl2], freeze-thawed >5 times in liquid nitrogen and 42°C water bath with intermittent vortexing, and then extruded 11 times through a 0.05-μm polycarbonate filter (Avanti Polar Lipids).
Cryo-EM/tomography sample preparation
For AP-1core cryoEM/ET samples, 2 μM AP-1core, 8 μM Arf1myr, 8 μM Nefmyr, 125 nM ARF nucleotide-binding site opener, and 200 μM GTP were incubated with extruded liposomes (0.25 mg/ml) for 1 hour at 25°C in HKM buffer before vitrification. For AP-1βFL cryoEM/ET samples, 0.5 μM AP-1βFL, 2 μM Arf1myr, 2 μM Nefmyr, 125 nM ARNO, 200 μM GTP, and 250 nM clathrin were incubated with extruded liposomes (0.125 mg/ml) for 1 hour at 25°C in HKM buffer before vitrification. Ten-nanometer bovine serum albumin–gold fiducial marker was buffer exchanged into HKM buffer and concentrated to 10× and was added 1:10 to the sample just before sample application on the grid. The reaction plus fiducial (3.5 μl) was applied to glow-discharged lacey carbon grids (200 mesh, thick; Ted Pella) and incubated at 100% relative humidity and 25°C for 1 min before double-sided blotting for 4 to 5 s and plunge freezing into liquid ethane using a Vitrobot.
Tilt series data acquisition
Bidirectional tilt series (3° increment, ± 60° tilt range) were collected from vitrified samples containing AP-1core, Arf1myr, Nefmyr, and liposomes incorporated with MHC-Icyto-lipopeptide. Data were collected on an FEI Titan Krios electron microscope operated at 300 kV using a Gatan Quantum energy filter with a slit width of 25 eV and a K3 direct detector operated in correlated-double sampling mode. The total exposure of ~120 e−/Å2 was equally distributed between 61 tilts. Three to four frame movies were acquired for each tilt. The details of data collection are given in table S1. The selection of acquisition areas was guided by suitability for high-resolution tomographic data collection (i.e., vitreous ice quality, lack of crystalline ice contaminations, and intactness of the carbon support).
Dose-symmetrical tilt series acquisition (62) was collected on cryo-preserved grids containing AP-1βFL, Arf1myr, Nefmyr, clathrin, and liposomes incorporated with MHC-Icyto-lipopeptide. Data were collected on an FEI Titan Krios electron microscope operated at 300 kV using a Gatan Quantum energy filter with a slit width of 25 eV and a K3 direct detector operated in CDS mode. The total exposure of ~120 e−/Å2 was equally distributed between 41 tilts. Three to four frame movies were acquired for each tilt. The details of data collection are given in table S1. The selection of acquisition areas was guided by suitability for high-resolution tomographic data collection (i.e., vitreous ice quality, lack of crystalline ice contaminations, and intactness of the carbon support) and obvious presence of tubulated membranes.
Tomogram reconstruction
Image preprocessing and tomogram reconstruction were performed essentially as described in (65). The IMOD v. 4.10.3 package (66) was used to align frames in raw movies and correct for detector gain. Tilt series were excluded from further analysis based on large tracking errors or significant beam-induced sample movements. Defective high-tilt images (due to tracking error, large objects like a grid bar, or contaminations coming in the field of view) were also removed before tilt series alignment. Tilt series were aligned using gold fiducial markers in the IMOD package. CTFPLOTTER (within IMOD) was used to estimate the defocus. Aligned and unbinned tilt series were low-pass–filtered according to cumulative dose (67), three-dimensional (3D) contrast-transfer function-corrected by phase flipping using strip widths of 15 nm, and reconstructed into tomograms by back projection using NovaCTF (68). Reconstructed tomograms were binned by factors of 2 and 4 using antialiasing in IMOD. Bin4 tomograms were Simultaneous Iterative Reconstruction Technique-like filtered and used for tube annotation in Dynamo (69). Tomograms were denoised with IsoNet for illustration purposes only (70).
Subtomogram averaging
Subtomogram alignment and averaging were done in Dynamo (1.1.532). MATLAB scripts were adapted and/or written in-house. Centers of coated tubules were manually traced in bin4, filtered tomograms, and their diameters were recorded in Dynamo (1.1.532). All tubules were annotated regardless of apparent diameter. The positions of subtomograms were defined on the surface of tubes with uniform radial sampling and uniform separation between rings. Initial subtomogram orientations were set to be normal to the membrane surface. The full dataset of annotated tubes was partitioned into 10-nm windows (10 to 20 nm, 20 to 30 nm, 30 to 40 nm, etc.), and each subset was individually processed to identify correlations in AP-1 lattice structure and tube diameter.
For each subset, 2000 random subtomograms were extracted from bin4, unfiltered tomograms and averaged according to their initial orientations, producing an initial reference. The subtomograms were then aligned to this reference, allowing both shifts and angular search. A saddle-shaped mask was applied, passing through the protein layer and the membrane. Eight iterations of such alignment were performed with an angular search increment of 20° to 2° and an appropriate low-pass filter. A second round of alignments was performed using two noisy versions of the resulting average plus low-pass filtering to parse γ-γ lattice–centered subtomograms and β1-β1 lattice–centered subtomograms. The resulting two averages were shifted and rotated to more precisely center the γ-γ or β1-β1 tracks before further processing.
The resulting averages (γ-γ–centered lattice or β1-β1–centered lattice) were used as templates to align the complete 10-nm window datasets, using alignment and sampling parameters as above. Overlapping subtomograms resulting from oversampling at the initial extraction stage were removed by selecting the subtomogram with the highest cross-correlation score within a distance threshold equivalent to a third of the box size. Subtomograms were then split into odd and even half datasets for further processing. Subsequent alignments were performed independently on the odd and even half-sets. The search space and angular increments were gradually decreased, and the low-pass filter was gradually moved toward a higher resolution. After each round of alignments, subtomogram cross-correlation values were reweighted as a function of the θ Euler angle, as in (65), on a tube-by-tube basis. Cross-correlation cutoff values were determined from visual inspection of the subtomogram real-space coordinates colored by cross-correlation values using the chimera PlaceObject plugin. Thresholds were applied globally to both half-sets. Subvolumes located close to the edge of tomograms were also removed. At the end of each iteration, the resolution of a given structure was estimated by Fourier shell correlation of the independent half-maps in Dynamo.
Subboxing was performed in Dynamo (1.1.532). Coordinates for individual dimers or specific interfaces mapped to positions on the γ-γ centered lattice or β1-β1 centered lattice and used extract to subtomograms, initially from bin4 tomograms. Subtomogram alignment and averaging of each subboxed region were performed as described above. The global resolution of each EM map was estimated in Relion-3.1 (71). Local resolution was measured using relion_postprocess from the Relion 3.1 package (71). The local resolution ranges for each map are listed in table S2. The reported global resolution is based on FSC 0.143 values. Final half-maps were cropped to remove density corresponding to areas residing outside the mask used for subtomogram alignment. The final sharpened maps were prepared using local resolution filtering and denoising implemented in Relion 3.1.
Model building
Rigid-body fitting and molecular visualization were performed in Chimera (1.14) and ChimeraX (1.3) (72). To build models of the asymmetric unit on narrow tubes, a single protomer from the trimeric AP-1:Arf1:BST2-Nef (PDB: 6CM9) assembly was fit to the STA map. To model the MHC-Icyto peptide bound to Nef, the crystal structure of μ1:Nef-MHC-I (PDB: 4EN2) was fit to the same STA map, and a composite atomic model was created by replacing the MHC-I peptide and Nef of 4EN2 with the corresponding models in 6CM9. To model the N terminus of Arf1 (2 to 16), the full-length, human Arf1 model produced by AlphaFold2 (UniProt: P84077) was aligned to Arf1γ in the new composite model, and the N-terminal residues were manually adjusted to fit the STA map density in ISOLDE. The modeled N terminus was then cropped and stitched to the Arf1γ from 6CM9 in Coot (version 0.9.1). The full-length modeled Arf1 replaced Arf1β1 and Arf1γ models in the new composite structure. To build the asymmetric unit on wide tubes, the asymmetric unit from narrow tubes was fit to the STA map corresponding to the asymmetric unit on wide tubes. One additional Arf1 molecule from the narrow asymmetric unit was manually placed in the density associated with Arf1γ′. Composite maps were generated for visualization purposes only. Overlapping regions of the γ-γ and β1-β1 lattices were overlaid and combined in Chimera (1.14) or ChimeraX (1.3) using the vop command. Helical parameters for the lattice were manually calculated using the composite maps. Models were real-space–refined against merged, locally filtered maps by global minimization in Phenix (1.20-4459) and global FSC 0.143 as the resolution cutoff (73).
Modeling the open AP-1:Arf1 lattice
Previously, a model of an open AP-1:Arf1 hexagonal lattice compatible with a hexagonal clathrin coat was generated by combining open AP-1:Arf trimers (56) and crystal structure of an Arf1β-linked dimer (32). We revisited this model on the basis of the membrane-associated Arf1 dimers visualized in this study. Assembly of AP-1:Arf1 trimers linked by “narrow” tube Arf1γ:γ dimeric interfaces results in a pentameric arrangement of AP-1 molecules that matches the spatial distribution of clathrin hubs at pentagonal faces of clathrin cages (fig. S5). Assembly with Arf1γ:γ dimeric contacts like those in the “wide” tubular lattice results in a flatter and wider arrangement of AP-1 molecules such that a hexagon results instead (fig. S5). The binding site for the third Arf1 molecule remains intact as observed in the wide tube lattice and the dimensions of the hexagon again match the dimensions of the clathrin hexagonal face (fig. S5).
Sequence alignments
Primary sequences encoding Arf and AP proteins were aligned using the COBALT, Clustal Omega, and/or Tcoffee/Mcoffee/Espresso Server. Alignments were formatted in BoxShade.
DNA plasmids and siRNA for cell imaging
The pcDNA5 NA7-Nef-HALO vector was generated by fusing the codon-optimized NA7-Nef sequence (protein ID: ABB51086.1), SGGTG linker–HALO tag DNA sequence, and the pcDNA5 vector backbone via Gibson assembly. The pCI NL4-3-Nef-HALO vector was generated by fusing the pCI NL4-3 Nef and STSGGTG linker–HALO tag DNA sequence via Gibson assembly. pEGFPN1 AP-1-GFP was generated by inserting the Hs AP1M1 sequence with GSGS linker into a pEGFPN1 backbone through double restriction digest with Xho I and Hind III. Clathrin knockdown was conducted with SMARTPool ON-TARGETplus Human CLTC siRNA purchased from Dharmacon.
Tissue culture
All live-cell imaging was conducted using HeLa cells obtained from the University of California, Berkeley Cell Culture Facility. Cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium) F12 medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (PenStrep) and grown at 37°C and 5% CO2. Twenty-four hours before live-cell imaging, cells were plated onto eight-well chambered cover glass (Cellvis C8-1.5H-N) and incubated with 100 nM JF635 dye. Before imaging, cells were transferred to imaging medium consisting of DMEM F12–phenol red supplemented with 10% FBS and PenStrep. Cells were allowed to recover for at least 10 min in the incubator before movie acquisition.
DNA transfection and siRNA knockdown
For knockdowns, HeLa cells were cultured to ~60% confluency in six-well plates and transfected with Lipofectamine 3000 (Invitrogen L3000) following directions provided by the manufacturer. The final concentration of siRNA used for knockdown was 20 nM. After 1 day, cells were split into 12-well plates for transfection with DNA plasmids. The following day, 1 μg of each DNA plasmid (AP-1-GFP, NA7-Nef-HALO, and/or NL4-3-Nef-HALO) was transfected as desired using the TransIT-LT1 reagent (MIR2304) following directions provided by the manufacturer. Last, cells were plated onto the eight-well chambered cover glass for imaging.
Cell lysis and Western blots
HeLa cells, grown in 12-well plates, were washed with phosphate-buffered saline (PBS), trypsinized, and resuspended in 24 μl of lysis buffer, consisting of 50 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 1% NP40, PhosSTOP phosphatase inhibitor (Roche, 04906845001), and protease inhibitor (Roche, 1183617001). Cells were lysed for 15 min on ice with gentle agitation every 5 min. Lysed cells were then centrifuged at 4°C and 13,000 rpm for 15 min, and 18 μl of the supernatant was collected as lysate. The lysate was mixed with Laemmli reducing buffer with 5% β-ME, and 10 μl of the final solution was loaded onto a 10% acrylamide gel for SDS-PAGE analysis. The SDS-PAGE gel was transferred onto nitrocellulose membrane for immunoblotting via wet transfer. Blots were blocked with 5% milk in tris-buffered saline (TBS) and incubated with primary antibody. The antibodies used for Western blotting were anti-cltc [1:1000 in 0.1% Tween 20 and 0.05% milk in PBS, room temperature (RT) for 1 hour; Abcam, ab21679] and anti–glyceraldehyde-3-phosphate dehydrogenase [1:10,000 in TBS with Tween 20 (TBST), RT for 1 hour; Abcam, ab9485]. Blots were then incubated with IRDye 800/680–conjugated antibodies (1:10,000 in 5% milk TBST, RT 1 hour, 926-32212 926-68071) and imaged by the LICOR Odyssey scanner.
Cell imaging
Live-cell imaging was conducted on a Zeiss LSM900 with Airyscan 2.0 detection in SR-2Y mode, using the ZEN (blue edition) software. All imaging was conducted in an incubation chamber at 37°C and 5% CO2. Ten-micrometer Z-stack movies with 140-nm intervals were acquired for all analyzed cells. During multichannel imaging, all channels were frame-sequentially acquired before moving to the next Z slice to minimize cross-talk and time offset. Images were batch processed by ZEN Blue software for Airyscan reconstruction using filter strength setting of 5 and 3D processing.
Image analysis
To test for colocalization between Nef and AP-1 signal, both channels from the same slice in a Z stack was analyzed. Both channels were first background corrected using the 15-pixel median filtering as described above and thresholded using the “Auto Threshold” Otsu and Li thresholds for the AP-1 and Nef channels, respectively, in ImageJ. Particles less than 0.10 μm2 were filtered out. AP-1 signal was then classified as “spheroid” if particles had circularity between 0.75 and 1.00 or as “tubule” if particles had circularity between 0 and 0.75 according to the “Analyze Particles” function. Using the thresholded images of AP-1 and Nef, the Manders correlation coefficient was calculated for each type of AP-1 structure using the JACoP plugin in ImageJ (74). Specifically, the fraction of AP-1 pixels overlapping with Nef pixels was reported for colocalization. To confirm that colocalization of AP-1 and Nef was not random, the Van Steensel CCF function was generated using JACoP as well. The ImageJ scripts used for AP-1 tubule analysis can be found at https://github.com/yuichiro-iwamoto/AP1_tubule_quantification.
Statistical analysis
Statistical analysis of AP-1/Nef colocalization was performed using the GraphPad Prism 9 software. In all cases, the Kruskal-Wallis test was used to make one-way comparisons across all conditions. Not significant (ns), P > 0.05; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Acknowledgments
We thank J. Hutchings and G. Zanetti for providing a dataset for workflow validation, D. Toso and J. Remis for cryo-EM operational support, C. Buffalo for critical reading of the manuscript and for producing Fig. 7E, and D. Drubin for use of the Zeiss LSM900 microscope.
Funding: This research was supported by NIH grants F32 AI152971 (R.M.H.), R01 AI120691 (J.H.H.), and P50 AI150476 (J.H.H.).
Author contributions: R.M.H. and J.H.H. conceived and designed research; R.M.H., Y.I., D.T., and X.R. carried out research; J.H.H. supervised research; R.M.H. and J.H.H. wrote the first draft, and all authors edited the manuscript.
Competing interests: J.H.H. is a cofounder and shareholder of Casma Therapeutics and receives research funding from Casma Therapeutics, Genentech, and Hoffmann-La Roche. The other authors declare that they have no competing interests.
Data and materials availability: Coordinates and density maps have been deposited in the RCSB and EMDB, respectively, with accession numbers RCSD 7UX3, 8D4C, 8D4D, 8D4E, 8D4F, and 8D4G, and EMDB EMD-26853, EMD-27181, EMD-27182, EMD-27183, EMD-27184, EMD-27185, EMD-27186, EMD-27187, EMD-27188, EMD-27189, EMD-27190, and EMD-27191. The scripts for tubule quantification have been deposited with Dryad at https://doi.org/10.6078/D1S72k. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Tables S1 to S4
REFERENCES AND NOTES
- 1.Dell’Angelica E. C., Bonifacino J. S., Coatopathies: Genetic disorders of protein coats. Annu. Rev. Cell Dev. Biol. 35, 131–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faini M., Beck R., Wieland F. T., Briggs J. A., Vesicle coats: Structure, function, and general principles of assembly. Trends Cell Biol. 23, 279–288 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Kovtun O., Leneva N., Bykov Y. S., Ariotti N., Teasdale R. D., Schaffer M., Engel B. D., Owen D. J., Briggs J. A. G., Collins B. M., Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janvier K., Bonifacino J. S., Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell 16, 4231–4242 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V., Deschamps C., Raposo G., Benaroch P., Benmerah A., Chavrier P., Niedergang F., AP-1 and ARF1 control endosomal dynamics at sites of FcR mediated phagocytosis. Mol. Biol. Cell 18, 4921–4931 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haberg K., Lundmark R., Carlsson S. R., SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J. Cell Sci. 121, 1495–1505 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Vowels J. J., Payne G. S., A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 17, 2482–2493 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J., Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164, 1065–1076 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirst J., Bright N. A., Rous B., Robinson M. S., Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell 10, 2787–2802 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dell’Angelica E. C., Mullins C., Bonifacino J. S., AP-4, a novel protein complex related to clathrin adaptors. J. Biol. Chem. 274, 7278–7285 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Jackson L. P., Kümmel D., Reinisch K. M., Owen D. J., Structures and mechanisms of vesicle coat components and multisubunit tethering complexes. Curr. Opin. Cell Biol. 24, 475–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Weijer M. L., Luteijn R. D., Wiertz E. J., Viral immune evasion: Lessons in MHC class I antigen presentation. Semin. Immunol. 27, 125–137 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Collins D. R., Collins K. L., HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLOS Pathog. 10, e1003851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekaban G. A., Dikeakos J. D., HIV-I Nef inhibitors: A novel class of HIV-specific immune adjuvants in support of a cure. AIDS Res. Ther. 14, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buffalo C. Z., Iwamoto Y., Hurley J. H., Ren X., How HIV nef proteins hijack membrane traffic to promote infection. J. Virol. 93, e01322-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauter D., Kirchhoff F., Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev. 40, 3–12 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Ramirez P. W., Sharma S., Singh R., Stoneham C. A., Vollbrecht T., Guatelli J., Plasma membrane-associated restriction factors and their counteraction by HIV-1 accessory proteins. Cell 8, 1020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staudt R. P., Alvarado J. J., Emert-Sedlak L. A., Shi H., Shu S. T., Wales T. E., Engen J. R., Smithgall T. E., Structure, function, and inhibitor targeting of HIV-1 Nef-effector kinase complexes. J. Biol. Chem. 295, 15158–15171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Gall S., Erdtmann L., Benichou S., Berlioz-Torrent C., Liu L., Benarous R., Heard J. M., Schwartz O., Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8, 483–495 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Roeth J. F., Williams M., Kasper M. R., Filzen T. M., Collins K. L., HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol. 167, 903–913 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasper M. R., Roeth J. F., Williams M., Filzen T. M., Fleis R. I., Collins K. L., HIV-1 Nef disrupts antigen presentation early in the secretory pathway. J. Biol. Chem. 280, 12840–12848 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Lubben N. B., Sahlender D. A., Motley A. M., Lehner P. J., Benaroch P., Robinson M. S., HIV-1 Nef-induced down-regulation of MHC class I requires AP-1 and clathrin but not PACS-1 and is impeded by AP-2. Mol. Biol. Cell 18, 3351–3365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wonderlich E. R., Leonard J. A., Kulpa D. A., Leopold K. E., Norman J. M., Collins K. L., ADP ribosylation factor 1 activity is required to recruit AP-1 to the major histocompatibility complex class I (MHC-I) cytoplasmic tail and disrupt MHC-I trafficking in HIV-1-infected primary T cells. J. Virol. 85, 12216–12226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer M. R., Wonderlich E. R., Roeth J. F., Leonard J. A., Collins K. L., HIV-1 Nef targets MHC-I and CD4 for degradation via a final common beta-COP-dependent pathway in T cells. PLOS Pathog. 4, e1000131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blagoveshchenskaya A. D., Thomas L., Feliciangeli S. F., Hung C. H., Thomas G., HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111, 853–866 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Janvier K., Craig H., Hitchin D., Madrid R., Sol-Foulon N., Renault L., Cherfils J., Cassel D., Benichou S., Guatelli J., HIV-1 Nef stabilizes the association of adaptor protein complexes with membranes. J. Biol. Chem. 278, 8725–8732 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Dirk B. S., Pawlak E. N., Johnson A. L., van Nynatten L. R., Jacob R. A., Heit B., Dikeakos J. D., HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci. Rep. 6, 37021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavares L. A., de Carvalho J. V., Costa C. S., Silveira R. M., de Carvalho A. N., Donadi E. A., daSilva L. L. P., Two functional variants of AP-1 complexes composed of either γ2 or γ1 subunits are independently required for major histocompatibility complex class I downregulation by HIV-1 Nef. J. Virol. 94, e02039-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen D. J., Collins B. M., Evans P. R., Adaptors for clathrin coats: Structure and function. Annu. Rev. Cell Dev. Biol. 20, 153–191 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Jackson L. P., Kelly B. T., McCoy A. J., Gaffry T., James L. C., Collins B. M., Höning S., Evans P. R., Owen D. J., A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141, 1220–1229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canagarajah B. J., Ren X., Bonifacino J. S., Hurley J. H., The clathrin adaptor complexes as a paradigm for membrane-associated allostery. Protein Sci. 22, 517–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren X., Farias G. G., Canagarajah B. J., Bonifacino J. S., Hurley J. H., Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 152, 755–767 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly B. T., Graham S. C., Liska N., Dannhauser P. N., Höning S., Ungewickell E. J., Owen D. J., Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science 345, 459–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L., Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114, 299–310 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Stamnes M. A., Rothman J. E., The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell 73, 999–1005 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Traub L. M., Ostrom J. A., Kornfeld S., Biochemical dissection of AP-1 recruitment onto Golgi membranes. J. Cell Biol. 123, 561–573 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi C. E., Dell’Angelica E. C., Bonifacino J. S., ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol. 142, 391–402 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boehm M., Aguilar R. C., Bonifacino J. S., Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs). EMBO J. 20, 6265–6276 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noviello C. M., Benichou S., Guatelli J. C., Cooperative binding of the class I major histocompatibility complex cytoplasmic domain and human immunodeficiency virus type 1 Nef to the endosomal AP-1 complex via its mu subunit. J. Virol. 82, 1249–1258 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia X., Singh R., Homann S., Yang H., Guatelli J., Xiong Y., Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat. Struct. Mol. Biol. 19, 701–706 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia X., Weber E., Tokarev A., Lewinski M., Rizk M., Suarez M., Guatelli J., Xiong Y., Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. eLife 3, e02362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris K. L., Buffalo C. Z., Stürzel C. M., Heusinger E., Kirchhoff F., Ren X., Hurley J. H., HIV-1 nefs are cargo-sensitive AP-1 trimerization switches in tetherin downregulation. Cell 174, 659–671.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heldwein E. E., Macia E., Wang J., Yin H. L., Kirchhausen T., Harrison S. C., Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. U.S.A. 101, 14108–14113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grzesiek S., Bax A., Clore G. M., Gronenborn A. M., Hu J. S., Kaufman J., Palmer I., Stahl S. J., Wingfield P. T., The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 3, 340–345 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Bresnahan P. A., Yonemoto W., Ferrell S., Williams-Herman D., Geleziunas R., Greene W. C., A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 8, 1235–1238 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Janvier K., Kato Y., Boehm M., Rose J. R., Martina J.´. A., Kim B. Y., Venkatesan S., Bonifacino J. S., Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes. J. Cell Biol. 163, 1281–1290 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Q. T., Ren X., Zhang R., Lee I. H., Hurley J. H., HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science 350, aac5137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson M. S., Cloning and expression of gamma-adaptin, a component of clathrin-coated vesicles associated with the Golgi apparatus. J. Cell Biol. 111, 2319–2326 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Futter C. E., Gibson A., Allchin E. H., Maxwell S., Ruddock L. J., Odorizzi G., Domingo D., Trowbridge I. S., Hopkins C. R., In polarized MDCK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. J. Cell Biol. 141, 611–623 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang F., Nesterov A., Carter R. E., Sorkin A., Trafficking of yellow-fluorescent-protein-tagged mu1 subunit of clathrin adaptor AP-1 complex in living cells. Traffic 2, 345–357 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Guo X., Mattera R., Ren X., Chen Y., Retamal C., González A., Bonifacino J. S., The adaptor protein-1 μ1B subunit expands the repertoire of basolateral sorting signal recognition in epithelial cells. Dev. Cell 27, 353–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krauss M., Jia J. Y., Roux A., Beck R., Wieland F. T., de Camilli P., Haucke V., Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J. Biol. Chem. 283, 27717–27723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beck R., Prinz S., Diestelkötter-Bachert P., Röhling S., Adolf F., Hoehner K., Welsch S., Ronchi P., Brügger B., Briggs J. A. G., Wieland F., Coatomer and dimeric ADP ribosylation factor 1 promote distinct steps in membrane scission. J. Cell Biol. 194, 765–777 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., Wieland F., Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. U.S.A. 105, 11731–11736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bottanelli F., Kilian N., Ernst A. M., Rivera-Molina F., Schroeder L. K., Kromann E. B., Lessard M. D., Erdmann R. S., Schepartz A., Baddeley D., Bewersdorf J., Toomre D., Rothman J. E., A novel physiological role for ARF1 in the formation of bidirectional tubules from the Golgi. Mol. Biol. Cell 28, 1676–1687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diestelkoetter-Bachert P., Beck R., Reckmann I., Hellwig A., Garcia-Saez A., Zelman-Hopf M., Hanke A., Nunes Alves A., Wade R. C., Mayer M. P., Wieland F., Structural characterization of an Arf dimer interface: Molecular mechanism of Arf-dependent membrane scission. FEBS Lett. 594, 2240–2253 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Kovtun O., Dickson V. K., Kelly B. T., Owen D. J., Briggs J. A. G., Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles. Sci. Adv. 6, eaba8381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paraan M., Mendez J., Sharum S., Kurtin D., He H., Stagg S. M., The structures of natively assembled clathrin-coated vesicles. Sci. Adv. 6, eaba8397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenberg M., DeTulleo L., Rapoport I., Skowronski J., Kirchhausen T., A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8, 1239–1242 (1998). [DOI] [PubMed] [Google Scholar]
- 60.Mangasarian A., Piguet V., Wang J. K., Chen Y. L., Trono D., Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J. Virol. 73, 1964–1973 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coleman S. H., Hitchin D., Noviello C. M., Guatelli J. C., HIV-1 Nef stabilizes AP-1 on membranes without inducing ARF1-independent de novo attachment. Virology 345, 148–155 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Dodonova S. O., Aderhold P., Kopp J., Ganeva I., Röhling S., Hagen W. J. H., Sinning I., Wieland F., Briggs J. A. G., 9Å structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments. eLife 6, e26691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melero A., Boulanger J., Kukulski W., Miller E. A., Ultrastructure of COPII vesicle formation characterised by correlative light and electron microscopy. bioRxiv 10.1101/2022.03.13.484130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nickel W., Wieland F. T., [41] Receptor-dependent formation of COPI-coated vesicles from chemically defined donor liposomes. Methods Enzymol. 329, 388–404 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Hutchings J., Stancheva V., Miller E. A., Zanetti G., Subtomogram averaging of COPII assemblies reveals how coat organization dictates membrane shape. Nat. Commun. 9, 4154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant T., Grigorieff N., Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turoňová B., Schur F. K. M., Wan W., Briggs J. A. G., Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4Å. J. Struct. Biol. 199, 187–195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castano-Diez D., Kudryashev M., Arheit M., Stahlberg H., Dynamo: A flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178, 139–151 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Liu Y.-T., Zhang H., Wang H., Tao C.-L., Bi G.-Q., Zhou Z. H., Isotropic reconstruction of electron tomograms with deep learning. bioRxiv 10.1101/2021.07.17.452128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolte S., Cordelières F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Tables S1 to S4