Abstract
The level of hepatitis B virus (HBV) DNA in serum reflects the replicative activity of HBV. To compare serum HBV DNA levels in different states of hepatitis B, 47 sera of patients with HBeAg-positive chronic hepatitis B, 4 sera of patients with HBeAg-negative chronic hepatitis B, 40 samples of patients after HBeAg seroconversion during alpha interferon treatment, 57 sera of inactive HBsAg carriers, and 42 sera of patients who had recovered from chronic hepatitis B more than 12 months prior to blood collection were checked for the presence of HBV DNA with the Amplicor HBV Monitor Test. In patients with HBeAg-positive chronic hepatitis B, the median of serum HBV DNA levels (8.3 × 108 copies/ml) was significantly higher than that for patients after HBeAg seroconversion (6.2 × 103 copies/ml) and than that for inactive HBsAg carriers (5.6 × 103 copies/ml). None of the patients who had recovered from hepatitis B had detectable HBV DNA in serum. Quantitative PCR proved to be a valuable tool for identification of different states of HBV infection. This technique was found to be a good method for determination of serum HBV DNA levels both for patients with HBeAg seroconversion and for inactive carriers who showed low viremia not detectable by conventional hybridization assays.
Monitoring of hepatitis B virus (HBV) DNA in serum has become the standard method of assessing the replicative activity of HBV. The clinical importance of this method has been reported for the assessment, management, and antiviral treatment of patients with chronic HBV infection (8, 15). Hybridization assays for detection of serum HBV DNA have been used since the beginning of the 1980s. Introduction of PCR for detection of serum HBV DNA resulted in a significant improvement of sensitivity over that of hybridization techniques (1, 2, 4). Home-brew PCR-based assays employed radioactive or nonradioactive hybridization techniques (7, 20). Those assays, however, lack standardization and reproducibility, as has been shown by the results of the EUROHEP proficiency study, where more than 50% of participating laboratories failed either the sensitivity or the specificity criteria (14, 20).
The standardized quantitative Amplicor HBV Monitor Test (Roche Diagnostic Systems, Pleasanton, Calif.), which has been introduced recently, is based on a previously described quantitative PCR with colorimetric detection (7, 16). This assay includes a simplified sample preparation procedure, a PCR amplification, a hybridization step, and colorimetric detection on a microwell plate. The Amplicor HBV Monitor Test proved to be relatively easy to use, and the whole procedure can be carried out in 7 h (3, 6). The lower detection limit of this assay was found to be 102 to 103 HBV DNA copies/ml, thus making the assay 1,000 to 10,000 times more sensitive than a widely used commercially available hybridization assay (6). According to the manufacturer's package insert, the Amplicor HBV Monitor Test shows linearity from 4.0 × 102 (lower detection limit) to 4.0 × 107 HBV DNA copies per ml. The reproducibility data of the Amplicor HBV Monitor Test indicate that this test is very reliable, with coefficients of variation being mostly below 20% (6).
The aim of this study was to compare serum HBV DNA levels in different states of hepatitis B. Furthermore, sera of patients collected more than 12 months after remission of hepatitis B were tested for the presence of HBV DNA. All HBV DNA levels were measured with the Amplicor HBV Monitor Test.
A total of 190 sera from 92 (female/male ratio, 43/49; age range, 12 to 68 years) patients were studied. The serological profile of all patients is shown in Table 1. Patients who were repeat tested showed no change in their serological profile. Forty-seven serum samples were collected from 25 patients (22 of them repeat tested within 6 months) with chronic hepatitis B (CHB; group 1a), and four sera were collected from four patients with HBV e antigen (HBeAg)-negative CHB (group 1b). In all patients of groups 1a and 1b, serum alanine aminotransferase levels were >1.5 times the upper limit of normal range and none of the patients received anti-HBV treatment. Forty serum samples were obtained from five patients after HBeAg seroconversion during an alpha interferon treatment study (group 2). For all of them, serum for determination of the HBV DNA level was collected monthly. For participation in the treatment study, HBeAg positivity had been one of the inclusion criteria. All patients were treated at the Departments of Pediatrics and Internal Medicine, Karl Franzens University Graz. The study protocol was approved by the local Ethics Committee, and all patients or their legal representatives gave informed consent. Finally, 57 serum samples were obtained from 37 inactive carriers (20 of them repeat tested within 6 months), and 42 serum samples were collected from 21 patients (all of them repeat tested within 6 months) who had recovered from CHB.
TABLE 1.
Serological profile of patient groups
| Parameter | Group 1ab | Group 2c | Group 3d | Group 4e |
|---|---|---|---|---|
| HBsAg | Positive | Positive | Positive | Negative |
| Anti-HBs | Negative | Negative | Negative | Positive |
| Anti-HBc | Positive | Positive | Positive | Positive |
| HBeAg | Positivea/negativeb | Negative | Negative | Negative |
| Anti-HBe | Negative | Positive | Positive | Positive |
Patients with HBeAg-positive CHB.
Patients with HBeAg-negative CHB.
Patients after HBeAg seroconversion during an alpha interferon treatment study.
Inactive carriers.
Patients who had recovered from CHB more than 12 months prior to blood collection.
Within 60 min of blood draw, blood collection tubes were centrifuged at 15,000 × g for 20 min at room temperature. After centrifugation, aliquots were prepared and were either tested within 6 h or immediately frozen at −70°C until testing.
Serological parameters included HBV surface antigen (HBsAg), anti-HBs, anti-HBc, HBeAg, and anti-HBe, which were measured by microparticle enzyme immunoassay (Abbott, North Chicago, Ill.).
Serum HBV DNA levels were tested with the Amplicor HBV Monitor Test, according to the manufacturer's package insert instructions. Briefly, virus was collected from 50 μl of serum by polyethylene glycol 8000 precipitation, lysed by dilute sodium hydroxide, and neutralized. For PCR, a set of primers which allows amplification of a 104-bp fragment of the pre-C and C region of the HBV genome was used. The Master Mix contained AmpErase (uracil-N-glycosylase) and internal standard (IS) DNA. Following amplification, two aliquots of the PCR mixture were pipetted into separate wells of a streptavidin-coated microwell plate. The biotinylated amplification products were allowed to bind to the streptavidin. After this, the DNA was denatured, and the nonbiotinylated DNA strand was removed by a washing step. To one well, the dinitrophenyl (DNP)-labeled probe, specific for the target HBV sequence, was added, and to the other well, a DNP-labeled probe specific for the IS DNA was added. Following hybridization to the immobilized amplification product, the DNP moiety of the probes was colorimetrically detected using an anti-DNP–alkaline phosphatase conjugate and p-nitrophenylphosphate substrate. The amount of HBV DNA in each specimen was calculated from the ratio of the optical density for the HBV-specific well to the optical density for the IS-specific well. The number of HBV DNA copies was calculated from a standard curve (0, 4.0 × 102, 4.0 × 103, 4.0 × 104, 4.0 × 105, and 4.0 × 106 copies/ml) prepared from each amplification run. If the result exceeded 4.0 × 107 HBV DNA copies/ml, serum was diluted and retested.
For statistical analysis, medians of each of the patient groups were calculated. For comparative analysis, the Mann-Whitney U test was used. A finding of P < 0.05 was considered statistically significant.
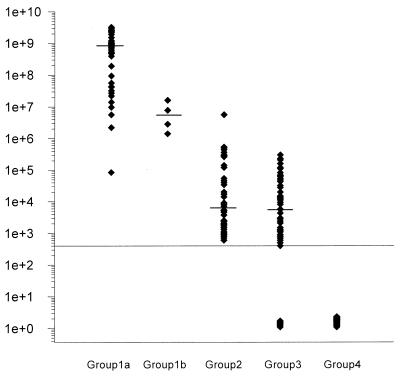
Serum HBV DNA levels of patient groups 1 to 4 are shown in Fig. 1. In patients with HBeAg-positive CHB (group 1a), the median serum HBV DNA load was 8.3 × 108 copies/ml (range, 8.4 × 104 to 3.2 × 109 copies/ml). The serum HBV DNA load in patients with HBeAg-negative CHB (group 1b) was 1.6 × 107, 7.7 × 106, 2.8 × 106, and 1.4 × 106 copies/ml. In patients after HBeAg seroconversion during the alpha interferon treatment study (group 2), the median serum HBV DNA load was 6.2 × 103 copies/ml (range, 6.0 × 102 to 5.6 × 106). In only two of the samples collected from patients with HBeAg-positive CHB, HBV DNA levels were lower (8.4 × 104 and 2.2 × 106 copies/ml) than the highest level measured in the group of patients after HBeAg seroconversion during the alpha interferon treatment study (5.6 × 106 copies/ml), and one sample showed an identical result. In inactive carriers (group 3), the median serum HBV DNA load was 5.6 × 103 copies/ml. Seven samples tested below the detection limit, and the highest HBV DNA level found in this group was 3.0 × 105 copies/ml. Only the lowest HBV DNA level (8.4 × 104 copies/ml) measured in one of the patients with CHB was within the range of HBV DNA levels measured in inactive carriers. In all patients who had recovered from CHB more than 12 months prior to blood collection (group 4), HBV DNA load was found to be below the detection limit of the employed molecular assay.
FIG. 1.
Serum HBV DNA levels (copies per milliliter) in patients with HBeAg-positive CHB (group 1a), in patients with HBeAg-negative CHB (group 1b), in patients after HbeAg seroconversion during alpha interferon treatment (group 2), in inactive carriers (group 3), and in patients who had recovered from CHB more than 12 months prior to blood collection (group 4).
In patients with HBeAg-positive CHB, median serum HBV DNA load was significantly higher than that in patients after HBeAg seroconversion (P < 0.001) and in inactive carriers (P < 0.001). Median serum HBV DNA loads of patients after HBeAg seroconversion and of inactive carriers were comparable.
The molecular assay employed in this study proved to be suitable for a laboratory specializing in molecular diagnostics. The whole procedure could be carried out in 7 h. The assay, however, lacked automation of the sample preparation, hybridization, and detection steps, requiring specialized personnel and limiting the number of samples processed in parallel. An automated version has recently been evaluated and found to be highly sensitive and reproducible (13).
Because of the improved sensitivity of PCR assays, it is possible to distinguish between inactive carrier and CHB states. In this study, serum HBV DNA levels in patients with HBeAg-positive CHB were found to be significantly higher than those in patients after HBeAg seroconversion during an alpha interferon treatment study and in inactive carriers. The median serum HBV DNA load in patients with HBeAg-negative CHB was lower than that in patients with HBe-positive CHB, but this did not have statistical importance because of the small number of samples. Results of the present study are in accordance with those of a recent retrospective study, which concluded that the asymptomatic carrier and CHB states could be distinguished by a serum HBV DNA concentration of 105 copies/ml (12). In this study, the borderline between patients with CHB on one side and both patients after HBeAg seroconversion and inactive carriers on the other was found to be at a serum HBV DNA concentration of 106 copies/ml.
It may, however, be very difficult to identify patients who may benefit from antiviral therapy and to evaluate the success of treatment. Before interferon treatment, no difference between HBV DNA levels of responders and nonresponders could be observed (11). If, after termination of interferon treatment, serum HBV DNA concentration was found to be less than 104 copies/ml, hepatitis did not occur thereafter (12). However, even after loss of HBsAg and anti-HBs seroconversion, which are considered absolute markers of viral clearance, residual viremia was detected by PCR for several patients for a maximum of 12 months following treatment termination (5, 9, 18). Similar observations were made for patients with spontaneous loss of HBsAg (9, 10, 19). Those results, however, were obtained by home-brew PCR assays and may well include false positives because of contamination problems. On the other hand, HBV may never be completely cleared. This has recently been supported indirectly by another study in which not only did a patient who had recovered from acute hepatitis B 23 years earlier still have circulating HBV-specific cytotoxic T cells but these cells displayed markers of activation showing evidence of recent contact with the virus (17). In this study, serum HBV DNA of all patients who had recovered from CHB more than 12 months prior to blood collection was found to be below the detection limit (4.0 × 102 HBV DNA copies/ml) of the employed assay. However, intermittent viremia which might be detected over a longer period of observation cannot be excluded.
In conclusion, testing with the Amplicor HBV Monitor Test could distinctly distinguish patients with CHB from patients after HBeAg seroconversion during alpha interferon treatment and from inactive carriers. Further prospective studies, however, are desirable to determine cutoffs for diagnostic and therapeutic decisions. For the routine diagnostic laboratory, the automated assay version might replace the manual version in future.
REFERENCES
- 1.Brechot C. Polymerase chain reaction for the diagnosis of viral hepatitis B and C. Gut. 1993;34:S39–S44. doi: 10.1136/gut.34.2_suppl.s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erhardt A, Schaefer S, Athanassiou N, Kann M, Gerlich W H. Quantitative assay of PCR-amplified hepatitis B virus DNA using a peroxidase-labelled DNA probe and enhanced chemiluminescence. J Clin Microbiol. 1996;34:1885–1891. doi: 10.1128/jcm.34.8.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerken G, Gomes J, Lampertico P, Colombo M, Rothaar T, Trippler M, Colucci G. Clinical evaluation and applications of the Amplicor HBV Monitor test, a quantitative HBV DNA PCR assay. J Virol Methods. 1998;74:155–165. doi: 10.1016/s0166-0934(98)00081-0. [DOI] [PubMed] [Google Scholar]
- 4.Heermann K H, Hagos Y, Thomssen R. Liquid-phase hybridization and capture of hepatitis B virus DNA with magnetic beads and fluorescence detection of PCR products. J Virol Methods. 1995;50:43–58. doi: 10.1016/0166-0934(94)90162-7. [DOI] [PubMed] [Google Scholar]
- 5.Jardi R, Buti M, Rodriguez-Frias F, Cortina M, Esteban R, Guardia J, Pascual C. The value of quantitative detection of HBV-DNA amplified by PCR in the study of hepatitis B infection. J Hepatol. 1996;24:680–685. doi: 10.1016/s0168-8278(96)80263-7. [DOI] [PubMed] [Google Scholar]
- 6.Kessler H H, Pierer K, Dragon E, Lackner H, Santner B, Stünzner D, Stelzl E, Waitzl B, Marth E. Evaluation of a new assay for HBA DNA quantitation in patients with chronic hepatitis B. Clin Diagn Virol. 1998;9:37–43. doi: 10.1016/s0928-0197(97)10008-3. [DOI] [PubMed] [Google Scholar]
- 7.Lehtovaara P, Uusi-Oukari M, Buchert P, Laaksonen M, Bengtström M, Ranki M. Quantitative PCR for hepatitis B virus with colorimetric detection. PCR Methods Appl. 1993;3:169–175. doi: 10.1101/gr.3.3.169. [DOI] [PubMed] [Google Scholar]
- 8.Lok A S F. Treatment of chronic hepatitis B. J Viral Hepatitis. 1994;1:105–124. doi: 10.1111/j.1365-2893.1994.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 9.Loriot M A, Marcellin P, Walker F, Boyer N, Degott C, Randrianatoavina I, Benhamou J P, Erlinger S. Persistence of hepatitis B virus DNA in serum and liver from patients with chronic hepatitis B after loss of HBsAg. J Hepatol. 1997;27:251–258. doi: 10.1016/s0168-8278(97)80168-7. [DOI] [PubMed] [Google Scholar]
- 10.Michalak T I, Pasquinelli C, Guilhot S, Chisari F V. Hepatitis B virus persistence after recovery from acute viral hepatitis. J Clin Investig. 1994;93:230–239. doi: 10.1172/JCI116950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagata I, Colucci G, Gregorio G V, Cheeseman P, Williams R, Mieli-Vergani G, Vergani D. The role of HBV DNA quantitative PCR in monitoring the response to interferon treatment in chronic hepatitis B virus infection. J Hepatol. 1999;30:965–969. doi: 10.1016/s0168-8278(99)80247-5. [DOI] [PubMed] [Google Scholar]
- 12.Niitsuma H, Ishii M, Miura M, Kobayashi K, Toyota T. Low level hepatitis B viremia detected by polymerase chain reaction accompanies the absence of HBe antigenemia and hepatitis in hepatitis B virus carriers. Am J Gastroenterol. 1997;92:119–123. [PubMed] [Google Scholar]
- 13.Noborg U, Gusdal A, Pisa E K, Hedrum A, Lindh M. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor Test. J Clin Microbiol. 1999;37:2793–2797. doi: 10.1128/jcm.37.9.2793-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawlowsky J M, Bastie A, Lonjon I, Remire J, Darthuy F, Soussy C J, Dhumeaux D. What technique should be used for routine detection and quantification of HBV DNA in clinical samples? J Virol Methods. 1997;65:245–253. doi: 10.1016/s0166-0934(97)02196-4. [DOI] [PubMed] [Google Scholar]
- 15.Perrillo R P, Schiff E R, Davis G L, Bodenheimer H C, Lindsay K, Payne J, Dienstag J L, O'Brien C, Tamburro C, Jacobson I M, Sampliner R, Feit D, Lefkowitch J, Kuhns M, Meschievitz C, Sanghvi B, Albrecht J, Gibas A the Hepatitis Interventional Therapy Group. A randomized, controlled trial of interferon alpha-2b alone and after prednisone withdrawal for the treatment of chronic hepatitis B. N Engl J Med. 1990;323:295–301. doi: 10.1056/NEJM199008023230503. [DOI] [PubMed] [Google Scholar]
- 16.Ranki M, Schätzl H M, Zachoval R, Uusi-Oukari M, Lehtovaara P. Quantification of hepatitis B virus DNA over a wide range from serum for studying viral replicative activity in response to treatment and in recurrent infection. Hepatology. 1995;21:1492–1499. [PubMed] [Google Scholar]
- 17.Rehermann B, Ferrari C, Pasquinelli C, Chisari F V. The hepatitis B virus persists for decades after patients' recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med. 1996;2:1104–1108. doi: 10.1038/nm1096-1104. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Moreno M, Otero M, Millan A, Castillo I, Cabrerizo M, Jimenez F J, Oliva H, Ramon y Cajal S, Carreno V. Clinical and histological outcome after hepatitis B e antigen to antibody seroconversion in children with chronic hepatitis B. Hepatology. 1999;29:572–575. doi: 10.1002/hep.510290230. [DOI] [PubMed] [Google Scholar]
- 19.Yotsuyanagi H, Yasuda K, Iino S, Moriya K, Shintani Y, Fujie H, Tsutsumi T, Kimura S, Koike K. Persistent viremia after recovery from self-limited acute hepatitis B. Hepatology. 1998;27:1377–1382. doi: 10.1002/hep.510270526. [DOI] [PubMed] [Google Scholar]
- 20.Zaaijer H L, ter Borg F, Cuypers H T M, Hermus M C A H, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]