Abstract
There is increasing evidence for inflammation as a determinant in the pathogenesis of Parkinson’s disease, but its role in parkinsonian neurodegeneration remains elusive. It is not clear whether inflammatory cascades are causes or consequences of dopamine neuron death. In the present study, we aim to perform an in-depth statistical investigation of the causal relationship between inflammation and Parkinson’s disease using a two-sample Mendelian randomization design.
Genetic instruments were selected using summary-level data from the largest genome-wide association studies to date (sample size ranging from 13 955 to 204 402 individuals) conducted on a European population for the following inflammation biomarkers: C-reactive protein, interleukin-6, interleukin 1 receptor antagonist and tumour necrosis factor α. Genetic association data on Parkinson’s disease (56 306 cases and 1 417 791 controls) and age at onset of Parkinson’s disease (28 568 cases) were obtained from the International Parkinson’s Disease Genomics Consortium. On primary analysis, causal associations were estimated on sets of strong (P-value < 5 × 10−8; F-statistic > 10) and independent (linkage disequilibrium r2 < 0.001) genetic instruments using the inverse-variance weighted method. In sensitivity analysis, we estimated causal effects using robust Mendelian randomization methods and after removing pleiotropic genetic variants. Reverse causation was also explored. We repeated the analysis on different data sources for inflammatory biomarkers to check the consistency of the findings.
In all the three data sources selected for interleukin-6, we found statistical evidence for an earlier age at onset of Parkinson’s disease associated with increased interleukin-6 concentration [years difference per 1 log-unit increase = −2.364, 95% confidence interval (CI) = −4.789–0.060; years difference per 1 log-unit increase = −2.011, 95% CI = −3.706 to −0.317; years difference per 1 log-unit increase = −1.569, 95% CI = −2.891 to −0.247]. We did not observe any statistical evidence for causal effects of C-reactive protein, interleukin 1 receptor antagonist and tumour necrosis factor α on both Parkinson’s disease and its age at onset. Results after excluding possible pleiotropic genetic variants were consistent with findings from primary analyses. When investigating reverse causation, we did not find evidence for a causal effect of Parkinson’s disease or age at onset on any biomarkers of inflammation.
We found evidence for a causal association between the onset of Parkinson’s disease and interleukin-6. The findings of this study suggest that the pro-inflammatory activity of the interleukin-6 cytokine could be a determinant of prodromal Parkinson’s disease.
Keywords: inflammation, Parkinson’s disease, Mendelian randomization, causal inference, age at onset of Parkinson’s disease
Bottigliengo et al. investigate the causal relationship between inflammation and Parkinson’s disease using a two-sample Mendelian randomization design. They show that higher IL-6 levels are associated with younger age at onset of Parkinson's disease, suggesting that elevated IL-6 levels could be part of the disease prodrome.
Introduction
In recent years, increasing biological evidence supporting a link between inflammatory processes and the pathogenesis of Parkinson’s disease has emerged.1–4 Inflammation origin has been hypothesized to be from the CNS through microglia activation, which might play a crucial role in the inflammatory cascades associated with death of dopaminergic neurons.5 In particular, activation of toxic microglia leads to the production of pro-inflammatory cytokines, such as interleukin (IL) 6 and tumour necrosis factor-α (TNF-α), which will ultimately produce damage to dopamine neurons.6,7 Furthermore, inflammation can also be triggered by mitochondrial stress through the release of damage-associated molecular patterns.8 Several studies found increased levels of pro-inflammatory cytokines and TNF-α in the CSF of Parkinson’s disease patients.9–13 Moreover, higher blood concentrations of IL-1β, IL-6, IL-10 and TNF-α were observed in Parkinson’s disease patients than controls, suggesting a possible involvement of the peripheral tissue in the origin of the inflammatory cascades.14,15 Despite the increasing number of findings pointing to a relationship between inflammation and Parkinson’s disease, the role of inflammatory processes remains controversial, as does its direction, possible causality and effect magnitude. For example, high cytokines levels in blood have been hypothesized to be the consequence of brain clearance processes through the glymphatic system.16 Moreover, lower plasma concentration levels of some cytokines have been found in Parkinson’s disease patients.17 The supporting evidence for the hypothesized link was mainly derived from observational studies, which are known to be prone to several sources of bias. Indeed, issues such as reverse causation (activation of microglia is the cause and not the consequence of neurodegeneration) and confounding (the association between inflammation and Parkinson’s disease is driven by other factors associated with both exposure and outcome) are often present in observational studies and can undermine reliability of results.
The causal role of modifiable exposures on outcomes, such as diseases, has been increasingly studied using Mendelian randomization (MR), a statistical framework that uses genetic variants as instruments for proxying the effect of exposures on the outcome of interest.18 Given the random inheritance at conception, genetic variant allocation is not influenced by environmental or lifestyle factors and, thus, MR estimates are usually less prone to confounding and reverse causation biases.19 The increasing availability of information on genetic associations for phenotypes and disease traits has greatly aided the use of MR methods for reliable causal inference. Indeed, genome-wide association studies (GWAS) data from large-scale consortia have the potential to improve the power of MR analysis for detecting causal effects.20
Few recent studies have evaluated the causal relationship of inflammation with Parkinson’s disease using MR methodology. A previous study investigated the causal relationship of C-reactive protein (CRP) with several diseases in the European population and did not find any statistical evidence of an effect on Parkinson’s disease.21 Conversely, another study found strong statistical evidence for a causal association of CRP with Parkinson’s disease with a phenome-wide MR on the UK Biobank and FinnGenn population.22 An MR approach was used to examine the role of long-term TNF-α inhibition on Parkinson’s disease risk and age at onset (AAO) in the European population, but no statistical evidence for a causal relationship was observed.23
In the present study, we aimed to further investigate the causal relationship of inflammation with Parkinson’s disease within a two-sample MR framework to reduce the risk of biases that are common in observational studies. In the context of disease prevention and therapy, the MR approach represents a valid alternative to randomized clinical trials where they cannot be conducted. Hence, findings can be used to inform and prioritize potential drug targets, which are more likely to be developed when supported by genetic evidence.24–27 We considered multiple additional well-known inflammatory biomarkers as exposures and we leveraged the information from different GWAS data sources to evaluate the robustness of findings and their replicability by checking the consistency of the MR estimates. Finally, to evaluate the direction of the causality and to exclude reverse causation, a bi-directional approach—considering inflammation biomarkers as exposures and Parkinson’s disease as the outcome and vice versa—was used on available data. The present study was conducted following the STROBE-MR guidelines for transparent reporting of MR studies.28
Materials and methods
Study design
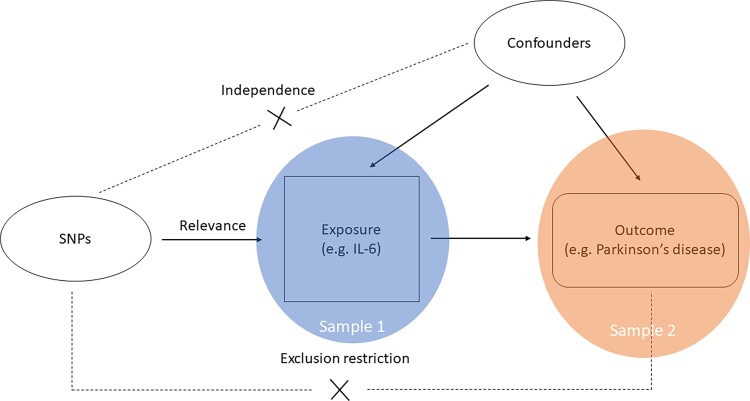
A two-sample MR design was followed, i.e. summary genetic data for exposures and outcome were retrieved from two independent samples avoiding bias due to overlapping.29 Multiple genetic variants were used as instruments when available, and both instrument selection and instrument-exposure estimates came from the same combined analysis of discovery and replication studies, that is, from the largest GWAS to date for maximum power. Moreover, when available, we retrieved more than one data source for each exposure to investigate the robustness and the replicability of the estimates. For each data source, we estimated the causal effects and evaluated their directions. We focused on studies conducted on ethnically homogeneous participants of European ancestry. A graphical overview of the general MR design is provided in Fig. 1.
Figure 1.
Graphical representation of the MR assumptions [(i) relevance; (ii) independence; (iii) exclusion restriction] in a two-sample MR design. The continuous lines represent the relationships that hold in MR analysis. Dashed lines depict the association that should not be present to satisfy the second and third assumptions. SNP-exposure associations are derived in Sample 1 (blue circle), and SNP-outcome associations in Sample 2 (orange circle).
Data sources
Details on the studies on the exposures are summarized in Table 1. We selected the following inflammation biomarkers as exposures: CRP, IL-6, IL-1 receptor antagonist (IL-1ra) and TNF-α. They were identified on the basis of public availability and sample size (at least 5000 participants) to ensure sufficient power for detecting a causal effect and to minimize the risk of small sample bias.30 A description of a more comprehensive list of GWAS conducted on common markers of inflammation is available in Supplementary Table 30. More information on exposure data sources can be found in the Supplementary material.
Table 1.
Description of the data sources of the study
| Trait | References | Sample size (discovery + replication) (number of studies) | Measurement | Statistical analysis |
|---|---|---|---|---|
| Exposure | ||||
| CRP | Ligthart et al.41 | 204 402 (88 studies) | Immune assay techniques as mg/l | Linear model on log-transformed CRP adjusting for sex, age and population structure |
| CCGC42 | 194 418 (47 studies) | High-sensitivity assay standardized on International Reference standards for CRP immunoassay | Linear model on log-transformed CRP adjusting for sex and ethnicity | |
| IL-6 | Ahluwalia et al.43 | 67 428 (38 studies) | Immunoassay techniques as pg/ml | Linear model on log-transformed IL-6 adjusting for age, sex, population structure and study site |
| IL6R consortium45 | 125 222 (46 studies) | Immunoassay techniques | Linear model on log-transformed IL-6 adjusting for age and sex | |
| Folkersen et al.44 (SCALLOP) | 30 931 (14 studies) | Olink Proximity Extension Assay CVD-I panel | Linear model on rank-based inverse normal transformed IL-6 adjusting for population structure and study-specific features | |
| IL-1ra | Herder et al.46 | 13 955 (11 studies) | Immunoassay techniques | Linear model on log-transformed IL-1ra adjusting for age, sex, body mass index, waist-to-hip ratio and smoking |
| Folkersen et al.44 (SCALLOP) | 30 931 (14 studies) | Olink Proximity Extension Assay CVD-I panel | Linear model on rank-based inverse normal transformed IL-6 adjusting for population structure and study-specific features | |
| TNF-α | Prins21 | 30 912 (25 studies) | Immunoassay techniques as pg/ml | Linear model on log-transformed TNF-α adjusting for a quadratic effect of age, sex, body mass index, study-specific covariates and relatedness |
| Outcome | ||||
| PD diagnosis | Nalls et al.31 | 37 688 cases, 18 618 proxy-cases, and 1 417 791 controls (17 studies) | Self-reporting or clinical ascertainment | Most studies used logistic regression on Parkinson’s disease status adjusting for AAO for cases and age at the most recent examination for controls, sex and the first six principal components of population structure. For each study, uniform summary statistics were generated and then pooled through fixed-effects meta-analysis |
| AAO | Blauwendraat et al.32 | 28 568 Parkinson’s disease case (18 studies) | Self-reporting of first Parkinson’s disease motor signs or age at diagnosis | Linear model on AAO adjusting for sex and the first five principal components of population structure. For each study, uniform summary statistics were generated and then pooled through fixed-effects meta-analysis |
Information on the traits, the sources, the sample size and the number of studies when results were meta-analysed, measurement and assessment of the traits, and statistical analysis are provided. CCGC = CRPC-reactive protein coronary heart disease genetics collaboration; CVD = cardiovascular disease; PD = Parkinson's disease; SCALLOP = systematic and combined analysis of Olink proteins.
We focused on two outcomes: the diagnosis of Parkinson’s disease and its AAO. In the former case, we retrieved genetic data from the most recent GWAS meta-analysis among 16 cohorts within the International Parkinson’s Disease Genomics Consortium (IPDGC) and 23andMe study, with a total of 37 688 cases, 18 618 proxy-cases (individuals without a diagnosis of Parkinson’s disease but with a first degree relative with Parkinson’s disease diagnosis) and 1 417 791 controls.31 For AAO, we extracted genetic association estimates from the largest GWAS conducted on 28 568 Parkinson’s disease cases by the IPDGC and 23andMe.32 More details on outcomes data sources are available in the Supplementary material.
Mendelian randomization analyses
We investigated causal effects of exposures on outcomes using two-sample MR methods. Before performing MR analyses, we selected genetic variants as instruments such that they were strongly associated with the exposure (P < 5 × 10−8 and F-statistic > 10) and independent [linkage disequilibrium (LD) r2 < 0.001], and we harmonized the datasets as previously described.33 Details on the selection of instruments and the data harmonization are in the Supplementary material. Depending on the number of selected genetic instruments, in primary MR analysis, we estimated causal effects using the Wald ratio estimator and inverse-variance weighted fixed or random effects methods. In secondary analyses, we used two-sample MR methods robust to the presence of pleiotropy (details in the Supplementary material).
To check the robustness of MR findings, we performed further sensitivity analyses. First, we removed from the initial set of instruments those genetic variants for which we found biological or statistical evidence of pleiotropy. Second, to avoid the risk of observing results driven by a few potentially pleiotropic variants, we selected genetic instruments with a more liberal LD clumping approach based on r2 < 0.1. Last, we performed a reverse causation investigation to understand whether an increased liability to Parkinson’s disease and increased AAO are causally associated with the selected biomarkers of inflammation. Details on the sensitivity analyses are included in the Supplementary material.
When considering AAO (continuous outcome), we summarized the causal estimates as the expected differences for a unit increase in exposure (logarithmic or rank-based inverse normal transformation scales, depending on the dataset). For Parkinson’s disease (binary outcome), we used the odds ratio (OR) to represent the causal effect. Given the increasing awareness towards the issues with statistical significance when interpreting findings of a study,34 we presented the results with point estimates and relative 95% CIs thus focusing on the magnitude, the direction and the associated uncertainty of a causal effect.35
Power analysis
An a priori power analysis has been conducted following the approach proposed by Brion et al.36 Different scenarios are shown in Supplementary Fig. 1. Assuming a sample size of 200 000 participants, an α level of 0.05 and a proportion of Parkinson’s disease cases of about 3%, we estimated a power of 80% to detect an OR of at least 1.15, when the exposure variability explained by the instruments is at least ∼5%.
Data availability
All the GWAS included in the present study obtained written informed consent from participants and were approved by ethics committees. No further ethical consents were required since our study is based on publicly available summary-level data.
Concerning exposure data, we extracted the data from the tables in the main paper or the supplementary material of the original publication, and from the repository (https://zenodo.org/record/2615265#.YS0CBt_OOUk). Regarding the outcomes, the summary-level data are available on the The International Parkinson Disease Genomics Consortium (IPDGC) consortium website (https://pdgenetics.org/resources).
All the analyses were performed using R software for statistical computing (v.4.1.1).37 Data manipulation and harmonization were implemented using the tidyverse meta-package (v.1.3.1)38 and MR analyses were performed using TwoSampleMR (v.0.5.6) and MendelianRandomization (v.0.5.1) R packages.39,40 The code used to perform all the steps of the analysis is available in an online Github repository (https://github.com/EuracBiomedicalResearch/mr.inflpd).
Results
Table 2 shows details on the selected instruments for MR analyses. Overall, one to five instruments were selected in most of the data sources, except for CRP where 31 instruments were identified in one meta-analysis. All genetic variants were strongly associated with the exposures, with the F-statistic values >25, and exposure variances explained by genetic instruments ranging from 0.2 to 5.3%.
Table 2.
Description of the genetic variants used as instrumental variables in the MR analysis for each dataset
| Exposure | Data source | Number of IVs | F-statistica | Overall R2 |
|---|---|---|---|---|
| CRP | Ligthart et al.41 | 31 | 49 (25; 2070.3) | 5.3% |
| CCGC42 | 1 | 311.2 | 0.7% | |
| IL-6 | Ahluwalia et al.43 | 2 | 37.8; 397.7 | 0.4% |
| IL6R consortium45 | 1 | 63.1 | 0.4% | |
| Folkersen et al.44 (SCALLOP) | 2 | 35.6; 198.5 | 1.7% | |
| IL-1ra | Herder et al.46 | 1 | 169 | 0.2% |
| Folkersen et al.44 (SCALLOP) | 5 | 170.2 (30.0; 384.8) | 4.9% | |
| TNF-α | Prins21 | 3 | 30.9 (28.4; 39.5) | 0.2% |
Information on the exposure, the dataset source, the number of instrumental variables (IVs), the strength of the SNP-exposure association (using F-statistic) and the overall proportion of exposure variance explained by the IVs (R2). CCGC = CRPC-reactive protein coronary heart disease genetics collaboration; SCALLOP = systematic and combined analysis of Olink proteins.
When only one IV was available, a single value is reported. For two instruments, both F-statistic values are shown. For >2 instruments, median (range) are reported.
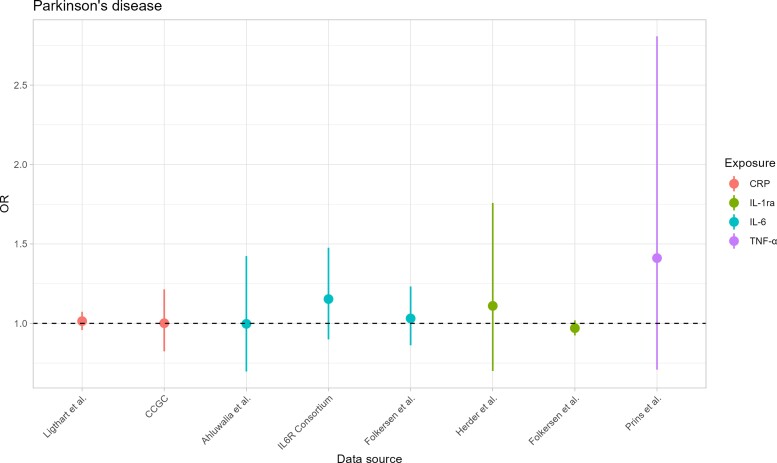
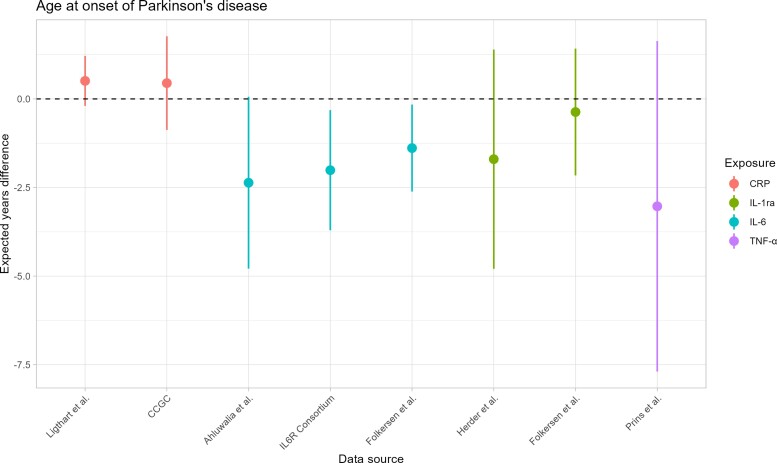
Results from primary MR analysis are shown in Table 3, Fig. 2 (for Parkinson’s disease) and Fig. 3 (for AAO). Regarding CRP, we did not find any statistical evidence for an association with the risk of Parkinson’s disease, both either Ligthart et al.41 [OR = 1.014; 95% confidence interval (CI) = 0.958–1.073] or CRP coronary heart disease genetics collaboration (CCGC) data42 (OR = 1.001; 95% CI = 0.824–1.215). Similarly, we did not observe any evidence of an effect on AAO in Ligthart et al.41 (0.509 years difference; 95% CI = −0.195 to 1.213) and CCGC42 (0.446 years difference; 95% CI = −0.875 to 1.767) datasets. In secondary analyses (Supplementary Tables 33 and 34), no effect was observed on Parkinson’s disease risk by any robust MR method, whereas we observed statistical evidence for later AAO associated with an increase of 1-unit of CRP on the log scale using MR-Egger (1.486 years difference; 95% CI = 0.470–2.501) and weighted mode (0.893 years difference; 95% CI = 0.052–1.734) methods. The exclusion of pleiotropic variants (Supplementary Table 45) and the selection of instruments with r2 < 0.1 (Supplementary Tables 35–42) led to results consistent with those previously obtained. Moreover, no evidence of reverse causation was observed (Supplementary Tables 43 and 44).
Table 3.
Results for primary MR analysis
| Exposure | Data source | PD OR (95% CI) | Estimated years difference (95% CI) |
|---|---|---|---|
| CRP | Ligthart et al.41 | 1.014 (0.958; 1.073) | 0.509 (−0.195; 1.213) |
| CCGC42 | 1.001 (0.824; 1.215) | 0.446 (−0.875; 1.767) | |
| IL-6 | Ahluwalia et al.43 | 0.997 (0.697; 1.424) | −2.364 (−4.789; 0.060) |
| IL6R consortium45 | 1.152 (0.900; 1.475) | −2.011 (−3.706; −0.317) | |
| Folkersen et al.44 (SCALLOP) | 1.031 (0.862; 1.232) | −1.387 (−2.615; −0.160) | |
| IL-1ra | Herder et al.46 | 1.109 (0.700; 1.757) | −1.700 (−4.793; 1.393) |
| Folkersen et al.44 (SCALLOP) | 0.970 (0.924; 1.102) | −0.370 (−2.159; 1.420) | |
| TNF-α | Prins21 | 1.410 (0.709; 2.807) | −3.030 (−7.692; 1.633) |
For Parkinson’s disease, results are reported as OR along with 95% CI for a unit increase in the exposure level. For AAO, results are reported as expected years’ difference of AAO along with 95% CI for a unit increase in the exposure level. SCALLOP = systematic and combined analysis of Olink proteins.
Figure 2.
MR estimates for Parkinson’s disease. On the x-axis, the data sources are depicted. On the y-axis, the ORs are shown. Points represent the OR estimate of the effect of the exposure on Parkinson’s disease, whereas the lines represent the 95% CIs of the point estimate. Each colour identifies an inflammatory biomarker. The dashed line represents the situation of absence of association between exposures and Parkinson’s disease, i.e. OR = 1.
Figure 3.
MR estimates for Parkinson’s disease-AAO. On the x-axis, the data sources are depicted. On the y-axis, the estimated Parkinson’s disease-AAO differences are shown. Points represent the estimated Parkinson’s disease-AAO differences of the effect of the exposure on Parkinson’s disease-AAO, whereas the lines represent the 95% CIs of the point estimate. Each colour identifies an inflammatory biomarker. The dashed line represents the situation of absence of association between exposures and Parkinson’s disease-AAO, i.e. estimated Parkinson’s disease-AAO difference = 0.
For IL-6, we did not observe any strong statistical evidence for an association with Parkinson’s disease risk using either Ahluwalia et al.43 (OR 0.997; 95% CI = 0.697–1.424), IL6R Genetics Consortium (OR 1.152; 95% CI = 0.900–1.475), or Folkersen et al.44 (OR 1.031; 95% CI = 0.862–1.232) datasets. However, we found evidence for younger AAO associated with increase IL-6 levels in IL6R Genetics Consortium45 (−2.011 years difference; 95% CI = −3.076 to −0.317) and Folkersen et al.44 (−1.387 years difference; 95% CI = −2.615 to −0.160) data, whereas less evidence was observed in Ahluwalia et al.43 (−2.364 years difference; 95% CI = −4.789 to 0.060) dataset. In sensitivity analyses on Ahluwalia et al.,43 one single-nucleotide polymorphism (SNP) (rs660895 in the HLA-DRB1 locus) associated with rheumatoid arthritis (Supplementary Table 47) was excluded. After SNP exclusion for possible pleiotropy, no strong evidence for increased risk of Parkinson’s disease associated with increased IL-6 levels was observed, whereas we found that higher IL-6 concentrations decreased the AAO (−2.785 years difference; 95% CI = −5.305 to −0.265) (Supplementary Tables 35 and 36). On the Folkersen et al.44 data, we excluded the SNP rs4959106 in HLA-DRB1 locus associated with rheumatoid arthritis and cholesterol (Supplementary Table 47) in sensitivity analyses. Consistently with primary MR analysis, we did not find evidence for an association between IL-6 and Parkinson’s disease risk (OR 1.117; 95% CI = 0.921–1.355), whereas we observed evidence for younger AAO associated with increased IL-6 levels (−1.569 years difference; 95% CI = −2.891 to −0.247). When genetic instruments were selected using r2 < 0.1, findings were consistent with previous analyses (Supplementary Tables 39–42). We did not find any evidence of reverse causation (Supplementary Tables 43 and 44).
Regarding IL-1ra, no association with the risk of Parkinson’s disease was observed in either Herder et al.46 (OR = 1.109; 95% CI = 0.700–1.757) or Folkersen et al.44 (OR = 0.970; 95% CI = 0.924–1.102) data. Lack of statistical evidence was also observed for AAO using both datasets (Herder et al.: −1.700 years difference; 95% CI = −4.793 to 1.393. Folkersen et al.44: −0.370 years difference; 95% CI = −2.159 to 1.420). In secondary analyses, no MR methods identified a causal effect of IL-1ra on both Parkinson’s disease risk and AAO (Supplementary Tables 33 and 34). No SNP was excluded for pleiotropy based on both statistical and biological criteria. Finally, no evidence of reverse causation was observed (Supplementary Tables 43 and 44) and results with a more liberal LD clumping threshold (r2< 0.1) were in agreement with those from primary and secondary analyses (Supplementary Tables 39–42).
For TNF-α, there was no statistical evidence that higher concentration levels affected either the risk of Parkinson’s disease (OR = 1.410; 95% CI = 0.709–2.807) or AAO (−3.030 years difference; 95% CI = −7.692–1.633). Excluding pleiotropic SNPs (Supplementary Tables 35 and 36) and using r2 < 0.1 for LD clumping (Supplementary Table 39–42) findings were confirmed. No data were available for TNF-α to investigate reverse causation.
Discussion
Previous observational studies pointed to a potential link between inflammation and neurodegeneration in Parkinson’s disease.9,14,47 However, observational studies can be prone to many sources of bias, such as confounding and reverse causation, that can undermine findings’ reliability. In the present study, we explored a possible causal relationship between inflammatory processes and Parkinson’s disease using an MR design, which is a statistical framework that provides valid causal inference on the effect of exposures on outcomes by leveraging genetic information. Connections between inflammation and Parkinson’s disease through that design have been explored by previous studies. We extended this evaluation by considering multiple well-established markers of inflammation. Moreover, we studied the causal effect of the exposures on datasets from different data sources to assess the consistency of the results. Based on the present findings, we did not observe any strong evidence for causality between inflammatory biomarkers and Parkinson’s disease except for IL-6 and AAO.
Regarding CRP, findings from previous studies are controversial. Borsche et al.48 compared inflammatory biomarker levels in serum between patients with monogenic or idiopathic Parkinson’s disease and controls, and did not find any statistical evidence for a difference of CRP levels between the two groups. Similar results were also observed in Prins et al.,21 suggesting no association between CRP and Parkinson’s disease, as we observed from our primary analysis results. Conversely, both the Qin et al.14 and Si et al.22 studies found evidence for a relationship between CRP and the disease. In the first study, after meta-analysing results from six studies, the authors observed that increased CRP levels were associated with increased Parkinson’s disease risk. Despite the strong evidence often supported by meta-analysis findings, results were pooled from observational studies, which might be affected by issues such as confounding. Opposite results were found in the second study, where the authors used MR designs to evaluate the relationship between CRP and several diseases using data from UK Biobank and FinnGenn cohorts. Strong evidence that elevated CRP levels decreased the risk of Parkinson’s disease was observed using UK Biobank data but the results were not replicated in the FinnGen cohort. These findings suggested that the relationship between CRP levels and Parkinson’s disease might be heterogeneous in different populations. Moreover, despite the large sample size that can enhance statistical power, the UK Biobank cohort may be affected by selection bias issues and not be representative of the general European population.49 In our study, findings from secondary analyses using MR-Egger and weighted mode methods indicated that higher CRP levels might delay the onset of the disease. However, the results were not robust after the exclusion of pleiotropic genetic instruments. The link between CRP and Parkinson’s disease remains elusive. Indeed, CRP is a general marker of inflammation and elevated levels signal a very broad range of inflammatory processes, some of which might not be involved in the parkinsonian neurodegeneration.
In our primary MR analysis, we did not observe any strong statistical evidence for a link between IL-6 and the risk of Parkinson’s disease. However, the results from IL6R Genetics Consortium and Folkersen et al. data suggested that increased IL-6 levels might play a role, even though there was no statistical evidence at a 95% level. Using Ahluwalia et al. data for instruments selection, there was no evidence for an association in primary MR analysis. In sensitivity analysis, we removed the SNP rs660895, which might induce pleiotropy issues given its association with rheumatoid arthritis, a risk factor for Parkinson’s disease,50 and we observed results similar to those obtained using the other datasets. Statistical evidence for increased IL-6 levels associated with earlier AAO was observed in IL6R Genetics Consortium and Folkersen et al.44 datasets. Using Ahluwalia et al.,43 strong evidence emerged only in sensitivity analysis after removal of the pleiotropic instrument in the HLA-DBR1 locus. Moreover, when performing LD clumping with a more liberal criterion (r2 < 0.1), we observed estimates in agreement with those from the previous analyses on both data sources, suggesting robustness of the results to different analysis specifications. The present findings are consistent with previous studies that pointed to a link between IL-6 and Parkinson’s disease.11,12,48,51 Indeed, pro-inflammatory cytokines production through microglia activation might be considered as a more relevant marker of the inflammatory cascades involved in neurodegeneration than CRP. More specifically, our findings suggested that higher IL-6 production might anticipate the onset of the disease. The lack of statistical evidence for the risk of Parkinson’s disease might be partially explained by the insufficient power of the present study to identify the effect of interest.
IL-1ra is involved in anti-inflammatory processes and acts as a natural inhibitor of the pro-inflammatory activities of IL-1α and IL-1β cytokines, whose levels in CSF and serum were found to be higher in Parkinson’s disease subjects than controls.13,15,52 In the present study, we did not find any evidence of an effect of IL-1ra on the risk and the onset of the disease, which we proposed to be decreased and postponed, respectively, for higher production of the anti-inflammatory cytokines.
Regarding TNF-α, our findings are consistent with the results from Kang et al.,23 who did not observe any effect on both risk and onset of Parkinson’s disease using data from Ligthart et al. on genetic variants in or near TNF receptor 1. Lack of association of TNF-α with Parkinson’s disease from MR analyses suggests that previous findings from observational studies might be affected by some degree of bias.11,13,14
In the present study, the absence of statistical evidence for an opposite relationship between inflammation and Parkinson’s disease, i.e. the disease affects the inflammatory processes, suggests that, probably, neurodegeneration is not a driver of the inflammatory cascades in Parkinson’s disease patients.
The use of MR design to minimize the risk of confounding and reverse causation is the main strength of the present study. We focused on multiple well-known biomarkers of inflammation and not just on a single exposure. Moreover, to improve the reliability and consistency of the results, we carefully selected genetic instruments with conservative strategies, applied robust MR methods and sensitivity analyses for reducing pleiotropic issues, and evaluated the causal effects using data from large-scale GWAS. Additionally, we restricted the investigation using data from studies conducted on European individuals to minimize the risk of population stratification bias.53
Our study has some limitations. First, MR analyses estimate the ‘lifetime’ effect of the exposure on the disease.54 Under this consideration, causal inference will be valid only if the exposure does not change over time, i.e. the levels of the inflammatory markers are constant across time. Second, we did not evaluate the presence of survival bias, which is an issue in MR studies, since conditioning on participants who survived long enough to be included in the study might distort the final estimates.55 A future direction for this research area could be to perform separate MR analyses across different groups of age and check the consistency of the results. Third, we selected data from studies that measured biomarker concentrations only in blood samples, even if evidence for increased inflammatory biomarkers levels in Parkinson’s disease patients were also identified in CSF samples.11–13,48 Indeed, such an evaluation would provide more insights into the role played by both neural and peripheral inflammation in the neurodegeneration of Parkinson’s disease. Fourth, we used the same exposure datasets, i.e. summary genetic data from meta-analyses of discovery and replication cohorts, to perform both the selection of instruments and retrieval of SNP-exposure association estimates, a procedure that might introduce the ‘winner’s curse’ bias.53,56 We used this approach for consistency on the type of exposure datasets, and for maximizing the statistical power. Fifth, despite the careful selection of genetic instruments to overcome issues related to weak instruments and LD, MR estimates were obtained using a few genetic variants in most of the situations, which might have lowered the power. Recently, more sophisticated MR methods that can handle hundreds of weak and correlations genetic variants have been proposed, even though their use in MR studies is still scarce.57–59 In future, results using these MR methods should be complemented with those obtained from the most popular strategies for a better bias-variance trade-off. Sixth, even though considering multiple markers of inflammation, we did not evaluate their adjusted causal effects using multivariable MR. However, given the absence of overlap among the genetic instruments selected for each biomarker, we hypothesized that standard univariable MR should be sufficient to identify an independent effect of inflammatory biomarkers without further adjustments.60 Furthermore, power is typically an issue in MR studies, since the genetic variants used as instruments often capture only a small fraction of the exposure variance, leading to power for rejecting the null hypothesis of no association below the conventional 80% level.61 In light of this, we performed an a priori analysis to understand the expected power for detecting a causal effect and we found that even using the largest available GWAS so far, the power was <80% for most of the present MR analyses. Finally, despite the definition of cases in Parkinson’s disease GWAS was assessed differently and AAO of Parkinson’s disease is a phenotype often difficult to measure rigorously and comprehensively, summary genetic data were generated uniformly by each study for the outcomes. Hence, it does not represent an issue in our MR study.
In summary, the present study suggests that increased IL-6 levels could anticipate the onset of Parkinson’s disease. Based on these findings, IL-6 concentration, together with clinical examinations, family information and genetic data, can help in raising awareness towards a clinical profile that could be prodromal of Parkinson’s disease. Despite IL-6 being a drug target, suggesting consideration of therapeutic strategies that inhibit the pro-inflammatory IL-6 activity, it is a pleiotropic cytokine that exerts several anti-inflammatories and molecular protective functions.62 Indeed, the consideration of lowering IL-6 strategies for disease prevention cannot disregard further careful investigations of IL-6 role in parkinsonian neurodegeneration.
Supplementary Material
Acknowledgements
The authors acknowledge the investigators of the original studies for sharing the GWAS data used in this project. The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the open access publication cost.
Abbreviations
- AAO
age at onset
- CRP
C-reactive protein
- GWAS
genome-wide association study
- IL
interleukin
- LD
linkage disequilibrium
- MR
Mendelian randomization
- SNP
single-nucleotide polymorphism
- TNF-α
tumour necrosis factor α
Contributor Information
Daniele Bottigliengo, Institute for Biomedicine, Eurac Research, Bolzano/Bozen, Italy.
Luisa Foco, Institute for Biomedicine, Eurac Research, Bolzano/Bozen, Italy.
Philip Seibler, Institute of Neurogenetics, University of Lübeck and University Hospital of Schleswig-Holstein, Lübeck, Germany.
Christine Klein, Institute of Neurogenetics, University of Lübeck and University Hospital of Schleswig-Holstein, Lübeck, Germany; Department of Psychiatry and Psychotherapy, University of Lübeck, Germany.
Inke R König, Institute of Medical Biometry and Statistics, University of Lübeck and University Hospital of Schleswig-Holstein, Lübeck, Germany.
Fabiola Del Greco M, Institute for Biomedicine, Eurac Research, Bolzano/Bozen, Italy.
Funding
This study was supported by grants from the German Research Foundation (Research Unit ProtectMove, FOR 2488).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Hirsch EC, Hunot S. Neuroinflammation in Parkinson’s disease: A target for neuroprotection? Lancet Neurol. 2009;8:382–397. [DOI] [PubMed] [Google Scholar]
- 2. Wang Q, Liu Y, Zhou J. Neuroinflammation in Parkinson’s disease and its potential as therapeutic target. Transl Neurodegener. 2015;4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Houser MC, Tansey MG. The gut-brain axis: Is intestinal inflammation a silent driver of Parkinson’s disease pathogenesis? NPJ Parkinson’s Dis. 2017;3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kline EM, Houser MC, Herrick MK, et al. Genetic and environmental factors in Parkinson’s disease converge on immune function and inflammation. Movement Disorders. 2021;36:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gundersen V. Parkinson’s disease: Can targeting inflammation be an effective neuroprotective strategy? Front Neurosci. 2021;14:580311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Main BS, Zhang M, Brody KM, et al. Type-1 interferons contribute to the neuroinflammatory response and disease progression of the MPTP mouse model of Parkinson’s disease. Glia. 2016;64:1590–1604. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Q-S, Heng Y, Yuan Y-H, Chen N-H. Pathological α-synuclein exacerbates the progression of Parkinson’s disease through microglial activation. Toxicol Lett. 2017;265:30–37. [DOI] [PubMed] [Google Scholar]
- 8. Sliter DA, Martinez J, Hao L, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Karpenko MN, Vasilishina AA, Gromova EA, Muruzheva ZM, Miliukhina IV, Bernadotte A. Interleukin-1β, interleukin-1 receptor antagonist, interleukin-6, interleukin-10, and tumor necrosis factor-α levels in CSF and serum in relation to the clinical diversity of Parkinson’s disease. Cell Immunol. 2018;327:77–82. [DOI] [PubMed] [Google Scholar]
- 10. Starhof C, Winge K, Heegaard NHH, Skogstrand K, Friis S, Hejl A. Cerebrospinal fluid pro-inflammatory cytokines differentiate parkinsonian syndromes. J Neuroinflammation. 2018;15:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schröder JB, Pawlowski M, Meyer zu Hörste G, et al. Immune cell activation in the cerebrospinal fluid of patients with Parkinson’s disease. Front Neurol. 2018;9:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lian TH, Guo P, Zuo LJ, et al. Tremor-dominant in Parkinson disease: The relevance to iron metabolism and inflammation. Front Neurosci. 2019;13:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iwaoka K, Otsuka C, Maeda T, et al. Impaired metabolism of kynurenine and its metabolites in CSF of Parkinson’s disease. Neuroscience Lett. 2020;714:134576. [DOI] [PubMed] [Google Scholar]
- 14. Qin X-Y, Zhang S-P, Cao C, Loh YP, Cheng Y. Aberrations in peripheral inflammatory cytokine levels in Parkinson disease: A systematic review and meta-analysis. JAMA Neurol. 2016;73:1316–1324. [DOI] [PubMed] [Google Scholar]
- 15. Kim R, Kim H-J, Kim A, et al. Peripheral blood inflammatory markers in early Parkinson’s disease. J Clin Neurosci. 2018;58:30–33. [DOI] [PubMed] [Google Scholar]
- 16. Iliff JJ, Wang M, Liao Y, et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci Transl Med. 2012;4:147ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rocha EM, De Miranda B, Sanders LH. Alpha-synuclein: Pathology, mitochondrial dysfunction and neuroinflammation in Parkinson’s disease. Neurobiol Dis. 2018;109(Pt B):249–257. [DOI] [PubMed] [Google Scholar]
- 18. Smith GD, Ebrahim S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 19. Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G. Mendelian Randomization: Using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. [DOI] [PubMed] [Google Scholar]
- 20. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, EPIC- InterAct Consortium . Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prins BP, Abbasi A, Wong A, et al. Investigating the causal relationship of C-reactive protein with 32 complex somatic and psychiatric outcomes: A large-scale cross-consortium Mendelian randomization study. PLoS Med. 2016;13:e1001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Si S, Li J, Tewara MA, Xue F. Genetically determined chronic low-grade inflammation and hundreds of health outcomes in the UK biobank and the FinnGen population: A phenome-wide Mendelian randomization study. Front Immunol. 2021;12:720876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang X, Ploner A, Pedersen NL, et al. Tumor necrosis factor inhibition and Parkinson disease: A Mendelian randomization study. Neurology. 2021;96:e1672–e1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. [DOI] [PubMed] [Google Scholar]
- 25. King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15:e1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hingorani AD, Kuan V, Finan C, et al. Improving the odds of drug development success through human genomics: Modelling study. Sci Rep. 2019;9:18911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gill D, Georgakis MK, Walker VM, et al. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thompson JR, Minelli C, Del Greco MF. Mendelian randomization using public data from genetic consortia. Int J Biostat. 2016;12:20150074. [DOI] [PubMed] [Google Scholar]
- 31. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blauwendraat C, Heilbron K, Vallerga CL, et al. Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov Disord. 2019;34:866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartwig FP, Davies NM, Hemani G, Smith GD. Two-sample Mendelian randomization: Avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45:1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amrhein V, Greenland S, McShane B. Scientists rise up against statistical significance. Nature. 2019;567:305–307. [DOI] [PubMed] [Google Scholar]
- 35. Wasserstein RL, Schirm AL, Lazar NA. Moving to a world beyond “p < 0.05”. Am Stat. 2019;73:1–19. [Google Scholar]
- 36. Brion M-JA, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. R Core Team . R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2021. https://www.R-project.org/.
- 38. Wickham H, Averick M, Bryan J, et al. Welcome to the tidyverse. J Open Source Software. 2019;4:1686. [Google Scholar]
- 39. Hemani G, Zheng J, Elsworth B, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yavorska OO, Burgess S. Mendelian randomization: An R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ligthart S, Vaez A, Võsa U, et al. Genome analyses of >200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am J Hum Genet. 2018;103:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Collaboration (CCGC) CRPCHDG . Association between C-reactive protein and coronary heart disease: Mendelian randomisation analysis based on individual participant data. BMJ. 2011;342:d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ahluwalia TS, Prins BP, Abdollahi M, et al. Genome-wide association study of circulating interleukin 6 levels identifies novel loci. Human Mol Genet. 2021;30:393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Folkersen L, Gustafsson S, Wang Q, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2:1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. IL6R Genetics Consortium Emerging Risk Factors Collaboration . Interleukin-6 receptor pathways in coronary heart disease: A collaborative meta-analysis of 82 studies. The Lancet. 2012;379:1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Herder C, Nuotio ML, Shah S, et al. Genetic determinants of circulating interleukin-1 receptor antagonist levels and their association with glycemic traits. Diabetes. 2014;63:4343–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nilsonne G, Lekander M. Circulating interleukin 6 in Parkinson disease. JAMA Neurol. 2017;74:607–608. [DOI] [PubMed] [Google Scholar]
- 48. Borsche M, König IR, Delcambre S, et al. Mitochondrial damage-associated inflammation highlights biomarkers in PRKN/PINK1 parkinsonism. Brain. 2020;143:3041–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tyrrell J, Zheng J, Beaumont R, et al. Genetic predictors of participation in optional components of UK biobank. Nat Commun. 2021;12:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li C, Ou R, Shang H. Rheumatoid arthritis decreases risk for Parkinson’s disease: A Mendelian randomization study. npj Parkinsons Dis. 2021;7:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6:a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams-Gray CH, Wijeyekoon R, Yarnall AJ, et al. Serum immune markers and disease progression in an incident Parkinson’s disease cohort (ICICLE-PD). Mov Disord. 2016;31:995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith GD, Hemani G. Mendelian Randomization: Genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Labrecque JA, Swanson SA. Interpretation and potential biases of Mendelian randomization estimates with time-varying exposures. Am J Epidemiol. 2019;188:231–238. [DOI] [PubMed] [Google Scholar]
- 55. Smit RAJ, Trompet S, Dekkers OM, Jukema JW, le Cessie S. Survival bias in Mendelian randomization studies: A threat to causal inference. Epidemiology. 2019;30:813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taylor AE, Davies NM, Ware JJ, VanderWeele T, Smith GD, Munafò MR. Mendelian Randomization in health research: Using appropriate genetic variants and avoiding biased estimates. Econ Hum Biol. 2014;13:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gkatzionis A, Burgess S, Conti DV, Newcombe PJ. Bayesian variable selection with a pleiotropic loss function in Mendelian randomization. Stat Med 2021;40:5025–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patel A, Ditraglia FJ, Zuber V, Burgess S. Selection of invalid instruments can improve estimation in Mendelian randomization. arXiv:210701513 [stat]. Published online July 3, 2021. Accessed September 3, 2021.https://arxiv.org/abs/2107.01513
- 60. Burgess S, Thompson SG. Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. IntJ Epidemiol. 2014;43:922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McElvaney OJ, Curley GF, Rose-John S, McElvaney NG. Interleukin-6: Obstacles to targeting a complex cytokine in critical illness. Lancet Respiratory Med. 2021;9:643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the GWAS included in the present study obtained written informed consent from participants and were approved by ethics committees. No further ethical consents were required since our study is based on publicly available summary-level data.
Concerning exposure data, we extracted the data from the tables in the main paper or the supplementary material of the original publication, and from the repository (https://zenodo.org/record/2615265#.YS0CBt_OOUk). Regarding the outcomes, the summary-level data are available on the The International Parkinson Disease Genomics Consortium (IPDGC) consortium website (https://pdgenetics.org/resources).
All the analyses were performed using R software for statistical computing (v.4.1.1).37 Data manipulation and harmonization were implemented using the tidyverse meta-package (v.1.3.1)38 and MR analyses were performed using TwoSampleMR (v.0.5.6) and MendelianRandomization (v.0.5.1) R packages.39,40 The code used to perform all the steps of the analysis is available in an online Github repository (https://github.com/EuracBiomedicalResearch/mr.inflpd).