Abstract
The endocannabinoid system is a highly conserved and ubiquitous signalling pathway with broad-ranging effects. Despite critical pathway functions, gene variants have not previously been conclusively linked to human disease.
We identified nine children from eight families with heterozygous, de novo truncating variants in the last exon of DAGLA with a neuro-ocular phenotype characterized by developmental delay, ataxia and complex oculomotor abnormality. All children displayed paroxysms of nystagmus or eye deviation accompanied by compensatory head posture and worsened incoordination most frequently after waking. RNA sequencing showed clear expression of the truncated transcript and no differences were found between mutant and wild-type DAGLA activity. Immunofluorescence staining of patient-derived fibroblasts and HEK cells expressing the mutant protein showed distinct perinuclear aggregation not detected in control samples.
This report establishes truncating variants in the last DAGLA exon as the cause of a unique paediatric syndrome. Because enzymatic activity was preserved, the observed mislocalization of the truncated protein may account for the observed phenotype. Potential mechanisms include DAGLA haploinsufficiency at the plasma membrane or dominant negative effect. To our knowledge, this is the first report directly linking an endocannabinoid system component with human genetic disease and sets the stage for potential future therapeutic avenues.
Keywords: endocannabinoid, episodic ataxia, paroxysmal tonic upgaze, nystagmus, developmental delay
Bainbridge et al. show that mutation of the endocannabinoid gene DAGLA causes a paediatric syndrome with developmental delay, ataxia and abnormal head and eye movements. DAGLA activity is preserved in affected individuals, suggesting that mislocalization of the truncated protein may account for the observed phenotype.
Introduction
The endocannabinoid (eCB) system is a modulatory system involved in cell and tissue homeostasis that is highly conserved among species.1 The pathway consists of G-protein coupled receptors (primarily CB1 and CB2), their endogenous ligands (eCBs) and the proteins involved in the synthesis and degradation of eCBs.2 The two main eCBs are N-arachidonoyl-ethanolamine (AEA; anandamide) and 2-arachidonoylglycerol (2-AG). AEA is synthesized by a complex system involving multiple enzymatic pathways.3 In contrast, 2-AG is synthesized primarily by a pair of enzymes in the brain, diacylglycerol lipase alpha and beta (DAGLA and DAGLB, respectively) encoded by the DAGLA and DAGLB genes.3
DAGLA is expressed in neurons and astrocytes throughout the brain, most prominently in cerebellum, hippocampus, ventral tegmental area and striatum.4 It is found in the plasma membrane of neuronal cells5 and contributes to intracellular 2-AG signalling.6 DAGLA may also be found in the nucleus or cytoplasm of a variety of cell types.7–9 DAGLB, on the other hand, is mostly expressed in the nervous system by microglia.10
In the developing brain, the eCB signalling pathway has important influences on neural development, signalling and brain repair.1,3 DAGLA activity is required within the developing nervous system for axonal growth and guidance.6 Once the nervous system is mature, eCBs modulate neuronal activity and network function2 where eCBs are released from postsynaptic neurons and act retrogradely on presynaptic CB1 and CB2, resulting in short- and long-term suppression of neurotransmitter release.6 This intricate system influences a wide array of pathophysiological functions, including: emotion, cognition, energy balance, pain sensation and neuroinflammation.11
DAGLA animal models display a range of neurological phenotypes. DAGLA knockdown zebrafish show morphologic abnormalities in mid- and hind-brain regions associated with abnormal locomotor and optokinetic behaviours.12Dagla knockout mice are viable (mousephenotype.org), display increased seizure susceptibility and severity,13,14 anxiety and depression-like behaviours,14 abnormalities in regions involved in learning and memory including altered cholinergic innervation of CA1 pyramidal cells of the hippocampus15 and compromised adult neurogenesis in the hippocampus and subventricular zone.14
We report nine children from eight families with heterozygous apparently de novo premature termination variants in DAGLA. Each displays a unique neuro-ocular phenotype characterized by global developmental delay and ataxia with superimposed paroxysms of worsening ataxia and complex oculomotor abnormality of nystagmus or eye deviation accompanied by compensatory head posture.
Subjects and methods
Ethical approval
Consent was obtained according to the respective institutional ethics guidelines and with approval of the local ethics committees in the participating study centres.
Patient ascertainment and review of clinical features
The index case was identified by trio genome sequencing. Genematcher16 was utilized to identify additional individuals with DAGLA variants and initiate an international collaboration of eight centres. For each case, medical records were reviewed. Anonymized data were provided via a standardized study questionnaire.
Subjects
Case 1 is an 11-year-old Hispanic male with global developmental delay, hypotonia, ataxia, dysarthria, congenital nystagmus and intellectual disability. From birth, the child displayed daily paroxysmal spells characterized by eye deviation and/or nystagmus (vertical>>horizontal) with abnormal head posture either alone or in combination with generalized ataxia. Spells are most prominent within 5–10 min of waking in the morning or after a nap. Symptoms typically persist until midday and are less prominent or entirely absent in the afternoon and evening. Abnormal eye movements are mostly upward eye deviation with chin down posture with less common episodes of downward eye deviation with upward head tilt and convergence movements. Simultaneously, he may display whole body titubation, dysarthria and/or wide based markedly unsteady gait. Over time, the child made forward developmental motor progress and demonstrated improved baseline ataxia. He also showed reduced frequency, duration and severity of daily spells, albeit without complete cessation. Parents note improvement in eye movements, balance and speech with acetazolamide and after eating foods high in carbohydrates. They also report reduced severity of morning episodes when cornstarch is consumed at bedtime. Brain MRIs were normal. EEG showed mild epileptiform discharges at age 23 months and mild slowing posterior dominant rhythm at nine years. Muscle biopsy supported a possible mitochondrial dysfunction (Supplementary Table 1). Blood and muscle mitochondrial sequencing revealed no pathogenic variants and mitochondrial depletion studies were normal. Genomic sequencing of proband and parents revealed a de novo variant in the DAGLA gene NM_006133.3:c.2440G>T, p.(Glu814X) (Supplementary Video 1).
Cases 2–9 are described in the Supplementary material and Supplementary Videos 2–4. All cases are summarized in Table 1 and Supplementary Table 1.
Table 1.
Genotype and phenotype of case cohort
| Case # | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 |
|---|---|---|---|---|---|---|---|---|---|
| Genetics | |||||||||
| DAGLA variant | c.2440G>T p.E814X |
c.2484delC p.E829RfsX6 | c.2551C>T p.Q851X |
c.2456_2457delAC p.H819PfsX5 | c.2484delC p.E829RfsX6 | c.2484delC p.E829RfsX6 | c.2370C>G p.Y790X |
c.2485G>T p.E829X |
c.2613dup p.S872QfsX6 |
| Inheritance | De novo | De novo | De novo | De novo | De novo a | De novo a | De novo | De novo | De novo |
| Phenotype | |||||||||
| Gender | Male | Male | Male | Male | Male | Female | Male | Male | Male |
| Age (year of birth) | 11 years (2010) | 9 years (2012) | 5 years (2016) | 7 years (2014) | 15 years (2006) | 12 years (2009) | 12 years (2009) | 4 years (2017) | 5 years (2016) |
| Motor development | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed |
| Language development | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed | Normal | Normal |
| Age at symptom onset | Birth | 6 months | Birth | Birth | 9 months | First weeks | 3 months | First month | 7 months |
| First symptom | Nystagmus (v,h) | Nystagmus (v,h) | Apnoeic spells; feeding difficulty | Clubfoot | Nystagmus (v) | Nystagmus | Poor head control | Poor head control; pendular nystagmus | Paroxysmal upward eye deviation |
| Nystagmus (v/h) | Y (v,h) | Y (v,h) | N | Y (v) | Y (v) | Y (v) | Y (v,h) | Y (v) | N |
| Nystagmus onset | Birth | 6 months | None | 7 months | 9 months | First weeks | 13 months | First weeks | None |
| Other eye movements | Amblyopia; Sac Pur | N | Sac Pur | N | Sac Pur | Sac Pur | Sac Pur; Abnl Sac | Sac Pur | Sac Pur |
| Dysarthria | Y | Y | Y | Y | Y | Y | Y | N | Y |
| Hypotonia | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Ataxia | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Intellectual disability | Y | Y | Too young to assess | N: low average IQ | Y | Y | Y | N | N |
| MRI brainb | Normal | Normal | Abnormal | Normal | Normal | Normal | Possibly anormal | Abnormal | Possibly anormal |
| Paroxysmal events | |||||||||
| Paroxysmal nystagmus | Y | Y | N | Y | Y | Y | Y | Y | N |
| Paroxysmal chin down posture | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Paroxysmal chin up posture | Y | N | N | N | Y | Y | N | Y | N |
| Paroxysmal upward eye deviation | Y | Y | Y | N | N | N | N | Y | Y |
| Paroxysmal ataxia | Y | Y | N | N | Y | Y | N | Y | N |
| Paroxysmal event triggers | Morning; post sleep; post nap | Morning; post nap; fever | n/a | n/a | Morning; post nap; fever | Morning; post nap; fever | Morning; post sleep; fatigue | Morning; post sleep; post nap | Morning; post sleep |
| Relieving factors | Acetazol; sweets | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Reverse diurnal fluctuation | Y | Y | N | N: possible diurnal | Y | Y | Y | Y | N |
| Spell trajectory over time | Improving | Slightly improved | Improving | Improving | Improving | Improving | Improving | Improving | Improving |
Abnl Sac = abnormal saccades; Acetazol = acetazolamide; h = horizontal; N = no; n/a = not available; Sac Pur = saccadic pursuits; v = vertical; Y = yes.
Probable parental mosaicism.
Details in Supplementary Table 1.
Laboratory methods
Genomic sequencing
Exome or genome sequencing was performed on proband and informative family members using standard methods (Supplementary material).
RNA sequencing
Blood was obtained from the index patient (Case 1) and parents. RNA was isolated using Qiagen Blood RNA kit (Cat. No. 762164). Total RNA [Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus (20040529)] and mRNA libraries (TruSeq Stranded mRNA Library Prep 20020595) were generated and sequenced on NovaSeq to produce ∼100 M reads per subject. RNA sequence reads were aligned and variants called using the DRAGEN v3.84 pipeline.
DAGLA activity evaluation
Cloning of full-length and mutant DAGLA (1–813)
Full-length Homo sapiens DAGLA cDNA was cloned into expression vector pcDNA3.1 with an N-terminal FLAG-tag as previously described.17 For mutant DAGLA, a stop codon was introduced at E814 using QuikChange site-directed mutagenesis.17
Recombinant expression of enzymes by transient transfection of HEK293T cells
Transient transfection of HEK293T cells with DAGLA constructs, either 2 µg of wild-type or mutant DAGLA DNA constructs, were conducted using (JetOPTIMUS-11701) as per the manufacturer’s instructions. Transfected cells were plated 24 h after transfection for experiments.
Assessment of DAGLA activity by gel-based activity-based protein profiling
Lysates from HEK293T cells expressing wild-type DAGLA or mutant DAGLA (1–813) (50 μl, 1 mg/ml) were treated with either FP-Rh (1 µM final concentration) or HT-01 (2 μM final concentration) for 30 min at 37°C. The reactions were quenched by adding 20 µl of 4× sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. After separation by SDS–PAGE, samples were visualized by in-gel fluorescence scanning using the ChemiDoc MP system (Bio-Rad). Band intensities were quantified with Image Lab (5.2.1) software (Bio-Rad).
Immunofluorescence staining
For immunofluorescence, 200 000 cells were plated on poly-D-lysine–coated coverslips (neuVitro GG-18-PDL) and incubated for 24 h. Cells were then washed with chilled 1× PBS and fixed in 4% paraformaldehyde (PFA) before staining with primary antibody against DAGLA (Invitrogen PA5-23765) in 1:200 dilution and incubated overnight. Cells were washed in 1× Tris-buffered saline with 0.1% Tween (TBST), incubated with secondary antibody (Invitrogen A32732) in 1:1000 dilution, and incubated for 40 min. ProLong-Gold antifade reagent with DAPI (Invitrogen P36935) was used as nuclear stain. Pictures were taken using Echo microscope.
Data availability
Supporting data for this study are available from the corresponding author upon request.
Results
Sequencing reveals distal truncating variants in DAGLA
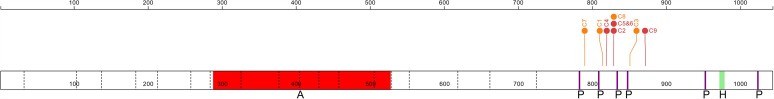
Seven unique apparently de novo DAGLA variants were identified in nine children from eight families. All variants lead to premature protein truncation within an 82 amino acid window. These truncating variants disrupt multiple predicted phosphorylation sites as well as a protein–protein interaction domain previously shown to interact with HOMER scaffolding proteins5 (Fig. 1). RNA was isolated from peripheral blood from the index patient and parents. Two libraries of total RNA and mRNA were generated for each subject. The truncating variant was identified in each library from the proband with 10/25 reads (40%), indicating that the transcript does not undergo nonsense-mediated decay (NMD). The library reads of the proband’s mother and father (26× and 30× coverage, respectively) showed only the wild-type allele.
Figure 1.
DAGLA variants and domains. Schematic of DAGLA (bottom box) with exon–exon boundaries (dotted lines) and codon position (top). Frameshift and nonsense variants [orange (C1,3,7,8) and red (C2, 4, 5, 6, 9) lollipops, respectively] disrupt HOMER motif (green, ‘H’) and phosphorylation sites (purple ‘P’) but leave the active domain (red ‘A’) intact.
Detailed phenotyping identifies a distinct DAGLA associated phenotype
Clinical features of individuals with pathogenic DAGLA variants are summarized in Table 1 and Supplementary Table 1. Eight of nine children are male. All displayed hypotonia and gross motor delay; seven also had language delay, but without autistic features. All children had baseline ataxia and oculomotor abnormalities including nystagmus or saccadic pursuits. In addition, all had prominent paroxysmal events during which there was worsened incoordination and eye movement abnormalities including worsened nystagmus, forced vertical versions or both, often accompanied by compensatory head postures. For seven of nine children, episodic events were most prominent in the morning after awakening or after a nap. For most, paroxysmal episodes reduced in frequency and severity as the child aged.
Functional studies
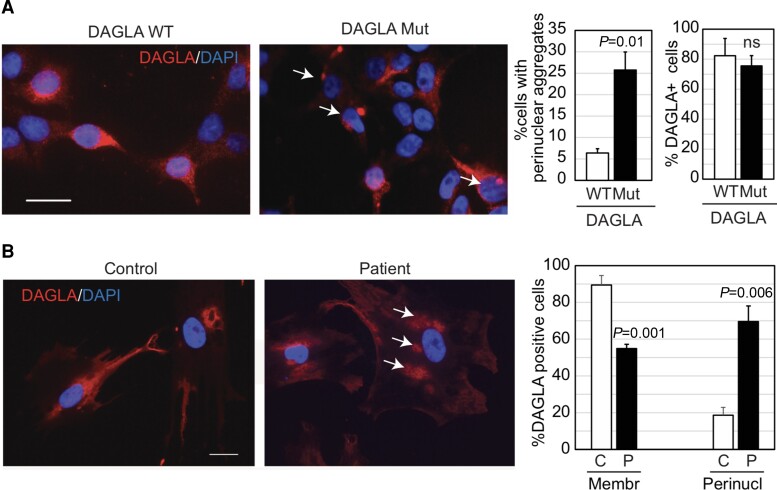
To characterize the effects of the truncating variant on DAGLA protein levels, activity and subcellular localization, fibroblasts from the index case were obtained and immortalized. RNA levels of DAGLA were comparable between patient-derived fibroblasts and cells from an unaffected control. Immunofluorescent staining showed no difference in the proportion of DAGLA-positive cells between patient-derived and control. However, immunofluorescent staining identified distinct perinuclear aggregates of DAGLA in the patient’s cells (Fig. 2A). To confirm this cellular characteristic, HEK293T cells were transfected with either wild-type DAGLA or DAGLA containing the p.Glu814X variant. Lysates from the HEK293T cells were assessed for DAGLA activity by Activity-Based Protein Profiling (ABPP),17 which showed no difference in DAGLA activity (Supplementary Fig. 1). The perinuclear aggregation of DAGLA observed in patient-derived fibroblasts was validated in HEK293T cells expressing the mutant truncated DAGLA, which had an ∼4-fold increase in the cell numbers with perinuclear aggregates relative to isogenic control cells transfected with wild-type DAGLA (P < 0.005, Fig. 2B). The total number of DAGLA-positive cells was comparable between mutant and wild-type-expressing HEK293T cells.
Figure 2.
Immunofluorescence studies. Representative photomicrographs of immunofluorescence for DAGLA in HEK293T cells transfected with either wild-type (WT) or mutant (Mut) DAGLA cDNA (A) and in patient-derived fibroblasts (P) and control (C) cells from an unaffected individual (B). DAPI was used for nuclear counterstain. Scale bars = 50 µm. Accompanying bar graphs represent quantification of the number of cells with perinuclear (perinucl) aggregation (arrows) or membrane-bound (membr) DAGLA. Graphs: bars indicate the mean and error bars the standard deviation. Two-tailed Student’s t-test determined P-values.
Discussion
We define neuro-ocular DAGLA-related syndrome (NODRS), a new clinical syndrome in nine children from eight families with apparently de novo premature termination variants in DAGLA. The neuro-ocular features of this cohort are unique and consistent across all individuals, characterized by ataxia and developmental delays with daily superimposed paroxysmal exacerbations. The episodic events cause worsened ataxia and/or increased oculomotor abnormalities (nystagmus and/or forced vertical versions) with compensatory head postures. The NODRS acronym reflects the distinctive chin-down posture common across the cohort. The majority report a striking pattern of exacerbation in the morning shortly after waking and after naps. The skewed gender ratio (8/9 male) is statistically unlikely, although whether this is a real phenomenon or a chance finding is unclear.
The NODRS phenotype overlaps, but does not conform with several previously defined conditions, including: non-progressive congenital ataxias, infantile nystagmus syndromes, episodic ataxias, and paroxysmal tonic upgaze (PTU). Of these conditions, NODRS most closely resembles complicated forms of PTU. PTU is characterized by episodic, sustained, conjugate upward eye deviation in association with downbeat nystagmus on attempted downgaze, sometimes with mild ataxia, and symptom resolution in a few months to years.18 It is a heterologous condition that, in its complicated form, may manifest additional persistent neurological symptoms including developmental delays, significant ataxia and oculomotor abnormalities.19,20 Episodic ataxia exacerbation, as seen in our cohort, is not generally considered to be part of the PTU phenotype, but several cases displaying this aspect have been reported. For example, one previously described complicated PTU case that was strikingly similar to our cohort demonstrated compensatory head postures and ataxia that were most severe after sleep.21 The molecular aetiology in this case is unknown. Other children with PTU and episodic ataxia resembling our cohort were found to carry pathogenic variants in the CACNA1A gene,20,22,23 although, in contrast to our cohort, PTU attacks in these individuals were relieved by sleep.
Several clinical aspects of NODRS may provide insight into disease pathophysiology. Anatomically, the prominent features of ataxia and eye movement abnormalities localize to the cerebellum and/or brain stem. Brain imaging identified mild inferior vermian hypoplasia for two children in our cohort. For the remainder, these regions were unremarkable, although subtle or non-anatomic changes in these regions cannot be excluded (Table 1 and Supplementary Table 1). Abnormalities in this region are consistent with high expression of DAGLA in cerebellar Purkinje cells both in human and rodent (www.proteinatlas.org).24,25 Most individuals report the unusual phenomenon of symptom onset after waking. Early morning symptoms may suggest impaired energy metabolism exacerbated by the overnight fast. Potential secondary mitochondrial impacts are supported by muscle biopsy abnormalities in Case 1 and myopathic changes on electromyogram in Case 7 (Supplementary Table 1). Worsening symptoms after a nap suggest links to circadian rhythm as opposed to fasting; considering this, the circadian regulation of eCB signalling and its light entrainment role in the rat is of potential relevance.26
The clustering of pathogenic DAGLA variants in our NODRS cohort may provide insight into the mechanism of disease. All variants are located proximally in the final exon of DAGLA, permitting the transcript to escape NMD. The termination variants may disrupt a variable number of known phosphorylation sites and all eliminate a HOMER binding domain, which is critical for the recruitment of DAGLA to the plasma membrane but not for 2-AG production5 (Fig. 1). In concordance with this, transfected cells expressing mutant DAGLA and mutant patient-derived fibroblasts revealed mislocalization of mutant DAGLA with no effect on DAGLA enzymatic activity (Fig. 2 and Supplementary Fig. 1). Variants in DAGLA that both disrupt the HOMER motif and escape NMD must fall within a length of 262 amino acids. The probability of identifying eight independent de novo variants within this constrained location is small (P < 1.78 × 10−5). Although the precise pathophysiological mechanisms underlying NODRS are uncertain, our findings suggest either a dominant negative effect of truncated and mislocalized DAGLA or effects of reduced DAGLA concentration at the plasma membrane. Given the broad and diverse role of eCBs, either or both mechanisms may be at play in distinct brain regions at different times in development, resulting in static and paroxysmal symptoms that may potentially evolve over time. Truncating variants in more proximal regions of DAGLA, predicted to lead to NMD, are vanishingly rare in public databases, implying that globally reduced levels of DAGLA are lethal.27 In contrast, our findings imply that reduced levels at the plasma membrane are compatible with life, although may be associated with disease.
Prior reports suggested that disruption of DAGLA were associated with various neurological conditions but were not conclusive. Autoantibodies to DAGLA were detected in the sera of patients with CNS syndromes who presented with cerebellitis, epilepsy and hippocampal sclerosis or brainstem encephalitis.28 One study implicated a duplication of 20 genes, including DAGLA, with adult-onset spinocerebellar ataxia, while another study reported rare missense variant enrichment in eCB system genes DAGLA and CNR1 within a cohort of individuals with a wide range of neurological phenotypes.14,29 Similarly, DAGLA animal models implicate DAGLA in development and function of the nervous system. These models show some similarity but do not fully recapitulate the NODRS phenotype. This may be due to the changing evoluationary importance of DAGLA in humans or that these models do not reproduce the mislocaltization described here.
In summary, we define NODRS, a novel childhood-onset neuro-ocular phenotype associated with premature termination variants in the last exon of DAGLA. We hypothesize that complete haploinsufficiency of DAGLA is lethal in humans, and that the syndrome described here is caused by reduced concentration of DAGLA at the plasma membrane or potentially a dominant negative effect of mislocalized mutant DAGLA. Development of animal models with truncating variants in the last DAGLA exon will be needed to understand the mechanism by which mislocalized DAGLA leads to disease. Additional studies will also be required to explore whether truncating DAGLA variants may be identified in other patients with a PTU-like phenotype as well as to develop targeted therapeutic options for these children. As the first report conclusively linking cannabinoid dysfunction to human disease, we are hopeful this work will play a pivotal role in better understanding the role of eCBs in health and disease and will also contribute to optimization of the use of cannabis-derived medications for a broad range of disorders.
Supplementary Material
Acknowledgements
We thank the patients and their caregivers, investigators, health care providers, research and laboratory staff that participated in this study. We thank Kimberly Greer ScienceDocs for editorial assistance with the manuscript relating to grammar, style and formatting. Dr Greer was funded by RCIGM.
Abbreviations
- eCB
endocannabinoid
- NODRS
neuro-ocular DAGLA-related syndrome
- PTU
paroxysmal tonic upgaze
Appendix 1
Undiagnosed Disease Network authors
Maria T. Acosta, Margaret Adam, David R. Adams, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Jimmy Bennet, Beverly Berg-Rood, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Brenna Boyd, Lauren C. Briere, Elly Brokamp, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Olveen Carrasquillo, Thomas Cassini, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, Matthew Coggins, F. Sessions Cole, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D’Souza, Hongzheng Dai, Surendra Dasari, Joie Davis, Jyoti G. Dayal, Matthew Deardorff, Esteban C. Dell’Angelica, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L. Doss, Emilie D. Douine, Laura Duncan, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Marni Falk, Liliana Fernandez, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, William A. Gahl, Ian Glass, Bernadette Gochuico, Rena A. Godfrey, Katie Golden-Grant, Madison P. Goldrich, Alana Grajewski, Irma Gutierrez, Don Hadley, Sihoun Hahn, Rizwan Hamid, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yong Huang, Wendy Introne, Rosario Isasi, Kosuke Izumi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Lefkothea Karaviti, Jennifer Kennedy, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N. Kobren, Isaac S. Kohane, Jennefer N. Kohler, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Susan Korrick, Mary Koziura, Seema R. Lalani, Byron Lam, Christina Lam, Grace L. LaMoure, Brendan C. Lanpher, Ian R. Lanza, Kimberly LeBlanc, Brendan H. Lee, Roy Levitt, Richard A. Lewis, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Bryan C. Mak, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Alexa T. McCray, Elisabeth McGee, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava, Paolo M. Moretti, Mariko Nakano-Okuno, Stan F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina GS. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips III, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey, Aaron Quinlan, Wendy Raskind, Archana N. Raja, Deepak A. Rao, Anna Raper, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Natalie Rosenwasser, Francis Rossignol, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Mario Saporta, C. Ron Scott, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, Vandana Shashi, Jimann Shin, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Edward C. Smith, Kevin S. Smith, Emily Solem, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C. Spillmann, Joan M. Stoler, Jennifer A. Sullivan, Kathleen Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Amelia L. M. Tan, Queenie K.-G. Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Brianna M. Tucker, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Stephanie Wallace, Nicole M. Walley, Melissa Walker, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Monika Weisz-Hubshman, Mark Wener, Tara Wenger, Katherine Wesseling Perry, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Kim Worley, Changrui Xiao, Shinya Yamamoto, John Yang, Diane B. Zastrow, Zhe Zhang, Chunli Zhao, Stephan Zuchner, Hugo Bellen, Rachel Mahoney.
Contributor Information
Matthew N Bainbridge, Rady Children’s Institute for Genomic Medicine (RCIGM), San Diego, CA 92123, USA.
Aloran Mazumder, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA 92037, USA.
Daisuke Ogasawara, The Scripps Research Translational Institute, The Scripps Research Institute, La Jolla, CA 92037, USA.
Rami Abou Jamra, Institute of Human Genetics, University Medical Center Leipzig, Leipzig 04103, Germany.
Geneviève Bernard, Department of Neurology and Neurosurgery, McGill University, Montreal, Canada; Department of Pediatrics and Human Genetics, McGill University, Montreal, Canada; Department of Human Genetics, McGill University, Montreal, Canada; Department Specialized Medicine, Division of Medical Genetics, McGill University Health Center, Montreal, Canada; Child Health and Human Development Program, Research Institute of the McGill University Health Center, Montreal, Canada.
Enrico Bertini, Unit of Neuromuscular and Neurodegenerative Disorders, Department of Neurosciences ‘Bambino Gesu’ Children’s Research Hospital, IRCCS, Rome, Italy.
Lydie Burglen, Centre de Référence Malformations et Maladies Congénitales du Cervelet, Département de génétique, AP-HP Sorbonne Université, Hôpital Trousseau, Paris, France; Developmental Brain Disorders Laboratory, Imagine Institute, INSERM UMR 1163, Paris, France.
Heidi Cope, Department of Pediatrics, Division Medical Genetics Durham, Duke University Medical Center, North Carolina, USA.
Ali Crawford, Illumina, San Diego, CA 92122, USA.
Alexa Derksen, Department of Neurology and Neurosurgery, McGill University, Montreal, Canada; Child Health and Human Development Program, Research Institute of the McGill University Health Center, Montreal, Canada.
Leon Dure, Division of Pediatric Neurology, Department of Pediatrics, University of Alabama at Birmingham, Birmingham, AL 35233, USA.
Emily Gantz, Division of Pediatric Neurology, Department of Pediatrics, University of Alabama at Birmingham, Birmingham, AL 35233, USA.
Margarete Koch-Hogrebe, Vestische Kinder- und Jugendklinik, 45711 Datteln, Germany.
Anna C E Hurst, Department of Genetics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
Sonal Mahida, Department of Neurology, Boston Children’s Hospital, Boston, Massachusetts, USA.
Paige Marshall, Department of Neurology, Boston Children’s Hospital, Boston, Massachusetts, USA.
Alessia Micalizzi, Translational Cytogenomics Research Unit, Bambino Gesù Children’s Hospital, IRCCS, Roma, Italy.
Antonio Novelli, Translational Cytogenomics Research Unit, Bambino Gesù Children’s Hospital, IRCCS, Roma, Italy.
Hongfan Peng, The Scripps Research Translational Institute, The Scripps Research Institute, La Jolla, CA 92037, USA.
Rady Children's Institute for Genomic Medicine, Rady Children’s Institute for Genomic Medicine (RCIGM), San Diego, CA 92123, USA.
Diana Rodriguez, Sorbonne Université, INSERM UMR 1141, AP-HP.SU, Centre de Référence Maladies Rares Malformations et Maladies Congénitales du Cervelet & Service de Neuropédiatrie, Hôpital Trousseau, Paris, France.
Shira L Robbins, Ratner Children’s Eye Center at the Shiley Eye Institute; Viterbi Family Department of Ophthalmology, University of California San Diego, La Jolla, CA 92093, USA.
S Lane Rutledge, Division of Pediatric Neurology, Department of Pediatrics, University of Alabama at Birmingham, Birmingham, AL 35233, USA; Department of Genetics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
Roberta Scalise, Department of Developmental Neuroscience, IRCCS Stella Maris Foundation, Pisa, Italy; Tuscan PhD Program of Neuroscience, University of Florence, Pisa and Siena, Florence, Italy.
Sophia Schließke, Institute of Human Genetics, University Medical Center Leipzig, Leipzig 04103, Germany.
Vandana Shashi, Department of Pediatrics, Division Medical Genetics Durham, Duke University Medical Center, North Carolina, USA.
Siddharth Srivastava, Department of Neurology, Boston Children’s Hospital, Boston, Massachusetts, USA.
Isabella Thiffault, Genomic Medicine Center, Children’s Mercy Hospital, Kansas City, Missouri, USA; Faculty of Medicine, University of Missouri Kansas City, Kansas City, Missouri, USA; Department of Pathology, Children’s Mercy Hospital, Kansas City, Missouri, USA.
Sarah Topol, The Scripps Research Translational Institute, The Scripps Research Institute, La Jolla, CA 92037, USA.
Leila Qebibo, Centre de Référence Malformations et Maladies Congénitales du Cervelet, Département de génétique, AP-HP Sorbonne Université, Hôpital Trousseau, Paris, France.
Dagmar Wieczorek, Institute of Human Genetics, Medical Faculty and University Hospital Düsseldorf, Heinrich-Heine-University Düsseldorf, 40225, Düsseldorf, Germany.
Benjamin Cravatt, The Scripps Research Translational Institute, The Scripps Research Institute, La Jolla, CA 92037, USA.
Svasti Haricharan, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA 92037, USA.
Ali Torkamani, The Scripps Research Translational Institute, The Scripps Research Institute, La Jolla, CA 92037, USA.
Jennifer Friedman, Rady Children’s Institute for Genomic Medicine (RCIGM), San Diego, CA 92123, USA; Division of Neurology, Rady Children’s Hospital San Diego, CA 92123, USA; Department of Neurosciences, University of California La Jolla, CA 92037, USA; Department of Pediatrics, University of California La Jolla, CA 92037, USA.
Undiagnosed Disease Network:
Maria T Acosta, Margaret Adam, David R Adams, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A Ashley, Mahshid S Azamian, Carlos A Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H Beggs, Edward Behrens, Gill Bejerano, Jimmy Bennet, Beverly Berg-Rood, Jonathan A Bernstein, Gerard T Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Brenna Boyd, Lauren C Briere, Elly Brokamp, Gabrielle Brown, Elizabeth A Burke, Lindsay C Burrage, Manish J Butte, Peter Byers, William E Byrd, John Carey, Olveen Carrasquillo, Thomas Cassini, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Gary D Clark, Terra R Coakley, Laurel A Cobban, Joy D Cogan, Matthew Coggins, F Sessions Cole, Heather A Colley, Cynthia M Cooper, Heidi Cope, William J Craigen, Andrew B Crouse, Michael Cunningham, Precilla D’Souza, Hongzheng Dai, Surendra Dasari, Joie Davis, Jyoti G Dayal, Matthew Deardorff, Esteban C Dell’Angelica, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L Doss, Emilie D Douine, Laura Duncan, Dawn Earl, David J Eckstein, Lisa T Emrick, Christine M Eng, Cecilia Esteves, Marni Falk, Liliana Fernandez, Elizabeth L Fieg, Paul G Fisher, Brent L Fogel, Irman Forghani, William A Gahl, Ian Glass, Bernadette Gochuico, Rena A Godfrey, Katie Golden-Grant, Madison P Goldrich, Alana Grajewski, Irma Gutierrez, Don Hadley, Sihoun Hahn, Rizwan Hamid, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M Hisama, Ingrid A Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yong Huang, Wendy Introne, Rosario Isasi, Kosuke Izumi, Fariha Jamal, Gail P Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Lefkothea Karaviti, Jennifer Kennedy, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N Kobren, Isaac S Kohane, Jennefer N Kohler, Deborah Krakow, Donna M Krasnewich, Elijah Kravets, Susan Korrick, Mary Koziura, Seema R Lalani, Byron Lam, Christina Lam, Grace L LaMoure, Brendan C Lanpher, Ian R Lanza, Kimberly LeBlanc, Brendan H Lee, Roy Levitt, Richard A Lewis, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K Loo, Joseph Loscalzo, Richard L Maas, Ellen F Macnamara, Calum A MacRae, Valerie V Maduro, Bryan C Mak, May Christine V Malicdan, Laura A Mamounas, Teri A Manolio, Rong Mao, Kenneth Maravilla, Ronit Marom, Gabor Marth, Beth A Martin, Martin G Martin, Julian A Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Alexa T McCray, Elisabeth McGee, Heather Mefford, J Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava, Paolo M Moretti, Mariko Nakano-Okuno, Stan F Nelson, John H Newman, Sarah K Nicholas, Deborah Nickerson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P Orengo, Laura Pace, Stephen Pak, J Carl Pallais, Christina G S Palmer, Jeanette C Papp, Neil H Parker, John A Phillips III, Jennifer E Posey, Lorraine Potocki, Barbara N Pusey, Aaron Quinlan, Wendy Raskind, Archana N Raja, Deepak A Rao, Anna Raper, Genecee Renteria, Chloe M Reuter, Lynette Rives, Amy K Robertson, Lance H Rodan, Jill A Rosenfeld, Natalie Rosenwasser, Francis Rossignol, Maura Ruzhnikov, Ralph Sacco, Jacinda B Sampson, Mario Saporta, C Ron Scott, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A Scott, Vandana Shashi, Jimann Shin, Edwin K Silverman, Janet S Sinsheimer, Kathy Sisco, Edward C Smith, Kevin S Smith, Emily Solem, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C Spillmann, Joan M Stoler, Jennifer A Sullivan, Kathleen Sullivan, Angela Sun, Shirley Sutton, David A Sweetser, Virginia Sybert, Holly K Tabor, Amelia L M Tan, Queenie K-G Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J Tifft, Camilo Toro, Alyssa A Tran, Brianna M Tucker, Tiina K Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P Vogel, Colleen E Wahl, Stephanie Wallace, Nicole M Walley, Melissa Walker, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F Wangler, Patricia A Ward, Daniel Wegner, Monika Weisz-Hubshman, Mark Wener, Tara Wenger, Katherine Wesseling Perry, Monte Westerfield, Matthew T Wheeler, Jordan Whitlock, Lynne A Wolfe, Kim Worley, Changrui Xiao, Shinya Yamamoto, John Yang, Diane B Zastrow, Zhe Zhang, Chunli Zhao, Stephan Zuchner, Hugo Bellen, and Rachel Mahoney
Funding
M.B. and J.F. receive funding from Rady Children’s Institute for Genomic Medicine (RCIGM) and support for this work was funded in part by Clinical and Translational Science Award (5 UL1 TR002550); S.S. receives funding from NIH-NINDS (K23NS119666). Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG007672 (Duke University). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health; G.B.’s research reported in this manuscript was supported by grants from the Canadian Institute of Health Research (377869, 426534) and her research was enabled in part by support provided by Compute Canada (www.computecanada.ca). G.B. received a Clinical Research Scholar Junior 1 award from the Fonds de Recherche du Quebec – Santé (FRQS) (2012–2016), the New Investigator Salary Award from the Canadian Institutes of Health Research (CIHR) (2017–2022) and a Clinical Research Scholar Senior award from the FRQS (2022–2025); G.B.’s research was funded by research grants from the Canadian Institutes for Health Research (project grant 426534 and 201610PJT- 377869) and her research was enabled in part by support provided by Compute Canada (www.computecanada.ca). A.D. is funded by CIHR, Fondation du Grand défi Pierre Lavoie and Healthy Brains for Healthy Lives (HBHL). A.T., S.T. and H.P. receive funding through NCATS UL1TR002550, Shaffer Family Foundation and the Anne and Henry Zarrow Foundation; IT work was made possible by the generous gifts to Children’s Mercy Research Institute and Genomic Answers for Kids program at Children’s Mercy Kansas City.
Competing interests
All authors declare no actual conflicts of interest related to this work. The following financial and personal relationships are disclosed: S.S. has received consulting fees from GLG, Guidepoint (which connected to a client, Fortress Biotech), Novartis, ExpertConnect, Orchard Therapeutics; G.B. is/was a consultant for Passage Bio Inc. (2020–2021) and Ionis (2019). She is/was a site investigator for the Alexander’s disease trial of Ionis (2021), Metachromatic leukodystrophy of Shire/Takeda (2020–2021), Krabbe and GM1 gene therapy trials of Passage Bio, and Adrenoleukodystrophy/Hematopoietic stem cell transplantation natural history study of Bluebird Bio (2019) and a site sub-investigator for the MPS II gene therapy trial of Regenxbio (2021). She has received an unrestricted educational grant from Takeda (2021). She serves on the scientific advisory board of the Pelizaeus-Merzbacher Foundation and is the Chair of the Medical Advisory Board of the United Leukodystrophy Foundation. She is on the editorial boards of Neurology Genetics, Frontiers in Neurology – Neurogenetics and Journal of Medical Genetics; A.C. is an employee and shareholder of Illumina, Inc.; J.F. conducts Clinical Trials with Biogen (Angelman’s Syndrome); J.F.’s spouse is Founder and Principal of Friedman Bioventure, which holds a variety of publicly traded and private biotechnology interests; A.T. declares ownership in geneXwell Inc., advisory for Vivid Genomics, Seqster and InsideTracker, and grant funding from Takeda.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Garcia-Arencibia M, Molina-Holgado E, Molina-Holgado F. Effect of endocannabinoid signalling on cell fate: Life, death, differentiation and proliferation of brain cells. Br J Pharmacol. 2019;176(10):1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu HC, Mackie K. Review of the endocannabinoid system. Biol Psychiatry Cogn Neurosci Neuroimaging. 2021;6(6):607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iannotti FA, Di Marzo V, Petrosino S. Endocannabinoids and endocannabinoid-related mediators: Targets, metabolism and role in neurological disorders. Prog Lipid Res. 2016;62:107–128. [DOI] [PubMed] [Google Scholar]
- 4. Murataeva N, Straiker A, MacKie K. Parsing the players: 2-Arachidonoylglycerol synthesis and degradation in the CNS. Br J Pharmacol. 2014;171(6):1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jung KM, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D. A key role for diacylglycerol lipase-α in metabotropic glutamate receptor-dependent endocannabinoid mobilization. Mol Pharmacol. 2007;72(3):612–621. [DOI] [PubMed] [Google Scholar]
- 6. Bisogno T, Howell F, Williams G, et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 2003;163(3):463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia del Caño G, Montaña M, Aretxabala X, et al. Nuclear phospholipase C-β1 and diacylglycerol LIPASE-α in brain cortical neurons. Adv Biol Regul. 2014;54(1):12–23. [DOI] [PubMed] [Google Scholar]
- 8. Nielsen JE, Rolland AD, Rajpert-De Meyts E, et al. Characterisation and localisation of the endocannabinoid system components in the adult human testis. Sci Rep. 2019;9(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang L, Kolaj M, Renaud LP. Endocannabinoid 2-AG and intracellular cannabinoid receptors modulate a low-threshold calcium spike-induced slow depolarizing afterpotential in rat thalamic paraventricular nucleus neurons. Neuroscience. 2016;322:308–319. [DOI] [PubMed] [Google Scholar]
- 10. Viader A, Ogasawara D, Joslyn CM, et al. A chemical proteomic atlas of brain serine hydrolases identifies cell type-specific pathways regulating neuroinflammation. Elife. 2016;5:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baggelaar MP, Maccarrone M, van der Stelt M. 2-Arachidonoylglycerol: A signaling lipid with manifold actions in the brain. Prog Lipid Res. 2018;71:1–17. [DOI] [PubMed] [Google Scholar]
- 12. Martella A, Sepe RM, Silvestri C, et al. Important role of endocannabinoid signaling in the development of functional vision and locomotion in zebrafish. FASEB J. 2016;30:4275–4288. [DOI] [PubMed] [Google Scholar]
- 13. Powell DR, Gay JP, Wilganowski N, et al. Diacylglycerol lipase α knockout mice demonstrate metabolic and behavioral phenotypes similar to those of cannabinoid receptor 1 knockout mice. Front Endocrinol (Lausanne). 2015;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith DR, Stanley CM, Foss T, Boles RG, McKernan K. Rare genetic variants in the endocannabinoid system genes CNR1 and DAGLA are associated with neurological phenotypes in humans. PLoS One. 2017;12(11):e0187926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keimpema E, Alpar A, Howell F, et al. Diacylglycerol lipase a manipulation reveals developmental roles for intercellular endocannabinoid signaling. Sci Rep. 2013;3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogasawara D, Deng H, Viader A, et al. Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc Natl Acad Sci USA. 2016;113(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ouvrier R, Billson F. Paroxysmal tonic upgaze of childhood—A review. Brain Dev. 2005;27(3 SPEC. ISS.):185–188. [DOI] [PubMed] [Google Scholar]
- 19. Hayman M, Harvey AS, Hopkins IJ, Kornberg AJ, Coleman LT, Shield LK. Paroxysmal tonic upgaze: A reappraisal of outcome. Ann Neurol. 1998;43(4):514–520. [DOI] [PubMed] [Google Scholar]
- 20. Humbertclaude V, Krams B, Nogue E, et al. Benign paroxysmal torticollis, benign paroxysmal vertigo, and benign tonic upward gaze are not benign disorders. Dev Med Child Neurol. 2018;60(12):1256–1263. [DOI] [PubMed] [Google Scholar]
- 21. Apak RA, Topçu M. A case of paroxysmal tonic upgaze of childhood with ataxia. Eur J Paediatr Neurol. 1999;3(3):129–131. [DOI] [PubMed] [Google Scholar]
- 22. Roubertie A, Echenne B, Leydet J, et al. Benign paroxysmal tonic upgaze, benign paroxysmal torticollis, episodic ataxia and CACNA1A mutation in a family. J Neurol. 2008;255(10):1600–1602. [DOI] [PubMed] [Google Scholar]
- 23. Blumkin L, Leshinsky-Silver E, Michelson M, et al. Paroxysmal tonic upward gaze as a presentation of de-novo mutations in CACNA1A. Eur J Paediatr Neurol. 2015;19(3):292–297. [DOI] [PubMed] [Google Scholar]
- 24. Oudin MJ, Hobbs C, Doherty P. DAGL-dependent endocannabinoid signalling: Roles in axonal pathfinding, synaptic plasticity and adult neurogenesis. Eur J Neurosci. 2011;34(10):1634–1646. [DOI] [PubMed] [Google Scholar]
- 25. Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 26. Sládek M, Liška K, Houdek P, Sumová A. Modulation of single cell circadian response to NMDA by diacylglycerol lipase inhibition reveals a role of endocannabinoids in light entrainment of the suprachiasmatic nucleus. Neuropharmacology. 2021;185:108455. [DOI] [PubMed] [Google Scholar]
- 27. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miske R, Scharf M, Hahn S, et al. Identification of DAGLA as an autoantibody target in cerebellar or hippocampal degeneration. In: 12th International Congress on Autoimmunity (ICA), Virtueller Kongress. 2021. [Google Scholar]
- 29. Knight MA, Hernandez D, Diede SJ, et al. A duplication at chromosome 11q12.2-11q12.3 is associated with spinocerebellar ataxia type 20. Hum Mol Genet. 2008;17(24):3847–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data for this study are available from the corresponding author upon request.