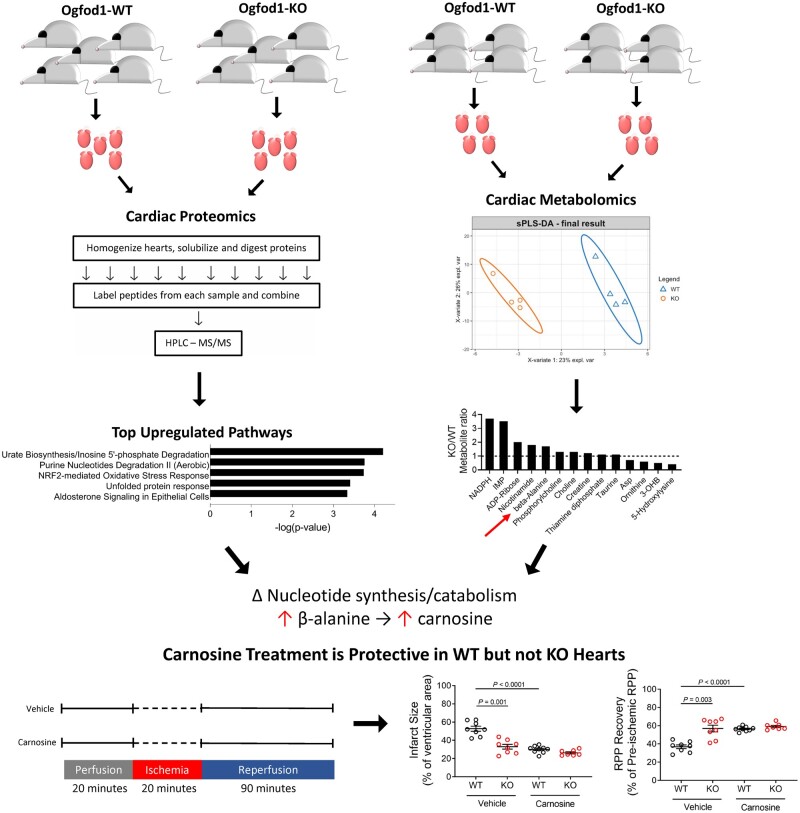
Abstract
Aims
Prolyl hydroxylation is a post-translational modification that regulates protein stability, turnover, and activity. The proteins that catalyze prolyl hydroxylation belong to the 2-oxoglutarate- and iron-dependent oxygenase family of proteins. 2-oxoglutarate- and iron-dependent oxygenase domain-containing protein 1 (Ogfod1), which hydroxylates a proline in ribosomal protein s23 is a newly described member of this family. The aims of this study were to investigate roles for Ogfod1 in the heart, and in the heart’s response to stress.
Methods and results
We isolated hearts from wild-type (WT) and Ogfod1 knockout (KO) mice and performed quantitative proteomics using tandem mass Tag labelling coupled to liquid chromatography and tandem mass spectrometry (LC-MS/MS) to identify protein changes. Ingenuity pathway analysis identified ‘Urate Biosynthesis/Inosine 5′-phosphate Degradation’ and ‘Purine Nucleotides Degradation II (Aerobic)’ as the most significantly enriched pathways. We performed metabolomics analysis and found that both purine and pyrimidine pathways were altered with the purine nucleotide inosine 5′-monophosphate showing a 3.5-fold enrichment in KO hearts (P = 0.011) and the pyrimidine catabolism product beta-alanine showing a 1.7-fold enrichment in KO hearts (P = 0.014). As changes in these pathways have been shown to contribute to cardioprotection, we subjected isolated perfused hearts to ischaemia and reperfusion (I/R). KO hearts showed a 41.4% decrease in infarct size and a 34% improvement in cardiac function compared to WT hearts. This protection was also evident in an in vivo I/R model. Additionally, our data show that treating isolated perfused WT hearts with carnosine, a metabolite of beta-alanine, improved protection in the context of I/R injury, whereas treating KO hearts with carnosine had no impact on recovery of function or infarct size.
Conclusions
Taken together, these data show that Ogfod1 deletion alters the myocardial proteome and metabolome to confer protection against I/R injury.
Keywords: Cardioprotection, Ischaemia–reperfusion injury, Nucleotide metabolism, Proteomics, Metabolomics
Graphical Abstract
Graphical Abstract.
Translational perspective
Heart disease is the leading cause of death in the USA. In characterizing the cardiovascular effects of deleting the prolyl hydroxylase Ogfod1 (2-oxoglutarate- and iron-dependent oxygenase domain-containing protein 1) and investigating its role in disease pathology, we found that deleting Ogfod1 protected hearts against ex vivo and in vivo ischaemia and reperfusion injury. Ogfod1-KO hearts showed significant metabolomic and proteomic changes that supported altered purine and pyrimidine nucleotide synthesis and turnover. Beta-alanine, a precursor of the anti-oxidant carnosine and a product of pyrimidine degradation, accumulated in KO hearts to help confer cardioprotection. Altogether, these data suggest a role for Ogfod1 down-regulation as a therapeutic strategy for heart disease.
1. Introduction
Ogfod1 (2-oxoglutarate- and iron-dependent oxygenase domain-containing protein 1) is a newly described member of a family of enzymes1,2 that utilize molecular oxygen to catalyze the hydroxylation of specific proline residues in proteins through a reaction that couples the decarboxylation of α-ketoglutarate to the generation of succinate and carbon dioxide.2,3 Oxygen is a substrate for these enzymes and many have Km values for oxygen that are in the range to sense physiological changes in oxygen.4 Hypoxia-inducible factor–prolyl hydroxylase domain enzymes (HIF-PHDs) are well-known members of this family which have been shown to regulate levels of hypoxia-inducible factor (HIF) by hydroxylation of a specific proline that targets HIF for degradation. Under conditions of low oxygen, HIF-PHDs are inhibited leading to stabilization and activation of HIF and transcription of HIF targets.
Ogfod1 catalyzes hydroxylation of Pro-62 in the small ribosomal protein s23 (Rps23),5–7 a component of the 40S ribosomal subunit. Ogfod1 knockdown or ablation has been shown under some conditions to lead to phosphorylation of eIF2α,6,8,9 translation repression,9–11 and stress granule formation.6 Ogfod1 knockdown was also shown to protect cultured Nalm6 cells from stress-induced cell death.12 Ogfod1 is expressed in the heart and we have shown previously that its expression levels decrease during differentiation.10 However, as the role of Ogfod1 in adult cardiomyocytes is unknown, the aim of this study is to characterize the function of Ogfod1 in the heart.
Previous studies have shown that changes in OGFOD1 levels can alter the proteomic and metabolic landscape and may therefore be involved in how cells respond to stress. To test this, we utilized a mouse in which Ogfod1 gene function is ablated, and subjected this mouse to ex vivo and in vivo ischaemia–reperfusion (I/R) injury. These studies provide the first in vivo characterization of Ogfod1 function. Here, we show that the cardiac proteome of Ogfod1 knockout (KO) mice exhibited changes in metabolic pathways, including nucleotide degradation. Ogfod1-KO hearts showed an increase in beta-alanine, which is a product of pyrimidine degradation13 and a precursor for histidine dipeptides such as carnosine. Carnosine has been shown to be cardioprotective,14,15 and over-expressing carnosine synthase, the enzyme that converts beta-alanine and histidine to carnosine, was sufficient to protect mouse hearts from I/R injury.14 In this study, we show that Ogfod1 deletion increases beta-alanine, which in turn increases carnosine and confers cardioprotection in I/R injury.
2. Methods
2.1 Mice
Ogfod1-KO mice were generated as previously described.6 All animal studies were performed in a manner consistent with the recommendations established by the Guide for the Care and Use of Laboratory Animals (National Institutes of Health), and all animal protocols were approved by the National Heart, Lung and Blood Institute’s Animal Care and Use Committee. Age-matched Ogfod1- wild-type (WT) and Ogfod1-KO male mice aged 2–6 months were used for this study. In accordance with approved guidelines, mice were euthanized by intraperitoneal injection of 125 mg/kg pentobarbital sodium salt (Sigma #P3761). Euthanasia was confirmed by failure of the animal to respond to firm toe pinch.
2.2 Ischaemia–reperfusion injury ex vivo
In brief, hearts were quickly dissected from mice, perfused in Langendorff mode, and subjected to 20 min of equilibration perfusion, 20 min of ischaemia, and 90 min of reperfusion as previously described.16 For carnosine treatment, L-carnosine (Sigma #C9625) was dissolved in Krebs buffer to a final concentration of 1 mM, and hearts were perfused in Krebs + carnosine for 20 min prior to beginning I/R, and hearts were also perfused in Krebs + carnosine throughout the reperfusion period. We measured recovery of left ventricular developed pressure (LVDP) and heart rate (HR) during reperfusion following ischaemia, and determined the recovery of rate pressure product (RPP = HR × LVDP) of contractile function. Following reperfusion, hearts were perfused with 1% 2,3,5-triphenyltetrazolium chloride (TTC), and incubated in TTC for 30 min at 37°C. TTC-treated hearts were fixed in 10% formaldehyde, cut into cross-sectional slices, and images were captured using a Leica MDZL3 dissecting microscope. Infarct size was calculated as a percentage of total cross-sectional area for each image. Average infarct size for each heart was calculated from six images.
2.3 In vivo left anterior descending coronary artery ligation
Mice were anaesthetized with 1–3% isofluorane and intubated prior to being kept on a ventilator. The thoracic cavity was accessed by first making an incision along the ventral midline, then cutting the intercostal muscle between the 3rd and 4th ribs. The ribs were retracted to expose the heart. Once the left atrial appendage was identified, a 7-0 silk braided suture was placed perpendicular to the long axis of the heart around the left anterior descending coronary (LAD) artery. A slipknot was used to ligate the artery for 40 min. At the end of the 40-min ischaemia period, the slipknot was released for 24 h of reperfusion. Directly following release of the slipknot, the lungs were fully expanded using airway pressure not exceeding 30 cm of water, and the ribs were closed using 5-0 silk braided sutures, 0.5% Bupivacaine (up to 2 mg/kg) was placed on the intercostal muscles along the site of the incision to relieve pain, and 5-0 sutures were used to close the chest. The skin was closed with surgical staples. After 24 h of reperfusion, mice were sacrificed as described above. Hearts were quickly excised, cannulated through the aorta, and perfused via Langendorff retrograde with Krebs buffer for 5 min to remove blood. The LAD artery ligation suture was retied in the same location as during the 40 min occlusion. The heart was perfused with 0.5 mL 10% Evans blue solution to delineate area at risk, sectioned, and incubated in 1% TTC at 37°C for 20 min to distinguish infarcted tissue from normal tissue. Ten to 14 images were captured of each heart using a Leica MDZL3 dissecting microscope, and the area at risk and infarct sizes for each heart were determined using Image J. Tissue processing, imaging, area at risk measurements, and infarct size measurements were done blinded.
2.4 Metabolite levels in ischaemic hearts
In brief, hearts were quickly dissected from mice, perfused in Langendorff mode, and subjected to 20 min of equilibration perfusion and 10 min of ischaemia. Hearts were then snap-frozen using liquid nitrogen-chilled clamps and ground in 4% perchloric acid in liquid nitrogen. The metabolite suspension was cleared using centrifugation, and the pellet was dried and weighed as an indicator of dry weight. The pH of the cleared metabolite solution was neutralized using 4M potassium carbonate. After clearing the lysate once again, inosine levels were determined using an inosine assay kit (Abcam #ab126286) according to the manufacturer’s protocol, and hypoxanthine/xanthine levels were determined using a xanthine/hypoxanthine assay kit (Abcam #ab155900) according to the manufacturer’s protocol.
2.5 Metabolomics and data processing
Hearts were dissected from mice, and immediately snap-frozen using liquid nitrogen-chilled clamps. Hearts were then submitted to Human Metabolome Technologies America, Inc. for capillary electrophoresis–mass spectrometry (CE-MS). Metabolites with >50% missing value were excluded from the analysis. For the rest of the metabolites, the missing values were imputed with half of the minimum measurement value of the respective metabolites measured and scaled. Sparse partial least square discriminant analysis (sPLS-DA) from mixOmics17 was used to identify metabolites with the variable importance in prediction (VIP) score >1.
2.6 Preparation of protein extracts for mass spectrometry
Hearts were dissected from mice, and immediately flash-frozen using liquid nitrogen-chilled clamps. Protocol details provided in Supplementary material online.
2.7 Offline HPLC peptide fractionation and mass spectrometry
Protocol details provided in Supplementary material online.
2.8 Mass spectrometry data processing
Protocol details provided in Supplementary material online.
2.9 Statistics
Results are shown as fold change (KO/WT), percent difference from WT, or as mean ± standard error of the mean (SEM). A Student’s t-test with Welch’s correction or a Mann–Whitney test was used to compare two groups depending upon the normalcy of the data spread. Brown–Forsythe and Dunnett’s T3 multiple comparisons tests were used to compare more than two groups. A P-value of <0.05 was considered significant.
3. Results
3.1 Ablating Ogfod1 alters proteins in nucleotide metabolism
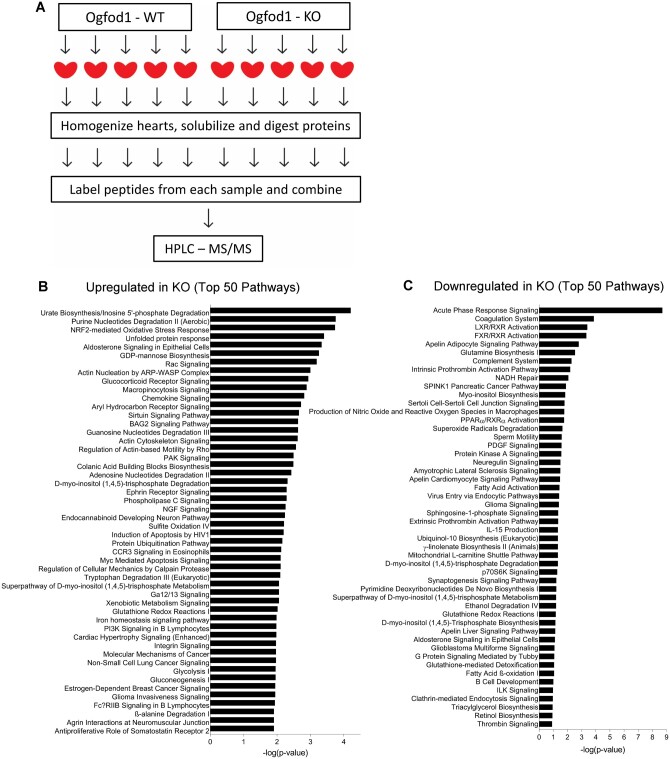
To identify changes in the cardiac proteome that resulted from Ogfod1 ablation, we performed proteomic analysis using tandem mass spectrometry on hearts from 5 Ogfod1-KO and 5 Ogfod1-WT mice. Ogfod1-KO mice were generated as described by Singleton et al.6 and were viable and fertile. We tagged tryptic peptides from each mouse heart with a unique isobaric mass tag, combined tagged tryptic peptides from all 10 mouse hearts, and performed off-line high-performance liquid chromatography followed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Figure 1A). We found that 203 of the 4810 identified proteins were significantly altered in the KO hearts. Of these, 138 (68%) were increased and 65 (32%) were decreased (Figure 1B and C, Supplementary material online, Table S1). Ingenuity pathway analysis showed that ‘Urate Biosynthesis/Inosine 5′-phosphate Degradation’ and ‘Purine Nucleotides Degradation II (Aerobic)’ (P = 0.000062 and P = 0.00017, respectively) were the most significantly enriched pathways. Of note, inosine monophosphate dehydrogenase 2 (Impdh2), xanthine dehydrogenase/oxidase (Xdh/Xo), and hypoxanthine phosphoribosyl transferase, three key enzymes in purine metabolism, were increased in the Ogfod1-KO hearts (7.9% increase P = 0.015, 8.9% increase P = 0.039, and 18.1% increase P = 0.056, respectively). These changes support alterations in purine metabolism in KO hearts.
Figure 1.
Ogfod1 ablation alters the cardiac proteome. (A) Schematic of workflow in isolating hearts from Ogfod1-WT and Ogfod1-KO hearts for proteomics. (B and C) Ingenuity® Pathway Analysis (IPA®) analysis on the 138 significantly increased proteins (B), and the 65 significantly decreased proteins (C) identified by mass spectrometry. Bar length represents the significance with which each pathway was identified on the –log(P-value) scale. n = 5 biological replicates per group. Student’s t-test was used to determine significance between groups. A P-value of <0.05 was considered significant. HPLC-MS/MS, high-performance liquid chromatography tandem mass spectrometry.
3.2 Ablating Ogfod1 alters the metabolic profile
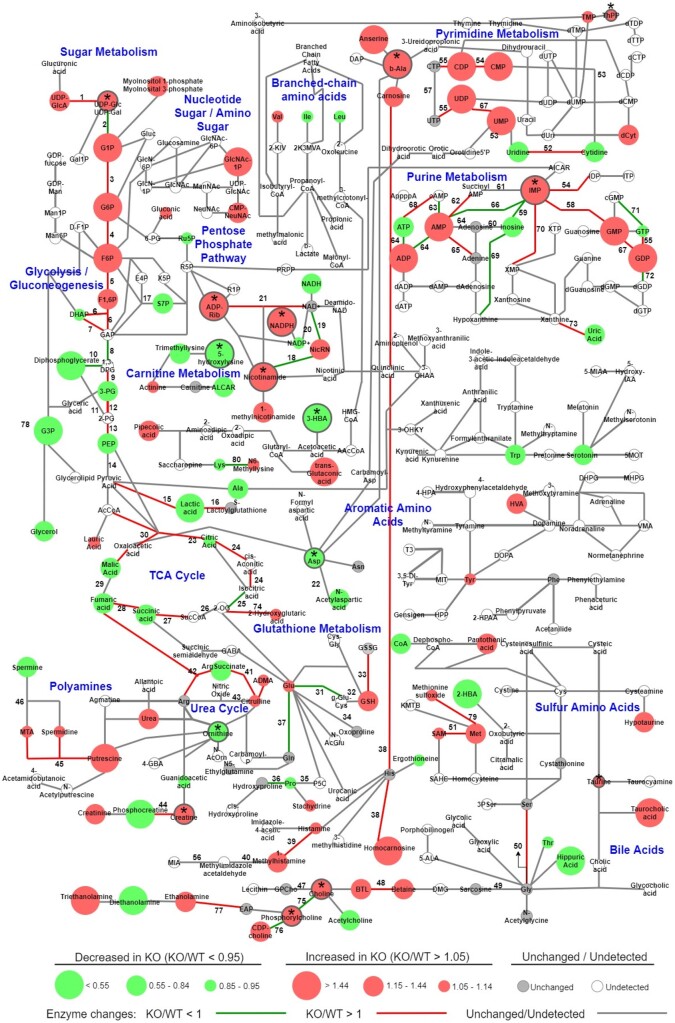
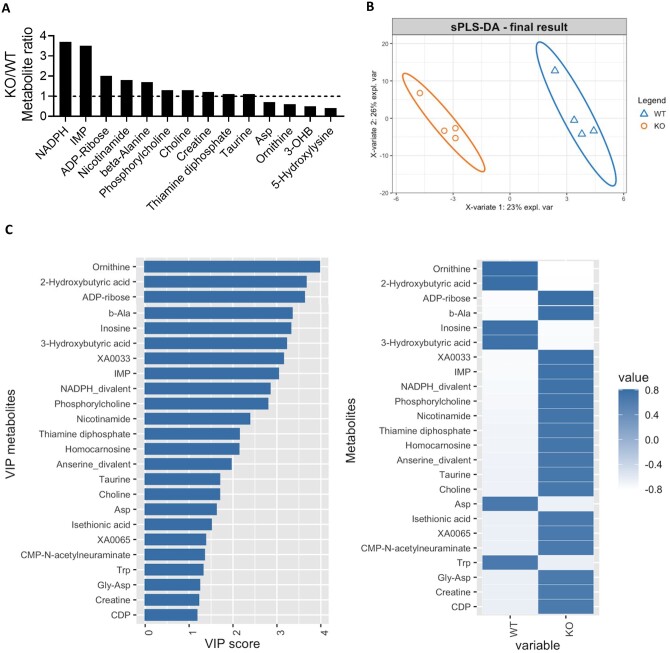
In the proteomic study, we identified changes in pathways involved in purine degradation and inosine 5′-monophosphate (IMP) metabolism. To determine if these proteomic changes led to metabolic changes, we performed a metabolomic analysis. We isolated hearts from WT and KO mice and processed these hearts for CE-MS. We then overlayed the metabolomic and proteomic studies to determine how the two datasets correlated. As shown in Figure 2, we identified metabolites belonging to purine metabolism, pyrimidine metabolism, glycolysis, gluconeogenesis, carnitine metabolism, and tricarboxylic acid cycle, among other pathways. To correlate the proteomic and metabolomic datasets, changes in the abundances of enzymes identified in the proteomics were overlayed with the metabolite changes (Supplementary material online, Tables S2 and S3). Focusing on the significant metabolite changes, which are reported in arbitrary units (a.u.), the purine nucleotide IMP, which is a key component of the purine synthesis and salvage pathways, showed one of the largest changes in KO hearts with a 3.5-fold increase over WT (WT: 3.7 × 10−4 ± 1.2 × 10−4 a.u., KO: 1.3 × 10−3 ± 1.9 × 10−4 a.u., P = 0.011) (Figure 3A). This change was second only to NADPH, which is produced in several metabolic pathways including the pentose phosphate shunt (WT: 1.6 × 10−4 ± 8.1 × 10−5 a.u., KO: 6.1 × 10−4 ± 1.3 × 10−4 a.u., P = 0.035). We performed sPLS-DA on the metabolomics data (Figure 3B), and assigned the metabolites VIP scores that represented their importance in predicting the group to which the metabolite belonged. Beta-alanine (WT: 7.2 × 10−4 ± 1.1 × 10−4 a.u., KO: 1.2 × 10−3 ± 4.3 × 10−5 a.u., P = 0.014), a by-product of pyrimidine catabolism, as well as inosine (WT: 5.2 × 10−4 ± 3.8 × 10−4 a.u., KO: 3.3 × 10−4 ± 7.5 × 10−5 a.u., P = 0.661) and IMP in purine synthesis and salvage, were all among the top VIP’s (Figure 3C), making them and their respective pathways primary in distinguishing WT and KO hearts. These data are consistent with the alteration in nucleotide metabolism that was identified in the proteomic analysis.
Figure 2.
Ogfod1 deletion leads to changes in the cardiac metabolome. Metabolomics results (circles) from Ogfod1-WT and Ogfod1-KO hearts correlated with the proteomics results (lines connecting the circles) for enzymes responsible for the indicated metabolite connections. Colour and size of circles indicates direction and magnitude of change, while colour of lines indicates the directionality of the protein change as described in the key at the bottom of the figure. Asterisks combined with grey outline of red or green circles indicate significant metabolite changes. n = 4 biological replicates per group. Student’s t-test was used to determine significance between groups. A P-value of <0.05 was considered significant.
Figure 3.
Ogfod1-KO hearts show significant metabolite changes. (A) Significant metabolite changes in KO/WT hearts. KO/WT = 1 is shown as a horizontal dotted line. (B) Final results of sparse partial least squares-discriminant analysis (sPLS-DA) on the metabolomics data. (C) Left panel: Variable importance in prediction (VIP) scores for metabolites. Right panel: Mean relative expression for the indicated metabolite in each of the two groups. n = 5 biological replicates per group. Student’s t-test was used to determine significance between groups. A P-value of <0.05 was considered significant.
3.3 Ogfod1 ablation leads to protection against ischaemia–reperfusion injury
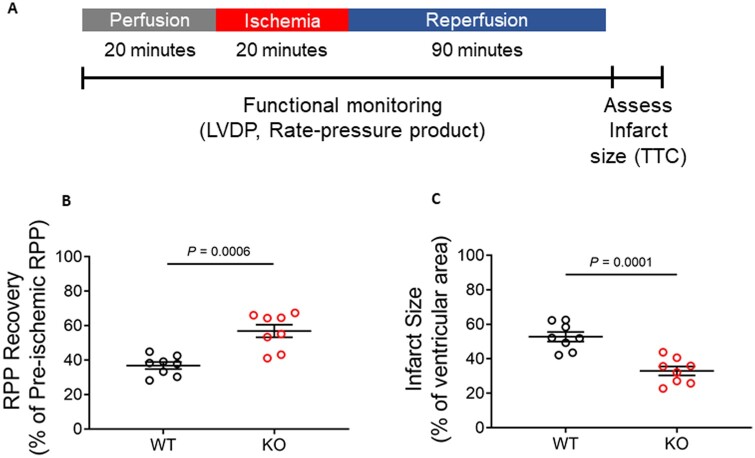
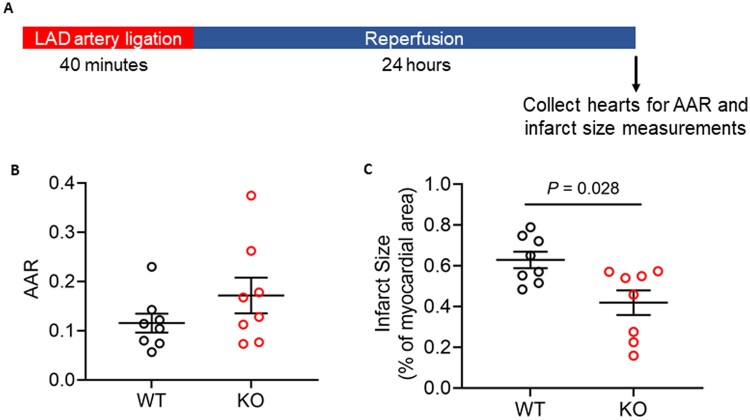
We next examined whether these proteomic and metabolite alterations resulted in phenotypic changes in the heart. At baseline, Ogfod1-KO hearts showed no signs of functional defects (Supplementary material online, Figure S1). To determine if ablation of Ogfod1 alters the response to cell stress, we subjected WT and KO ex vivo perfused hearts to I/R, and measured recovery of LVDP and HR during reperfusion following ischaemia, and determined the recovery of contractile function rate pressure product (RPP = HR × LVDP) (Figure 4A). We also measured infarct size at the end of reperfusion. As shown in Figure 4B and C, loss of Ogfod1 is protective against I/R injury in mice, with Ogfod1-KO hearts showing a 34% improvement in cardiac function (measured as RPP) (Figure 4B) and a 41.4% reduction in infarct size compared to Ogfod1-WT (Figure 4C). The improvement in RPP was due to a difference in LVDP, as there were no differences in hemodynamic parameters such as HR and flow rate (Supplementary material online, Table S4). To determine whether this protection was reflected in vivo, we subjected WT and KO mice to LAD coronary artery ligation for 40 min followed by 24 h of reperfusion (Figure 5A). At the end of the reperfusion period, we assessed area at risk and infarct size and found that Ogfod1 deletion led to a significant reduction in infarct size (Figure 5B and C). These results indicate that Ogfod1 loss is protective against the injury that occurs following both ex vivo and in vivo I/R in the heart.
Figure 4.
Ogfod1 ablation leads to protection against ischaemia–reperfusion injury ex vivo. (A) Langendorff-perfused hearts from WT and KO mice were subjected to ischaemia and reperfusion protocol as shown in the schematic. (B) Rate pressure product (RPP) recovery following ischaemia and reperfusion. (C) Infarct size measured at the end of the ischaemia–reperfusion period. Data shown as mean ± standard error. n = 8 biological replicates per group. Student’s t-test with Welch’s correction was used to determine significance between groups. A P-value of <0.05 was considered significant. LVDP, left ventricular developed pressure; TTC, 2,3,5-triphenyltetrazolium chloride.
Figure 5.
Ogfod1 ablation leads to protection against damage resulting from in vivo LAD artery ligation. (A) In vivo ischaemia reperfusion injury was induced by left anterior descending (LAD) coronary artery ligation in WT and KO mice as shown in the schematic. (B) Area at risk (AAR) for WT and KO hearts subjected to LAD artery ligation. (C) Infarct size shown as a percentage of the heart. Data shown as mean ± standard error. n = 8 biological replicates per group. Student’s t-test with Welch’s correction was used to determine significance between groups. A P-value of <0.05 was considered significant.
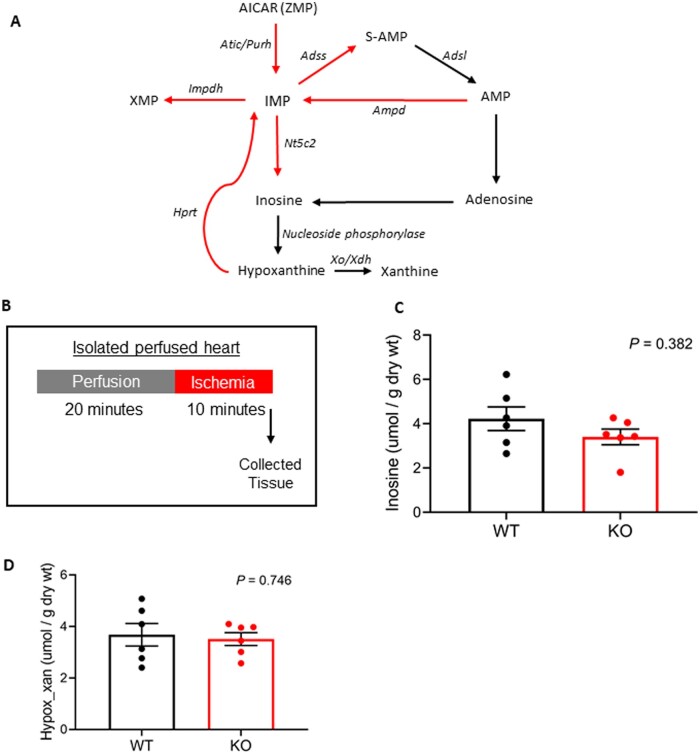
3.4 Baseline IMP accumulation in Ogfod1-KO hearts does not impact levels of inosine or hypoxanthine during ischaemia
Several molecular changes occur during ischaemic injury, with one of the key changes being severe ATP depletion.18,19 Due to this high energy phosphate depletion, re-synthesizing ATP is important in recovering cardiac function following ischaemia.20 Inosine, adenosine, hypoxanthine, and xanthine all accumulate in ischaemic tissues21 but are quickly lost upon post-ischaemic reperfusion. The purine salvage pathway can function to retain nucleotide bases such as these which are needed for the re-synthesis of ATP.22 Because IMP was higher in the KO mice and plays a role in the purine salvage pathway used for the rapid re-synthesis of ATP, we postulated that if IMP was diverted towards S-AMP and AMP for ATP resynthesis, the IMP degradation products inosine, hypoxanthine, and xanthine would be decreased in ischaemic KO hearts as indicated by the purine nucleotide degradation and salvage pathway (Figure 6A). We induced ischaemia for 10 min in WT and KO hearts, and collected hearts to assess metabolite levels (Figure 6B). Interestingly, both inosine (WT: 4.2 ± 0.53 umol/g, KO: 3.4 ± 0.35 umol/g, P = 0.382) and hypoxanthine/xanthine (WT: 3.7 ± 0.44 umol/g, KO: 3.5 ± 0.25 umol/g, P = 0.746) were unchanged in ischaemic KO hearts compared to WT hearts (Figure 6C and D). These results indicated that changes in the purine nucleotide salvage pathway are not responsible for the protection observed in the KO hearts.
Figure 6.
Investigating the potential for the purine nucleotide pathway to be altered in ischaemic Ogfod1-KO hearts. (A) Schematic showing purine nucleotide degradation and salvage. (B) Brief protocol showing the timepoint in ischaemia when hearts were collected for metabolite analysis. (C) Inosine levels (umol/g dry wt) in ischaemic WT and KO hearts. (D) Hypoxanthine and xanthine levels (umol/g dry wt) in ischaemic WT and KO hearts. n = 6 biological replicates per group. Student’s t-test with Welch’s correction was used to determine significance between groups. A P-value of <0.05 was considered significant.
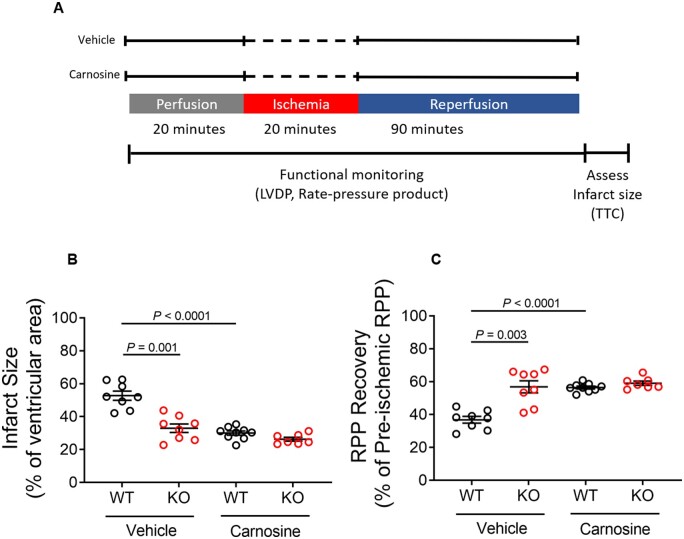
3.5 Histidine dipeptide accumulation in Ogfod1-KO hearts leads to protection against ischaemia–reperfusion injury
From the metabolomics results, both purine and pyrimidine pathways were altered. Beta-alanine, which is significantly up-regulated in KO hearts, is a by-product of pyrimidine catabolism. Beta-alanine is also the rate-limiting amino acid precursor in synthesizing the histidine dipeptide carnosine, which shows a 40% increase in KO hearts (WT: 6.2 × 10−4 ± 5.3 × 10−5 a.u., KO: 8.6 × 10−4 ± 1.1 × 10−4 a.u., P = 0.117). Carnosine has been shown to protect hearts from I/R injury,14,15 so we hypothesized that the increase in beta-alanine and histidine dipeptides such as carnosine might mediate protection in the KO hearts. To test this hypothesis, we treated WT and KO hearts with carnosine prior to ischaemia and during reperfusion (Figure 7A). If the hypothesis was correct, we would expect carnosine treatment to confer I/R protection in WT, but not KO hearts. Perfusing hearts with carnosine prior to I/R reduced infarct size by 44% (Figure 7B) and improved recovery of cardiac function by 55% (Figure 7C) in WT hearts, but failed to improve cardiac function or reduce infarct size in KO hearts. These results support the hypothesis that Ogfod1 loss leads to an increase in the carnosine precursor beta-alanine, which protects the myocardium against I/R injury.
Figure 7.
Carnosine treatment protects WT, but not KO hearts from ischaemia–reperfusion injury. (A) Schematic showing the ischaemia–reperfusion protocol as well as the carnosine and vehicle treatments. (B) Infarct size measured at the end of the ischaemia–reperfusion period. (C) Rate pressure product (RPP) recovery following ischaemia and reperfusion. Data shown as mean ± standard error. n = 7–9 biological replicates per group. Brown–Forsythe and Dunnett’s T3 multiple comparisons tests were used to compare more than two groups. A P-value of <0.05 was considered significant. LVDP, left ventricular developed pressure; TTC, 2,3,5-triphenyltetrazolium chloride.
4. Discussion
Ogfod1 catalyzes the prolyl hydroxylation of Rps23, a component of the 40S ribosomal subunit, on proline 62 which is located in the decoding centre of the ribosome. Hydroxylation of Rps23 has been reported to regulate translation.6,9–11,23–25 This is consistent with post-translational modifications on other ribosomal proteins, which have been shown to regulate translation,26 ribosome biogenesis,27 and targeted mRNA silencing.28 Ablating Ogfod1 reduces translation and activates translational stress pathways in a context-dependent manner.6 We previously reported that deleting OGFOD1 in human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) leads to changes in the expression of translation initiation factors, mRNA splicing factors, and ribosomal proteins—which all function in regulating protein levels.10 Therefore, we hypothesized that Ogfod1 might be a metabolic switch that alters the response of the cell to stress, and investigated whether Ogfod1 might modulate the translational landscape in adult heart and whether this might alter cardiac physiology.
We performed proteomic and metabolomic analysis on hearts from Ogfod1-KO mice and found alterations in metabolic enzyme levels, particularly those involved in regulating purine and pyrimidine turnover. These proteomic results were supported by accumulated IMP and NADPH, the latter is generated by the pentose phosphate pathway that produces phosphoribosyl pyrophosphate for de novo purine synthesis. As changes in these pathways have been shown to contribute to cardioprotection, we subjected isolated perfused hearts to I/R injury and found that KO hearts showed a 41.4% decrease in infarct size and a 34% improvement in functional recovery compared to WT hearts.
To determine the contribution of purine and pyrimidine pathway regulation to the cardioprotection in I/R injury, we assessed the levels of purine salvage pathway metabolites that resulted from IMP degradation and found no differences that indicated they contributed to the protection. Next, we investigated the contribution of pyrimidine metabolism to I/R injury protection. Beta-alanine, a product of nucleotide turnover and a precursor for carnosine synthesis, is significantly up-regulated in KO hearts, with anserine, carnosine, and homocarnosine all trending towards increases. Upon treating WT and KO isolated perfused hearts with carnosine, WT hearts were protected from I/R injury to an extent comparable to KO hearts without carnosine. However, KO hearts treated with carnosine showed no improvement in protection from I/R injury over WT hearts.
Previous studies have shown that alterations in transcriptional and translational machinery can alter metabolism. In studies in which RNA polymerase III-mediated transcription was increased, it was shown to lead to increased metabolic demand, increased protein catabolism, increased nucleotide levels to meet the demand for increased transcription, and changes in both purine and pyrimidine nucleotide synthesis and turnover.29 RNA polymerase III transcribes 5S ribosomal RNA, tRNA, and small non-coding RNAs,30 so changes in its activity also impact translation.31,32 Therefore, increasing transcription through this polymerase ultimately links broad polymerase activity to specific protein and metabolic changes.29 Similarly, deleting the ribosomal prolyl hydroxylase Ogfod1, which regulates translation and ribosome biology, increased protein catabolism, increased levels of several enzymes functioning in nucleotide metabolism, and altered levels of purines, pyrimidines, and related metabolites, such as IMP, NADPH, and beta-alanine.
Altogether, deleting Ogfod1 leads to specific changes in the proteomic and metabolomic landscapes that protect the heart against I/R injury both ex vivo and in vivo, suggesting the need for further investigation of Ogfod1 for its therapeutic potential. This strategy of inhibiting PHD’s for therapeutic benefit is currently being utilized in other clinical applications as Roxadustat (FG-4592), a PHD inhibitor, has been approved for clinical use in China,33 Japan, and Chile and is under review for approval in the US Roxadustat is intended to be used as a HIF-PHD inhibitor that induces HIF accumulation. HIF accumulation then activates erythropoietin expression34,35 to treat anaemia in patients with chronic kidney disease.36 Interestingly, it is also a potent Ogfod1 inhibitor, and thus this inhibitor could have clinical effects that have yet to be fully characterized.37 This study illuminates the potential for OGFOD1 inhibitors, to be used as a preventative treatment for individuals prone to ischaemic episodes. It also highlights the possibility that inhibitors such as Roxadustat could have off-target effects that need to be investigated.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
M.H., J.S., and L.M.K. did experiments and assisted in revising the manuscript. J.S. did ex vivo mouse studies, K.K. did in vivo LAD studies, and D.S. did echocardiogram studies. A.A. and M.G. performed and supervised liquid chromatography and tandem mass spectrometry. K.S. and M.P. did bioinformatics. M.E.C. was instrumental in providing Ogfod1-KO mice. E.M. and L.M.K. designed the study and wrote the manuscript.
Funding
This work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health Intramural Research Program (ZO1-HL002066). The work was also supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001501); the UK Medical Research Council (FC001501); and the Wellcome Trust (FC001501).
Supplementary Material
Contributor Information
Michael Harris, Cardiovascular Branch, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Junhui Sun, Cardiovascular Branch, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Karen Keeran, Animal Surgery and Resources Core Facility, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Angel Aponte, Proteomics Core Facility, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Komudi Singh, Bioinformatics and Computational Biology Core Facility, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Danielle Springer, Murine Phenotyping Core Facility, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Marjan Gucek, Proteomics Core Facility, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Mehdi Pirooznia, Bioinformatics and Computational Biology Core Facility, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Matthew E Cockman, The Francis Crick Institute, London, UK.
Elizabeth Murphy, Cardiovascular Branch, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Leslie M Kennedy, Cardiovascular Branch, National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Data availability
The data used to generate the figures in this article are available in the supplementary material.
References
- 1. Loenarz C, Schofield CJ.. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 2008;4:152–156. [DOI] [PubMed] [Google Scholar]
- 2. Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 2004;39:21–68. [DOI] [PubMed] [Google Scholar]
- 3. Hanauske-Abel HM, Günzler V.. A stereochemical concept for the catalytic mechanism of prolylhydroxylase: applicability to classification and design of inhibitors. J Theor Biol 1982;94:421–455. [DOI] [PubMed] [Google Scholar]
- 4. Kaelin WG Jr, Ratcliffe PJ.. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008;30:393–402. [DOI] [PubMed] [Google Scholar]
- 5. Ge W, Wolf A, Feng T, Ho CH, Sekirnik R, Zayer A, Granatino N, Cockman ME, Loenarz C, Loik ND, Hardy AP, Claridge TDW, Hamed RB, Chowdhury R, Gong L, Robinson CV, Trudgian DC, Jiang M, Mackeen MM, McCullagh JS, Gordiyenko Y, Thalhammer A, Yamamoto A, Yang M, Liu-Yi P, Zhang Z, Schmidt-Zachmann M, Kessler BM, Ratcliffe PJ, Preston GM, Coleman ML, Schofield CJ.. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol 2012;8:960–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singleton RS, Liu-Yi P, Formenti F, Ge W, Sekirnik R, Fischer R, Adam J, Pollard PJ, Wolf A, Thalhammer A, Loenarz C, Flashman E, Yamamoto A, Coleman ML, Kessler BM, Wappner P, Schofield CJ, Ratcliffe PJ, Cockman ME.. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci USA 2014;111:4031–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horita S, Scotti JS, Thinnes C, Mottaghi-Taromsari YS, Thalhammer A, Ge W, Aik W, Loenarz C, Schofield CJ, McDonough MA.. Structure of the ribosomal oxygenase OGFOD1 provides insights into the regio- and stereoselectivity of prolyl hydroxylases. Structure 2015;23:639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wehner KA, Schutz S, Sarnow P.. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol Cell Biol 2010;30:2006–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katz MJ, Acevedo JM, Loenarz C, Galagovsky D, Liu-Yi P, Perez-Pepe M, Thalhammer A, Sekirnik R, Ge W, Melani M, Thomas MG, Simonetta S, Boccaccio GL, Schofield CJ, Cockman ME, Ratcliffe PJ, Wappner P.. Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc Natl Acad Sci USA 2014;111:4025–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stoehr A, Kennedy L, Yang Y, Patel S, Lin Y, Linask KL, Fergusson MM, Zhu J, Gucek M, Zou J, Murphy E.. The ribosomal prolyl-hydroxylase OGFOD1 decreases during cardiac differentiation and modulates translation and splicing. JCI Insight 2019;4:e128496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM.. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 2006;26:5237–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saito K, Adachi N, Koyama H, Matsushita M.. OGFOD1, a member of the 2-oxoglutarate and iron dependent dioxygenase family, functions in ischemic signaling. FEBS Lett 2010;584:3340–3347. [DOI] [PubMed] [Google Scholar]
- 13. Wasternack C, Lippmann G, Reinbotte H.. Pyrimidine-degrading enzymes. Purification and properties of beta-ureidopropionase of Euglena gracilis. Biochim Biophys Acta 1979;570:341–351. [DOI] [PubMed] [Google Scholar]
- 14. Zhao J, Conklin DJ, Guo Y, Zhang X, Obal D, Guo L, Jagatheesan G, Katragadda K, He L, Yin X, Prodhan MAI, Shah J, Hoetker D, Kumar A, Kumar V, Wempe MF, Bhatnagar A, Baba SP.. Cardiospecific overexpression of ATPGD1 (carnosine synthase) increases histidine dipeptide levels and prevents myocardial ischemia reperfusion injury. J Am Heart Assoc 2020;9:e015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee JW, Miyawaki H, Bobst EV, Hester JD, Ashraf M, Bobst AM.. Improved functional recovery of ischemic rat hearts due to singlet oxygen scavengers histidine and carnosine. J Mol Cell Cardiol 1999;31:113–121. [DOI] [PubMed] [Google Scholar]
- 16. Sun J, Nguyen T, Aponte AM, Menazza S, Kohr MJ, Roth DM, Patel HH, Murphy E, Steenbergen C.. Ischaemic preconditioning preferentially increases protein S-nitrosylation in subsarcolemmal mitochondria. Cardiovasc Res 2015;106:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rohart F, Gautier B, Singh A, Le Cao KA.. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 2017;13:e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jennings RB, Hawkins HK, Lowe JE, Hill ML, Klotman S, Reimer KA.. Relation between high energy phosphate and lethal injury in myocardial ischemia in the dog. Am J Pathol 1978;92:187–214. [PMC free article] [PubMed] [Google Scholar]
- 19. Jennings RB, Reimer KA, Hill ML, Mayer SE.. Total ischemia in dog hearts, in vitro. 1. Comparison of high energy phosphate production, utilization, and depletion, and of adenine nucleotide catabolism in total ischemia in vitro vs. severe ischemia in vivo. Circ Res 1981;49:892–900. [DOI] [PubMed] [Google Scholar]
- 20. Reimer KA, Hill ML, Jennings RB.. Prolonged depletion of ATP and of the adenine nucleotide pool due to delayed resynthesis of adenine nucleotides following reversible myocardial ischemic injury in dogs. J Mol Cell Cardiol 1981;13:229–239. [DOI] [PubMed] [Google Scholar]
- 21. Van Bilsen M, van der Vusse GJ, Coumans WA, de Groot MJ, Willemsen PH, Reneman RS.. Degradation of adenine nucleotides in ischemic and reperfused rat heart. Am J Physiol 1989;257:H47–H54. [DOI] [PubMed] [Google Scholar]
- 22. Fujii K, Kubo A, Miyashita K, Sato M, Hagiwara A, Inoue H, Ryuzaki M, Tamaki M, Hishiki T, Hayakawa N, Kabe Y, Itoh H, Suematsu M.. Xanthine oxidase inhibitor ameliorates postischemic renal injury in mice by promoting resynthesis of adenine nucleotides. JCI Insight 2019;4:e124816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henri J, Rispal D, Bayart E, van Tilbeurgh H, Seraphin B, Graille M.. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J Biol Chem 2010;285:30767–30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, Mackeen MM, McDonough MA, Cockman ME, Kessler BM, Ratcliffe PJ, Wolf A, Schofield CJ.. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci USA 2014;111:4019–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stoehr A, Yang Y, Patel S, Evangelista AM, Aponte A, Wang G, Liu P, Boylston J, Kloner PH, Lin Y, Gucek M, Zhu J, Murphy E.. Prolyl hydroxylation regulates protein degradation, synthesis, and splicing in human induced pluripotent stem cell-derived cardiomyocytes. Cardiovasc Res 2016;110:346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imami K, Milek M, Bogdanow B, Yasuda T, Kastelic N, Zauber H, Ishihama Y, Landthaler M, Selbach M.. Phosphorylation of the ribosomal protein RPL12/uL11 affects translation during mitosis. Mol Cell 2018;72:84–98.e9. [DOI] [PubMed] [Google Scholar]
- 27. Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, Pende M.. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 2014;33:474–483. [DOI] [PubMed] [Google Scholar]
- 28. Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL.. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 2003;115:187–198. [DOI] [PubMed] [Google Scholar]
- 29. Willis IM, Moir RD, Hernandez N.. Metabolic programming a lean phenotype by deregulation of RNA polymerase III. Proc Natl Acad Sci USA 2018;115:12182–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turowski TW, Tollervey D.. Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem Soc Trans 2016;44:1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet 2008;24:622–629. [DOI] [PubMed] [Google Scholar]
- 32. Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A.. The expanding RNA polymerase III transcriptome. Trends Genet 2007;23:614–622. [DOI] [PubMed] [Google Scholar]
- 33. Dhillon S. Roxadustat: first global approval. Drugs 2019;79:563–572. [DOI] [PubMed] [Google Scholar]
- 34. Nangaku M, Eckardt KU.. Hypoxia and the HIF system in kidney disease. J Mol Med (Berl) 2007;85:1325–1330. [DOI] [PubMed] [Google Scholar]
- 35. Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ.. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001;292:468–472. [DOI] [PubMed] [Google Scholar]
- 36. Hasegawa S, Tanaka T, Nangaku M.. Hypoxia-inducible factor stabilizers for treating anemia of chronic kidney disease. Curr Opin Nephrol Hypertens 2018;27:331–338. [DOI] [PubMed] [Google Scholar]
- 37. Yeh TL, Leissing TM, Abboud MI, Thinnes CC, Atasoylu O, Holt-Martyn JP, Zhang D, Tumber A, Lippl K, Lohans CT, Leung IKH, Morcrette H, Clifton IJ, Claridge TDW, Kawamura A, Flashman E, Lu X, Ratcliffe PJ, Chowdhury R, Pugh CW, Schofield CJ.. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem Sci 2017;8:7651–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to generate the figures in this article are available in the supplementary material.