ABSTRACT
Candida auris has emerged globally as a multidrug-resistant pathogen causing outbreaks in health care facilities. Whole genome sequencing (WGS) analysis has identified four major clades, while earlier WGS data from a single Iranian isolate suggested the existence of a potential fifth clade. Here, we confirm the existence of this fifth clade by providing WGS data of another four Iranian isolates. These clade V isolates differed less than 100 single-nucleotide polymorphisms (SNPs) between each other, while they were separated from the other clades by more than 200,000 SNPs. Two of these isolates were resistant to fluconazole and were found to harbour mutations in the TAC1b and ERG11 genes.
KEYWORDS: Candida auris, clade V, genotyping, whole genome sequencing, microsatellite typing, Iran
Introduction
Candida auris emerged as a multidrug-resistant yeast in the last decade in over 50 countries [1]. This yeast colonizes human skin, hospital environments and causes nosocomial outbreaks and invasive candidiasis [2]. The majority of C. auris isolates are resistant to fluconazole and are responsible for a high mortality rate in intensive care units [3]. C. auris has first been identified in Japan from a human external ear canal in 2009 [4]. Since then cases of systemic infections have been reported globally in other Asian countries, the Americas, Africa and Europe [5]. Initial genetic analysis identified four major clades, named after the geographical location: South-Asia, East-Asia, Africa, South America, respectively clade I, II, III and IV [6,7]. Between any two clades, isolates differ at least ten thousand single nucleotide polymorphisms (SNPs). Previous whole genome sequencing (WGS) data from a single Iranian isolate suggested the existence of a potential fifth clade separated by >200,000 SNPs from the other encountered clades [8].
More than 90% of C. auris isolates from clade I and III and about half of the isolates from clade IV are resistant against fluconazole [3,6,9]. In addition antifungal resistance against polyenes and echinocandins has been reported in these clades. Typically, clade II isolates are susceptible to azoles and other common antifungals [10,11]. Amino acid substitutions in ERG10 and ERG11, essential components of the ergosterol biosynthesis pathway, are associated with fluconazole resistance [12,13]. Fluconazole resistance in C. auris is associated with amino acid substitutions in ERG11 at position F126L, Y132F and K143R, while I466M and Y501H might contribute to reduced susceptibility [14]. The prevalence of these mutations differ between clades [12]. In addition, recent findings of ergosterol biosynthesis pathway mutations indicate that an increased expression of efflux pump CDR1 corresponds with mutations in TAC1b, which also coincide with triazole resistance [15,16]. Moreover, the separate clades present differences in their biochemical profiles. Clade II isolates are not able to utilize N-acetyl glucosamine [17], while only clade III isolates assimilate L-rhamnose which correlates with the presence of a 7-gene cluster involved in L-rhamnose utilization [18]. The first reported potential clade V isolate was reported to also utilize L-rhamnose and contains the L-rhamnose gene cluster [19].
In 2018, the first C. auris isolate from Iran (IFRC2087) was reported in a patient from Babol with C. auris otomycosis [20]. Subsequent WGS indicated that this isolate did not localize to any known clade and might constitute a potential fifth clade [8]. Since then, four more patients with C. auris have been reported in Iran [21–24]. All isolates were analysed by short tandem repeat (STR) genotyping. The STR profiles indicated that one isolate belonged to clade I and the other isolates to the potential clade V. Two of these potential clade V isolates were isolated from patients with otomycosis, one originating from Babol (IFRC 4050), and the other from Isfahan (MRL40) [22,23]. Furthermore, in 2021 C. auris was isolated from skin and ear (TMML616 and TMML617) from a girl with ichthyosis from Shiraz [24]. Only the first isolate from Babol (IFRC2087)was susceptible to fluconazole but the other two clade V isolates from Babol and Isfahan were resistant. Antifungal susceptibility profiles from the Shiraz isolates were not reported [24]. All analysed isolates from Iran were susceptible to other triazoles, amphotericin B and echinocandins. Limited data regarding the fifth clade are available compared to the other four major clades. Herein, we present WGS data from all reported potential fifth clade C. auris isolates, confirm the existence of the fifth clade and describe some differences in antifungal resistance patterns within the fifth clade.
Materials and methods
Isolates
Five C. auris isolates from Iran and eight C. auris isolates from Canada, India, Pakistan, South-Africa, the United States and Venezuela representing four clades, were used (Supplementary Table S1). Isolates were stored at −80°C according to standard procedures. All isolates were identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as previously described [25].
Culture and DNA extraction
C. auris isolates were retrieved from −80°C storage and grown on Sabouraud (SAB) agar plates (Oxoid) at 30°C. For STR genotyping and WGS a single colony was resuspended in 500 µL MagNA Pure bacteria lysis buffer and MagNA Lyser green beads. The cells were mechanically lysed for 30 s at 6500 rpm using the MagNA Lyser system (all Roche Diagnostics GmbH, Mannheim, Germany). DNA was extracted and purified with the MagNA Pure 96 instrument and the MagNA Pure DNA and Viral NA Small Volume Kit (Roche Diagnostics), according to the recommendations of the manufacturer. For WGS, all samples were subsequently treated with RNase at a final concentration of 5 µg/µL for one hour at room temperature, after which DNA was extracted and purified as described earlier. Purified DNA was measured with a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) using the double-stranded DNA (dsDNA) High sensitivity option.
STR typing
Short tandem repeat (STR) typing of C. auris isolates was performed as described previously [26]. C. auris STRs were amplified by four multiplex PCR reactions, amplifying STR targets with a repeat size of two, three or nine nucleotides. Corresponding copy numbers were determined using GeneMapper Software (Applied Biosystems, Foster City, CA, USA), and analysed using BioNumerics 7.6.1. (Applied Maths, Kortrijk, Belgium) by employing the unweighted pair group method with arithmetic mean averages (UPGMA) as described previously [27].
Whole-genome sequencing pipeline validation
Read data of 23 C. auris isolates were downloaded from the SRA database and uploaded to the Galaxy tool [28] (Supplementary Table S2). Additionally, FastQC was used to assess the quality of the read data, no trimming was performed. Read data were aligned against the genome of C. auris B11205 (Genbank assembly accession number GCA_016772135.1) using BWA-MEM [29]. PCR duplicates were removed using RmDup, local realignment was performed with BamLeftAlign, and unpaired reads were removed with BAM filter. Mapped reads with a MAPQ score <60 were removed. Variants were detected with Freebayes using the default settings except for population model option (ploidy: 1) and allelic scope options (ignore indels, multiple nucleotide polymorphisms, and complex events) [30]. Variants in the resulting VCF file were removed with a read depth (DP) of <30, a quality (QUAL) of <100, an allele frequency (AO) of <0.15 x DP, and an AO >0.90 x DP. Subsequently variants in the first and last 5% of each chromosome were removed. Phylogenetic analysis was performed with VCF2PopTree, using the genetic drift algorithm and a MEGA distance based matrix was developed [31]. The matrix was uploaded to MEGA11 and a phylogenetic tree was generated using the Neighbour-joining Tree method [32].
Whole-genome sequencing variant calling
Genomic libraries of the Iranian isolates were prepared and sequenced with Illumina technology (Illumina, San Diego, CA, USA) with 150-bp paired-end-read mode at Eurofins Genomics (Ebersberg, Germany). All raw read data in this study have been submitted to the National Center for Biotechnology Information’s Sequence Read Archive (BioProject ID PRJNA816104). Read data of the remaining eight isolates from four different clades was accessed with the NCBI SRA toolkit (Supplementary Table S1). Read data and variant calling was performed as described earlier. Read data were aligned against the genome of C. auris B11221 (GenBank assembly accession number GCA_002775015.1) using BWA-MEM, with a mean coverage of 148.52 ± 12.63 [29]. Alignments around genes ERG10, ERG11 and TAC1b, involved in fluconazole resistance, and the L-rhamnose gene cluster, were visually inspected using JBrowse v1.16.11 [33]. To provide a clear overview isolates 10-11-10-18 (IFRC2087), 10-13-08-58 (IFRC4050), 10-13-10-90 (RML40), 10-13-10-56 (TMML616), 10-13-10-57 (TMML617) were indicated as Babol-1, Babol-2, Isfahan, Shiraz-1 and Shiraz-2, respectively.
Biochemical analysis and antifungal susceptibility testing (AFST)
12 C. auris isolates were grown on SAB agar plates before testing (Supplementary Table S1). Biochemical profile analysis with a Vitek 2XL (BioMérieux, Marcy I’Etoile, France) was performed according to the manufacturer’s instructions by using a Vitek 2 YST ID card and the latest software update of the Vitek 2XL identification system (version 9.02). All isolates were analysed in triplo with Vitek 2XL. Minimal inhibitory concentrations (MICs) of amphotericin B (AMB), fluconazole (FLU), itraconazole (ITR), voriconazole (VOR), posaconazole (POS), isavuconazole (ISA), anidulafungin (AFG) and micafungin (MFG) were determined by broth microdilution according to the Clinical and Laboratory Standard Institute (CLSI) M27-A3 standard [34].
Results
STR genotyping
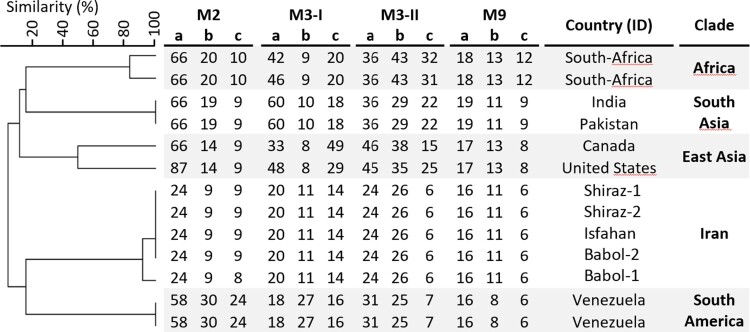
Genotyping of the five C. auris isolates from Iran is shown in Supplementary Table S1. The Iranian isolates clustered in a separate clade, apart from isolates from the other four clades, which were included as a comparison (Figure 1). Among the Iranian isolates two genotypes were found. The Iranian isolates differed in one STR marker.
Figure 1.
STR genotypes of 13 C. auris isolates. Cluster analysis showed that the five different clades form distinct clusters based on STR profiles. The UPGMA dendrogram was generated with BioNumerics.
WGS SNP calling
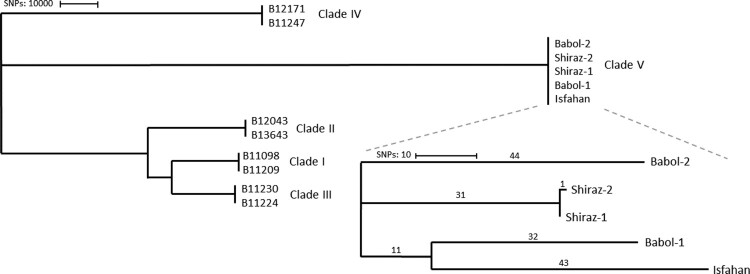
Next, we investigated whether it was possible to further distinguish the five Iranian isolates using WGS analysis. For this purpose, we first validated our variant calling pipeline by analysing 23 C. auris isolates previously published for validation purposes by the Centers for Disease Control and Prevention (CDC) [35] (Supplementary Table S2). Phylogenetic analysis showed that all branches of the phylogenetic tree correspond to those inferred in the original tree (Supplementary Figure S1). In total we found 229 single nucleotide polymorphisms (SNPs) as compared to 268 SNPs reported with the CDC workflow, leading to an acceptable difference (14.6% less SNPs) between both pipelines. Having validated our pipeline, reads from the Iranian strains were mapped to the B11221 reference genome and SNPs between isolates were determined. All isolates from Iran differed >200,000 SNPs compared to isolates from the other four clades that were included as a comparison (Figure 2). Isolates from Iran differed one to 98 SNPs from each other. The two samples that differed one SNP from each other were from the skin and ear of the same patient suffering from ichthyosis.
Figure 2.
Phylogenetic tree based on SNPs of 13 C. auris. Numbers above the branch indicate the number of SNPs. Isolates from clade V differed >200,000 SNPs from other clades. The tree was generated with MEGA11 using the neighbour-joining tree method.
Biochemical analysis
Biochemical profile analysis with Vitek 2 demonstrated that all C. auris isolates from clades III and V displayed assimilation of L-rhamnose. Among isolates from the three other clades no assimilation of L-rhamnose was observed. The reported L- rhamnose gene cluster was found in all clade III and V samples, as determined with WGS data analysis. All isolates, including those from clade V, were able to utilize N-acetyl glucosamine except for isolates from clade II.
Fluconazole resistance
MICs according to CLSI standards for all clade V isolates were determined (Table 1). Both isolates from Shiraz showed wildtype MICs of all tested antifungals while the MICs of the remaining isolates were concordant with previously reported MICs [20,22,23]. To further analyse the mechanism of fluconazole resistance in Babol-2 (IFRC 4050) and Isfahan (MRL40) isolates, genes ERG10, ERG11 and TAC1b were assessed with JBrowse for potential mutations. The Babol-2 (IFRC4050) isolate demonstrated a TAC1b D599G and ERG11 Y132F mutation, which were not present in the other isolates. An ERG11 I466L mutation was found in the isolate from Isfahan. In genes ERG10, ERG11 and TAC1b the resistant isolates did not exhibit any unique mutation compared to the susceptible isolates.
Table 1.
MIC values of clade V isolates. Breakpoints determined according to CLSI microbroth dilution (mg/L).
| ID | City | AMB | FLU | ITC | VOR | POS | ISA | AFG | MFG |
|---|---|---|---|---|---|---|---|---|---|
| IFRC2087 | Babol | 0.25 | 8 | 0.031 | 0.063 | 0.031 | 0.016 | 0.031 | 0.031 |
| IFRC4050 | Babol | 0.25 | 64 | 0.016 | 1 | 0.016 | 0.016 | 0.031 | 0.063 |
| MRL40 | Isfahan | 0.5 | 64 | 0.125 | 1 | 0.031 | 0.016 | 0.063 | 0.031 |
| TMML616 | Shiraz | 0.25 | 1 | 0.016 | 0.016 | 0.016 | 0.016 | 0.063 | 0.031 |
| TMML617 | Shiraz | 0.25 | 1 | 0.016 | 0.016 | 0.016 | 0.016 | 0.063 | 0.031 |
Discussion
STR genotyping of the five Iranian C. auris clade V isolates demonstrated that they form a separate cluster. WGS confirmed that this cluster was separated from the other four major clades with >200,000 SNPs. Compared to the other C. auris clades, clade V appears at a basally branching position due to the large number of SNPs. Our WGS results together with previous research suggest the fifth clade diverged before the separation of the other clades [19].
Isolates within the fifth clade were separated with less than 100 SNPs. Remarkable, the isolates of patients from Babol (IFRC2087) differed 87 SNPs, while only 75 SNPs were found between Babol-2 (IFRC4050) and the Isfahan (MRL40) isolates [20,22,23]. These isolates (Babol-2; IFRC4050 and Isfahan; MRL40) were resistant against fluconazole with the Babol-2(IFRC4050) isolate demonstrating a TAC1b D599 mutation and an ERG11 Y132F mutation and the Isfahan isolate an ERG11 I466L mutation. The occasional presence of different mutations suggests a local resistance development. All Iranian isolates were able to utilize L-rhamnose, a trait shared with clade III, and this corresponded with the presence of the L-rhamnose gene cluster in all clade V isolates [18]. Previously it has been hypothesized that this gene cluster is missed in clades I, II an IV rather than a gain in clades III an V [19]. The presence of this gene cluster in all studied fifth clade isolates strengthens this hypothesis.
The ERG11 Y132F as found in the Babol-2 (IFRC4050) and previously in clades I and IV isolates, is known to coincide with fluconazole resistance [9,12]. To our knowledge, the ERG11 I466L mutation as found in the Isfahan (MRL40) isolate has not been reported earlier, although the I466M mutation was found earlier in clade IV isolates and, together with other mutations, likely contributes to reduced susceptibility [14]. Moreover, in C. albicans the mutation I471T, which is equivalent to I466 in C. auris, was reported in two resistant C. albicans isolates [36,37]. Therefore, it is likely that ERG11 I466L contributes to fluconazole resistance. Finally, Babol-2 (IFRC4050) also harboured a TAC1b D599G mutation, which might contribute to its resistance to fluconazole, as different mutations in TAC1b are associated with fluconazole resistance as they cause overexpression of the efflux pump CDR1 [15,16].
Remarkably, all four Iranian patients with clade V isolates suffered from otomycosis, which is also observed for clade II isolates. Furthermore, only two out of five isolates were resistant against fluconazole, while they were all susceptible to other triazoles, amphotericin B and echinocandins. The majority of reported isolates from clades I, III and IV are resistant to fluconazole, and have elevated MICs for amphotericin B and echinocandins [3,6]. Most clade II isolates are susceptible for fluconazole and other common antifungals [10,11]. Finally, no outbreaks with clade V isolates have yet been reported, while isolates from clades I, III and IV were involved in multiple outbreaks. Thus, isolates from clade II and V share similar characteristics, for which the underlying mechanism is still unknown. Regarding outbreaks, a previous report indicated that the outbreak-causing clades have a highly syntenic genome compared to clade II isolates, which have many chromosomal rearrangements and large sub-telomeric deletions, possibly due to a nonsense mutation in DCC1 [19]. The first Iranian isolate, Babol-1 (IFCR2087), also demonstrated a highly syntenic genome [19], and we did not find nonsense mutations in DCC1 for any clade V isolate, suggesting that genome instability does not play a role in the prevalence of outbreaks. Furthermore, clade II isolates are known to harbour mutations and deletions in genes related to biofilm formation and cell wall proteins MNT4 and adhesion-like genes which may impact the ability to cause invasive infections [19,38–41]. Further investigations towards differences in cell wall and biofilm associated genes between the five clades might reveal differences between outbreak causing clades and clades which mostly cause ear infections.
As of to date C. auris is rarely detected in Iran which may be due to the limited availability of diagnostic options. The number of patients affected is likely underestimated. The isolates originated from the cities Babol, Isfahan and Shiraz, which are more than 600 km apart from each other. This suggests that clade V is endemic in Iran and might also be present in neighbouring countries. While the origin of C. auris remains elusive, it has been hypothesized that C. auris existed as a plant saprobe in wetlands and acquired the characteristics to cause infections and outbreaks through climate change, the use of antiseptics in hospitals and environmental stress [42]. The isolation of environmental C. auris isolates from the Andaman Islands fits this hypothesis [43]. Following this hypothesis, it is unlikely that the isolates from Shiraz and Isfahan originated from these regions, as these cities are located in the desert which have a more arid climate compared to the humid climate of Babol, close to the Caspian Sea. It is more likely that the fifth clade originated from wetlands like the ones around the Persian Gulf or the Caspian Sea from where it migrated to cities like Shiraz and Isfahan. However, more surveillance and research in these regions are required to investigate this hypothesis.
Supplementary Material
Funding Statement
This work was supported by Canisius Wilhelmina Hospital for the Centre of Expertise in Mycology RadboudUMC/CWZs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Jeffery-Smith A, Taori SK, Schelenz S, et al. . Candida auris: a review of the literature. Clin Microbiol Rev. 2018;31(1):e00029–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du H, Bing J, Hu T, et al. . Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10):e1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lockhart SR. Candida auris and multidrug resistance: defining the new normal. Fungal Genet Biol. 2019;131:103243. [DOI] [PubMed] [Google Scholar]
- 4.Satoh K, Makimura K, Hasumi Y, et al. . Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44. [DOI] [PubMed] [Google Scholar]
- 5.Rhodes J, Fisher MC.. Global epidemiology of emerging Candida auris . Curr Opin Microbiol. 2019;52:84–89. [DOI] [PubMed] [Google Scholar]
- 6.Chow NA, Muñoz JF, Gade L, et al. . Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio. 2020;11(2):e03364–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lockhart SR, Etienne KA, Vallabhaneni S, et al. . Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow NA, de Groot T, Badali H, et al. . Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary A, Prakash A, Sharma C, et al. . A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73(4):891–899. [DOI] [PubMed] [Google Scholar]
- 10.Sekizuka T, Iguchi S, Umeyama T, et al. . Clade II Candida auris possess genomic structural variations related to an ancestral strain. PLoS One. 2019;14(10):e0223433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon YJ, Shin JH, Byun SA, et al. . Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol. 2019;57(4):e01624–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaabane F, Graf A, Jequier L, et al. . Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front Microbiol. 2019;10:2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Wang Y, Hu W, et al. . Application of machine learning classifier to Candida auris drug resistance analysis. Front Cell Infect Microbiol. 2021;11:742062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Healey KR, Kordalewska M, Jiménez Ortigosa C, et al. . Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother. 2018;62(10):e01427–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rybak JM, Muñoz JF, Barker KS, et al. . Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio. 2020;11(3):e00365–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Coste AT, Liechti M, et al. . Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother. 2021;65(5):e02663–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhary A, Kumar VA, Sharma C, et al. . Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis. 2014;33(6):919–926. [DOI] [PubMed] [Google Scholar]
- 18.Ambaraghassi G, Dufresne PJ, Dufrense SF, et al. . Identification of Candida auris by use of the updated Vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J Clin Microbiol. 2019;57(11):e00884–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz JF, Welsh RM, Shea T, et al. . Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics. 2021;218(1):iyab029. doi: 10.1093/genetics/iyab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abastabar M, Maghani I, Ahangarkani F, et al. . Candida auris otomycosis in Iran and review of recent literature. Mycoses. 2019;62(2):101–105. [DOI] [PubMed] [Google Scholar]
- 21.Mirhendi H, Charsizadeh A, Aboutalebian S, et al. . South Asian (clade I) Candida auris meningitis in a paediatric patient in Iran with a review of the literature. Mycoses. 2022;65(2):134–139. [DOI] [PubMed] [Google Scholar]
- 22.Safari F, Madani M, Badali H, et al. . Chronic autochthonous fifth clade case of Candida auris Otomycosis in Iran. Mycopathologia. 2022;187(1):121–127. [DOI] [PubMed] [Google Scholar]
- 23.Armaki MT, Omran SM, Kiakojuri K, et al. . First fluconazole-resistant Candida auris isolated from fungal otitis in Iran. Curr Med Mycol. 2021;7(1):51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Youssefian L, Khodavaisy S, et al. . Chronic mucocutaneous candidiasis due to Candida auris and non-albicans Candida species in a family with a mild TP63-associated ectodermal dysplasia. [abstract]. J Invest Dermatol. 2022;142(8):S84. [Google Scholar]
- 25.Prakash A, Sharma C, Singh A, et al. . Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect. 2016;22(3):277. e1-9. [DOI] [PubMed] [Google Scholar]
- 26.de Groot T, Puts Y, Berrio I, et al. . Development of Candida auris short tandem repeat typing and Its application to a global collection of isolates. mBio. 2020;11(1):e02971–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Valk HA, Meis JF, Curfs IM, et al. . Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J Clin Microbiol. 2005;43(8):4112–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afgan E, Baker D, Batut B, et al. . The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R.. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrison E, Marth G.. Haplotype-based variant detection from short-read sequencing. Preprint at ArXiv. 2012: 1207–3907. [Google Scholar]
- 31.Subramanian S, Ramasamy U, Chen D.. VCF2PopTree: a client-side software to construct population phylogeny from genome-wide SNPs. Peer J. 2019;7:e8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Stecher G, Kumar S.. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skinner ME, Uzilov AV, Stein LD, et al. . JBrowse: a next-generation genome browser. Genome Res. 2009;19(9):1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CLSI. Reference method for broth dilution antifungal susceptibility testing of yeasts: Approved Standard 3rd Edition. CLSI Document M27-A3. Clinical and Laboratory Standards Institute Wayne, PA; 2008
- 35.Welsh RM, Misas E, Forsberg K, et al. . Candida auris whole-genome sequence benchmark dataset for phylogenomic pipelines. J Fungi (Basel). 2021;7(3):214. doi: 10.3390/jof7030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kakeya H, Miyazaki Y, Miyazaki H, et al. . Genetic analysis of azole resistance in the Darlington strain of Candida albicans. Antimicrob Agents Chemother. 2000;44(11):2985–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Chen L, Li C.. Susceptibility of clinical isolates of Candida species to fluconazole and detection of Candida albicans ERG11 mutations. J Antimicrob Chemother. 2008;61(4):798–804. [DOI] [PubMed] [Google Scholar]
- 38.Hall RA, Gow NA.. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol Microbiol. 2013;90(6):1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruno M, Kersten S, Bain JM, et al. . Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat Microbiol. 2020;5(12):1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabaldon T, Martin T, Marcet-Houben M, et al. . Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kean R, Delaney C, Sherry L, et al. . Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere. 2018;3(4):e00334–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadevall A, Kontoyiannis DP, Robert V.. On the emergence of Candida auris: climate change, azoles, swamps, and birds. mBio. 2019;10(4):e01397–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arora P, Singh P, Wang Y, et al. . Environmental isolation of Candida auris from the coastal wetlands of Andaman islands, India. mBio. 2021;12(2):e03181–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.