Abstract
Autoimmune diseases and coronavirus disease 2019 (COVID-19) share many similarities. Concerns have arisen that autoimmune diseases may increase the susceptibility and severity of COVID-19. We used Mendelian randomization to investigate whether liability to autoimmune diseases is related to COVID-19 susceptibility and severity. Genetic instruments for 8 autoimmune diseases, including type 1 diabetes mellitus, rheumatoid arthritis, systemic lupus erythematosus, psoriasis, multiple sclerosis, primary sclerosing cholangitis, primary biliary cirrhosis and juvenile idiopathic arthritis, were obtained from published genome-wide association studies. Two-sample Mendelian randomization analyses of the associations of liability to each autoimmune disease with COVID-19 infection, hospitalized COVID-19, and very severe COVID-19 were performed using the latest publicly available genome-wide association study for COVID-19. Genetic liability to each of the autoimmune diseases was largely not associated with COVID-19 infection, hospitalized COVID-19, or very severe COVID-19 after accounting for multiple comparison. Sensitivity analysis excluding genetic variants in the human leukocyte antigen gene, which has an important role in the immune response, showed similar results. The autoimmune diseases examined were largely not genetically associated with the susceptibility or severity of COVID-19. Further investigations are warranted.
Subject terms: Genetic association study, Viral infection, Autoimmune diseases
Introduction
Similarities in clinical manifestations and immune response are evident for autoimmune diseases and coronavirus disease 2019 (COVID-19)1 suggesting a close relation between these diseases. Furthermore, medications for treatment of autoimmune diseases have been used in severe cases of COVID-192. High type I interferon in systemic lupus erythematosus patients may involve a hyperinflammatory response in COVID-19, suggesting an increase in COVID-19 severity by inducing a hyperinflammatory response3. Some studies have reported higher risk of COVID-19 infection and severity of disease among people with pre-existing autoimmune conditions, such as rheumatoid arthritis (RA)4, while other studies have not5,6. A meta-analysis based on case–control studies found that the risk of COVID-19 infection was higher among autoimmune disease patients than controls, and also found a positive association of glucocorticoid use with the risk of COVID-197. However, observational studies are vulnerable to confounding from other factors, such as immunosuppressant treatment for autoimmune diseases. Whether autoimmune diseases causally increase the susceptibility and severity of COVID-19 remains unknown, which may involve the priority for COVID-19 prevention and management strategies.
Here, we conducted a two-sample Mendelian randomization (MR) study to assess the associations of liability to each major autoimmune disease with susceptibility and severity of COVID-19. Ethical approval is not applicable given the summary statistics used are all publicly available.
Results
Selection of genetic instruments for autoimmune diseases
We obtained strong (p-values < 5 × 10–8) and independent (r2 < 0.01) genetic instruments for 8 autoimmune diseases after excluding palindromic SNPs (Supplementary Table S1). The median F-statistic for the exposures ranged from 37.9 to 56.6. The genetic variants explained 18.8% to 43.5% of the variances of the exposures.
MR effect of each autoimmune disease on COVID-19 susceptibility and severity
Genetic liability to each autoimmune disease was largely not associated with COVID-19 infection, hospitalized COVID-19, or very severe COVID-19 in the analyses using the IVW method (Fig. 1). The estimates were generally similar using MR-Egger and the weighted median methods (Supplementary Tables S2–S4).
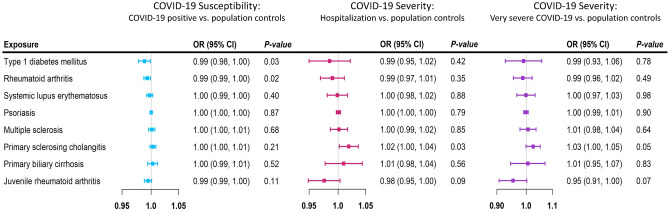
Figure 1.
Mendelian randomization (MR) estimates for each autoimmune diseases and COVID-19. Forest plot of MR estimates of 8 autoimmune diseases with COVID-19 infection, hospitalized COVID-19 and very severe COVID-19 using the multiplicative random-effects inverse variance weighting method. The black color represents COVID-19 infection, the blue color represents hospitalized COVID-19 and the red color represents very severe COVID-19. MR estimates are reported as odds ratios per unit of log-odds of each autoimmune disease examined.
Type 1 diabetes mellitus and rheumatoid arthritis were negatively associated with COVID-19 infection in the general population (Fig. 1; p-values for IVW = 0.031, 0.019, respectively), but were not related to hospitalized or very severe COVID-19. Primary sclerosing cholangitis, however, was positively associated with COVID-19 infection in the MR-Egger analysis (Supplementary Table S2; p = 0.009; p for intercept = 0.024) and was positively associated with hospitalized COVID-19 and very severe COVID-19 (Fig. 1; p-values for IVW = 0.010, 0.026, respectively). Juvenile rheumatoid arthritis was negatively associated with hospitalized COVID-19 and very severe COVID-19 in the weighted median (Supplementary Tables S3–S4; p-values = 0.007, 0.0003, respectively). However, none of the associations were significant after accounting for multiple comparisons in the main analysis.
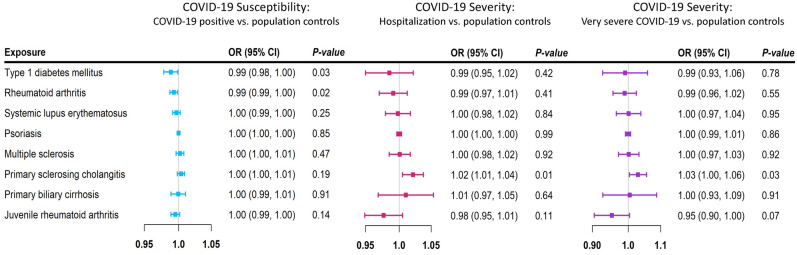
Sensitivity analysis was performed by excluding the SNPs in the HLA gene region. The results were largely similar to those of the primary analyses (Fig. 2; Supplementary Tables S5–S7).
Figure 2.
Mendelian randomization (MR) estimates of sensitivity analysis for each autoimmune disease and COVID-19. Forest plot of Mendelian randomization (MR) estimates of 8 autoimmune diseases with COVID-19 infection, hospitalized COVID-19 and very severe COVID-19 using the multiplicative random-effects inverse variance weighting method after excluding genetic variants in the human leukocyte antigen region. The black color represents COVID-19 infection, the blue color represents hospitalized COVID-19 and the red color represents very severe COVID-19. MR estimates are reported as odds ratios per unit of log-odds of each autoimmune disease examined.
Discussion
Our study provides genetic evidence with minimally confounded estimates that the genetic predisposition to autoimmune diseases examined are largely not genetically associated with the susceptibility or severity of COVID-19, which is consistent with previous findings in patients with autoimmune disorders8.
Potential confounders and selection bias could be major concerns in previous observational studies. A cohort study showed a positive association of RA with COVID-19 infection and hospitalization4. However, body mass index (BMI), which is a risk factor for both RA4 and COVID-199 and thus a potential confounder for the association, was not adjusted for in that study. Many of the relevant studies were selected from people in middle or older age on lifelong immunosuppressant treatment4,5,7. Age is significantly associated with higher COVID-19 mortality and severity10, while autoimmune diseases affect life expectancy11. As such, selection of people who survived genetic liability to or autoimmune diseases and competing risk of COVID-19 may generate selection bias for examining the relation between them12. Another source of selection bias is from participants taking long-term immunosuppressant treatment for autoimmune diseases who were generally vulnerable to COVID-19, masking the association of autoimmune disease and COVID-197. However, previous findings suggest that autoimmunity per se does not increase the susceptibility to COVID-19, independent of immunosuppressive treatment8.
The negative association of type 1 diabetes mellitus with COVID-19 susceptibility observed in our study was consistent with a previous MR study9, where significance changed due to the use of the latest GWAS for COVID-19. However, after excluding SNPs that were negatively associated with BMI, we did not find a significant association of type 1 diabetes mellitus with COVID-19 infection. An MR study showed a positive association of RA with interleukin-6 receptor (IL6R) antagonist13. Another MR study further showed a protective effect of IL6R inhibition on COVID-19 infection, but not on severe COVID-19, where IL6R may increase the risk of pneumonia14. Findings from these two studies suggest a slight positive association of RA with COVID-19 infection. However, the small effects may not be clinically significant as shown in our study.
Primary sclerosing cholangitis might be associated with higher risk of COVID-19. However, most primary sclerosing cholangitis patients after liver transplantation require lifelong immunosuppressant treatment, where liver transplantation is the only known cure for advanced disease15, making them vulnerable to infectious diseases16. A high mortality rate17 may also contribute to potential selection bias because the GWAS was selected on the survival18. Further investigation is warranted.
Some autoimmune antibodies were found in the COVID-19 patients, such as antiphospholipid antibodies19, while some studies have reported an increased risk of autoimmune diseases, such as type 1 diabetes mellitus after COVID-19 infection20. However, many of the GWAS available for autoimmune diseases were much less densely genotyped than the COVID-19 GWAS, making the reverse MR analysis less reliable. Moreover, given the auto-immune GWAS were conducted before the COVID-19 pandemic, MR studies only give pleiotropic effects of auto-immune diseases on COVID-19. Whether COVID-19 triggers the onset of autoimmune diseases requires further investigation.
Despite these findings, this study has some limitations. First, some autoimmune diseases were not included due to lack of relevant GWAS, such as Guillain–Barre syndrome. Second, we cannot fully test the relevance and exclusion restriction assumptions. However, most SNPs used were not related to potential confounders, such as BMI, and did not affect the outcome via other pathways, such as interleukin-6. Sensitivity analysis excluding SNPs in the HLA gene also showed consistent results. Third, potential selection bias may arise from selection on participants with a certain autoimmune disease that needs long-term immunosuppressive treatment and has a low survival rate17. In addition, people with autoimmune diseases may have taken more precautions to avoid COVID-19 infection than other people, which could bias towards the null. However, liability to autoimmune disease does not equate to overt disease, which may reduce this bias. MR studies are also open to selection bias, particularly for exposures that affect survival, where the sample is inevitably selected on surviving the exposure or its genetic predictors and the outcome or a competing risk of the outcome. As such a substantial proportion of deaths before recruitment from the relevant auto-immune disease could bias towards the null or even reverse the estimate. Fourth, sex-specific GWAS for autoimmune diseases and COVID-19 were not available, where sex-specific examination could be helpful as women tend to have higher risk of autoimmune diseases21 while men have higher risks of COVID-19 infection and severity22. Fifth, by examining the genetic predisposition to autoimmune diseases with COVID-19 susceptibility and severity, the study provides genetic evidence rather than the direct causal association of autoimmune diseases with COVID-19. Sixth, limited by data availability, the GWAS used mainly concern people of European descent. Caution should be taken when applying these findings to other populations.
Methods
Study design
This is a two-sample MR study design based on instrumental variable analysis using genetic instruments, here single-nucleotide polymorphisms (SNPs), to predict each selected autoimmune disease to test the causal relation with COVID-19. MR provides a less confounded estimate by taking advantage of the random allocation of genetic variants at conception, and is less susceptible to reverse causation23, but remains susceptible to selection bias24. MR relies on three assumptions, i.e., the genetic instruments are associated with the exposure (relevance) but are not associated with any confounder of exposure on outcome (independence) and can affect the outcome only via the exposure (exclusion-restriction)25.
Data sources
Genetic associations with autoimmune diseases and genetic instrument selection
We included 8 autoimmune diseases as exposures, i.e., type 1 diabetes mellitus26, rheumatoid arthritis (RA)27, systemic lupus erythematosus28, psoriasis29, multiple sclerosis30, primary sclerosing cholangitis18, primary biliary cirrhosis31 and juvenile idiopathic arthritis (JIA)32. We used the largest relevant publicly available genome-wide association study (GWAS) to obtain genetic variants that were strongly (p < 5 × 10–8) and independently (r2 < 0.01) associated with each autoimmune disease18,26,28–31, except for RA and JIA, where the genetic variants for RA27 and JIA32 were obtained from previous well-established studies. The GWAS used were largely based on European descent individuals. Details of the GWASs, such as the number of cases and sample size, are listed in the Supplementary Table S1.
Genetic associations with COVID-19
Summary statistics data for COVID-19 were obtained from the COVID-19 Host Genetics Initiative GWAS meta-analysis round six (updated June 2021; without the 23andMe study), which included 112,612 COVID-19 cases versus 2,474,079 population controls, 24,274 hospitalized COVID-19 cases versus 2,061,529 population, and 8,779 very severe respiratory confirmed COVID-19 cases versus 1,001,875 population controls adjusted for age, sex, and ancestry33. A COVID-19 case was defined by laboratory based on RNA and/or serology, by physician, or by self-report.
Statistical analysis
An approximated F statistic was used to assess instrument strength, where the square of beta for the SNP-exposure association is divided by its variance34.
We used inverse variance weighting (IVW) with multiplicative random effects35 as the main analysis. We used MR-Egger and the weighted median methods as sensitivity analysis, where MR-Egger assumes the instrument strength is independent of the direct effect (InSIDE)36, and the weighted median provides valid estimates when more than 50% of the information comes from valid instruments37. To avoid potential effects of the human leukocyte antigen (HLA), we also performed a sensitivity analysis by excluding the SNPs in the HLA gene given the important role of the HLA for both infectious diseases and autoimmune disorders38.
Analyses were conducted in R (version 4.0.3; R Foundation for Statistical Computing, Vienna, Austria). The TwoSampleMR R package (version 0.5.5) was used to extract genetic predictors and the MendelianRandomization R package (version 0.4.2) to obtain MR estimates based on the IVW, MR-Egger and weighted median methods. Bonferroni correction was used to counteract the issue of multiple comparisons (p = 0.05/8 = 0.006).
Conclusion
Genetic liability to the autoimmune diseases examined was largely not related to susceptibility to or severity of COVID-19. Further investigations of the relation of immunosuppressive treatments and other autoimmune diseases with susceptibility to and severity of COVID-19 is warranted. Whether COVID-19 infection induces autoimmune disease requires further studies.
Supplementary Information
Acknowledgements
We thank the MR-base and the COVID-19 Host Genetics Initiative.
Abbreviations
- BMI
Body mass index
- COVID-19
Coronavirus disease 19
- GWAS
Genome-wide association study
- HLA
Human leukocyte antigen
- IL6R
Interleukin-6 receptor
- IVW
Inverse variance weighting
- JIA
Juvenile idiopathic arthritis
- MR
Mendelian randomization
- RA
Rheumatoid arthritis
- SNP
Single-nucleotide polymorphisms
Author contributions
S.L. designed, analyzed and interpreted the data, and was a major contributor in writing the manuscript. S.Y. provided advice for the analysis, helped with the figures, and revised the manuscript. C.M.S. contributed to the design of the study, checked the study results, and revised the manuscript. S.C.L. supervised the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by Uppsala University.
Data availability
The exposure datasets generated during the current study are available in the MR-base repository, [https://gwas.mrcieu.ac.uk/datasets/], where the MR-base ID of each exposure were listed in the Supplementary Table S1. Specifically, “ebi-a-GCST005536” for type 1 diabetes mellitus, “ebi-a-GCST003156” for systemic lupus erythematosus, “ebi-a-GCST005527” for psoriasis, “ieu-b-18” for “multiple sclerosis”, “ieu-a-1112” for “primary sclerosing cholangitis”, “ebi-a-GCST005581” for primary biliary cirrhosis. The outcome datasets generated during the current study are available in the COVID19-hg GWAS, [https://www.covid19hg.org/results/r6/], where we used “COVID19_HGI_C2_ALL_leave_23andme_20210607.b37.txt” for COVID-19 infection in general population, “COVID19_HGI_B2_ALL_leave_23andme_20210607.b37.txt” for hospitalized COVID-19 and “COVID19_HGI_A2_ALL_leave_23andme_20210607.b37.txt” for very severe COVID-19. The datasets analyzed during the current study are available in the Autoimmune disease and COVID-19 repository, [https://osf.io/sxjyc/].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-22711-1.
References
- 1.Liu Y, Sawalha AH, Lu Q. COVID-19 and autoimmune diseases. Curr. Opin. Rheumatol. 2021;33:155–162. doi: 10.1097/bor.0000000000000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esmaeilzadeh A, Elahi R. Immunobiology and immunotherapy of COVID-19: A clinically updated overview. J. Cell Physiol. 2021;236:2519–2543. doi: 10.1002/jcp.30076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Ruiz R, Paredes JL, Niewold TB. COVID-19 in patients with systemic lupus erythematosus: Lessons learned from the inflammatory disease. Transl. Res. 2021;232:13–36. doi: 10.1016/j.trsl.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.England BR, et al. Risk of COVID-19 in rheumatoid arthritis: A national veterans affairs matched cohort study in at-risk individuals. Arthritis Rheumatol. 2021;73:2179–2188. doi: 10.1002/art.41800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barzegar M, et al. COVID-19 among patients with multiple sclerosis: A systematic review. Neurol. Neuroimmunol. Neuroinflamm. 2021 doi: 10.1212/nxi.0000000000001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakthiswary R, Chuah HY, Chiang KS, Liew YS, Muhammad Aizat NA. COVID-19 in systemic lupus erythematosus: A pooled analysis and systematic review of case reports and series. Lupus. 2021;30:1946–1954. doi: 10.1177/09612033211045057. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama S, Hamdeh S, Micic D, Sakuraba A. Prevalence and clinical outcomes of COVID-19 in patients with autoimmune diseases: A systematic review and meta-analysis. Ann. Rheum. Dis. 2020 doi: 10.1136/annrheumdis-2020-218946. [DOI] [PubMed] [Google Scholar]
- 8.Santacroce G, et al. Impact of COVID-19 in immunosuppressive drug-naïve autoimmune disorders: Autoimmune gastritis, celiac disease, type 1 diabetes, and autoimmune thyroid disease. Pediatr. Allergy Immunol. 2022;33(Suppl 27):105–107. doi: 10.1111/pai.13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leong A, et al. Cardiometabolic risk factors for COVID-19 susceptibility and severity: A Mendelian randomization analysis. PLoS Med. 2021;18:e1003553. doi: 10.1371/journal.pmed.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Driscoll M, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 11.Lens-Pechakova LS. Centenarian rates and life expectancy related to the death rates of multiple sclerosis, asthma, and rheumatoid arthritis and the incidence of type 1 diabetes in children. Rejuvenation Res. 2016;19:53–58. doi: 10.1089/rej.2015.1690. [DOI] [PubMed] [Google Scholar]
- 12.Schooling CM, et al. Use of multivariable mendelian randomization to address biases due to competing risk before recruitment. Front. Genet. 2020;11:610852. doi: 10.3389/fgene.2020.610852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan S, Li X, Lin A, Larsson SC. Interleukins and rheumatoid arthritis: bi-directional Mendelian randomization investigation. Semin. Arthritis Rheum. 2022;53:151958. doi: 10.1016/j.semarthrit.2022.151958. [DOI] [PubMed] [Google Scholar]
- 14.Larsson SC, Burgess S, Gill D. Genetically proxied interleukin-6 receptor inhibition: Opposing associations with COVID-19 and pneumonia. Eur. Respir. J. 2021;57:2003545. doi: 10.1183/13993003.03545-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zecher BF, et al. Prevalence of COVID-19 in patients with autoimmune liver disease in Europe: A patient-oriented online survey. United Eur. Gastroenterol. J. 2021;9:797–808. doi: 10.1002/ueg2.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez MDP, Martin P, Simkins J. Infectious complications after liver transplantation. Gastroenterol. Hepatol. 2015;11:741–753. [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone M, et al. Primary sclerosing cholangitis: burden of disease and mortality using data from the national rare diseases registry in Italy. Int. J. Environ. Res. Public Health. 2020;17:3095. doi: 10.3390/ijerph17093095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji SG, et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat. Genet. 2017;49:269–273. doi: 10.1038/ng.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascolini S, et al. COVID-19 and immunological dysregulation: Can autoantibodies be useful? Clin. Transl. Sci. 2021;14:502–508. doi: 10.1111/cts.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett CE, et al. Risk for newly diagnosed diabetes >30 days after SARS-CoV-2 infection among persons aged <18 years: United States, March 1, 2020-June 28, 2021. MMWR Morb. Mortal Wkly. Rep. 2022;71:59–65. doi: 10.15585/mmwr.mm7102e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angum F, Khan T, Kaler J, Siddiqui L, Hussain A. The prevalence of autoimmune disorders in women: A narrative review. Cureus. 2020;12:e8094–e8094. doi: 10.7759/cureus.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peckham H, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith GD, Ebrahim S. 'Mendelian randomization': Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson E, et al. Mendelian randomization. Nat. Rev. Methods Primers. 2022;2:6. doi: 10.1038/s43586-021-00092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patsopoulos NA, et al. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365:6460. doi: 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JZ, et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Isac E, et al. Combined genetic analysis of juvenile idiopathic arthritis clinical subtypes identifies novel risk loci, target genes and key regulatory mechanisms. Ann. Rheum. Dis. 2020;80:321–328. doi: 10.1136/annrheumdis-2020-218481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The COVID-19 Host Genetics Initiative a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden J, et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016;45:1961–1974. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortiz-Fernández L, et al. Genetic analysis with the immunochip platform in Behçet disease: Identification of residues associated in the HLA class I region and new susceptibility loci. PLoS ONE. 2016;11:e0161305. doi: 10.1371/journal.pone.0161305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The exposure datasets generated during the current study are available in the MR-base repository, [https://gwas.mrcieu.ac.uk/datasets/], where the MR-base ID of each exposure were listed in the Supplementary Table S1. Specifically, “ebi-a-GCST005536” for type 1 diabetes mellitus, “ebi-a-GCST003156” for systemic lupus erythematosus, “ebi-a-GCST005527” for psoriasis, “ieu-b-18” for “multiple sclerosis”, “ieu-a-1112” for “primary sclerosing cholangitis”, “ebi-a-GCST005581” for primary biliary cirrhosis. The outcome datasets generated during the current study are available in the COVID19-hg GWAS, [https://www.covid19hg.org/results/r6/], where we used “COVID19_HGI_C2_ALL_leave_23andme_20210607.b37.txt” for COVID-19 infection in general population, “COVID19_HGI_B2_ALL_leave_23andme_20210607.b37.txt” for hospitalized COVID-19 and “COVID19_HGI_A2_ALL_leave_23andme_20210607.b37.txt” for very severe COVID-19. The datasets analyzed during the current study are available in the Autoimmune disease and COVID-19 repository, [https://osf.io/sxjyc/].