Abstract
Gliomas typically have unfavorable prognosis, due to late detection and interventions. However, effective biomarkers for early glioma diagnosis based on 5-hydroxymethylcytosines (5 hm C) in circulating cell-free DNA (cfDNA) are not currently available. 5 hm C profiles in GSE132118 set were subjected for establishment of diagnostic model using the LASSO (least absolute shrinkage and selection operator) algorithm. The 5 hm C-based models demonstrated great potency in differentiating healthy subjects from gliomas, with area under the curves (AUCs) > 0.91 in the training and validation sets. Moreover, the indicator performed well in combination with clinicopathological characteristics to differentiate glioblastomas (GBMs) from lower grade glioma (LGGs). Enrichment analysis on 5 hm C profiles displayed great correlation with glioma pathophysiology. The 5 hm C-derived biomarker might act as an effective and non-invasive measure in glioma screening.
Keywords: Diagnostic biomarker, cfDNA, 5 hm C, Glioma, LASSO
Diagnostic biomarker; cfDNA, 5 hm C; Glioma; LASSO.
1. Introduction
Gliomas, a common primary intracranial tumor, which are diagnosed over 260,000 patients worldwide annually, have a poor prognosis and lack effective treatments [1]. Despite extensive research over the past several decades, the 5-year overall survival (OS) rates for glioblastoma (GBM) patients only remain about 5%, and about 25% for World Health Organization (WHO) III glioma [2], because of the remarkable glioma heterogeneity, immunosuppression microenvironment and high levels of anaplasia and aggressiveness [3, 4]. It is evident that the clinical benefit of screening program for early detection of glioma. Better methods for early detection and identification of tumor recurrence are in urgent need to develop of novel treatment strategies and thus improve clinical outcomes.
Traditionally, the imaging examinations make it possible to detect and diagnose a great variety of brain disorders, as well as to predict response to chemotherapy and the patient survival [5]. Meanwhile, recent research demonstrates targeted calculation of microvascular characteristics and oxygen tension of tissues based on physiological magnetic resonance imaging (MRI) as an early biomarker of angiogenesis activity, providing valuable guidance for diagnosis of recurrent WHO III glioma [6]. However, there is a phenomenon, called pseudoprogression, occurs in 10–30% of patients with GBM of brain MRI scans within the first 3 months after clinical treatments. As a contrast-enhancing lesions, pseudoprogression may be induced by tumor progression, but may also be produced by post-radiotherapy changes, that may spontaneously regent, which may put sand to the wheels of its clinical application for the misleading potentiality [7].
Liquid biopsy, which is less invasive, safer, and quicker to obtain than tissue biopsy, has been applied to figure out isocitrate dehydrogenase (IDH) mutation status and other circulating signatures, for example, microRNAs in blood, cerebrospinal fluid (CSF) of gliomas [8, 9, 10]. In spite of the promising results, there are still severe limitations for the clinical application of the established biomarkers to detect and diagnose glioma owing to the insufficient sensitivity and specificity [7, 11, 12]. Due to these challenges, there is a great need to develop technologies to augment standard of diagnostic and monitoring approaches to optimally manage glioma patients. Circulating cell-free DNA (cfDNA) is defined as a short fragments DNA (150–200 bp), which could be derived through apoptosis, necrosis, and active secretion of normal and cancer cells and released into the peripheral plasma, or CSF [13]. Physiologically, the plasma concentration of cfDNA usually keep in a low level (10 ng/ml) [14]. Under the pathological circumstances, the significantly upregulated levels of cfDNA could be detected, including cancer, trauma, stroke, and etc. [15]. Notably, the cfDNA concentration in patients with malignancies is about 50 times that of normal individuals, laying a solid foundation for research on cfDNA-associated biomarker. 5-hydroxymethylcytosine (5 hm C) signature is defined as the oxidation product of 5-methylcytosine (5 mC) by the ten-eleven translocation enzymes, which has been recognized as a significant marker of several malignancies and occurs as early changes during tumorigenesis [16]. And with the advances and development of sequencing technologies, the application of cell-free methylated DNA immunoprecipitation and high-throughput bisulfite-free sequencing (cfMeDIP-seq) make it possible to carry out the analysis of more tissue-specific and cancer-specific epigenetic alterations of 5 hm C profiles in cfDNA [17].
As a noninvasive screening approach, recent research demonstrated an epigenetics-based test on 5 hm C profiles, which could significantly overcome tumor heterogeneity, could in all possibility provide an accurate and reliable approach for the cancer early detection and even evaluation of clinical treatment response and prognosis. Hu etal. reported a diagnostic model including 37 5 hm C features, which demonstrate high potentiality to differentiate lung cancer subjects from normal individuals, with area under curves (AUCs) of 0.90 and 0.84 in the two included sets [18]. Zhou etal. established a 5 hm C-LncRNA diagnostic score able to accurately separate human pan-cancers from healthy controls with AUCs of 0.96 and 0.91 in the training and internal validation cohorts, respectively [19]. In glioma, recent research has demonstrated that the 5 hm C signatures of gliomas have higher clinical sensitivity than the traditional molecular marker such as IDH, in precisive diagnosis of gliomas (AUC = 0.84), GBM patients (AUC = 0.84), WHO II-III glioma patients (AUC = 0.86), pioneering in detecting glioma using 5 hm C profiles [20]. However, given the inherent heterogeneity of glioma, the 5 hm C signatures with great sensitivity and accuracy remains to be explored.
In this study, a comprehensive diagnostic model was developed based on the 5 hm C profiles in cfDNA to distinguish gliomas from the normal subjects. Moreover, we explored the diagnostic correlation of the 5 hm C biomarkers with IDH mutation status. Our findings may act as the solid foundation for further research on a strong and potent liquid biopsy-based approach to detect and screen gliomas.
2. Materials and methods
2.1. Study participants
222 samples, including age-, sex-matched 111 primary gliomas and 111 non-tumor tissues, were obtained from GEO database (GSE132118). No sample received radiotherapy before biopsy. Lower-grade gliomas (LGGs) were referred to as WHO II and III gliomas. Among the gliomas, there were 47 subjects diagnosed as LGGs, and 64 samples were diagnosed as glioblastomas (GBMs). Then, we randomly divided the glioma patients and normal controls into independent training, testing cohorts, with a proportion of 7:3, into a training set (LGGs, n = 33; GBMs, n = 51; non-tumors (NTs), n = 71), and a testing one (LGGs, n = 14; GBMs, n = 13; NTs, n = 36). Meanwhile, for validation of the results, we used all the 222 individuals in the GSE132118 cohort (LGGs, n = 47; GBMs, n = 64; NTs, n = 111) as a set to examine the reproductivity of the diagnostic model. Notably, no remarkable differences were obtained in the light of sex, age, and IDH status among the three included sets (Table S1). Three RNA-seq 5 hm C datasets of solid tumors were also acquired from the GEO database and designed to test the predictive performance in other malignant diseases and the specificity of diagnostic model constructed in the present research. In detail, GSE112679 set, included 1402 non-tumor subjects and 1023 hepatocellular carcinoma (HCC) [21]; GSE89570 set contained 604 samples, including 166 colon cancer, 25 liver cancer, 34 pancreatic cancer, 165 gastric carcinoma, 46 thyroid cancer and 167 normal controls [22]. And for GSE152137 set, there were 338 samples, consisting of 243 healthy individuals and 95 patients with pancreatic cancer [23]. Recent research proposed that genes with differential 5 hm C are more likely to be brain-specific, between gliomas and the normal and have relatively higher 5 hm C modification relative to other tissues. Then, the 5 hm C data from 19 types normal tissues were retrieved for the GSE144530 set for testing the tissue specificity of the 5 hm C-Seal data from cfDNA. The workflow of the present research was displayed in Figure S1.
2.2. Development and specificity assessment of GDScore
The 5 hm C raw count data in the above four sets: GSE112679, GSE89570, GSE152137 and GSE132118 were handled based on R package GDCRNATools, for performance of TMM normalization and Voom transformation [24]. For establishment of diagnostic score for glioma, we first detected the differentially 5 hm C modified regions (DhMPs) with the cutoffs of false discovery rate (FDR) < 0.01 and |log2FC| > 0.5, using the Wilcoxon rank-sum test in GSE132118 set. Then, a separate 5 hm C-based diagnostic model to distinguish glioma from non-glioma subjects was constructed on based on two-step procedures-least absolute shrinkage and selection operator (LASSO) and logistic regression analysis. LASSO regression was carried out to achieve the selection of differentially modified 5 hm C signatures using R package glmnet. Then, 5 hm C markers cross-validated in all iterations was selected for multivariable logistic regression analysis. And the 5 hm C signatures with statistical significance (P < 0.05) were retained for diagnostic model. The calculation of the diagnostic score was performed based on the following formula: glioma diagnostic score (GDScore) = , where is the coefficient from the final multivariable logistic regression for the specific marker, and Genei is the 5 hm C values of the signatures. The predicting accuracy for GDScore was calculated by AUC with 95% CI. Spearman test and Wilcoxon rank-sum test was conducted to evaluate the correlation of the diagnostic score and the demographic characteristics, such as age, sex, IDH mutation status of gliomas.
2.3. Functional relevance of differentiating 5 hm C genes
To gain in-depth understanding into the possible mechanisms underlying the 5 hm C modification patterns in the glioma-associated genomic features, we conducted the enrichment analysis of the differentially modified 5 hm C candidates between glioma and non-tumor samples using the DAVID website (version 6.8) for analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO). GO biological processes and KEGG pathways with adjusted p values < 0.05 were retained and subjected to further interpretation.
2.4. Statistical analysis
Continuous variables are presented as mean ± SD and the categorical variables were analyzed on the root of x2 test. When comparing variables between two groups, statistical significance was calculated by Wilcoxon rank-sum test, and when over two subgroups, the statistical differences were tested based on Kruskal-Wallis test. R package ROCR was utilized to draw receiver operating characteristic (ROC) curve and calculate the area under the curve (AUC) with 95% CI to examine the diagnostic performance of the established model. All heatmaps were achieved by the R package pheatmap. All statistical analyses were carried out on R project (v4.0.1). All tests were two-sided and P < 0.05 was considered significant.
3. Results
3.1. Demographics and clinical characteristics of the participants
The mean age of the 111 healthy controls was 47.26 ± 12.90 year and in the diagnosed 111 gliomas, the mean age of was 47.17 ± 12.60 year. Meanwhile, in the both groups, the ratio of male and female patients was 0.67:0.33. For the all gliomas, there were 64 GBM samples included in our research, among which, 58 GBMs were IDH wildtype and 6 GBMs IDH mutated. In the LGG group, there were 47 samples included, among which, 12 LGGs were IDH wildtype, and 35 ones were IDH mutant (Table S2).
3.2. Differential expression of 5 hm C features in glioma
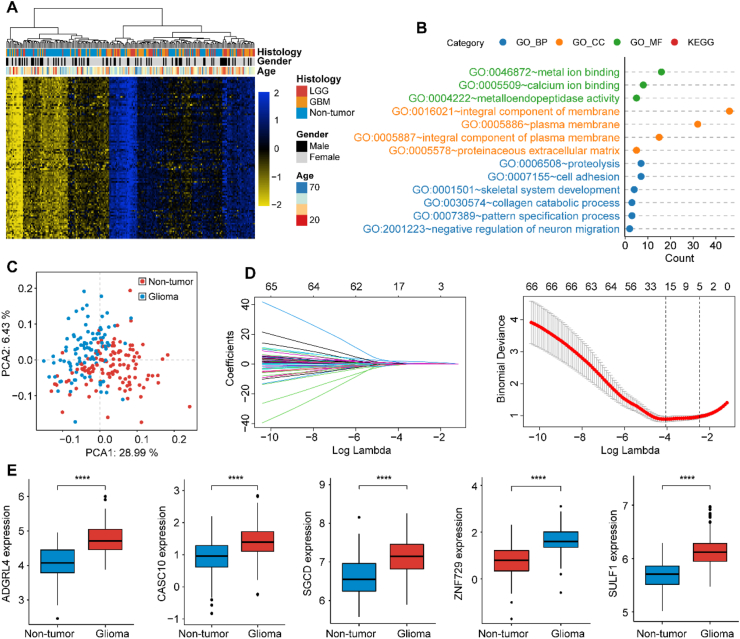
In Comparison with the non-tumor subjects, glioma samples demonstrated higher 5 hm C levels (P < 0.001; Figure S2A). Differential analyses identified 100 expression-dysregulated genes with the cutoffs of FDR < 0.01 and |log2 (fold change [FC])| > 0.5. And all the 100 potential markers were upregulated in the glioma samples, compared with non-tumor samples (Table S3). Meanwhile, the heatmap further demonstrated that the differential 5 hm C genes were more possibly to be brain-derived in 5 hm C levels across different normal samples, providing evidence for their glioma correlation (Figure S2B). Figure 1A demonstrated the differentially 5 hm C-modified features alone could separate gliomas into 2 major clusters using hierarchical clustering analysis. Enrichment analysis of 100 differential 5 hm C modified gene bodies revealed the dysregulated genes were implicated in tumorigenesis and progression of glioma such as the collagen catabolic process, metalloendopeptidase activity and cell adhesion, all of which had been identified to be associated with glioma pathogenesis or malignancy (Figure 1B, Table S4).
Figure 1.
Enrichment analysis and feature selection. Heatmap of differentially modified 5-hydroxymethylcytosine (5 hm C) gene bodies between gliomas and non-tumor samples (A). Scatterplot of enrichment analysis using DAVID database, the count in x-axis represented the genes involved in correspond terms (B). Principal component analysis (PCA) on differentially modified 5-hydroxymethylcytosine (5 hm C) gene bodies and demonstrated the first two principal components (PCA1 and PCA2) (C). LASSO regression analysis for dimensionality reduction (D). The six gene bodies were all upregulated in glioma in 5 hm C-Seal data (E). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Abbreviations: LGG, lower grade glioma; GBM, glioblastoma; LASSO, least absolute shrinkage and selection operator.
3.3. Integrative diagnostic models for gliomas
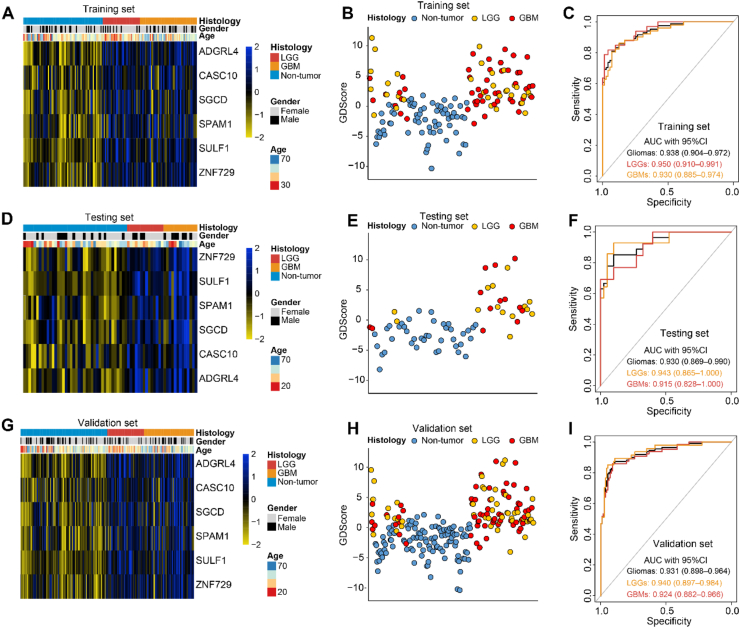
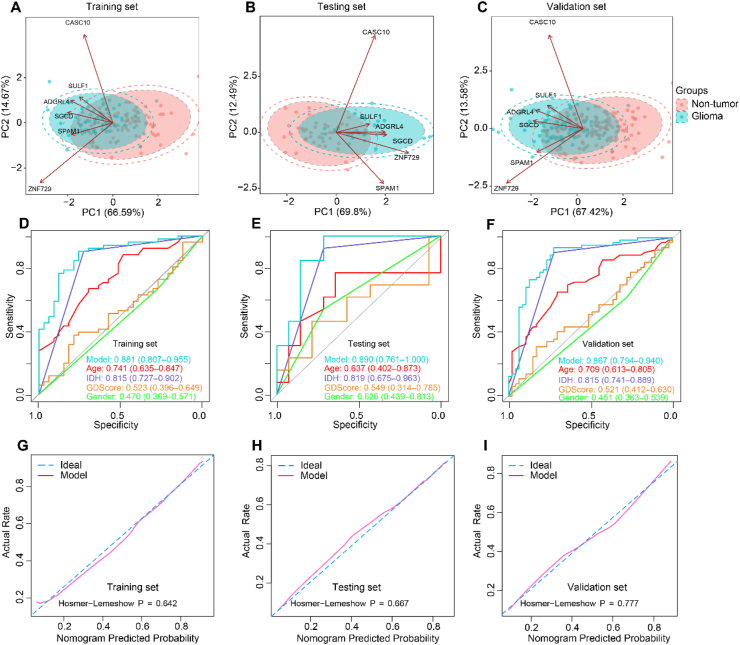
Principal components analysis (PCA) indicated obvious distribution differences between gliomas and non-tumor samples could be obtained based on the 100 differentially 5 hm C-modified genes (Figure 1C). For dimensionality reduction, we conducted least absolute shrinkage and selection operator (LASSO) regression analysis. First, total 222 samples were randomly divided into two sets: a training set of 155 samples (gliomas, n = 84; controls, n = 71) and a testing set of 67 samples (gliomas, n = 27; controls, n = 40), with balanced distributions of age and sex between the two sets, and all the 222 samples were applied as a validation set to confirm the performance of our findings. Then, following feature selection in the training set, six signature genes, including ADGRL4 (Adhesion G Protein-Coupled Receptor L4), CASC10 (Cancer Susceptibility Candidate 10), SGCD (Sarcoglycan Delta), SPAM1 (Sperm Adhesion Molecule 1), SULF1 (Sulfatase 1), ZNF729 (Zinc Finger Protein 729) were selected from the training set (Figure 1D and Table 1). Figure 1E revealed all the six 5 hm C signatures revealed evaluated modifications between gliomas and normal controls. The GDScore was obtained on the basis of the 6 genes. As shown in Figure 2A, 2D and 2G, there were obvious differences of the candidates between glioma and non-tumors. And an increase of GDScore was observed among normal, LGG and GBM samples (Figure 2B, 2E and 2H). Meanwhile, the GDScore demonstrated the great potentiality to tell apart normal people and gliomas (training cohort: AUC = 0.938, 95% CI: 0.904–0.972; testing cohort: AUC = 0.930, 95% CI: 0.869–0.990, validation cohort: AUC = 0.931, 95% CI: 0.898–0.964), GBMs (training cohort: AUC = 0.930, 95% CI: 0.885–0.974; testing cohort: AUC = 0.915, 95% CI: 0.828–1.000, validation cohort: AUC = 0.924, 95% CI: 0.882–0.966; LGGs (training cohort: AUC = 0.950; 95% CI: 0.910–0.991; testing cohort: AUC = 0.943, 95% CI: 0.865–1.000; validation cohort: AUC = 0.940, 95% CI: 0.897–0.984) (Figure 2C, 2F, and 2I). Figure 3A–C demonstrated the PCA, which takes six markers into consideration, also revealed obvious differences between the non-tumors and gliomas in the three cohorts. Meanwhile, the GDScore performed well in differentiating the healthy from IDH wildtype gliomas (training cohort: AUC = 0.929, 95% CI: 0.885–0.972; testing cohort: AUC = 0.930, 95% CI: 0.857–1.000 and validation cohort: AUC = 0.925, 95% CI: 0.885–0.965), and IDH mutant gliomas (training cohort: AUC = 0.955, 95% CI: 0.915–0.996; testing cohort: AUC = 0.930, 95% CI: 0.833–1.000 and validation cohort: AUC = 0.942, 95% CI: 0.896–0.989, Figure S3A-F). However, the diagnostic score achieved the AUC of 0.523 with 95% CI of 0.396–0.649 in training set, AUC = 0.549 with 95% CI of 0.314–0.785 in testing set and AUC = 0.521 95% CI of 0.412–0.630 in validation set in differentiating GBMs from LGGs (Figure S4A-C). In the context of combination with clinicopathological features such as age, sex, and IDH status and GDScore, the integrated model achieved the AUC of 0.881 (0.807–0.955), 0.890 (0.761–1.000) and 0.867 (0.794–0.940) in the separated three sets, outperforming clinicopathological characteristics or GDScore alone in differentiating GBMs from LGGs (Figure 3D–F). Meanwhile, the calibration curves in the three sets demonstrated the small deviations between predicted value and the actual value (Figure 3G–I).
Table 1.
Candidate features selected by LASSO-logistic regression.
| Symbol | log2foldchange | FDR | Chr | Start | End |
|---|---|---|---|---|---|
| ADGRL4 | 0.71 | 1.33E-26 | 1 | 79355449 | 79472403 |
| SGCD | 0.52 | 8.41E-13 | 5 | 1.55E+08 | 1.56E+08 |
| CASC10 | 0.51 | 3.01E-11 | 10 | 21781587 | 21786191 |
| SPAM1 | 0.50 | 5.4E-12 | 7 | 1.24E+08 | 1.24E+08 |
| SULF1 | 0.55 | 3.47E-25 | 8 | 70378859 | 70573150 |
| ZNF729 | 0.82 | 9.48E-17 | 19 | 22469210 | 22499978 |
Abbreviations: FDR, false discovery rate; Chr, chromosome.
Figure 2.
Diagnostic value of GDScore. Heatmap of 6 features selected in GDScore calculation. The expression values of candidate genes were normalized from −2 to 2 (A, D and G). Scatterplot of GDScore distribution among samples in the training, testing and validation sets (B, E, and H). AUC values demonstrated GDScore was a potential biomarker for early detection of glioma, GBM and LGG in the three sets (C, F and I). Abbreviations: AUC, area under curve.
Figure 3.
Nomogram performance assessment. PCA on the six signatures demonstrated the gliomas and non-tumors had a distinct distribution feature (A–C). The integrated model including clinicopathological features and GDScore outperformed the single variate in differentiating GBMs and LGGs by calculating AUC values in the three sets (D–F). The calibration curves for the integrated model. The x-axis represents the nomogram-predicted probability and y-axis represents the actual probability of glioma grades (G–I). Abbreviations: IDH, isocitrate dehydrogenase, AUC, area under curve.
3.4. Association of GDScore with clinical characteristics
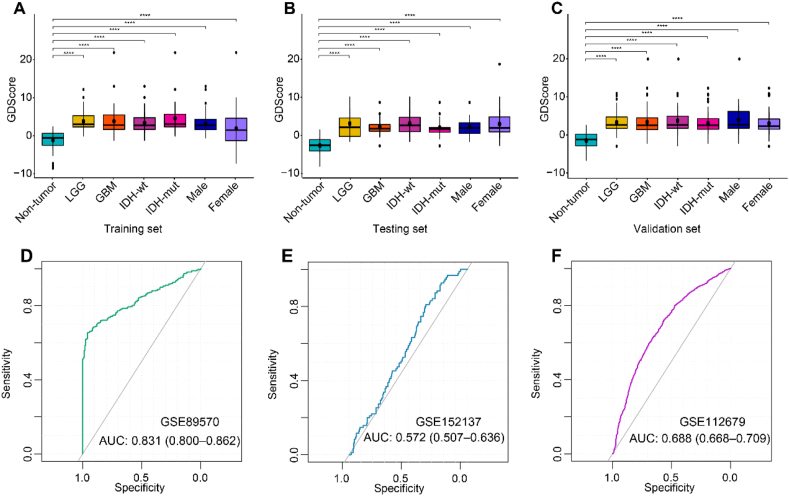
The correlation analysis was conducted of the GDScore with WHO classification, IDH mutation status, age and sex. As shown in Figure 4A–CA, the diagnostic scores increased with WHO grade of glioma, in comparison with the non-tumor controls (P < 0.001). Notably, gliomas also exhibited significantly higher diagnostic score than normal subjects, when taking IDH mutation and sex into account. However, no remarkable differences of diagnostic score were acquired between LGGs and GBMs, or IDH wildtype and IDH mutation, or female and male. Meanwhile, there were significant correlation of GDScore and the diagnostic age of glioma patients (Figure S4D-F).
Figure 4.
Association of GDScore with clinicopathological features and AUC calculation. Boxplots of correlation analysis between diagnostic score and clinical and pathological characteristics in the training (A), testing (B) and validation sets (C). ROC analyses of the performance of GDScore in GSE89570 (D), GSE152137 (E), GSE112679 (F) datasets. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Abbreviations: wt, wildtype; mut, mutant.
3.5. Diagnostic specificity of GDScore in glioma
We evaluated whether the GDScore based on 5 hm C modifications in cfDNA between glioma patients and healthy individuals could be glioma-specific. The GDScore were also computed in the three human solid tumor datasets including available in the GEO database. The predicting ability was assessed by ROC analysis. The results demonstrated in GSE89570, a set containing 5 types of malignant tumors in stomach, colon, pancreas, thyroid and liver, the AUC value with 95% CI was 0.831 (0.800–0.862). In GSE112679 HCC set, the AUC value with 95% CI was 0.688 (0.668–0.709) and in GSE152137 pancreatic cancer set, the AUC value with 95% CI was 0.572 (0.507–0.636).
Of note, comparing with AUC value of GDScore in glioma, GDScore demonstrated poor performance in differentiating other human tumors from healthy individuals, displaying high specificity of GDScore in diagnosing gliomas (Figure 4D–F).
3.6. Methylation analysis of the six hub genes
DiseaseMeth 3.0 database was used to analyze the methylation levels between tissues of glioma and normal samples of the six genes [25]. Differential methylation analysis demonstrated that two markers selected in diagnostic score construction-ADGRL4 and SPAM1 were methylation-upregulated in glioma tissues (Figure S5A and S5B, P < 0.05, respectively), however, CASC10, SGCD, SULF1, ZNF729 revealed no remarkable DNA methylation alterations in tissue levels (Figure S5C-F, P > 0.05, respectively).
4. Discussion
A great number of tumors, including glioma, demonstrate remarkable heterogeneity, which might restrict the capability of a single, tissue biopsy to acquire a full molecular information of the cancer. Liquid biopsies, by contrast, can detect tumor DNA at multiple sites within the tumor, providing a more comprehensive landscape of the cancer genome [26]. Recent research reports that aberrant 5 hm C levels in cfDNA obtained by plasma biopsies are involved in dysregulation of gene expression, tumorigenesis and tumor progression [27, 28, 29]. Meanwhile, growing evidence of abnormal 5 hm C data demonstrates tissue-specific and tumor-specific distribution characteristics, indicating the great probability of application of 5 hm C in cancer diagnostics [26, 30, 31]. With the development of cfMeDIP-seq technology of 5 hm C profiles in plasma, the comprehensive analysis of diagnostic value of 5 hm C are brought to light [32, 33, 34]. For example, recent research indicates the high potentiality of liquid biopsy of cfDNA in bodily fluids as a non-invasive, sensitive and accurate approach in screening and diagnosis of non–small cell lung cancer (NSCLC), and cancers of breast, and ovarian [35, 36, 37]. However, biomarkers based on 5 hm C in cfDNA in distinguishing gliomas from healthy people with great sensitivity and specificity have not been fully explored. In the present research, a cfDNA-based and non-invasive approach was established, which would offer a candidate method in the clinical detection of patients with glioma.
In this study, our research recruited age- and sex-matched 222 samples, including 111 gliomas and 111 healthy objects from the GEO database. Then, all the samples involved in our research were randomly separated with a proportion of 7:3 for accurate performance of the diagnostic model. The upregulated DhMPs between gliomas and normal samples were subjected to enrichment analysis, which demonstrated DhMPs were significantly enriched in cell adhesion, proteolysis, metalloendopeptidase (MMP) activity and extracellular matrix remodeling. Cell adhesion is an essential process for cell migration, abnormal change of which has been recognized as a common and essential process in tumor invasion and metastasis [38]. The MMP, a family of zinc-dependent endopeptidases, plays a major and important role in cell differentiation, proliferation, collagen remodeling, and angiogenesis, and contributes to the malignant behaviors such as tumorigenesis, and cancer invasion and progression [39, 40]. The consequences of enrichment analysis demonstrated that the aberrant 5 hm C profiles might reflect the pathological and physiological changes in carcinogenesis of brain cancer.
PCA on all the abnormal 5 hm C modified genes demonstrated there was a significantly different distributive characteristics between gliomas and normal samples. And the groups could be remarkably separated, which demonstrated the great probability of application of the differentially 5 hm C modified features into filtering patients with glioma out of healthy individuals. Then, to quantify the predictive ability of 5 hm C profiles, a model was constructed by introduction of machine-learning algorithms, including LASSO and logistic regression analyses. And a cfDNA-based, noninvasive epigenetic approach was developed here, in which there were six genes included: ADGRL4, CASC10, SGCD, SPAM1, SULF1, ZNF729. For the six signatures, findings related to SGCD dysregulation in glioma are not reported and deficiency of SGCD could contribute to the development of muscular dystrophy [41]. Dysregulated ADGRL4, an effective anti-angiogenic target for gliomas, has been recognized to be a crucial factor in glioma progression by abnormally activating JAK/STAT3/HIF-1alpha signaling pathway [42, 43]. CASC10 has been found to be relatively upregulated in ciliated glioma stem cells (GSCs) (log2FC = 1166, p < 0.05), however, the in-depth mechanism remains to be explored [44]. One report displayed genomic variations of SPAM1 are implicated in glioma development in Chinese patients [45]. SULF, one of heparan sulfate biosynthesis-related genes, has been illustrated to be significantly overexpressed in glioma, and the level of which positively correlated with glioma pathological grade and revealed a subtype-specific expression characteristics-elevated in proneural GBM, and relatively reduced in classical GBM [46]. Recent research demonstrated ZNF729 aberration in genomic level has a critical impact on dietary fat perception, while no literature has been reported related to glioma pathology. Increasing evidence reveals that dysfunction of lipid metabolism could result in oncogenesis of brain cancer [47]. And we hypothesized that ZNF729 dysregulation might be involved in lipid metabolic reprogramming, leading to the enhancement of malignant behaviors of glioma, which need further study for validation.
Then, for assessment of the predicting accuracy, the AUC was introduced here to calculate the performance in differentiating gliomas (or GBMs, LGGs) from the normal. As a result, the model revealed high diagnostic accuracy in discerning normal population from gliomas: (AUC = 0.938, 95 CI: 0.904–0.972), GBMs (AUC = 0.930, 95% CI: 0.885–0.974), LGGs (AUC = 0.950; 95% CI: 0.910–0.991) in the training set; and gliomas (AUC = 0.930, 95% CI: 0.869–0.990), GBM patients (AUC = 0.915, 95% CI: 0.828–1.000), LGGs (AUC = 0.943, 95% CI: 0.865–1.000) in the testing set; and gliomas (AUC = 0.931, 95% CI, 0.898–0.964), GBMs (AUC = 0.924, 95% CI: 0.882–0.966), LGGs (AUC = 0.940, 95% CI: 0.897–0.984) in the validation set of 222 samples. Meanwhile, the lower calculated AUC values GDScore in other human cancers confirmed the high specificity of GDScore in diagnosing glioma. In comparation with previous research on construction of diagnostic models for glioma, the diagnostic score here we calculated had an outstanding performance in early glioma detect. For example, a diagnostic indicator for gliomas, by scholar Cai, was developed based on 5 hm C profiles and demonstrated less accurate performance than our diagnostic model in distinguishing healthy individuals from gliomas (AUC = 0.84, 95% CI: 0.74–0.93); GBMs (AUC = 0.84; 95% CI: 0.74–0.94); and LGGs (AUC = 0.86; 95% CI: 0.76–0.96) [20]. Meanwhile, A recent study on glioma based on the spectroscopic blood biopsy also reaches a lower performance than our model (AUC = 0.80) [48]. Despite IDH mutation frequencies vary remarkably according to WHO grade of brain cancer, laying a scientific diagnostic foundation for distinguishing LGGs from GBMs, it is usually difficult to obtain due to the unbearable and unacceptable trauma of tumor biopsies [49]. Therefore, when taking the IDH mutation status into consideration, the model established here remained a potent diagnostic indicator for glioma, with IDH wildtype gliomas (training set: AUC = 0.929, 95% CI: 0.885–0.972; testing set: AUC = 0.930, 95% CI: 0.857–1.000 and validation set: AUC = 0.925, 95% CI: 0.885–0.965), and IDH mutant gliomas (training set: AUC = 0.955, 95% CI: 0.915–0.996; testing set: AUC = 0.930, 95% CI: 0.833–1.000 and validation set: AUC = 0.942, 95% CI: 0.896–0.989). The findings revealed the predicting performance of diagnostic score was independent of IDH mutation status, providing insights into distinguishing GBMs from LGGs by combining with IDH mutation. Therefore, our findings displayed the diagnostic model constructed here based on 5 hm C signatures, which could not only discriminate gliomas from healthy individuals but also perform well in distinct IDH mutation status, providing an alternative approach for screening and diagnosing gliomas. Conventional and traditional methods such as MRI and/or computed tomography (CT) contribute a lot to the clinical practice in glioma diagnosis. It is often difficult for radiologists to detect reliably, even if they have extensive experience in the field, owing to the different imaging examinations, and the sophisticated characteristics of glioma morphology [50]. Combination of techniques (eg, GDScore in combination with imaging methods) might allow for improved sensitivity and specificity. We also conducted the comparison of epigenetic patterns of the six biomarkers involved in construction of the diagnostic model in glioma tissues. Our data suggested that the two of the six genes-ADGRL4 and SPAM1 used in diagnostic biomarker establishment were methylation-changed in glioma tissue samples, while CASC10, SGCD, SULF1, ZNF729 demonstrated no significant changes in DNA methylation levels. The above findings might be interpreted with reasons that distinct sequencing platforms were introduced into the analysis and the methylation array could not tell 5 hm C and 5 mC apart, as well as differences because of sample categories (tissue and plasma).
Our findings in gliomas explored the potentiality of the new approach to be developed into a future screening approach, using only plasma examinations. However, some shortages should be focused and resolved in the further research. Firstly, comparison analysis between 5 hm C profiles in cfDNA and paired glioma subjects might contribute to illustrate the tissue or tumor correlation of plasma-extracted 5 hm C signatures. Secondly, external large-scale validation set including gliomas and normal subjects and more comprehensive pathological information would make our findings more scientific. Finally, our research was conducted based on the Chinese cohort, and calculation of the generalizability of the diagnostic score in other populations are essential. Therefore, our findings here remained further investigations for validation into glioma diagnostics.
5. Conclusions
In summary, a noninvasive diagnostic model was developed for patients with glioma by comprehensively analyzing of 5 hm C profiles. The 5 hm C-based diagnostic biomarker act in all probability as a sensitive and accurate approach for early detection and diagnosis of gliomas.
Declarations
Author contribution statement
Chunyu Zhang, Chunlong Zhong, Wei Zhou and Yinqiu Tan: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Daofeng Tian: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Medical Discipline Construction Project of Pudong Health Committee of Shanghai (PWYgy 2021-07) and the Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai (PWR12018-07).
Data availability statement
Data associated with this study has been deposited at GEO database under the accession number [GSE132118, GSE112679, GSE89570, GSE152137, and GSE144530]. (https://www.ncbi.nlm.nih.gov/geo).
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Daofeng Tian, Email: tiandaofeng@whu.edu.cn.
Chunlong Zhong, Email: drchunlongzhong@tongji.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Omuro A., DeAngelis L.M. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 3.Markwell S.M., Ross J.L., Olson C.L., Brat D.J. Necrotic reshaping of the glioma microenvironment drives disease progression. Acta Neuropathol. 2022;143(3):291–310. doi: 10.1007/s00401-021-02401-4. [DOI] [PubMed] [Google Scholar]
- 4.Mair M.J., Geurts M., van den Bent M.J., Berghoff A.S. A basic review on systemic treatment options in WHO grade II-III gliomas. Cancer Treat. Rev. 2021;92 doi: 10.1016/j.ctrv.2020.102124. [DOI] [PubMed] [Google Scholar]
- 5.Schwarzenberg J., Czernin J., Cloughesy T.F., Ellingson B.M., Pope W.B., Geist C., Dahlbom M., Silverman D.H., Satyamurthy N., Phelps M.E., et al. 3'-deoxy-3'-18F-fluorothymidine PET and MRI for early survival predictions in patients with recurrent malignant glioma treated with bevacizumab. J. Nucl. Med. 2012;53(1):29–36. doi: 10.2967/jnumed.111.092387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadlbauer A., Heinz G., Oberndorfer S., Zimmermann M., Kinfe T.M., Buchfelder M., Dörfler A., Kremenevski N., Marhold F. Physiological MRI of microvascular architecture, neovascularization activity, and oxygen metabolism facilitate early recurrence detection in patients with IDH-mutant WHO grade 3 glioma. Neuroradiology. 2022;64(2):265–277. doi: 10.1007/s00234-021-02740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller Bark J., Kulasinghe A., Chua B., Day B.W., Punyadeera C. Circulating biomarkers in patients with glioblastoma. Br. J. Cancer. 2020;122(3):295–305. doi: 10.1038/s41416-019-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silantyev A.S., Falzone L., Libra M., Gurina O.I., Kardashova K.S., Nikolouzakis T.K., Nosyrev A.E., Sutton C.W., Mitsias P.D., Tsatsakis A. Current and future trends on diagnosis and prognosis of glioblastoma: from molecular biology to proteomics. Cells. 2019;8(8) doi: 10.3390/cells8080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zachariah M.A., Oliveira-Costa J.P., Carter B.S., Stott S.L., Nahed B.V. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol. 2018;20(9):1155–1161. doi: 10.1093/neuonc/noy074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopkova A., Sana J., Machackova T., Vecera M., Radova L., Trachtova K., Vybihal V., Smrcka M., Kazda T., Slaby O., et al. Cerebrospinal fluid MicroRNA signatures as diagnostic biomarkers in brain tumors. Cancers. 2019;11(10) doi: 10.3390/cancers11101546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J., Jiang Y., Liu L., Zuo Z., Zeng C. Circulating MicroRNAs as promising diagnostic biomarkers for patients with glioma: a meta-analysis. Front Neurol. 2021;11 doi: 10.3389/fneur.2020.610163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueroa J.M., Carter B.S. Detection of glioblastoma in biofluids. J. Neurosurg. 2018;129(2):334–340. doi: 10.3171/2017.3.JNS162280. [DOI] [PubMed] [Google Scholar]
- 13.Mandel P., Metais P. Nuclear acids in human blood plasma. Comptes Rendus Seances Soc. Biol. Ses Fil. 1948;142(3-4):241–243. [PubMed] [Google Scholar]
- 14.Volik S., Alcaide M., Morin R.D., Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol. Cancer Res.: MCR. 2016;14(10):898–908. doi: 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 15.Kubiritova Z., Radvanszky J., Gardlik R. Cell-free nucleic acids and their emerging role in the pathogenesis and clinical management of inflammatory bowel disease. Int. J. Mol. Sci. 2019;20(15) doi: 10.3390/ijms20153662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo H., Wei W., Ye Z., Zheng J., Xu R.H. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol. Med. 2021;27(5):482–500. doi: 10.1016/j.molmed.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Shen S.Y., Singhania R., Fehringer G., Chakravarthy A., Roehrl M.H.A., Chadwick D., Zuzarte P.C., Borgida A., Wang T.T., Li T., et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563(7732):579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 18.Hu X., Luo K., Shi H., Yan X., Huang R., Zhao B., Zhang J., Xie D., Zhang W. Integrated 5-hydroxymethylcytosine and fragmentation signatures as enhanced biomarkers in lung cancer. Clin. Epigenet. 2022;14(1):15. doi: 10.1186/s13148-022-01233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou M., Hou P., Yan C., Chen L., Li K., Wang Y., Zhao J., Su J., Sun J. Cell-free DNA 5-hydroxymethylcytosine profiles of long non-coding RNA genes enable early detection and progression monitoring of human cancers. Clin. Epigenet. 2021;13(1):197. doi: 10.1186/s13148-021-01183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J., Zeng C., Hua W., Qi Z., Song Y., Lu X., Li D., Zhang Z., Cui X., Zhang X., et al. An integrative analysis of genome-wide 5-hydroxymethylcytosines in circulating cell-free DNA detects noninvasive diagnostic markers for gliomas. Neuro-oncol. Adv. 2021;3(1):vdab049. doi: 10.1093/noajnl/vdab049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai J., Chen L., Zhang Z., Zhang X., Lu X., Liu W., Shi G., Ge Y., Gao P., Yang Y., et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68(12):2195–2205. doi: 10.1136/gutjnl-2019-318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., Zhang J., Nie J., Zheng W., Xu D., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27(10):1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guler G.D., Ning Y., Ku C.J., Phillips T., McCarthy E., Ellison C.K., Bergamaschi A., Collin F., Lloyd P., Scott A., et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat. Commun. 2020;11(1):5270. doi: 10.1038/s41467-020-18965-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R., Qu H., Wang S., Wei J., Zhang L., Ma R., Lu J., Zhu J., Zhong W.-D., Jia Z. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34(14):2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- 25.Xing J., Zhai R., Wang C., Liu H., Zeng J., Zhou D., Zhang M., Wang L., Wu Q., Gu Y., et al. DiseaseMeth version 3.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2022;50(D1):D1208–d1215. doi: 10.1093/nar/gkab1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui X.L., Nie J., Ku J., Dougherty U., West-Szymanski D.C., Collin F., Ellison C.K., Sieh L., Ning Y., Deng Z., et al. A human tissue map of 5-hydroxymethylcytosines exhibits tissue specificity through gene and enhancer modulation. Nat. Commun. 2020;11(1):6161. doi: 10.1038/s41467-020-20001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen L., Song C.X., He C., Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasanthakumar A., Godley L.A. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. 2015;208(5):167–177. doi: 10.1016/j.cancergen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Thomson J.P., Lempiäinen H., Hackett J.A., Nestor C.E., Müller A., Bolognani F., Oakeley E.J., Schübeler D., Terranova R., Reinhardt D., et al. Non-genotoxic carcinogen exposure induces defined changes in the 5-hydroxymethylome. Genome Biol. 2012;13(10):R93. doi: 10.1186/gb-2012-13-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Globisch D., Münzel M., Müller M., Michalakis S., Wagner M., Koch S., Brückl T., Biel M., Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S.G., Jiang Y., Qiu R., Rauch T.A., Wang Y., Schackert G., Krex D., Lu Q., Pfeifer G.P. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71(24):7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guler G.D., Ning Y., Ku C.-J., Phillips T., McCarthy E., Ellison C.K., Bergamaschi A., Collin F., Lloyd P., Scott A., et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat. Commun. 2020;11(1):5270. doi: 10.1038/s41467-020-18965-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeifer G.P., Xiong W., Hahn M.A., Jin S.G. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res. 2014;356(3):631–641. doi: 10.1007/s00441-014-1896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., Zhang J., Nie J., Zheng W., Xu D., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27(10):1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crosby D. Delivering on the promise of early detection with liquid biopsies. Br. J. Cancer. 2022;126(3):313–315. doi: 10.1038/s41416-021-01646-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwo L., Jenna A. The promise of liquid biopsies for cancer diagnosis. Evid. base Oncol. 2021;27(7):SP261–SP262. [Google Scholar]
- 37.Mathios D., Johansen J.S., Cristiano S., Medina J.E., Phallen J., Larsen K.R., Bruhm D.C., Niknafs N., Ferreira L., Adleff V., et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021;12(1):5060. doi: 10.1038/s41467-021-24994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basu S., Cheriyamundath S., Ben-Ze'ev A. Cell-cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Research. 2018;7 doi: 10.12688/f1000research.15782.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassiouni W., Ali M.A.M., Schulz R. Multifunctional intracellular matrix metalloproteinases: implications in disease. FEBS J. 2021;288(24):7162–7182. doi: 10.1111/febs.15701. [DOI] [PubMed] [Google Scholar]
- 40.Payne L.S., Huang P.H. The pathobiology of collagens in glioma. Mol. Cancer Res. : MCR. 2013;11(10):1129–1140. doi: 10.1158/1541-7786.MCR-13-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabharwal R., Chapleau M.W. Autonomic, locomotor and cardiac abnormalities in a mouse model of muscular dystrophy: targeting the renin-angiotensin system. Exp. Physiol. 2014;99(4):627–631. doi: 10.1113/expphysiol.2013.074336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., Shen J., Wang Z., Xu H., Wang Q., Chai S., Fu P., Huang T., Anas O., Zhao H., et al. ELTD1 facilitates glioma proliferation, migration and invasion by activating JAK/STAT3/HIF-1α signaling axis. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-50375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziegler J., Pody R., Coutinho de Souza P., Evans B., Saunders D., Smith N., Mallory S., Njoku C., Dong Y., Chen H., et al. ELTD1, an effective anti-angiogenic target for gliomas: preclinical assessment in mouse GL261 and human G55 xenograft glioma models. Neuro Oncol. 2017;19(2):175–185. doi: 10.1093/neuonc/now147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goranci-Buzhala G. Universität zu Köln; 2020. Characterization of Primary Cilia in Patient-derived Glioma Stem Cells. PhD Thesis. [Google Scholar]
- 45.Li Y., Wang D., Wang L., Yu J., Du D., Chen Y., Gao P., Wang D.M., Yu J., Zhang F., et al. Distinct genomic aberrations between low-grade and high-grade gliomas of Chinese patients. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0057168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ushakov V.S., Tsidulko A.Y., de La Bourdonnaye G., Kazanskaya G.M., Volkov A.M., Kiselev R.S., Kobozev V.V., Kostromskaya D.V., Gaytan A.S., Krivoshapkin A.L., et al. Heparan sulfate biosynthetic system is inhibited in human glioma due to EXT1/2 and HS6ST1/2 down-regulation. Int. J. Mol. Sci. 2017;18(11) doi: 10.3390/ijms18112301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masui K., Cavenee W.K., Mischel P.S., Shibata N. The metabolomic landscape plays a critical role in glioma oncogenesis. Cancer Sci. 2022;113(5):1555–1563. doi: 10.1111/cas.15325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron J.M., Brennan P.M., Antoniou G., Butler H.J., Christie L., Conn J.J.A., Curran T., Gray E., Hegarty M.G., Jenkinson M.D., et al. Clinical validation of a spectroscopic liquid biopsy for earlier detection of brain cancer. Neuro-oncol. Adv. 2022;4(1):vdac024. doi: 10.1093/noajnl/vdac024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen A.L., Holmen S.L., Colman H. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 2013;13(5):345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upadhyay N., Waldman A.D. Conventional MRI evaluation of gliomas. Br. J. Radiol. 2011;84(Spec Iss 2):S107–111. doi: 10.1259/bjr/65711810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at GEO database under the accession number [GSE132118, GSE112679, GSE89570, GSE152137, and GSE144530]. (https://www.ncbi.nlm.nih.gov/geo).