Abstract
The main fatty acids at the sn-1 position of phospholipids (PLs) are saturated or monounsaturated fatty acids such as palmitic acid (C16:0), stearic acid (C18:0), and oleic acid (C18:1) and are constantly replaced, like unsaturated fatty acids at the sn-2 position. However, little is known about the molecular mechanism underlying the replacement of fatty acids at the sn-1 position, i.e., the sn-1 remodeling. Previously, we established a method to evaluate the incorporation of fatty acids into the sn-1 position of lysophospholipids (lyso-PLs). Here, we used this method to identify the enzymes capable of incorporating fatty acids into the sn-1 position of lyso-PLs (sn-1 lysophospholipid acyltransferase [LPLAT]). Screenings using siRNA knockdown and recombinant proteins for 14 LPLATs identified LPLAT7/lysophosphatidylglycerol acyltransferase 1 (LPGAT1) as a candidate. In vitro, we found LPLAT7 mainly incorporated several fatty acids into the sn-1 position of lysophosphatidylcholine (LPC) and lysophosphatidylethanolamine (LPE), with weak activities toward other lyso-PLs. Interestingly, however, only C18:0-containing phosphatidylcholine (PC) and phosphatidylethanolamine (PE) were specifically reduced in the LPLAT7-mutant cells and tissues from knockout mice, with a concomitant increase in the level of C16:0- and C18:1-containing PC and PE. Consistent with this, the incorporation of deuterium-labeled C18:0 into PLs dramatically decreased in the mutant cells, while deuterium-labeled C16:0 and C18:1 showed the opposite dynamic. Identifying LPLAT7 as an sn-1 LPLAT facilitates understanding the biological significance of sn-1 fatty acid remodeling of PLs. We also propose to use the new nomenclature, LPLAT7, for LPGAT1 since the newly assigned enzymatic activities are quite different from the LPGAT1s previously reported.
Supplementary key words: Phospholipid, sn-1, sn-2, fatty-acid remodeling, LPGAT1, palmitic acid, stearic acid, oleic acid, LPLAT
Abbreviations: acyl-CoA, acyl-coenzyme A; AGPAT, 1-acylglycerol-3-phosphate-O-acyltransferase; C16:0, palmitic acid; C17:0, margaric acid; C18:0, stearic acid; C18:1, oleic acid; C18:2, linoleic acid; C18:3, linolenic acid; C20:0, arachidic acid; C20:4, arachidonic acid; C20:5, icosapentaenoic acid; C22:0, behenic acid; C22:6, docosahexaenoic acid; C16:0-d31, 31-deuterium-labeled palmitic acid; C18:0-d35, 35-deuterium-labeled stearic acid; C18:1-d9, 9-deuterium-labeled oleic acid; CL, cardiolipin; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; G3P, glycerol 3-phosphate; GPAT, G3P acyltransferase; GPC, glycerophosphocholine; HBSS, Hank's Balanced Salt Solution; HPLC, high-performance liquid chromatography; KD, knockdown; KO, knockout; LC-MS/MS, liquid chromatography-tandem mass spectrometry; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPG, lysophosphatidyglycerol; LPGAT1, lysophosphatidylglycerol acyltransferase 1; LPI, lysophosphatidyinositol; lyso-PL, lysophospholipid; LPS, lysophosphatidylserine; LPAAT, LPA acyltransferase; LPEAT, LPE acyltransferase; LPLAT, lysophospholipid acyltransferase; LPSAT, LPS acyltransferase; MBOAT, membrane bound O-acyl transferase; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PL, glycerophospholipids; PLA1, phospholipase A1; PLA2, phospholipase A2; PS, phosphatidylserine; sgRNA, single guide RNA; siRNA, small interfering RNA; sn, stereospecifically numbered
Two fatty acids with various structures are bound at the sn-1 and sn-2 positions of PLs, resulting in a wide variety of PL fatty acid species (PL species). Recent development of mass spectrometry techniques has led to the identification of more than 1000 different PL species in mammalian cells (1). The biological significance of this wide variety of PL species has been unclear and has attracted much attention in recent years. In PLs, the two fatty acids are distributed asymmetrically. Namely, saturated fatty acids such as palmitic acid (C16:0) and stearic acid (C18:0) are mainly distributed in the sn-1 position. In contrast, polyunsaturated fatty acids such as linoleic acid (C18:2), arachidonic acid (C20:4), and docosahexaenoic acid (C22:6) are mainly distributed at the sn-2 position (2, 3). Monounsaturated fatty acids, such as oleic acid (C18:1), are distributed both in the sn-1 and sn-2 positions (2, 3). The asymmetry between the sn-1 and sn-2 positions is thought to form in two sequential reactions. These sequential reactions, also known as the PL remodeling reactions or Lands’ cycle, include excision of a fatty acid catalyzed by phospholipase A1 (PLA1) or PLA2 (the first reaction) and incorporation of a new fatty acid into the resulting lysophospholipid catalyzed by lysophospholipid acyltransferases (LPLATs) (the second reaction) (4). The PLA1/PLA2 molecules involved in the first-step reaction are unknown, but several LPLATs responsible for the second-step reaction have been identified (4).
In the last two decades, about a dozen of LPLATs were classified as either 1-acylglycerol-3-phosphate-O-acyltransferases (AGPATs) or membrane-bound O-acyl transferases (MBOATs) (4). Among them, LPCAT3/LPLAT12 and LPIAT1/LPLAT11 were shown to be responsible for the incorporation of arachidonic acid (C20:4) into the sn-2 positions of phosphatidylcholine (PC) (5) and phosphatidylinositol (PI) (6), respectively. Knockout (KO) mice of these LPLATs had decreased C20:4 content in PC and PI and several abnormal phenotypes, including abnormalities in intestinal structure and triacylglycerol absorption (LPLAT12) (7, 8) and neuronal development and liver function (LPLAT11) (9, 10). Other enzymes involved in the initial fatty acid incorporating reactions for the de novo synthesis of PLs and neutral lipids such as triacylglycerols include glycerol-3-phosphate (G3P) acyltransferases (GPATs) and lysophosphatidic acid (LPA) acyltransferases (LPAATs) (11). These enzymes introduce a fatty acid into G3P and LPA producing LPA and phosphatidic acid (PA), respectively. It should be emphasized that the positions of the glycerol backbone into which GPATs and LPAATs introduce a fatty acid are unclear. In addition, most of the enzymes involved in sn-1 remodeling and their biological significance remain to be identified.
One of the problems for the delay in understanding the molecular mechanism of the sn-1 fatty acid remodeling has been the lack of a versatile biochemical method to precisely evaluate the introduction of fatty acids into the sn-1 position. To do this, lyso-PLs with a fatty acid at the sn-2 position, 1-hydroxy-2-acyl-lysophospholipids (sn-2 lyso-PLs), are required as a substrate. However, sn-2 lyso-PLs are very unstable under neutral pH conditions and, especially, alkaline conditions because the fatty acids bound at the sn-2 position can easily migrate to the sn-1 position by an intramolecular acyl migration reaction, resulting in the formation of sn-1 lyso-PLs. Therefore, even if we prepare sn-2 lyso-PLs by PLA1 reaction, most of them would be converted quickly to the corresponding sn-1 lyso-PLs, resulting in a mixture of sn-1 lyso-PL and sn-2 lyso-PL isomers. We mention here that commercially available sn-1 lyso-PLs are a mixture of sn-1 lyso-PLs and sn-2 lyso-PLs with contamination of 5%–10% sn-2 lyso-PLs (12, 13). Therefore, when we use such commercially available sn-1 lyso-PLs to evaluate LPLAT activity, fatty acids can be incorporated into both sn-1 and sn-2 positions in theory. We previously showed that the intramolecular acyl migration reaction rarely occurred, ie, sn-2 lyso-PLs were stable under low pH conditions (13). Furthermore, we showed that the preparation of highly pure sn-2 lyso-PLs was possible by rapidly lowering the pH of the solvent to 4.0 after the PLA1 reaction, which made it possible to evaluate the incorporation of fatty acids into the sn-1 position (14).
Another problem for the delayed progress in elucidating the molecular mechanism of the sn-1 fatty acid remodeling reaction is the lack of methods to determine the position of the glycerol backbone to which a fatty acid is incorporated. For example, consider the LPLAT reaction in which we use oleoyl (C18:1) lysophosphatidylcholine (LPC) as an acyl acceptor and stearoyl (C18:0)-CoA as an acyl donor. In this reaction, the products would be a mixture of 1-oleoyl-2-stearoyl-GPC (glycerophosphocholine) and 1-stearoyl-2-oleoyl-GPC because oleoyl LPC used as an acyl acceptor is a mixture of 1-oleoyl-2-hydroxy-GPC and 1-hydroxy-2-oleoyl-GPC as stated above. However, it is difficult to detect the two asymmetric PC products separately. We recently showed that these two isomers could be distinguished after they were converted to LPC by a PLA2 reaction, which helped determine the precise glycerol sn positions to which a fatty acid is incorporated (14).
In the present study, we employed the two abovementioned methods to search for LPLATs capable of incorporating a fatty acid into the sn-1 position, which we call “sn-1 LPLATs.” First, we detected potent sn-1 LPLAT activities, comparable to sn-2 LPLAT activities incorporating arachidonic acid (C20:4), in several mouse tissues and cell lines. We performed screening using siRNAs and recombinant proteins for several LPLATs and identified LPLAT7/LPGAT1 as a candidate for sn-1 LPLATs. We performed biochemical analyses combined with lipidomics of LPLAT7 KO mice and cells. Accordingly, we found that LPLAT7/LPGAT1, which was previously assigned other biochemical properties, was a major sn-1 LPLAT selectively incorporating stearic acid (C18:0) into LPC and lysophosphatidylethanolamine (LPE). Our present data suggested that the name LPGAT1 did not reflect the enzymes' actual biochemical activities and functions. Thus, we propose to call the enzyme LPLAT7, according to the recently updated nomenclature proposal for LPLAT molecules. Thus, henceforth, the new nomenclature will be used throughout this paper.
Materials and methods
Reagents
Dulbecco's modified Eagle's medium (DMEM) was purchased from Nissui Pharmaceutical (Tokyo, Japan). Fetal bovine serum (FBS), Opti-MEM, Lipofectamine RNAiMAX, and Lipofectamine 2000 were obtained from Thermo Fisher Scientific (Waltham, MA). Honeybee venom PLA2, lipase from Rhizomucor miehei (intrinsic PLA1), stearic-d35 acid (C18:0-d35), and palmitic-d31 acid (C16:0-d31) were obtained from Merck (Darmstadt, Germany). 13C16 Palmitoyl (13C16 16:0) coenzyme A and 13C18 stearoyl (13C18 18:0) coenzyme A were purchased from Taiyo Nippon Sanso (Tokyo, Japan). All glycerophospholipids, lysophospholipids, nonlabeled acyl-coenzyme As (acyl-CoAs), and oleic-d9 acid (C18:1-d9) were purchased from Avanti Polar Lipids (Alabaster, AL). LC-MS-grade methanol and acetonitrile were purchased from Kanto Chemical (Tokyo, Japan). Chloroform, formic acid, and ammonium formate were purchased from Fujifilm-Wako (Osaka, Japan).
Preparation of sn-2-rich and sn-1-rich lysophospholipids
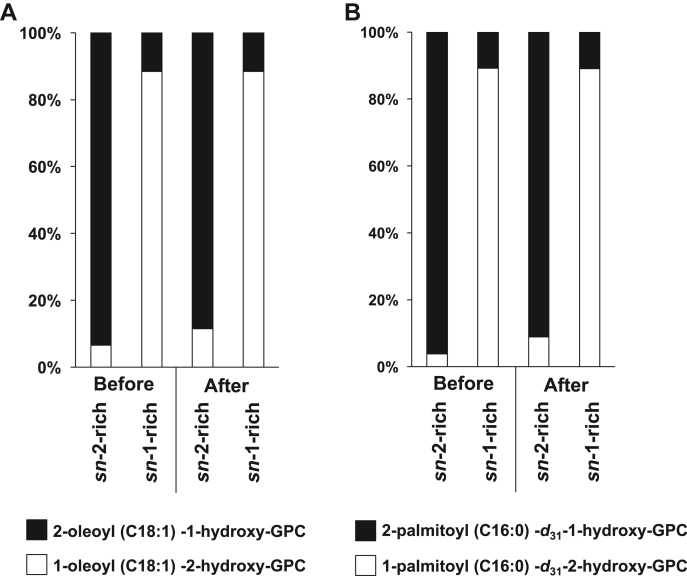
We prepared sn-2-rich LPC with oleic acid (C18:1) and palmitic-d31 acid (C16:0-d31) from dioleoyl (diC18:1) PC and dipalmitoyl-d31 PC, respectively, by PLA1 reaction as described previously (14). We used Rhizomucor miehei lipase as PLA1. The sn-1/sn-2 ratio and the concentration of the resulting sn-2-rich LPCs were determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The sn-2-rich C18:1 LPC and C16:0-d31 LPC preparations predominantly contained LPC with C18:1 and C16:0-d31, respectively, at the sn-2 position (> 90%) (Fig. 1). We stored the sn-2-rich LPCs at −80°C until use. Under this condition, sn-2-rich lysophospholipids (lyso-PLs) were stable for at least a year. The sn-1-rich C18:1 LPC and sn-1-rich C16:0 d31 LPC were prepared from the corresponding sn-2-rich LPC isomers using the spontaneous acyl-migration reaction. Since the acyl-migration reaction is fast in alkaline pH (pH 9.0), the stock sn-2-rich LPCs were dissolved in a weak alkaline solution (100 mM Tris-HCl [pH 8.9]) and were incubated for 2 h at 37°C before stopping the reaction by adding the acid methanol (pH 4.0). The sn-1-rich C18:1 LPC and C16:0-d31 LPC preparations predominantly contained LPC with C18:1 and C16:0-d31 at the sn-1 position (> 90%) (Fig. 1). sn-2-rich C18:1 LPE, sn-2-rich lysophosphatidylserine (LPS), sn-2-rich lysophosphatidylglycerol (LPG), and sn-2-rich LPA were prepared similarly from corresponding diC18:1 PLs. All these sn-2-rich C18:1 lyso-PLs predominantly contained lyso-PLs with C18:1 at the sn-2 position (> 90%, data not shown). Similarly, sn-2-rich LPCs with various acyl chains, including linoleic acid (C18:2), arachidonic acid (C20:4), and docosahexaenoic acid (C22:6), were prepared from corresponding diacyl PCs. In the case of lysophosphatidylinositol (LPI), sn-2-rich LPI was produced from soy PI (predominantly contained 1-C16:0-2-C18:2-glycerophosphoinositol). sn-2-rich LPI predominantly contained LPI with C18:2 at the sn-2 position (>90%, data not shown).
Fig. 1.
sn-2-rich and sn-1-rich LPCs used in this study. The ratio of sn-1 and sn-2 LPC isomers in sn-1-rich and sn-2-rich preparations which were prepared from dioleoyl (diC18:1) PC (A) and dipalmitoyl (diC16:0)-d31 PC (B) as described in Materials and methods. The ratio of sn-1 isomers (1-oleoyl (C18:1) -2-hydroxy-GPC or 1-palmitoyl (C16:0) -d31-2-hydroxy-GPC) and sn-2 isomers (2-oleoyl (C18:1) -1-hydroxy-GPC or 2-palmitoyl (C16:0) -d31-1-hydroxy-GPC) in each preparation were determined by LC-MS/MS. The ratio both before and after the reaction (37°C, 10 min, in a pH 7.4 Tris-HCl -based LPLAT assay buffer (see MATERIALS AND METHODS)) was shown.
Plasmids and vectors
cDNA for human LPLAT7 (hLPGAT1 isoform1; NCBI accession number NM_014873) was amplified by PCR using PrimeStar HS Polymerase (TAKARA BIO Inc) and HEK293A cell cDNA as a template. The PCR primers used were as follows.
Forward 5′- GCCACCATGGCTATAACTTTGGAAGAAGC-3′
Reverse 5′- GCGCTCGAGCTAAAACAGGCAATGGTAAAAATAC-3′
The amplified cDNA fragments were inserted into the pCAGGS vector (a gift from J. Miyazaki, Osaka University) with an N-terminal FLAG-tag. The nucleotide sequences of the prepared plasmids were checked by DNA sequencing (FASMAC).
siRNAs for human LPLATs
Silencer Select small interfering RNAs (siRNAs) for 14 human LPLATs were purchased from Thermo Fisher Scientific (Waltham, MA). The catalog numbers of siRNA used in this study are shown in the following: negative control #1 (4390843), LPLAT1/AGPAT1 (s7 and s9), LPLAT2/APGAT2 (s20702 and s20703), LPLAT3/AGPAT3 (s32329 and s32330), LPLAT4/AGPAT4 (s32331 and s32332), LPLAT5/AGPAT5 (s30735 and s30736), LPLAT6/LCLAT1 (s48420 and s48421), LPLAT7/LPGAT1 (s19258 and s19259), LPLAT8/LPCAT1 (s36575 and s36576), LPLAT9/LPCAT2 (s29823 and s29824), LPLAT10/LPCAT4 (s48551 and s48552), LPLAT11/MBOAT7 (s35614 and s35616), LPLAT12/LPCAT3 (s19799 and s19800), LPLAT13/MBOAT2 (s43424 and s43425), and LPLAT14/MBOAT1 (s45847 and s45848).
Cell culture and transfection
HeLa cells and HEK293A cells (purchased from Thermo Fisher Scientific) were maintained in DMEM supplemented with 10% FBS and penicillin-streptomycin-glutamine (Thermo Fisher Scientific) at 37°C in the presence of 5% CO2 gas. Transfection was performed using lipofection reagent (Lipofectamine 2000 for plasmid transfection and Lipofectamine RNAiMAX for siRNA transfection) and OptiMEM. Typically, HEK293A cells were seeded in a 24-well culture (for lipid analysis) plate or 10-cm dish (for preparing membrane fractions) at a cell density of 0.5-1 × 105 cells per mL and cultured for 1 day. Transfection was performed according to the manufacturer's instructions; typically, plasmid content was 200 ng per well in a 24-well plate, and two siRNAs targeting transcripts of the same gene were mixed at a final siRNA concentration of 10 nM. Twenty-four hours after the plasmid transfection and 48 h after the siRNA transfection, membrane fractions were prepared. In some cases, the cells were subjected to lipid analysis.
Generation of LPLAT7 KO cells
LPLAT7 KO HEK293A cells were generated from wild-type HEK293A cells (parent cells) using the CRISPR/Cas9 system, as previously reported (15). Single-guide RNA (sgRNA) constructs targeting the LPLAT7 genes were designed based on the online software CRISPR direct (https://crispr.dbcls.jp). The designed sgRNA target sequence was 5′- TTGGTGAATCATCAGGCAACAGG -3' (protospacer adjacent motif sequence is in bold and underline). The designed sgRNA-targeting sequences with homology arms were inserted into the BbsI site of the pSpCas9 (BB)-2A-GFP (PX458) vector (Addgene plasmid number #48138). Correctly inserted sgRNA-encoding sequences were verified by DNA sequencing (FASMAC). The PX458 plasmid encoding the sgRNA showed high activity in short assessments and corresponded to all splice variants of LPLAT7 identified at the NCBI. Parent cells were transfected with the PX458 plasmid and detached after 3 days and fractionated for EGFP-positive cells (Cas9 and the sgRNA expressed cells) by using a fluorescence-activated cell sorter (SH800, Sony). After cloning and expansion of cells by limiting dilution, mutations of the target gene were analyzed by DNA sequencing.
Preparation of membrane fractions
HEK293A cells or HeLa cells were recovered into ice-cold TSC buffer (20 mM Tris-HCl [pH 7.4], 300 mM sucrose and cOmplete protease inhibitor cocktail [Roche, Mannheim, Germany]). Minced mouse tissues (20–100 mg) were homogenized in ice-cold TSC buffer using Physcotron homogenizer (Microtec Co Ltd, Chiba, Japan). Cell and mouse tissues were sonicated three cycles of four times for 10 s using a probe sonicator (Microtec, Chiba, Japan). After the homogenates were centrifuged at 800 g for 10 min, the supernatants were further centrifuged at 100,000 g for 1 h. The resultant pellets were resuspended in TSE buffer (20 mM Tris-HCl [pH 7.4], 300 mM sucrose and 1 mM EDTA). After the BCA protein assay kit (Thermo Fisher Scientific) determined the protein concentration, the resulting membrane fractions were stored at −80°C until LPLAT assay.
LPLAT assay
LPLAT assays were performed as previously described (14). Briefly, lyso-PLs solution in a test tube was dried up in a glass tube. Then, assay mixtures containing 100 mM Tris-HCl (pH 7.4), 0.03% Tween-20, and acyl-CoAs (the concentrations of lyso-PLs and acyl-CoAs are indicated in figure legends) were added to the tube. The components were suspended by vortex and sonication. The reaction was initiated by adding a membrane fraction, maintained at 37°C for 10 min. Reactions were stopped by the addition of chloroform/methanol (v/v: 1/2). After adding corresponding internal standards (dilauryl PC, PE, PS, PG, and PA), lipids were extracted by the Bligh and Dyer method. After organic solvents were dried up using a centrifugal evaporator, and lipids were reconstituted in methanol. The resulting PLs were measured by LC-MS/MS.
Determination of the position into which fatty acids are incorporated
We determined the glycerol positions (sn-1 or sn-2 or both) into which LPLAT7 incorporated a fatty acid using a previously reported method (14). We first performed the LPLAT assay using lyso-PLs and acyl-CoA with different fatty acids in this method. Then, the products were subjected to PLA2 reaction, and the resulting LPCs were analyzed by LC-MS/MS. To distinguish the LPCs from the endogenous LPCs in the membrane fractions used as enzyme sources, we used a C16:0-d31 LPC (sn-2-rich) as an acceptor and a 13C1818:0-CoA as a donor. In addition, to remove C16:0-d31 LPC from the products, the reaction mixtures were applied to a Bond Elut C8 solid-phase cartridge column (Agilent Technologies) before the PLA2 treatment. The products (13C1818:0-C16:0-d31-GPC), C16:0-d31 LPC, and 13C1818:0-CoA were separately eluted with 5 mM ammonium formate in 99.5% (v/v) methanol from the cartridge column. The eluted fraction containing the product (13C1818:0-C16:0-d31-GPC) was concentrated by evaporation and dissolved in 0.1% Triton X-100-100 mM Tris-HCl buffer (pH 8.9). PLA2 reaction was performed by adding 0.1 unit bee venom PLA2 to the concentrated product and incubating it at 37°C for 10 min. After stopping the PLA2 reaction by adding acidic methanol (pH 4.0) containing 1 μM C17:0 LPC as an internal standard, the samples were subjected to LC-MS/MS analysis to determine the concentrations of sn-1-C16:0-d31 LPC and sn-1-13C18C18:0 LPC.
Incorporation of deuterium-labeled fatty acids into PLs in culture cells
HEK293A cells in 24-well plates were added with deuterium-labeled fatty acids (stearic-d35-acid [C18:0-d35, 10 μM], palmitic-d31-acid [C16:0-d31, 10 μM], and oleic-d9-acid [C18:1-d9, 25 μM]) and cultured for 2 h. The cells were collected and applied for PL analyses using LC-MS/MS. LPLAT7 KO HEK293A cells (clones #20, #58, and #116), and HEK293 cells overexpressing human LPLAT7 were analyzed. To overexpress LPLAT7, HEK293A cells were transfected with a human LPLAT7 vectors or an empty vector in 24-well culture plates.
Sample preparation for LC-MS/MS lipid analysis of culture cells and mouse tissues
Cells in 24-well plates were washed three times with HBSS (Hank's balanced salt solution) or OptiMEM and then added with ice-cold acidic methanol containing 17:0 LPC (1 μM), 17:0 LPA (100 nM), dilauryl PC (5 μM), dilauryl-PE (1 μM), dilauryl-PS (1 μM), dilauryl-PG (500 nM), dilauryl-PA (500 nM), and dioctanoyl PI (500 nM) as internal standards. After 10 min of incubation at room temperature, lipids dissolved in methanol were collected, passed through a filter (0.2 μm pore size, 4 mm inner diameter; YMC), and subjected to LC-MS/MS. Minced mouse tissues (∼20 mg) were homogenized in ice-cold acid methanol using a micro smash homogenizer (TOMY, Tokyo, Japan) with zirconia beads (3000 rpm and 4°C for 2 min). After centrifugation (21,500 g, 5 min), the supernatant was diluted with methanol containing internal standards (described above). The resulting samples were passed through a filter and subjected to LC-MS/MS analyses.
LC-MS/MS analyses
Lyso-PLs were detected and quantified as previously described (14) with minor modifications. The LC-MS/MS system consisted of Vanquish HPLC (high-performance liquid chromatography) and TSQ Altis Triple-Stage Quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with a heated-electrospray ionization-II (HESI-II) source. For HPLC, samples were separated on an L-column2 (100 mm × 2 mm, 3 μm particle size; CERI), using a gradient elution of solvent A (5 mM ammonium formate in water, pH 4.0) and solvent B (5 mM ammonium formate in 95% (v/v) acetonitrile, pH 4.0) at 200 μl/min. Diacyl PLs were detected and quantified by a similar method (14). The LC-MS/MS system consisted of UltiMate 3000 HPLC system and TSQ Quantiva Triple-Stage Quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with a HESI-II source or Vanquish- TSQ Altis system described above. HPLC separation was performed using a reverse-phase column (Capcell Pak C8 UG120, 150 mm × 1.5 mm, 5 μm particle size; Osaka soda) with a gradient elution of solvent A and solvent B (described above) at 400 μl/min. Lyso-PLs and PLs were detected by selected reaction monitoring in the positive ion mode. At MS1, the m/z values of [M+H] + ions for LPC, PC, PE, and PS and [M+NH4] + ions for PI, PG, and PA were selected. At MS3, phosphocholine fragments (m/z = 184.1) were detected for LPC and PC. DAG fragments were detected for PE, PS, PE, PG, and PA. Selected reaction monitoring was performed in the negative ion mode to characterize the acyl-chain composition of PL species. At MS1, [M+HCOO] – ions for PC and [M-H] – ions for other PLs were selected. At MS3, the m/z values of 255.2 for species containing palmitate, 281.2 for species containing oleate, 283.2 for those containing stearate, 286.4 for species containing palmitate-d31, 290.3 for species containing oleate-d9, and 318.5 for those containing stearate-d35.
Animals
All animal experiments were approved and performed under the guidelines of the animal experimentation committee of the University of Tokyo (approved number P31-1). All experiments on gene recombination were approved by and performed under the guidelines of the University of Tokyo Biosafety Committee (approval number 3–17). Lplat7/Lpgat1-KO mice (Strain Name: C57BL/6NJ-Lpgat1em1(IMPC)J/Mmjax, Stock Number:042167-JAX) were purchased from Mutant Mouse Resource and Research Centers (MMRRC). Mice were genotyped by PCR using primers annealing to the following genomic regions.
WT forward, GCTTTAGTTTCTGTCACTTGCC;
WT reverse, ATTACAACTTAGATGTTTACCAACAGG
KO forward, AGATTGAGTGCTTCCACGAGC;
KO reverse, CAAAATCAACAGTTCAAACACTGGC;
The expected product sizes are 207 bp for the WT allele and 242 bp for the KO allele.
Data analysis
The apparent Km and Vmax values were calculated from the Michaelis-Menten equation using GraphPad Prism 8.4.3 (GraphPad Software, Inc, San Diego, CA). Unless indicated otherwise, data are presented as mean ± SD. For statistical analysis, unpaired t-tests were used to compare two groups. After one-way or two-way ANOVA, multiple comparisons were performed with Bonferroni’s multiple comparison tests depending on the combinations of comparisons. All analyses were done with GraphPad Prism 8.4.3 and Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA). Statistically significant differences are marked with asterisks: ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; and “ns” indicates not significant.
Results
sn-1 LPLAT activities are detected in a wide range of tissues and cells
We first attempted to detect the endogenous LPLAT activities in tissues and cells incorporating fatty acids into the sn-1 position (sn-1 LPLAT activity) of LPC. For this, we used LPCs containing a high proportion of sn-2 isomers as acyl acceptors, which we call sn-2-rich LPCs in this study. The present sn-2-rich LPCs contain more than 90% of sn-2 isomers (2-oleoyl (C18:1)-1-hydroxy-GPC and 2-palmitoyl-d31 (C16:0-d31)-1-hydroxy-GPC) with less than 10% of sn-1 isomers (1- C18:1-2-hydroxy-GPC and 1- C16:0-d31-2-hydroxy-GPC) as judged by LC-MS/MS (Fig. 1). We also prepared the sn-1-rich LPCs from corresponding sn-2-rich LPCs (see Materials and methods). We confirmed that most of the sn-2-rich LPCs used as acyl acceptors remained as sn-2 isomers, even after incubating them in a pH 7.4 Tris-HCl-based LPLAT assay buffer (see Materials and methods) for 10 minutes (Fig. 1).
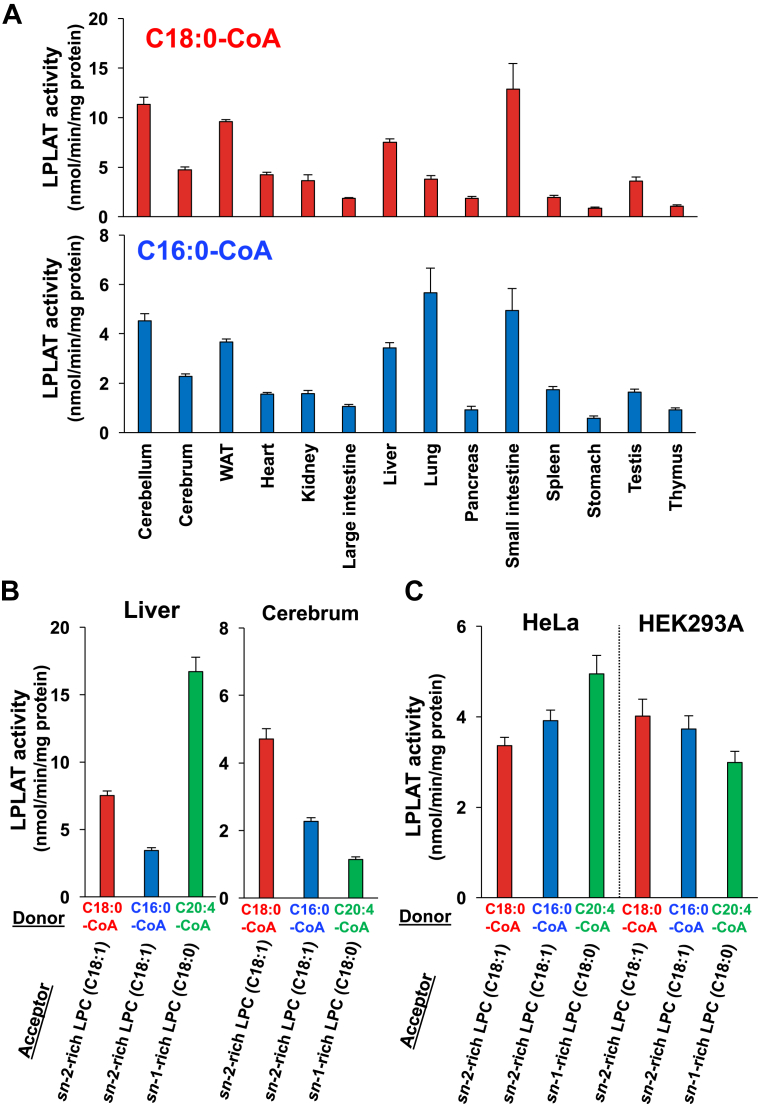
We performed the LPLAT assay using the sn-2-rich C18:1 LPC as an acyl acceptor, C18:0-CoA and C16:0-CoA as acyl donors, and membrane fractions prepared from various mouse tissues and two cultured cell lines (HeLa and HEK293) as enzyme sources. LPLAT activities for the two acyl donors were clearly detected (Fig. 2A). The activity with 18:0-CoA was about twice higher than that with 16:0-CoA. The tissue distribution patterns of activities with 18:0-CoA and with 16:0-CoA were similar but not necessarily identical. For example, the activity with 16:0-CoA was the strongest in the lung, while that with 18:0-CoA was not (Fig. 2A). This suggests that multiple LPLATs are responsible for these activities, especially in the lung. We also found that the sn-1 LPLAT activity with C18:0 was comparable to sn-2 LPLAT activity incorporating arachidonic acid (C20:4) into the sn-2 position of sn-1-rich LPC (C18:0 LPC) in both tissues (the liver and cerebrum) (Fig. 2B) and cell lines (HeLa and HEK293A) (Fig. 2C). These results indicate that sn-1 LPLAT activities for LPC incorporating C18:0 and C16:0 are ubiquitously and abundantly expressed in a wide range of tissues and cells.
Fig. 2.
sn-1 LPLAT activities are detected in a wide range of cells and tissues. A–C, sn-1 LPLAT activities in various mouse tissues and cells. A: LPLAT assays were performed using sn-2-rich LPC (oleoyl (C18:1) LPC (Fig. 1A)) as acyl acceptors, stearoyl-CoA (C18:0-CoA, upper), and palmitoyl-CoA (C16:0-CoA, lower) as acyl donors and membrane fractions of various mouse tissues as enzyme sources. B, C, LPLAT assays were performed using sn-2-rich LPC (C18:1 LPC) and sn-1-rich LPC (stearoyl (C18:0) LPC) as acyl acceptors, C18:0-CoA, C16:0-CoA and arachidonoyl (C20:4)-CoA as acyl donors and membrane fractions of mouse tissues (liver and cerebrum, B) and culture cells (HeLa and HEK293A, C) as enzyme sources. Data are shown as the mean ± SD of three data points. The data are representative of two independent experiments with similar results.
siRNA screening identified LPLAT7 as a candidate for sn-1 LPLAT
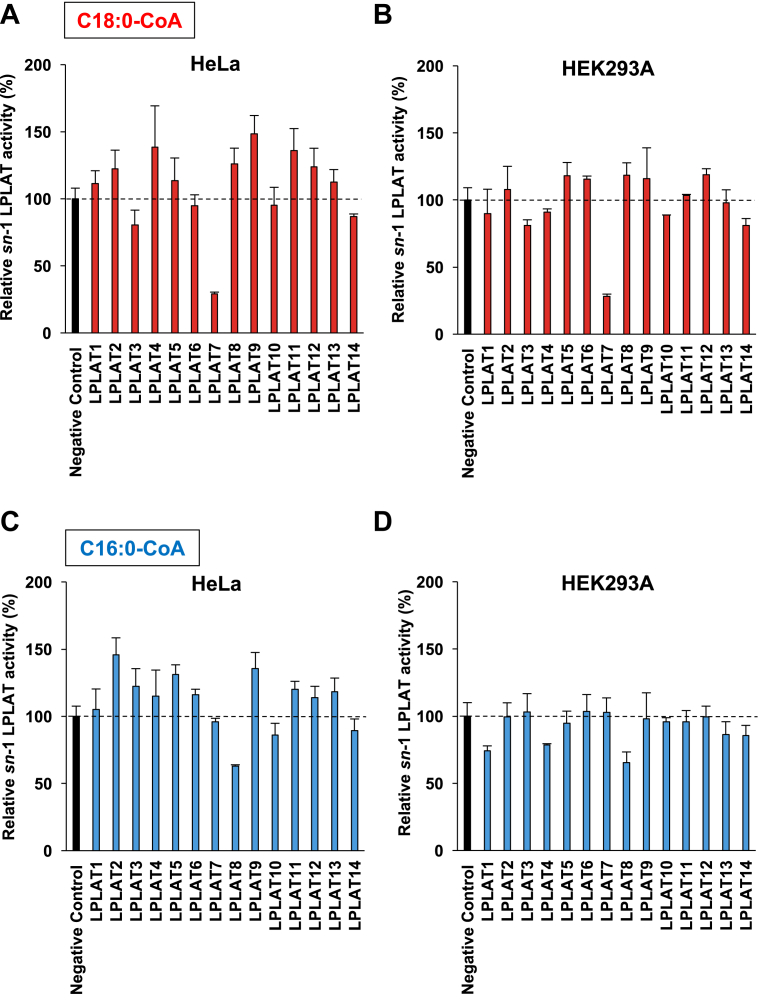
We examined 14 LPLATs belonging to either 1-acylglycerol-3-phosphate-O-acyltransferase (AGPAT) or MBOAT families as candidates of sn-1 LPLATs by screening with their siRNAs and recombinant proteins. HeLa and HEK293A cells were treated with siRNAs for the 14 LPLATs, and the resulting membrane fractions were tested for sn-1 LPLAT activities using sn-2-rich C18:1 LPC (Fig. 1A) as an acyl acceptor and C18:0- and C16:0-CoAs as acyl donors. Experiments using the two cell lines gave similar results: LPLAT7 (LPGAT1) was identified as a candidate for sn-1 LPLATs when C18:0-CoA was used (Fig. 3A, B), and LPLAT8 (LPCAT1) was identified as a candidate when C16:0-CoA was used (Fig. 3C, D). Since we previously showed that LPLAT8 had activities to incorporate C16:0 into both sn-1 and sn-2 positions of PC (14), we focused on LPLAT7.
Fig. 3.
siRNA screening for sn-1 LPLATs. HeLa and HEK293A cells were treated with either siRNA for each LPLAT (LPLAT1 (AGPAT1), LPLAT2 (AGPAT2), LPLAT3 (AGPAT3), LPLAT4 (AGPAT4), LPLAT5 (AGPAT5), LPLAT6 (LCLAT1), LPLAT7 (LPGAT1), LPLAT8 (LPCAT1), LPLAT9 (LPCAT2), LPLAT10 (LPCAT4), LPLAT11 (MBOAT7), LPLAT13 (MBOAT2), and LPLAT14 (MBOAT1) or negative control siRNA. The resulting membrane fractions were used for sn-1 LPLAT assays using sn-2-rich LPC (oleoyl (C18:1) LPC, Fig. 1A) as an acyl acceptor and stearoyl-CoA (C18:0-CoA, A: HeLa, B: HEK293A) and palmitoyl-CoA (C16:0-CoA, C: HeLa, D: HEK293A) as acyl donors. Data are shown as the mean ± SD of three data points. The data are representative of two independent experiments with similar results.
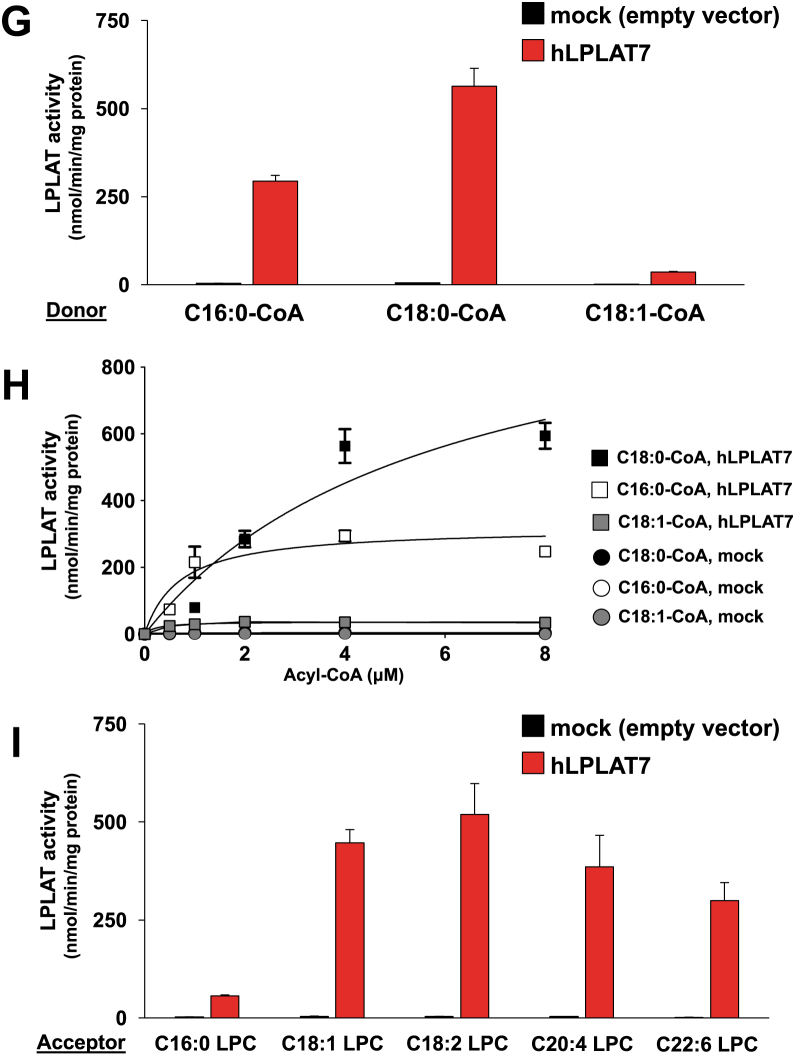
Biochemical characterization of LPLAT7
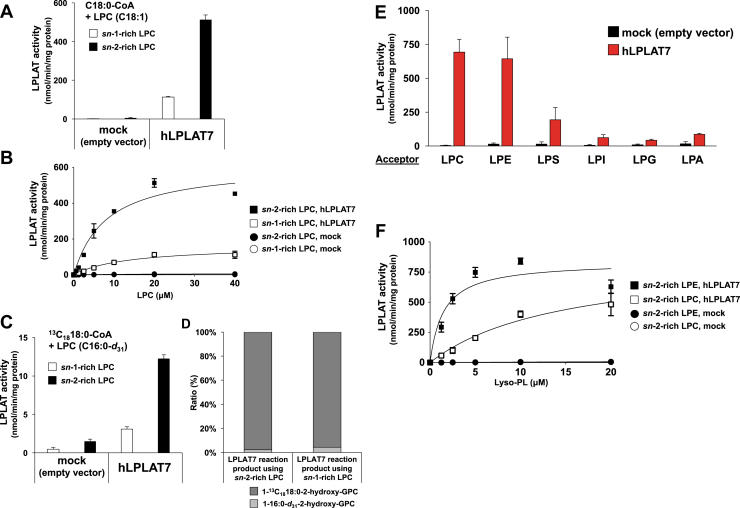
To characterize LPLAT7 biochemically, we overexpressed a human LPLAT7 protein in HEK293A cells and used the resulting membrane fraction as an enzyme source. A membrane fraction from HEK293A cells transfected with an empty vector was used as a negative control. When sn-2-rich LPCs (C18:1 LPC) were used as the acyl acceptors and C18:0-CoA was used as the acyl donor, the membrane fraction from LPLAT7-transfected HEK293A cells showed about 100 times more LPLAT activity than the control (Fig. 4A, filled bar). Of note, the sn-2-rich LPC was found to be a better substrate than the sn-1-rich LPC (Fig. 4A). It should be mentioned here that LPLAT7 showed significant activity against the sn-1-rich LPC with its activity about a quarter of that against the sn-2-rich LPC (Fig. 4A). The kinetic parameter experiments using different concentrations of acyl acceptors confirmed that the Vmax values were markedly greater for sn-2-rich LPC than for sn-1-rich LPC (∼600 vs. ∼150 nmol/min/mg protein). Interestingly, however, the apparent affinity (Km values) calculated from the curves were 9.8 ± 2.7 and 21.1 ± 8.8 μM for sn-2-rich and sn-1-rich LPCs, respectively, showing that the affinity for sn-2-rich LPC was slightly greater than that for sn-1-rich LPC (Fig. 4B).
Fig. 4.
Biochemical characterization of LPLAT7. A, B, Preference of LPLAT7 for sn-2 lysophospholipids. A: LPLAT activities of LPLAT7 against LPC. Oleoyl (C18:1) LPCs (both sn-2-rich and sn-1-rich, 20 μM) and stearoyl (C18:0)-CoA (4 μM) were used as acyl acceptors and an acyl donor, respectively. Membrane fractions from HEK293A cells transfected with a human LPLAT7 expression vector or an empty vector (Mock) were used as enzyme sources. B: Kinetic analyses of LPTAT7 activities. Substrate concentration dependence of LPLAT7 was determined with the indicated concentrations of LPCs (both sn-2-rich and sn-1-rich) in the presence of C18:0-CoA (4 μM). C, D, Determination of the glycerol sn positions into which LPLAT7 introduces a fatty acid. C: LPLAT activities of LPLAT7 against LPC using a set of labeled LPCs and a labeled-acyl-CoA. Palmitoyl (C16:0)-d31 LPCs (both sn-2-rich and sn-1-rich, 10 μM) and 13C1818:0-CoA (2 μM) were used as acyl acceptors and an acyl donor, respectively. D: The ratio of two labeled fatty acids at the sn-1 position of the LPLAT7 reaction products. The reaction products of LPLAT7 in (C) were subjected to a PLA2 reaction. The amount of the resulting two sn-1-acyl LPCs (1–13C1818:0-2-hydroxy-GPC and 1-16:0-d31-2-hydroxy-GPC) was determined by LC-MS/MS. E: LPLAT activities of LPLAT7 toward acyl acceptors with different head groups. Various sn-2-rich C18:1 lyso-PLs (LPC, LPE, LPS, LPG and LPA) and soy LPI, each 20 μM and C18:0-CoA (4 μM) were used. F: Kinetic analyses of LPTAT7 activities. Substrate concentration dependence of LPLAT7 was determined with the indicated concentrations of LPCs or LPEs in the presence of C18:0-CoA (4 μM). G: LPLAT activities of LPLAT7 toward the three acyl donors, determined using C16:0-CoA, C18:1-CoA, or C18:0-CoA (4 μM) with sn-2-rich C18:1 LPC (20 μM). H: Kinetic analyses of LPTAT7 activities. Substrate concentration dependence of LPLAT7 was determined with the indicated concentrations of acyl-CoAs (C18:0-, C18:1- and C16:0-CoAs) in the presence of sn-2-rich C18:1 LPC (20 μM). I: LPLAT activities of LPLAT7 toward various acyl acceptors with different fatty acids. Several LPCs (C16:0, C18:1, linoleoyl (C18:2), arachidonoyl (C20:4), and docosahexaenoyl (C22:6), each 20 μM) and C18:0-CoA (4 μM) were used.
To examine the position in the glycerol backbone into which LPLAT7 incorporated a fatty acid, we prepared another type of sn-2-rich LPC (C16:0-d31 LPC) and the corresponding sn-1-rich LPC (Fig. 1B). Note that both types of LPC were actually a mixture of 1-C16:0-d31-2-hydroxy-GPC (1-C16:0-d31-GPC) and 2-C16:0-d31-1-hydroxy-GPC (2-C16:0-d31-GPC). We obtained virtually the same result using a different set of substrates (C16:0-d31 LPCs and 13C1818:0-CoA) (Fig. 4C) that we obtained with C18:1 LPC and C18:0-CoA (Fig. 4A). Interestingly, when the LPLAT7 products, i.e. 13C1818:0-C16:0-d31-GPC (a mixture of 1–13C1818:0-2-C16:0-d31-GPC and 1-C16:0-d31-2-13C1818:0-GPC), were subjected to PLA2 digestion and the resulting LPC were analyzed by LC-MS/MS, the detected product from PLA2 digestion was almost exclusively 1–13C1818:0-2-hydroxy-GPC (1–13C1818:0 LPC) with a very slight amount of 1-C16:0-d31-2-hydroxy-GPC (1-C16:0-d31 LPC), even when the sn-1-rich C16:0-d31 LPC was used as an acyl acceptor (Fig. 4D), showing that LPLAT7 incorporated 13C1818:0 into the sn-1 position of 2-C16:0-d31-GPC which is contaminated in the present sn-1-rich LPC. This clearly demonstrated that LPLAT7 incorporated a fatty acid predominantly into the sn-1 position of the glycerol backbone.
We also measured the LPLAT7 activities using various substrates, i.e., lyso-PLs with different head (Fig. 4E) and acyl groups (Fig. 4I), and various acyl-CoAs (Fig. 4G and supplemental Fig. S1). With respect to the head groups, the rank order potencies of LPLAT7 were LPE ≥ LPC > lysophosphatidylserine (LPS) >> LPI = LPG = LPA (Fig. 4E). A precise kinetic parameter experiment using varying concentrations of LPC and LPE showed that the enzyme had a higher affinity for LPE than for LPC (the Km values were 9.8 ± 2.7 for LPC and 1.7 ± 0.3 for LPE), although the Vmax values (∼600 and ∼800 nmol/min/mg protein for LPC and LPE, respectively) were apparently similar (Fig. 4F). Among the three acyl-CoAs (C16:0-, C18:0-, and C18:1-CoAs), LPLAT7 showed the highest activity (Vmax = ∼600 nmol/min/mg protein) for C18:0-CoA and the highest Km value (i.e., the highest affinity) for C16:0-CoA (Fig. 4H). C18:1-CoA (Fig. 4G, H) and other acyl-CoAs (supplemental Fig. S1) were also found to be poor substrates. LPLAT7 showed roughly similar activities for various sn-2-rich LPCs including C18:1, linoleoyl (C18:2), arachidonyl (C20:4), and docosahexaenoyl (C22:6) LPCs (Fig. 4I). It should be mentioned that sn-2-rich 16:0 LPC was a relatively poor substrate (Fig. 4I).
The biochemical characterization of LPLAT7 showed that it had LPLAT activities to incorporate fatty acids into the sn-1 position of lyso-PLs, mainly LPC and LPE. Other lyso-PLs including LPI, LPG, and LPA were found to be poor substrates. LPS was a moderate substrate. Thus, LPLAT7 appears to be involved in the fatty acid remodeling of PLs, mainly PC, PE, and PS, rather than PI and PG. The fact that LPA was not a good substrate for LPLAT7 also suggests that LPLAT7 is not involved in the de novo PL synthesis.
LPLAT7 is the main sn-1 LPLAT in tissues and cells
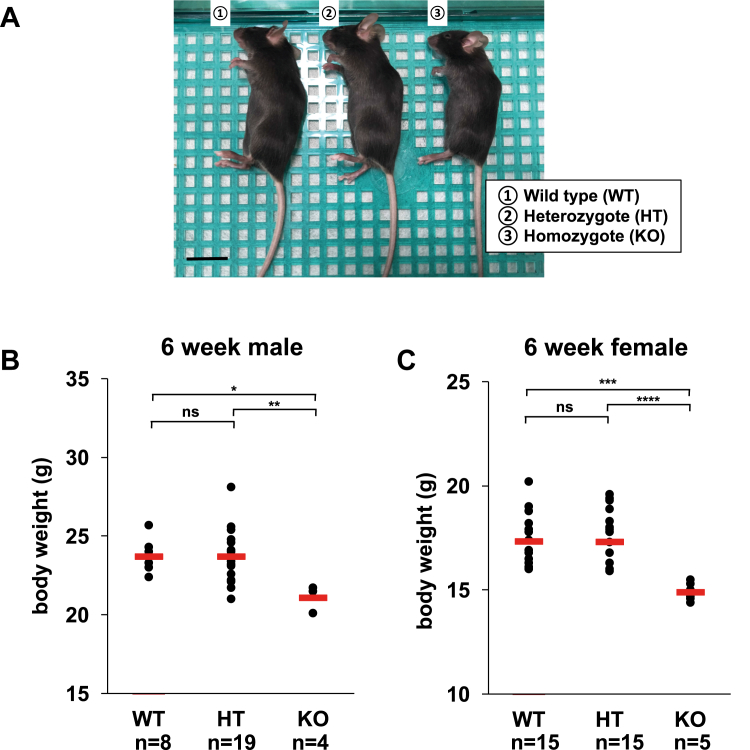
sn-1 LPLAT activities for sn-2-rich LPCs were detected in a wide range of tissues and cells (Fig. 2). Furthermore, LPLAT7 showed potent sn-1 LPLAT activities for various lyso-PLs (Fig. 4E). To examine the contribution of LPLAT7 to these sn-1 LPLAT activities, we evaluated the sn-1 LPLAT activities in Lplat7 KO mice and LPLAT7-deficient cells. Lplat7 KO mice generated by the Knockout Mouse Phenotyping Program (KOMP) using CRISPR technology lacked exon 3, which contains the AGPAT motif required for LPLAT activities, and the resulting premature stop codon (supplemental Fig. S2). Under our breeding conditions, about a half of the KO mice caused sudden death by 6 weeks of age and were present at a much lower rate than expected by the Mendelian rule (Table 1). Furthermore, at the same age, the KO mice had significantly lower body weights than the control wild-type and heterozygous littermates (Fig. 5A–C). We also established three lines of LPLAT7-deficient HEK293A cells using CRISPR technology, which also had mutations around the AGPAT motif (supplemental Fig. S3).
Table 1.
Number of offspring and genotype frequencies (% of total number) from heterozygote mating (6 weeks)
| Wild-type (Lplat7+/+) | Heterozygote (Lplat7+/−) | Homozygote (Lplat7−/−) | |
|---|---|---|---|
| Female | 111 (30.6%) | 213 (58.7%) | 39 (10.7%) |
| Male | 111 (31.3%) | 197 (55.5%) | 47 (13.2%) |
| Total | 222 (30.9%) | 410 (57.1%) | 86 (12.0%) |
Fig. 5.
Lplat7-deficient mice show abnormal growth. A: Gross appearance of 6-week-old female mice (Wild type, hetero Lplat7-deficient and homo Lplat7-deficient). Scale bar, 2 cm. B, C, Body weight of 6-weeks-old male mice (B) and female mice (C) (Wild type (WT), hetero Lplat7-deficient (HT) and homo Lplat7-deficient (KO)) Red bar show mean of each group. The number of mice are indicated below each genotype. Statistically significant differences (WT vs. HT, WT vs. KO and HT vs. KO) are marked with asterisks indicating P-values. ∗P<0.05; ∗∗P<0.01; ∗∗∗P<0.001; ∗∗∗∗P<0.0001; ns indicates not significant; one-way ANOVA, Bonferroni’s multiple comparison test.
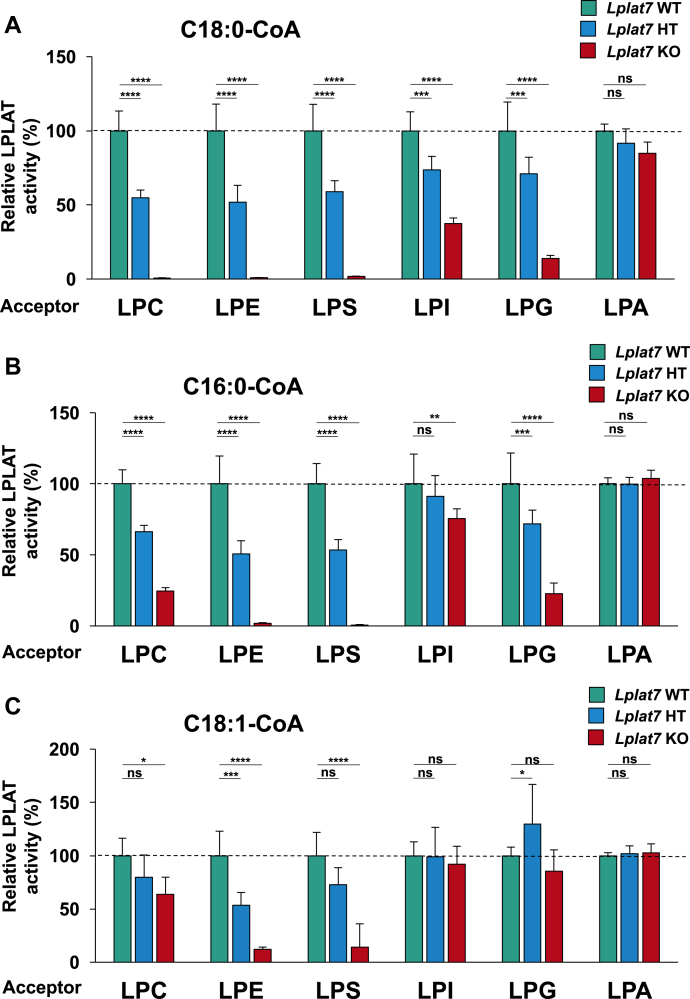
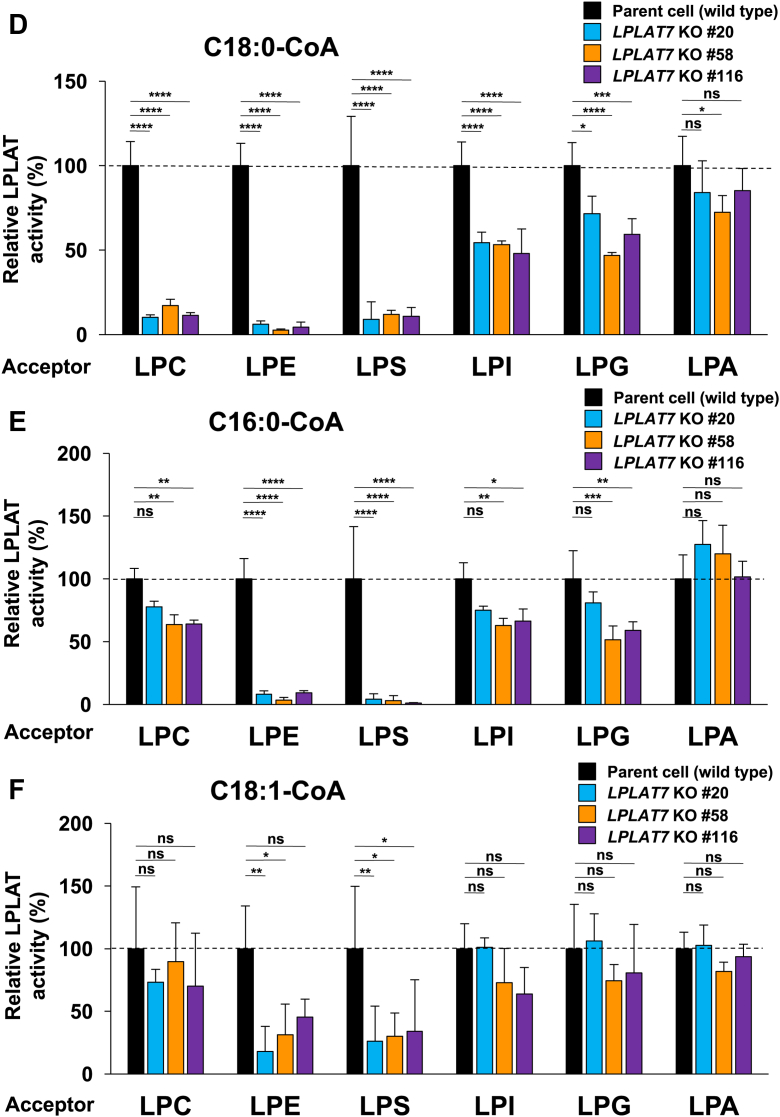
We prepared membrane fractions from the mouse liver and measured their sn-1 LPLAT activities using several combinations of acyl donors (C18:0-CoA, C16:0-CoA, and C18:1-CoA) and acceptors (sn-2-rich C18:1 lyso-PLs [LPC, LPE, LPS, and LPA] and soy LPI). Some sn-1 LPLAT activities dramatically disappeared in Lplat7 KO mice (Fig. 6A–C). For example, in the liver, LPLAT activities for LPC, LPE, and LPS almost completely disappeared when C18:0-CoA was used as the acyl donor. When C16:0-CoA was used, LPLAT activities for LPE and LPS were not detected. When C18:1-CoA was used, LPLAT activities for LPE and LPS were significantly decreased. On the other hand, the sn-1 LPLAT activities for LPC using C16:0-CoA or C18:1-CoA decreased but still left in the KO mice. The sn-1 LPLAT activities for LPI and LPG were also lower but still present in the KO mice. The activity for LPA was hardly reduced in the KO mice (Fig. 6A–C). Heterozygous deletion resulted in moderate activities (Fig. 6A–C). Similar changes in the sn-1 LPLAT activities were observed in the membrane fraction prepared from LPLAT7-KO HEK293A cells (Fig. 6D–F) and siRNA-treated cells (supplemental Fig. S4). One exception was the LPLAT activity detected when LPC and C16:0-CoA were used. In the liver of KO mice, the activity was reduced by ∼70%, whereas in KO cells, it was reduced by ∼30%. Thus, LPLAT7 is mainly responsible for the incorporation of C18:0 into LPC, LPE, and LPS and for the incorporation of C16:0 into LPE and LPS. LPLAT7 did not contribute to an LPLAT activity for LPA, suggesting that it contributes to the fatty acid remodeling of PLs rather than to the de novo synthesis of PLs. It should be also stressed that some sn-1 LPLAT activities for sn-2-rich lyso-PLs still remained in the KO mice liver and KO cells, implying that other sn-1 LPLATs, possibly LPLAT8/LPCAT1, act as sn-1 LPLATs for LPC and C16:0-CoA (14), and LPLAT6/LYCAT, as sn-1 LPLATs for LPI and LPG and C18:0-CoA (16).
Fig. 6.
LPLAT7 is the major sn-1 LPLAT in the mouse liver and human culture cells. (A, B and C) Membrane fractions of the liver from wild-type (green bars), Lplat7 heterozygous (blue bars) and Lplat7 homozygous (red bars) mice were tested for sn-1 LPLAT assays using various sn-2-rich oleoyl (C18:1) lysophospholipids (except for soy LPI) as acyl acceptors and stearoyl (C18:0)-CoA (A), palmitoyl (C16:0)-CoA (B) or oleoyl (C18:1)-CoA (C) as acyl donors. The relative LPLAT activities of wild-type mice being 100% are shown. Data are shown as the mean ± SD of 4–5 mice sample for each group. The data are representative of two independent experiments with similar results. (D, E and F) Membrane fractions of the HEK293 A parent cells (wild-type, black bars), LPLAT7 KO clone#20 (light blue bars), LPLAT7 KO clone#58 (orange bars) and LPLAT7 KO clone#116 (purple bars) were measured for sn-1 LPLAT assays using various sn-2-rich C18:1 lysophospholipids (except for soy LPI) as acyl acceptors and C18:0-CoA (D) or C16:0-CoA (E) or 18:1-CoA (F) as acyl donors. The relative LPLAT activities of parent cells (wild-type) being 100% are shown. Data are shown as the mean ± SD of three data points. The data are representative of two independent experiments with similar results. Statistically significant differences (WT vs. HT and WT vs. KO or Parent vs. KO #20, KO #58 and KO #116) are marked with asterisks indicating P-values. ∗P<0.05; ∗∗P<0.01; ∗∗∗P<0.001; ∗∗∗∗P<0.0001; ns indicates not significant; two-way ANOVA, Bonferroni’s multiple comparison test.
LPLAT7 is responsible for production of C18:0-containing PLs in tissues and cells
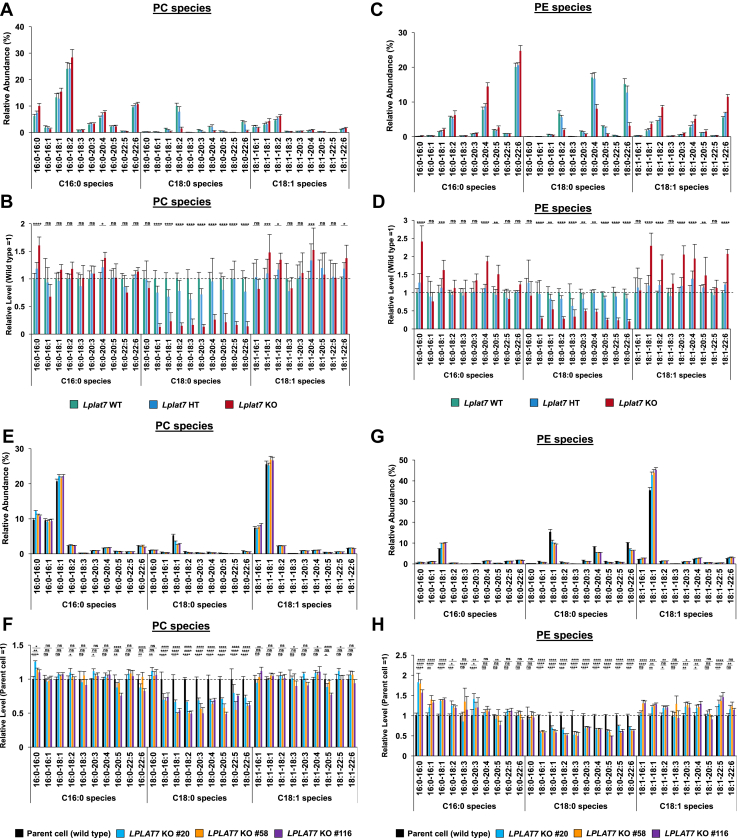
The substrates and products identified by in vitro experiments sometimes differ in vivo. To examine the validity of substrate specificities of LPLAT7 in vitro (Fig. 4), we analyzed the lipidomes of Lplat7 KO mice or LPLAT7 KO HEK293A cells. The analyses, which determined the fatty acid composition of all the PL classes, revealed a marked decrease in C18:0-containing PLs by the deletion of LPLAT7 in both mouse liver (Figs. 7A–D and S5) and HEK293A cells (Figs. 7E–H and S5). The reduction was evident in PC and PE species in both mouse liver and the cells (Fig. 7). It was also evident that C16:0- and C18:1-containing PC and PE species were increased, especially in the liver (Fig. 7A–D). Of note, a marked decrease in C18:0-containing PS species and a marked increase in C16:0- and 18:1-containing PS species were also detected in both the liver and cells (supplemental Fig. S5A, B, I, J). We detected only minor changes in the levels of 18:0-containing PG, PI, and PA (supplemental Fig. S5C–H, K–P). Thus, LPLAT7 is not likely to be involved in de novo PL synthesis. Rather, it affects the level of 18:0-containing PC, PE, and PS species by directly participating in the remodeling pathway. The lipidomic analyses revealed that LPLAT7 contributed to the production of C18:0-containing PL species including PC, PE, and PS. They also revealed that LPLAT7 determines the balance between C18:0-, C16:0-, and C18:1-containing PLs.
Fig. 7.
LPLAT7 determined the ratio of stearic acid (C18:0), palmitic acid (C16:0), and oleic acid (C18:1) in PC and PE in mouse liver and human culture cells. PL levels were determined using LC-MS/MS in negative mode. A–D, show results for mice and E-H for cells. (A, C, E, and G) The ion intensities of each PL species (PC (A, E) or PE (C, G)) were divided by the sum of the ion intensities of all PC or PE species and shown as “Relative Abundance (%)”. (B, D, F, and H) The relative PL levels of PC and PE species when the wild type (WT) values are “1” in the respective PL species graphs in A, C, E, and G. Mouse data were obtained from 8-10-weeks-old mice; WT (green bars), HT (blue bars), and KO (red bars). Results are expressed as the mean ± SD of 5–7 samples for each group. Cell data were obtained from HEK293 A cells; WT (black bars), KO clone#20 (light blue bars), KO clone#58 (orange bars), and KO clone#116 (purple bars). Results are expressed as the mean ± SD of four data points. Statistically significant differences (WT vs. KO or Parent vs. KO #20, KO #58 and KO #116) are marked with asterisks indicating P-values. ∗P<0.05; ∗∗P<0.01; ∗∗∗P<0.001; ∗∗∗∗P<0.0001; ns indicates not significant; two-way ANOVA, Bonferroni’s multiple comparison test. The results of other phospholipids (phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylglycerol (PG), and phosphatidic acid (PA)) data were shown in supplemental Fig. S5.
LPLAT7 incorporates C18:0 but not C16:0 or C18:1 into PLs
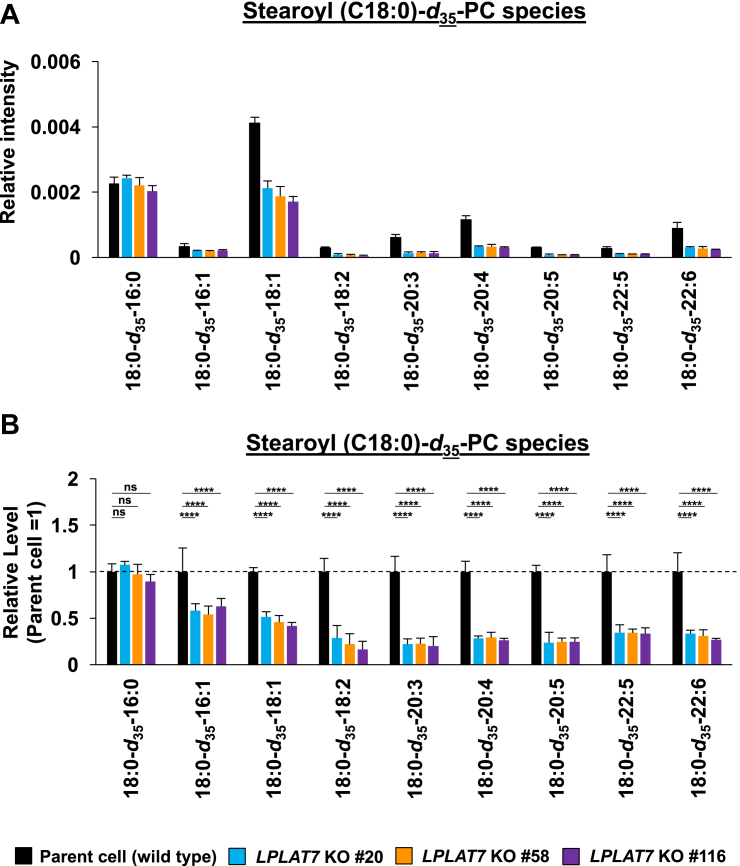
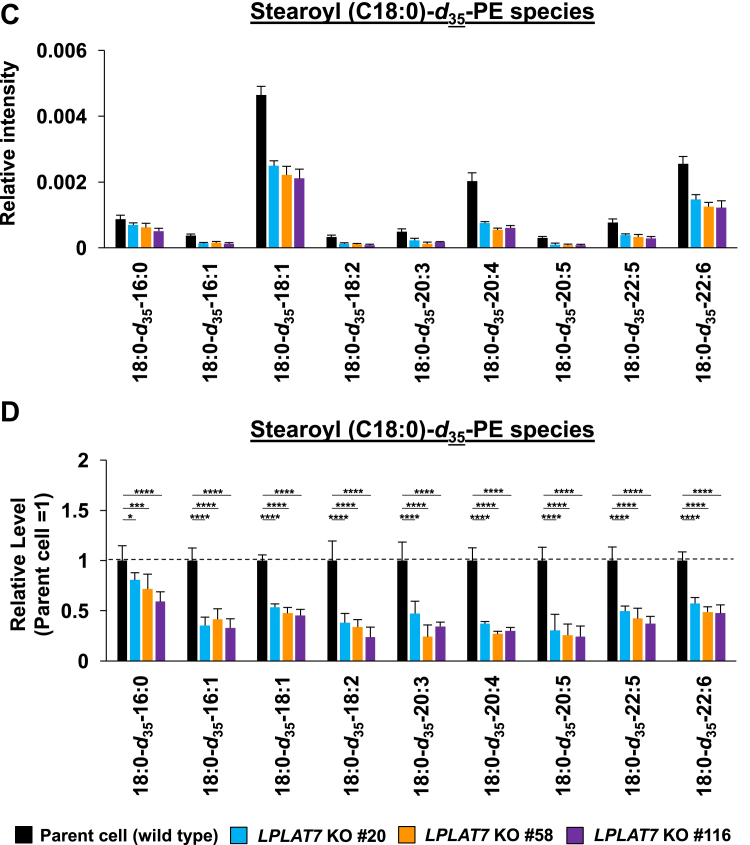
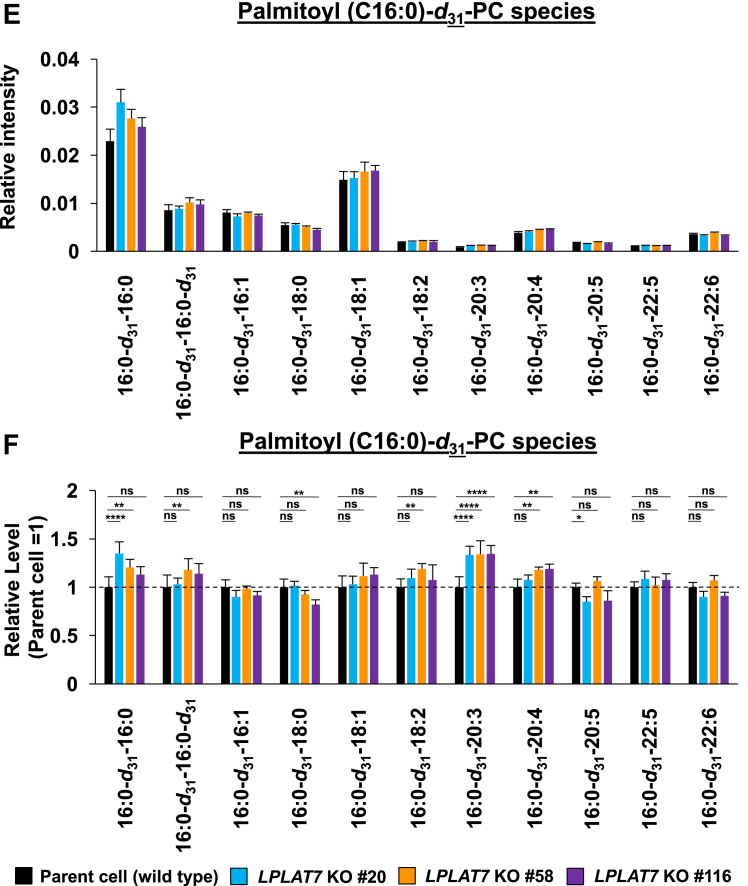
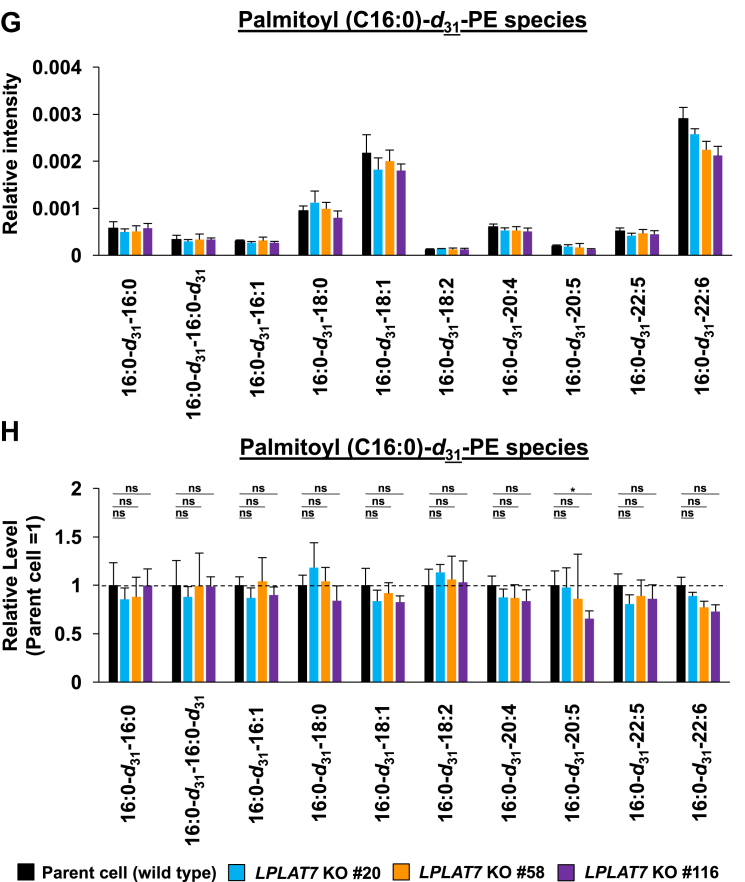
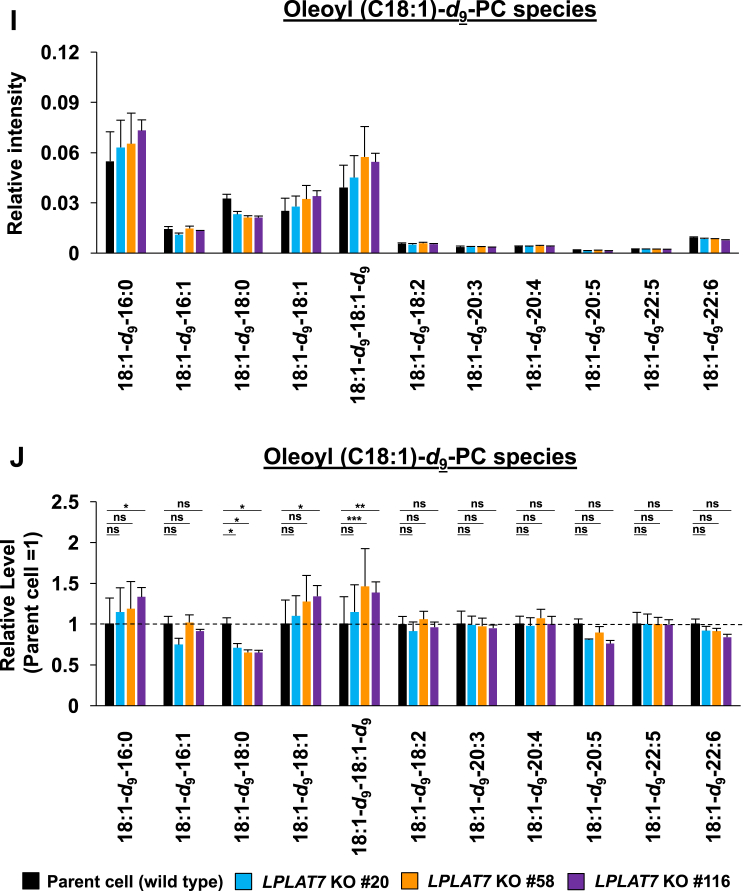
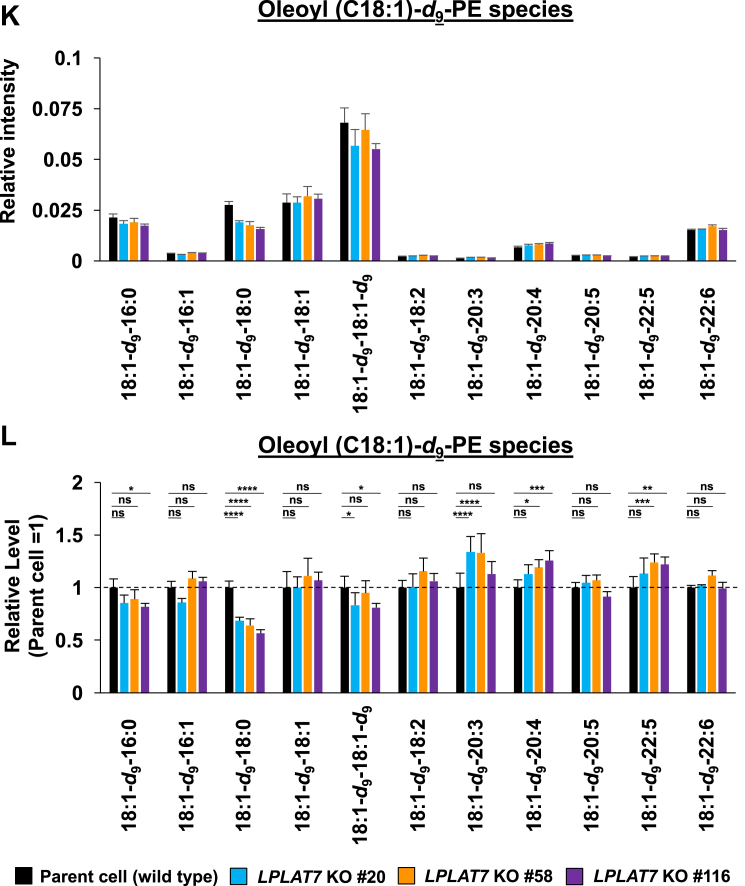
We then evaluated the direct incorporation of fatty acids into PLs by LPLAT7. To do this, we treated HEK293A cells with deuterium-labeled C18:0 (C18:0-d35), C16:0-d31, or C18:1-d9 for a short period (120 min) and then evaluated the incorporation of each deuterium-labeled fatty acid into the PLs by LC-MS/MS. We compared LPLAT7 KO cells and control parent cells and cells overexpressing-LPLAT7 and empty vector-transfected cells. One hundred twenty minutes after the treatment of C18:0-d35, the formations of most C18:0-d35-containing PL species (PC (Fig. 8A, B), PE (Fig. 8C, D), and PS (supplemental Fig. S6A, B) were significantly suppressed in each of the three LPLAT7 KO cell lines. The formations of most of the other C18:0-d35-containing PL species (PI, PG, and PA) were not affected in LPLAT7 KO cells, although formation of some C18:0-d35-containing PL species was also suppressed (eg C18:0-d35-C18:2-GPG (glycerophosphoglycerol), supplemental Fig. S6C–H), reflecting the substrate specificity observed in in vitro assay (Fig. 4). By contrast, the formation of C16:0-d31-containing PL species (PC (Fig. 8E, F) and PE (Fig. 8G, H)) and the formation of C18:1-d9-containing PL species (PC (Fig. 8I, J) and PE (Fig. 8K, L)) were unchanged except for C18:0-containing C18:1-d9-PL species (Fig. 8I–L).
Fig. 8.
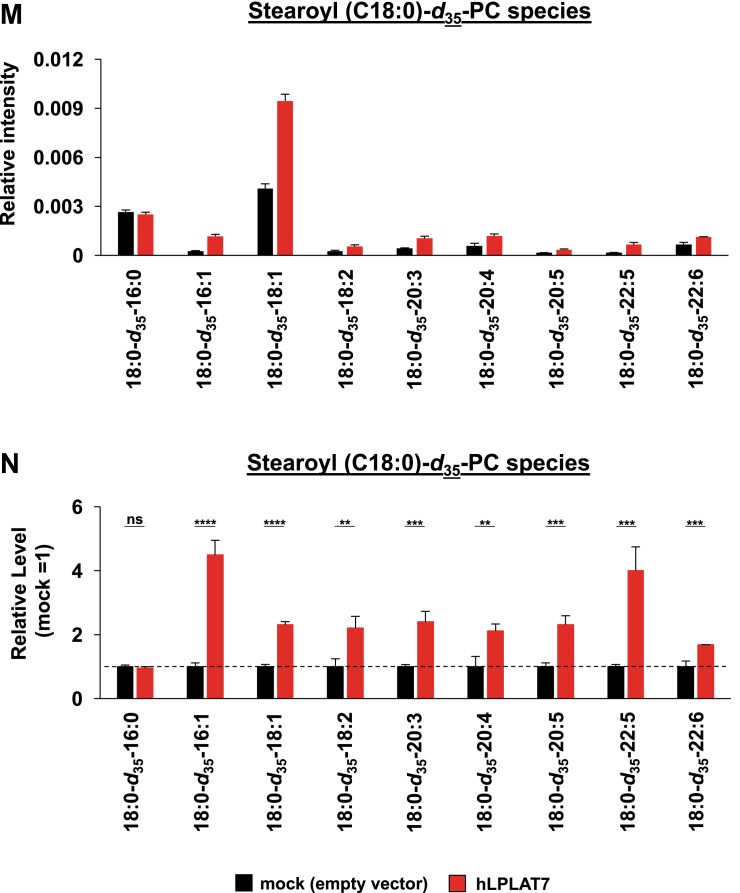
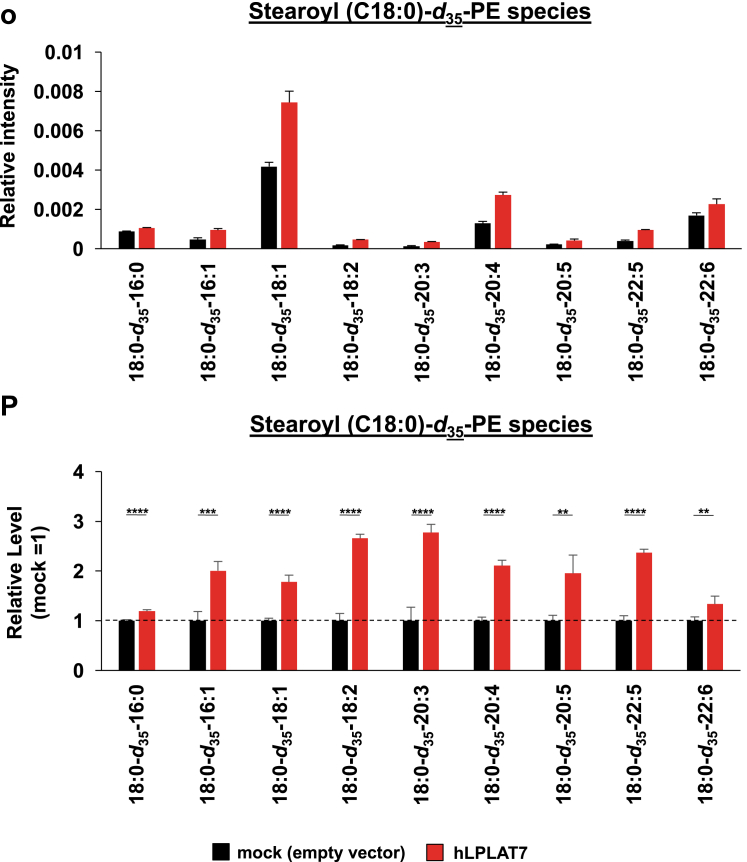
LPLAT7 is responsible for the incorporation of stearic acid (C18:0) into PC and PE in the human culture cells. Cells (LPLAT7 KO HEK293 A cells (A–L), HEK293 A cells overexpressing human LPLAT7 (M–P), and corresponding control cells) were treated with several deuterium-labeled fatty acids (stearic-d35 acid (C18:0-d35), palmitic-d31 acid (C16:0-d31) and oleic-d9 acid (C18:1-d9)) for two hours. The levels of C18:0-d35-labeled PC (A, B, M, and N), C18:0-d35-labeled PE (C, D, O, and P), C16:0-d31-labeled PC (E and F), C16:0-d31-labeled PE (G and H), C18:1-d9-labeled PC (I and J) and C18:1-d9-labeled PE (K and L) were determined by LC-MS/MS analyses in negative mode. (A, C, E, G, I, K, M, and O) The ion intensities of each PL species were normalized by the ion intensities of internal standard and presented as “Relative intensity”. (B, D, F, H, J, L, N, and P) The relative PL levels of deuterium-labeled PC and PE species, when the values of control cells (WT HEK293A cells for KO cells or HEK293A cells transfected with an empty vector) are “1” in the respective PL species graphs in A, C, E, G, I, K, M, and O. (A–L) WT cells (black bars), LPLAT7 KO clone#20 (light blue bars), LPLAT7 KO clone#58 (orange bars) and LPLAT7 KO clone#116 (purple bars). (M–P) Cells overexpressing human LPLAT7 (red bars) and control cells transfected with an empty vector (black bars). All data are expressed as the mean ± SD of three data points. Statistically significant differences (Parent vs. KO #20, KO #58 and KO #116 or mock vs. human LPLAT7) are marked with asterisks indicating P-values. ∗P<0.05; ∗∗P<0.01; ∗∗∗P<0.001; ∗∗∗∗P<0.0001; ns indicates not significant; two-way ANOVA, Bonferroni’s multiple comparison test for A-L and unpaired t test for M-P.
In contrast to the results using LPLAT7 KO cells (Fig. 8A–D), the formation of C18:0-d35-containing PL species, especially PC and PE, dramatically increased in HEK293 cells overexpressing LPLAT7 (Fig. 8M–P). The formation of C18:0-d35-containing other PL species and the formation of C16:0-d31- and C18:1-d9-containing PL species were not so affected by the overexpression of LPLAT7 (supplemental Figs. S6I–P and S7). It should be stressed here that C18:0 was incorporated into PC and PE in a similar time course (supplemental Figs. S8).
The results of these experiments using deuterium-labeled fatty acids were in good agreement with the results of lipidomic analyses at the mouse and cell level, but there were some discrepancies with the in vitro biochemical characters.
Discussion
LPLAT7 is an sn-1 specific LPLAT for LPC, LPE, and LPS and contributes to the production of stearic acid–containing PC, PE, and PS
One of the critical issues in determining the position of the glycerol backbone into which LPLAT introduces fatty acids is that lyso-PLs are always a mixture of sn-1 or sn-2 isomers, i.e., we cannot obtain 100% pure each isomer. Thus, even if we use pure lyso-PLs, enzymes can incorporate fatty acids into both the sn-1 and sn-2 positions. In particular, commercially available sn-1 lyso-PLs are a mixture of about 90% sn-1 lyso-PLs and 10% sn-2 lyso-PLs. Thus, in the LPLAT assays in which such sn-1 lyso-PLs are used as acyl acceptors, we must determine the positions carefully. In this study, we used highly pure (> 90%) sn-2 lyso-PLs (sn-2-rich lyso-PLs) and PLA2 digestion followed by lyso-PL analyses. We determined the precise position into which LPLAT7 incorporated a fatty acid. Accordingly, we found that LPLAT7 incorporated a fatty acid almost exclusively into the sn-1 position. This property of LPLAT7 contrasts with that of LPLAT8/LPCAT1, which introduces a given fatty acid into both the sn-1 and sn-2 positions (14). Using this method, we can determine the exact positions into which LPLATs and GPATs incorporate a fatty acid.
In the present study, we identified three biochemical preferences of LPLAT7: (1) sn-2-rich LPCs with unsaturated fatty acids as acyl acceptors (Fig. 4I), (2) choline and ethanolamine as head groups of lyso-PLs (Fig. 4E), and (3) C18:0-CoA and C16:0-CoA as acyl donors (Fig. 4G). In addition, using the membrane fraction from KO mice and KO cells as enzyme sources, LPLAT7 was found to be responsible for the majority of sn-1 LPLAT activities for LPC, LPE, and LPS detected in both tissues and cells (Fig. 6). More than half a century ago, Lands et al. found LPLAT activity toward sn-2-acyl LPC in the rat liver (17, 18). Also, Thompson et al. reported LPLAT activity toward sn-2-acyl LPS in the pig brain (19). LPLAT7 is assumed to be the LPLAT enzyme responsible for these activities. Of note, LPLAT7 is a major sn-1 LPLAT for both C18:0- and C16:0-CoAs. However, lipidomic analyses of the liver from Lplat7 KO mice and LPLAT7 KO cells suggested that LPLAT7 contributes to the incorporation of not C16:0 but C18:0 in cells and tissues, unlike the biochemical characters determined in vitro.
Comparison of the present results with those of previous reports
In 2004, Yang et al. first reported the biochemical properties of LPLAT7/LPGAT1. They used the cell lysates of insect cells (Sf9) infected with human LPLAT7/LPGAT1-carrying baculovirus and COS-7 cells transfected with human LPLAT7/LPGAT1 expression vector. LPLAT7/LPGAT1 had an LPG-specific LPLAT activity. They detected no significant LPLAT activities against various lyso-PLs, including LPC, LPE, LPI, and LPS. They also found that the enzyme recognized various acyl-CoAs and LPG as substrates but suggested that it had a clear preference for long-chain saturated fatty acyl-CoAs and C18:1-CoA as acyl donors (20). From these biochemical characteristics, Yang et al. proposed the name LPGAT1 for this enzyme. Zhang et al. (the same group of Yang et al.) performed lipidomic analyses of Lplat7/Lpgat1 KO and reported the alteration of several species of PG and CL (cardiolipin) in Lplat7/Lpgat1 KO mouse liver (21). Thus, the enzyme has been implicated in the metabolism of PG/CL (20, 21). Of note, Zhang et al. also reported C18:0-containing multiple PL species decreased in Lplat7/Lpgat1 KO mouse liver. However, they did not fully explain the reasons for the reduction of the C18:0-containing PLs (21).
Very recently, Xu et al. reported the biochemical properties of LPLAT7/LPGAT1. Lipidomic analyses of Lplat7/Lpgat1 KO mice demonstrated that C18:0-containing PC and PE species selectively decreased while C16:0-containing PC and PE species increased. Thus, they concluded that LPLAT7/LPGAT1 determines the stearate-to-palmitate ratio of PE and PC. They also demonstrated that LPLAT7/LPGAT1 preferred sn-2-acyl LPE over sn-1-acyl LPE using a recombinant enzyme expressed in E. coli. They claimed that LPLAT7/LPGAT1 had no LPLAT activity against LPC (22). The result is incompatible with our present results, which we discuss below.
More recently, Shibata et al. established zebrafish mutants of LPLAT7/LPGAT1 and showed that they had decreased levels of C18:0-containing PC and PE and increased levels of C16:0-containing PC and PE. PG species were not affected in the lplat7/lpgat1 mutant zebrafish (23).
Our present results agree with the recent developments by Xu et al. and Shibata et al. but not with those by Yang et al. and Zhang et al. Xu et al., and our present study share two conclusions: (1) the levels of C18:0-containing PC and PE are significantly reduced in LPLAT7 mutants, while C16:0-containing PC and PE levels increased in a complementary manner, and (2) LPLAT7 incorporated C18:0 into the sn-1 position of PE in vitro. However, as mentioned above, one distinct conclusion was that, while Xu et al. claimed that LPLAT7 did not use LPC as a substrate and thus was an LPE-specific sn-1 LPLAT, our results showed that LPLAT7 had a robust sn-1 LPLAT activity against LPC. Xu et al. used a purified His-tagged murine LPLAT7 expressed in Sf9 insect cells and LPLAT7 protein fused with maltose-binding protein (MBP) expressed in E. coli. On the other hand, we used the membrane fraction of HEK293A mammalian cells expressing human LPLAT7 as the enzyme source. For acyl acceptors, we used various LPC species (C16:0 LPC, C18:1 LPC, C18:2 LPC, C20:4 LPC, and C22:6 LPC), while Xu et al. used only C16:0 LPC, which we found to be a very poor substrate in our study (Fig, 4I). The differences in these enzyme sources and acyl acceptors are the possible cause of the differences in the results of Xu et al. and our study. Xu et al. showed that C18:0-containing PC levels were significantly reduced in Lpgat1 KO mice and speculated that the PC species come from C18:0-containing PE by a PE N-methyltransferase reaction. This hypothesis does not sufficiently explain that in various tissues where PE N-methyltransferase activity is weak, 18:0-containing PC and PE were also reduced. In our experiments, using deuterium-labeled fatty acids in HEK293A cells overexpressing LPLAT7, we found that C18:0-d35 was incorporated into PC and PE in an approximately similar time course (supplemental Fig. S8). We also showed that LPLAT7 was the major sn-1 LPLAT for LPC in the tissue (liver) and cells (HEK293A). Thus, although our results do not entirely rule out a pathway in which PE produced by LPLAT7 is converted to PC by methylation, our present results strongly indicated that LPLAT7 had the activity to incorporate directly C18:0 into LPC in addition to LPE.
In our present breeding condition, Lplat7 KO mice had reduced body weight, and about half of them died at the age of 1.5 months (Table 1 and Fig. 5). Most Lplat7 KO male mice died by the age of 4 months, and Lplat7 KO female mice lived much longer, but most of them died by the age of 5–6 months (data not shown). Xu et al. reported that Lplat7 KO mice were born according to the expected Mendelian ratio and showed no apparent weight fluctuations up to the age of 3 months. After 3.5 months, Lplat7 KO mice started to die, and the average life span of Lplat7 KO mice was reported to be around 5 months. On the other hand, Zhang et al. reported that Lplat7 KO mice were born according to the expected Mendelian ratio but showed reduced body weight immediately after birth to early adulthood. They also showed that weight loss was evident in male mice. They found that Lplat7 KO mice showed insulin resistance and hepatopathy but did not mention the lifespan of the Lplat7 KO mice. Thus, the phenotypes of Lplat7 KO mice, including the present ones, are similar but vary slightly. Interestingly, Xu et al. used Lplat7 KO mice of the same strain as ours (C57BL/6NJ-Lpgat1em1(IMPC)J/Mmjax from the Jackson Laboratory/MMRRC). Therefore, the phenotypic differences (weight loss and early death) of Lplat7 KO mice are probably due to the difference in the breeding conditions, such as the diet composition. The cause of weight loss and decreased survival rate is still unclear, and further work is needed to identify the cause of the phenotypes.
LPLAT7 determined the stearate/palmitate/oleate ratio PLs
As mentioned above, LPLAT7 was shown to have a role in determining the stearate-to-palmitate ratio in PL species (22). Our lipidomic analyses showed that loss of LPLAT7 increased not only C16:0-containing PLs but also C18:1-containing PLs (Fig. 7). Then, why does the loss of LPLAT7 lead to increased C16:0-containing and C18:1-containing PL levels?
One of the LPLATs that introduce C16:0 into the sn-1 position of PLs is LPLAT8/LPCAT1. Lplat8 KO mice have reduced levels of C16:0-containing PC species, including dipalmitoyl PC (24). Interestingly, Lplat8 KO mice had increased levels of C18:0-containing PC species. Therefore, there may be competition for sn-2-acyl LPCs between LPLAT7 and LPLAT8, resulting in a shunting phenomenon, in which loss of LPLAT7 causes increased availability of sn-2-acyl LPCs for LPLAT8. Similar competition may also occur between LPLAT7 and the putative sn-1 LPLATs for C18:1. Although the nature of the C18:1-preferring sn-1 LPLATs is not clear, the present study strongly suggests the presence of such sn-1 LPLATs for C18:1.
In general, C16:0 and C18:1 are incorporated into PLs faster than C18:0 in the de novo PL synthetic pathway. Indeed, this may also be the case in HEK293A cells in the present study based on the following observations: (1) Among C18:0-d35, C16:0-d31, and C18:1-d9 added to the cells, both C16:0-d31 and C18:1-d9 were incorporated into PC and PE approximately several times faster than C18:0-d35. In addition, (2) the LPLAT activities to incorporate C18:0, C16:0, and C18:1 into lyso-PLs were dramatically reduced, especially for LPE in Lplat7 KO mice and LPLAT7 KO HEK293A cells (Fig. 6), showing that LPLAT7 is the main sn-1 LPEAT and LPSAT for the three acyl-CoAs. Therefore, it is unlikely that other remodeling enzymes produced the deuterated PLs in the absence of LPLAT7. Thus, the majority of C16:0- and C18:1-containing PLs are produced by the de novo pathway rather than by a remodeling pathway. When we added C18:0-d35, C16:0-d31, and C18:1-d9 to LPLAT7-deficient HEK293A cells, the incorporation of C18:0-d35, but not C16:0-d31 and C18:1-d9, into PC and PE fractions was significantly attenuated. Thus, it is also possible that in the absence of LPLAT7, C16:0- and C18:1-containing PLs (mainly PC, PE, and PS) synthesized by the de novo pathway are not converted to C18:0-containing PLs, resulting in the accumulation of C16:0- and C18:1-containing PL species.
Proposal for a new nomenclature
Currently, 14 LPLATs (10 AGPATs and 4 MBOATs) are known, and they are highly conserved among vertebrates. Previous errors in determining substrate specificities of LPLATs may have led to their misnaming. Until now, the nomenclature LPXATn has been widely used for LPLAT enzymes, in which X indicates the polar head group of acyl acceptors and n indicates the order of discovery. This study showed that LPLAT7/LPGAT1 used various acyl acceptors (LPC, LPE, and LPS) and various acyl-CoAs (C18:0-, C16:0, C18:1-CoAs) in vitro. Our results also show that LPLAT7 regulates the level of C18:0-containing PC and PE in vivo, which indicates that it uses LPC and LPE as acyl acceptors and C18:0-CoA as an acyl donor. Thus, the nomenclature LPXATn apparently cannot cover the substrate specificities and the endogenous substrates. In addition, the previously proposed name of this enzyme, LPGAT1, does not reflect its properties. Recently, we proposed to use the new nomenclature LPLATn for mammalian LPLAT molecules, in which n is just the order number (4). Accordingly, we propose renaming LPGAT1 as LPLAT7.
Data availability
The raw mass spectrometric data will be made available from the corresponding author upon reasonable request.
Supplemental data
This article contains supplemental data.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank lab members for suggestions for improving current study. We thank the Laboratory of Molecular and Biochemical Research, Biomedical Research Core Facilities, Juntendo University Graduate School of Medicine for technical assistance.
Author contributions
H. K. designed and performed most of the experiments and wrote the manuscript. M. O. generated and analyzed LPLAT7-KO cells. T. Shibata, H. O., and Y. S. supported analyses of Lplat7-KO mice. K. K. supported mass spectrometry analyses. H. S., T. Shimizu, and N. K. provided technical assistance for some experiments and discussions. J. A. wrote and editing the manuscript with feedback from all of the authors; J. A. supervised all aspects of the study.
Funding and additional information
This work was supported by AMED-LEAP (21gm0010004h9905 for J. A.), AMED-CREST (22gm0910011 for H. S.) and KAKENHI (20K21379 for JA and 20K15984 and 22K15273 for H. K.).
Supplemental data
References
- 1.Harayama T., Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 2.Miller N.G., Hill M.W., Smith M.W. Positional and species analysis of membrane phospholipids extracted from goldfish adapted to different environmental temperatures. Biochim. Biophys. Acta. 1976;455:644–654. doi: 10.1016/0005-2736(76)90038-9. [DOI] [PubMed] [Google Scholar]
- 3.Amate L., Ramírez M., Gil A. Positional analysis of triglycerides and phospholipids rich in long-chain polyunsaturated fatty acids. Lipids. 1999;34:865–871. doi: 10.1007/s11745-999-0434-0. [DOI] [PubMed] [Google Scholar]
- 4.Valentine W.J., Yanagida K., Kawana H., Kono N., Noda N.N., Aoki J., et al. Update and nomenclature proposal for mammalian lysophospholipid acyltransferases, which create membrane phospholipid diversity. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2021.101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hishikawa D., Shindou H., Kobayashi S., Nakanishi H., Taguchi R., Shimizu T. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2830–2835. doi: 10.1073/pnas.0712245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., et al. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 2008;19:1174–1184. doi: 10.1091/mbc.E07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashidate-Yoshida T., Harayama T., Hishikawa D., Morimoto R., Hamano F., Tokuoka S.M., et al. Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. eLife. 2015;4 doi: 10.7554/eLife.06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rong X., Wang B., Dunham M.M., Hedde P.N., Wong J.S., Gratton E., et al. Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. eLife. 2015;4 doi: 10.7554/eLife.06557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H.C., Inoue T., Sasaki J., Kubo T., Matsuda S., Nakasaki Y., et al. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Mol. Biol. Cell. 2012;23:4689–4700. doi: 10.1091/mbc.E12-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka Y., Shimanaka Y., Caddeo A., Kubo T., Mao Y., Kubota T., et al. LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut. 2021;70:180–193. doi: 10.1136/gutjnl-2020-320646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi K., Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009;296:E1195–E1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plückthun A., Dennis E.A. Acyl and phosphoryl migration in lysophospholipids: importance in phospholipid synthesis and phospholipase specificity. Biochemistry. 1982;21:1743–1750. doi: 10.1021/bi00537a007. [DOI] [PubMed] [Google Scholar]
- 13.Okudaira M., Inoue A., Shuto A., Nakanaga K., Kano K., Makide K., et al. Separation and quantification of 2-acyl-1-lysophospholipids and 1-acyl-2-lysophospholipids in biological samples by LC-MS/MS. J. Lipid Res. 2014;55:2178–2192. doi: 10.1194/jlr.D048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawana H., Kano K., Shindou H., Inoue A., Shimizu T., Aoki J. An accurate and versatile method for determining the acyl group-introducing position of lysophospholipid acyltransferases. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:1053–1060. doi: 10.1016/j.bbalip.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami K., Yanagawa M., Hiratsuka S., Yoshida M., Ono Y., Hiroshima M., et al. Heterotrimeric Gq proteins act as a switch for GRK5/6 selectivity underlying β-arrestin transducer bias. Nat. Commun. 2022;13:487. doi: 10.1038/s41467-022-28056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imae R., Inoue T., Nakasaki Y., Uchida Y., Ohba Y., Kono N., et al. LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice. J. Lipid Res. 2012;53:335–347. doi: 10.1194/jlr.M018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lands W.E., Merkl I. Metabolism of glycerolipids. III. Reactivity of various acyl esters of coenzyme A with alpha'-acylglycerophosphorylcholine, and positional specificities in lecithin synthesis. J. Biol. Chem. 1963;238:898–904. [PubMed] [Google Scholar]
- 18.Lands W.E., Hart P. metabolism of glycerolipids. vi. specificities of acyl coenzyme a: phospholipid acyltransferases. J. Biol. Chem. 1965;240:1905–1911. [PubMed] [Google Scholar]
- 19.Thompson W., Belina H. Rapid accumulation of diacyl lipid in rat liver microsomes by selective acylation of 2-acyl-sn-glycero-3-phosphorylserine. Biochim. Biophys. Acta. 1986;876:379–386. doi: 10.1016/0005-2760(86)90023-8. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y., Cao J., Shi Y. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J. Biol. Chem. 2004;279:55866–55874. doi: 10.1074/jbc.M406710200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Zhang J., Sun H., Liu X., Zheng Y., Xu D., et al. Defective phosphatidylglycerol remodeling causes hepatopathy, linking mitochondrial dysfunction to hepatosteatosis. Cell Mol. Gastroenterol. Hepatol. 2019;7:763–781. doi: 10.1016/j.jcmgh.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Miller P.C., Phoon C., Ren M., Nargis T., Rajan S., et al. LPGAT1 controls the stearate/palmitate ratio of phosphatidylethanolamine and phosphatidylcholine in sn-1 specific remodeling. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata T., Kawana H., Nishino Y., Ito Y., Sato H., Onishi H., et al. Abnormal male reproduction and embryonic development induced by downregulation of a phospholipid fatty acid-introducing enzyme Lpgat1 in zebrafish. Sci. Rep. 2022;12:7312. doi: 10.1038/s41598-022-11002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harayama T., Eto M., Shindou H., Kita Y., Otsubo E., Hishikawa D., et al. Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 2014;20:295–305. doi: 10.1016/j.cmet.2014.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometric data will be made available from the corresponding author upon reasonable request.