Abstract
ADAMTS13, a plasma metalloprotease that cleaves von Willebrand factor, is crucial for normal hemostasis. Acquired autoantibody-mediated deficiency of plasma ADAMTS13 results in a potentially fatal blood disorder, immune thrombotic thrombocytopenic purpura (iTTP). Plasma ADAMTS13 protease appears to exist in multiple conformations. Under physiological conditions, plasma ADAMTS13 exists predominantly in its “closed” conformation (or latent form), which may be activated by lowering pH, ligand binding, and binding of an antibody against the distal domains of ADAMTS13. In patients with iTTP, polyclonal antibodies target at various domains of ADAMTS13. However, nearly all inhibitory antibodies bind the spacer domain, whereas antibodies that bind the distal C-terminal domains may activate ADAMTS13 through removing its allosteric inhibition. Additionally, the anti-C-terminal antibodies may alter the potency of inhibitory antibodies towards ADAMTS13 activity. This review summarizes some of the most recent knowledge about the ADAMTS13 conformation and its mechanism of inhibition by its autoantibodies.
Keywords: activation, ADAMTS13, autoantibody, inhibition, TTP/HUS
1 |. INTRODUCTION
Immune thrombotic thrombocytopenic purpura (iTTP) is an autoimmune disease, caused by autoantibody-mediated inhibition of a plasma metalloprotease ADAMTS13.1–5 The only known function of ADAMTS13 to date is to cleave the glycoprotein von Willebrand factor (VWF), which recruits platelets to sites of vascular injury for hemostasis.6,7 In the absence of functional ADAMTS13, ultra-large VWF multimers accumulate and cause a thrombotic microangiopathy that is potentially fatal unless being recognized early in the disease course and treated accordingly.8–10 Though significant progresses have been made over the past decades regarding the understanding of pathogenesis of iTTP, treatment options remain crude and nonspecific. Therapeutic plasma exchange has been part of the standard of care for iTTP, and it must be initiated early in the disease course to prevent high mortality associated with disseminated microvascular thrombosis.11–13 The monoclonal antibodies against CD20 on B-cells, rituximab and anti-VWF nanoboy, caplacizumab are the adjunct treatments that work indirectly to block the formation of autoantibodies and the recruitment of platelets to ultra-large VWF, respectively.14–23 To improve on our current therapeutic standards, there is still a need to explore the mechanism(s) by which autoantibodies cause iTTP. To do so, we must also learn more about how ADAMTS13 works. Recent work by our group and others suggests a complex landscape of conformations and functional states of ADAMTS13, both in the native context and when bound to antibodies.24–37 Both structural and functional studies have been used to gain insight into the function and antibody-mediated inhibition of ADAMTS13. We will breifly review the current state of knowledge of ADAMTS13 function and inhibition.
2 |. ADAMTS13 STRUCTURE
The partial and then completed primary structures of ADAMTS13 were first reported by several groups in 2001.1,38–41 Secreted ADAMTS13 protein consists of a metalloprotease (M) domain, a disintegrin (D) domain, the first thrombospondin type 1 (TSP1–1) repeat, the Cys-rich (C) and spacer (S) domains, followed by 7 more TSP1 repeats and two CUB domains, associated with complement components C1r/C1s, sea Urchin epidermal growth factor, Bone morphogenetic protein (Figure 1A). While the tertiary structure of a full-length ADAMTS13 protein has yet to be determined, the truncated versions of recombinant ADAMTS13 protein (e.g., MDTCS or its variants) have recently been described using a variety of techniques, including X-ray crystallography, small angle X-ray scattering, and mass spectrometry plus hydrogen/deuterium exchange, and cross-linking mass spectrometry.27,28,42,43 The MDTCS fragment contains all the necessary and sufficient domains of ADAMTS13 protein for substrate specificity and efficient cleavage.28,43,44,45 From these studies, coupled with functional assays, the spacer domain of ADAMTS13 has been identified to be crucial for substrate cleavage and specificity.45–48
FIGURE 1.
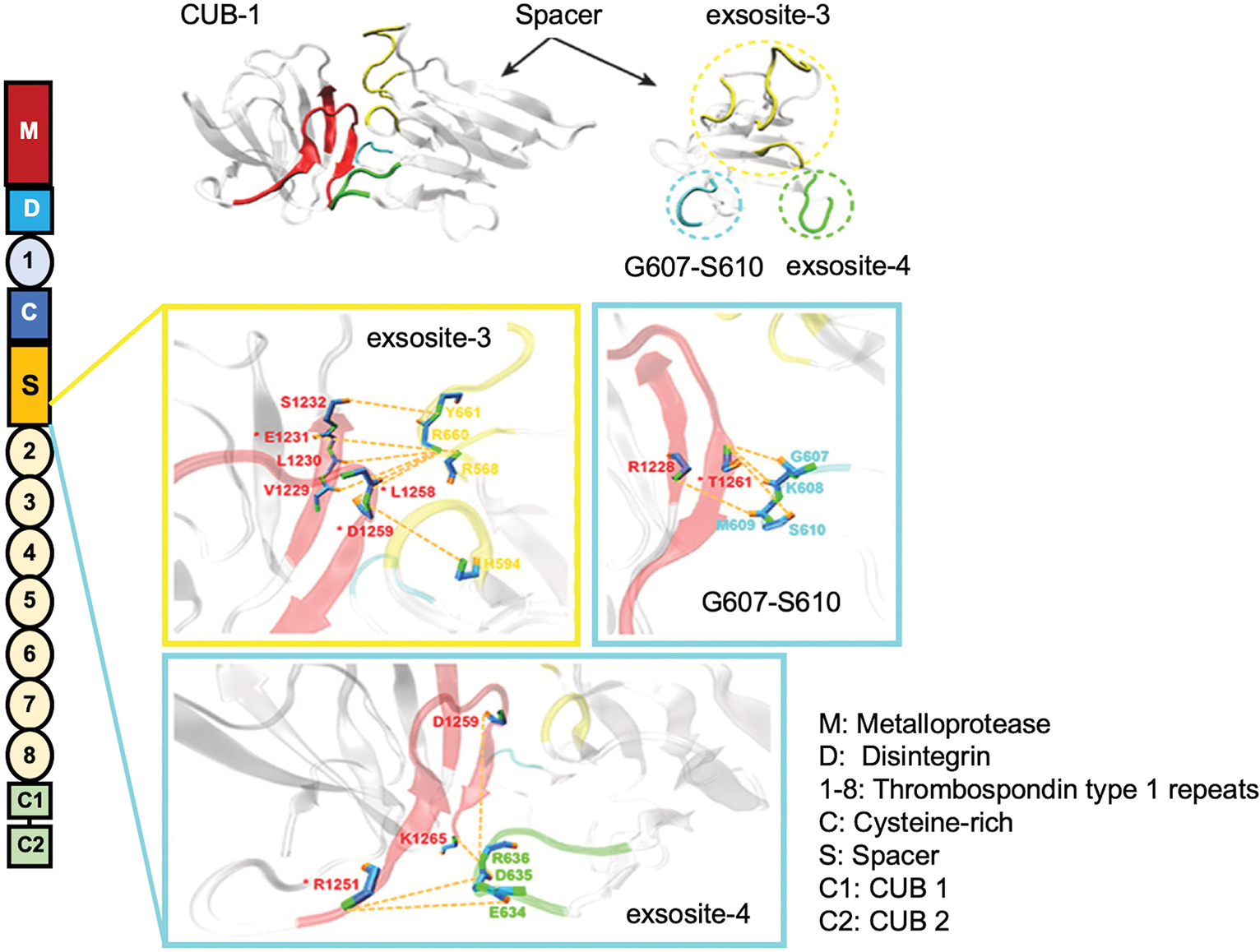
The primary domain structure of ADAMTS13 and proposed binding interaction between CUB and spacer domain. (A) ADAMTS13 consists of a metalloprotease, a disintegrin, the first TSP1 repeat, Cys-rich and spacer domain, followed by seven more TSP1 repeats, and two CUB domains. (B) Multiple binding regions in the Spacer interface including exosite 3 (R568, F592, R660, Y661, Y665), G607-S610, and exosite 4 (E634, D635, and R636) appear to interact with the CUB1 domain (E1231, R1251, L1258, D1259, and T1261) to mediate autoinhibition (Adapted from Yang et al.51).
The crystal structure of the DCTS fragment of ADAMTS13 was reported in 2009,44 which demonstrates that the spacer domain is highly structured, with 10 beta sheets forming a “sandwich” that exposes various loops. These loops are important epitopes for antibody binding, as we will review later. However, it was not until 2019 that the structure of MDTCS domains was determined,27 using a variant with the catalytic glutamate in the metalloprotease domain substituted for a glutamine (E225Q), bound with a mouse anti-human ADAMTS13 monoclonal antibody against the metalloprotease domain, which stabilizes the protein. The structure revealed that the active site is conformationally hidden, or buried, in this context. The extent to which MDTCS must be manipulated to obtain a crystal structure suggests that the wild-type native protein, as well as full-length recombinant ADAMTS13, may be somewhat disordered. This may also suggest that the metalloprotease domain may be flexible and plastic. This poses a significant challenge to structural biologists since ADAMTS13 was first discovered and cloned in 2001.1,38–41 Recently, cross-linking mass spectrometry was used to demonstrate that, when bound to a surrogate substrate (VWF73), the metalloprotease, disintegrin, cysteine-rich, and spacer domains undergo a significant rearrangement, with the first TSP1 repeat functioning as a “hinge” region, placing the spacer domain, which is the primary determinant of substrate specificity, in close proximity to the metalloprotease domain.43 This likely occurs via a series of low-affinity interactions with VWF at various exosites that causes dozens, if not hundreds, of small refolding events. The authors describe this as a “fuzzy complex” with a “dynamic zipper” mechanism, offering evidence that ADAMTS13 is likely an intrinsically disordered protein.43
3 |. ADAMTS13 CONFORMATIONS
The role of the other distal C-terminal domains of ADAMTS13 has started to become clearer in recent years. The CUB domains may associate with the spacer domain when the protein is in a less active state (e.g., before it is bound by a substrate).36,49 This likely prevents ADAMTS13 from indiscriminately cleaving VWF and possibly other proteins, suggesting an autoregulatory role of the distal domains. The crystal structure of the two CUB domains of human recombinant ADAMTS13 has been published recently, yielding important insights into the function of the CUB domains, which will be further discussed later.50
Though we begin to get a handle on what the CUB domains are doing, we have largely limited ourselves to the consideration of an intramolecular interaction between the distal C-terminal domains of ADAMTS13 and the proximal N-terminal domains. Yang et al. have performed molecular modeling and demonstrated that there may be three potential sites in the spacer domain—exosite 3 (R568, F592, R660, and Y661), G607-S610, and exosite 4 (E634, D635, and R636)—that may interact with the first CUB domain (Figure 1B).51 Mutations in the exosite 3 appear to disrupt the interaction between spacer and CUB, leading to an increase of ADAMTS13 activity.34,36,52
Recent data published by Rottensteiner et al. have demonstrated that there may be a role for the intermolecular ADAMTS13 interactions as well, with evidence that recombinant ADAMTS13 may form dimers or oligomers in nonreducing conditions.53 These oligomers may conceivably have interactions between the CUB domains on one monomer and the spacer domain on another monomer, for example. The implications of this oligomerization, as well as whether it is relevant in physiologic conditions, is an important area of ongoing investigation with tremendous implications regarding the function and inhibition of ADAMTS13.
Despite these significant advances, we still do not have any structural studies that are available for a full-length ADAMTS13. This is important because, though allosteric inhibition is implied by structural studies to some degree, functional studies suggest that allosteric changes can be communicated along the entire length of the protein.24,25,32,54 Our understanding of the mechanisms by which this allosteric regulation works is still evolving. More importantly, the study of anti-ADAMTS13 antibodies might have revealed surprising insights into the function and inhibition of ADAMTS13, as we will be discussing later.
4 |. ADAMTS13 INHIBITION: POTENTIAL NOVEL MECHANISM OF AUTOANTIBODY-MEDIATED INHIBITION
Most patients with iTTP have immunoglobulin G antibodies that bind the spacer domain to mediate their inhibition, although multiple other domains of ADAMTS13 may be targeted as well.55–64 To date, there is no published study to elucidate precisely how the anti-spacer (inhibitory) IgG work to prevent the VWF cleavage, with a widely held assumption that they somehow sterically interfere with substrate binding being unchallenged.33 Furthermore, a significant subset of patients may harbor antibodies that bind the C-terminal domains of ADAMTS13.26,56,65 Mouse monoclonal antibodies against the C-terminal domains have been developed, some of which are capable of stimulating ADAMTS13 activity instead of inhibiting it.24,25,66,67 This led to the hypothesis that these anti-C-terminal antibodies may allosterically create an “open” conformation of ADAMTS13 by relieving the autoinhibition of the CUB domains that are presumably binding to the spacer domain. This binding interaction may induce conformational changes that at the very least move the CUB domains away from spacer domain30 (Figure 2A).
FIGURE 2.
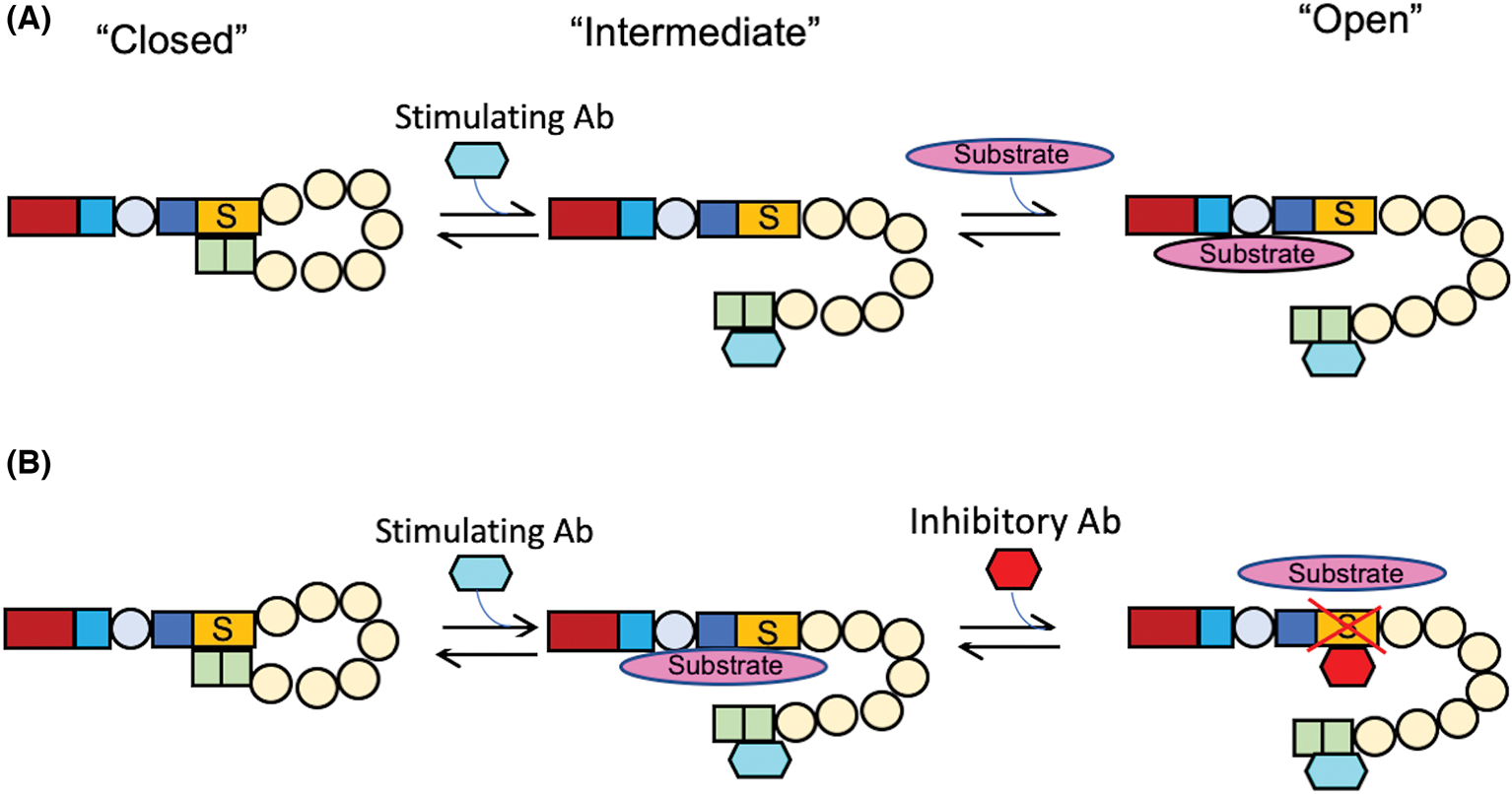
Conformational changes of ADAMTS13 induced by antibodies. (A) Transition from closed to open conformation of ADAMTS13 upon its binding with a human monoclonal anti-CUB IgG (scFv3–3) and its subsequent cleavage of VWF73 peptide. (B) The possible mechanism underlying the synergistic effect of a stimulating anti-CUB (scFv3–3) and an inhibitory anti-Spacer (i.e., scFv4–20) on proteolytic cleavage of VWF substrate (i.e., VWF73) by ADAMTS13 (Adapted from Halkidis and Zheng30)
To explore this hypothesis, Kim et al. used in silico docking simulations to identify the residues in the CUB domains that likely interact with the spacer domain. They then generated full-length variants of recombinant ADAMTS13 with mutations in putative interaction sites to test enzymatic activity in the presence of a truncated surrogate VWF substrate.50 They also assessed the activity of these variants in the presence and absence of an activating mouse monoclonal antibody (17G2). The hypothesis was that the “open” conformation would be favored in variants with decreased affinity between spacer and the CUB domains, and thus 17G2 should not increase ADAMTS13-mediated substrate cleavage. This was based on the assumption that the action of the stimulatory antibody is to decrease the CUB-mediated steric barrier to ADAMTS13-VWF binding. The authors then used a second round of docking simulations to test the hypothesis that the residues identified as abrogating the effects of 17G2 (W1245, W1250, K1252, R1326, E1387, and E1389) were important in the spacer-CUB interaction. The highest scoring pose using two different crystal structures of the spacer domain (PDB codes: 3GHM and 6QIG) and two different programs (ClusPro and HADDOCK) paired these CUB residues with spacer domain exosite residues previously shown to be important both in the function of ADAMTS13 and its inhibition by anti-ADAMTS13 antibodies. However, no in vitro binding data have yet been published to corroborate this hypothesis, and the mechanistic consequences are still not clear. For instance, it is not known if the spacer-CUB interaction sterically prevents VWF binding to ADAMTS13 or if other mechanism(s) are involved. The mechanism(s) by which 17G2 and other similar antibodies stimulate ADAMTS13 activity is still not known.
Our group recently used a human monoclonal antibody that binds to the C-terminal ADAMTS13 domains identified via phage display from a patient with iTTP,56 which showed strong activation of ADAMTS13.30 This is the first evidence of human monoclonal antibody-mediated stimulation of ADAMTS13 activity.30,56 We also showed that at physiologic pH, a condition in which ADAMTS13 activity is extremely low in vitro, our stimulatory antibody was capable of not only recovering activity equivalent to standard assay conditions, but in fact increased reaction velocity by threefold at saturating antibody concentrations.32,68,69 Presumably this may be mediated by promoting the “open” conformation of ADAMTS13. However, the physiological relevance of antibody-mediated activation of ADAMTS13 in patients with iTTP remains to be determined. Further in this review, we will revisit this data and discuss the potential implications.
Using a cryptic epitope of ADAMTS13 that is only recognized in the “open” conformation, it has recently been shown that the vast majority of patients with acute iTTP are in the “open” conformation, a seeming contradiction because these patients have little to no detectable ADAMTS13 activity.25,71 The group also showed that patients in remission who had ADAMTS13 in the “open” conformation were found to be more likely to develop a relapsed disease.
Combined with the structural insights reviewed previously, this paints a very complicated picture of ADAMTS13 biology in patients with iTTP, in which conformational accessibility of ADAMTS13 to bind VWF probably cannot explain exactly what is happening in these patients. For this reason, it is critical to identify whether anti-C-terminal and/or other “opening” antibodies are central to the pathogenesis of iTTP. If their mechanism of action can be elucidated, we can explore ways to rescue ADAMTS13 activity by similar mechanisms, or at the very least to prevent inhibitory anti-spacer domain antibodies from forming because of prolonged epitope exposure in their presence. Next, we will discuss recent mechanistic insights into ADAMTS13 using enzymology-based approaches.
5 |. KINETIC STUDIES: A STORY OF BINDING VS. ACTIVITY
The epitopes that inhibitory (anti-spacer domain) antibodies bind are also required for specific recognition of VWF.45,46,52,71 It was hypothesized that inhibitory anti-spacer antibodies may physically block the binding of ADAMTS13 to VWF, based on hydrogen exchange plus mass spectrometric analysis.33 As such, if this hypothesis is correct, the functional assays should reveal that the presence of inhibitory anti-spacer domain antibodies should lead to an increase in the substrate concentration that leads to the half-maximal reaction velocity. To test this hypothesis, most groups have relied on Michaelis–Menten kinetics-based experiments to determine the parameters kcat, KM, and kcat/KM of ADAMTS13.24,72 To wit, if the inhibitory antibodies cause problems with substrate binding, the KM should increase out of proportion to any change in kcat. If kcat is more affected than KM, it suggests that antibodies may affect the protein in a way that is independent of alterations in substrate binding.
One group has used a stimulatory mouse monoclonal antibody against the C-terminal domains of ADAMTS13 to determine the kcat, KM, and kcat/KM of recombinant ADAMTS13 in the presence of various concentrations of a surrogate substrate.24 Notably, they found that the kcat was more affected than the KM. This is consistent with an alternative hypothesis that the stimulatory antibody may not simply make ADAMTS13 easier to bind VWF by swinging the CUB domains away from the spacer domain as shown (Figure 2B), but somehow increases the catalytic turnover rate of ADAMTS13 by other unknown mechanisms. Clearly, this is potentially paradigm-shifting in the field, and it is a major focus of our group’s ongoing studies. This may help better elucidate ADAMTS13 function and regulation towards developing better diagnostic and therapeutic tools for management of iTTP in the future.
6 |. COOPERATIVITY: IMPLICATIONS FOR FUTURE RESEARCH IN iTTP
Cooperativity in protein chemistry is classically described by the example of the hetero-tetramer hemoglobin, in which binding of a ligand to one subunit of hemoglobin affects the ligand-binding affinity of adjacent subunits.73–75 It can also be seen in monomeric proteins, as in the other classic example of hexokinase, where stable intermediate conformations with different ligand binding affinities are more or less probable at different ligand concentrations.76
Our recent data using a human monoclonal anti-C-terminal antibody, isolated from a patient with iTTP, have shown that the antibody is not only capable of stimulating ADAMTS13 activity, but also exhibits a remarkable positive cooperativity.30 This is particularly evident when normal human plasma containing a wild-type ADAMTS13 is titrated with a stimulatory antibody under physiologic pH conditions. Even more strikingly, the simultaneous presence of stimulatory and inhibitory antibodies in physiologic conditions showed strong positive cooperativity. Our findings imply that ADAMTS13 may function as a multimeric protein in which the binding of one antibody to one monomer increases the probability of another antibody binding to an adjacent monomer. Alternatively, a monomeric ADAMTS13 may adapt multiple stable intermediate forms, only becoming more capable of binding to other antibodies when the ligand concentrations increase above a certain threshold.
Whether the cooperativity we observed occurs in the context of monomeric or multimeric ADAMTS13 is of critical importance because we do not know how ADAMTS13 would function optimally, poorly, or not at all as a monomer or a member of a complex. Identifying the factors that affect the multimer formation or the stability of multiple functional conformations of ADAMTS13, if they exist, would also enhance our understanding of the pathophysiology of iTTP. It may be also possible to exploit these properties to increase ADAMTS13 activity.
Despite the recent and historical elucidation of the role of ADAMTS13 in iTTP outlined here, which represents an impressive body of scientific achievement in a relatively short time frame, much work still needs to be done to elucidate the structure and function, as well as regulation of ADAMTS13 activity, the mechanism of antibody-mediated inhibition, and the pathophysiology of iTTP to improve our clinical management of this potentially fatal blood disease, and other inflammatory and thrombotic disorders that might be associated with imbalance of ADAMTS13/VWF axis.
ACKNOWLEDGMENTS
The study was supported in part by grants from NHLBI (HL126724, HL144552, HL157975–01A1 to X.L.Z.) and a 2020 Hemostasis and Thrombosis Research Society (HTRS) Mentored Research Award (to K.H.).
Footnotes
CONFLICT OF INTEREST
X.L.Z. was a speaker for Alexion and Sanofi, but continues serving a consultant for Alexion, Sanofi, and Takeda. X.L.Z. is also the co-founder of Clotsolution. K.H. has declared no relevant conflict.
REFERENCES
- 1.Zheng X, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276:41059–41063. [DOI] [PubMed] [Google Scholar]
- 2.Zheng X, Majerus EM, Sadler JE. ADAMTS13 and TTP. Curr Opin Hematol. 2002;9:389–394. [DOI] [PubMed] [Google Scholar]
- 3.George JN, Vesely SK. Thrombotic thrombocytopenic purpura-hemolytic uremic syndrome: diagnosis and treatment. Cleve Clin J Med 2001;68:857–878 [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Hutzler M, Li C, Pechet L. Thrombotic thrombocytopenic purpura (TTP) and hemolytic uremic syndrome (HUS): the new thinking. J Thromb Thrombolysis. 2001;11:261–272. [DOI] [PubMed] [Google Scholar]
- 5.Kelton JG. Thrombotic thrombocytopenic purpura and hemolytic uremic syndrome: will recent insight into pathogenesis translate into better treatment? Transfusion. 2002;42:388–392. [DOI] [PubMed] [Google Scholar]
- 6.Lian EC. Pathogenesis of thrombotic thrombocytopenic purpura: ADAMTS13 deficiency and beyond. Semin Thromb Hemost. 2005;31:625–632. [DOI] [PubMed] [Google Scholar]
- 7.Peyvandi F The role of ADAMTS13 in the new pathogenesis of TTP. Hematology. 2005;10(Suppl 1):47–48. [DOI] [PubMed] [Google Scholar]
- 8.Rock GA, Shumak KH, Buskard NA, et al. Comparison of plasma exchange with plasma infusion in the treatment of thrombotic thrombocytopenic purpura. Canadian Apheresis Study Group. N Engl J Med. 1991;325:393–397. [DOI] [PubMed] [Google Scholar]
- 9.Lara PN Jr, Coe TL, Zhou H, Fernando L, Holland PV, Wun T. Improved survival with plasma exchange in patients with thrombotic thrombocytopenic purpura-hemolytic uremic syndrome. Am J Med. 1999;107:573–579. [DOI] [PubMed] [Google Scholar]
- 10.Zheng XL, Kaufman RM, Goodnough LT, Sadler JE. Effect of plasma exchange on plasma ADAMTS13 metalloprotease activity, inhibitor level, and clinical outcome in patients with idiopathic and nonidiopathic thrombotic thrombocytopenic purpura. Blood. 2004;103:4043–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rock GA. A clinical research project to study plasma exchange and plasma infusion in treatment of thrombotic thrombocytopenic purpura (TTP). Prog Clin Biol Res. 1982;106:307–315. [PubMed] [Google Scholar]
- 12.Rock G, Shumak K, Nair R. A study of plasma exchange in TTP. The Canadian Apheresis Study Group. Prog Clin Biol Res. 1990;337:125–127. [PubMed] [Google Scholar]
- 13.Rock G Plasma exchange in the management of thrombotic thrombocytopenic purpura. Vox Sang. 2002;83(Suppl 1):141–143. [DOI] [PubMed] [Google Scholar]
- 14.Zheng X, Pallera AM, Goodnough LT, Sadler JE, Blinder MA. Remission of chronic thrombotic thrombocytopenic purpura after treatment with cyclophosphamide and rituximab. Ann Intern Med. 2003;138:105–108. [DOI] [PubMed] [Google Scholar]
- 15.Yassa SK, Blessios G, Marinides G, Venuto RC. Anti-CD20 monoclonal antibody (Rituximab) for life-threatening hemolytic-uremic syndrome. Clin Transplant. 2005;19:423–426. [DOI] [PubMed] [Google Scholar]
- 16.Cataland SR, Wu HM. Immunotherapy for thrombotic thrombocytopenic purpura. Curr Opin Hematol. 2005;12:359–363. [DOI] [PubMed] [Google Scholar]
- 17.Jestin M, Benhamou Y, Schelpe AS, et al. Preemptive rituximab prevents long-term relapses in immune-mediated thrombotic thrombocytopenic purpura. Blood. 2018;132:2143–2153. [DOI] [PubMed] [Google Scholar]
- 18.Owattanapanich W, Wongprasert C, Rotchanapanya W, Owattanapanich N, Ruchutrakool T. Comparison of the long-term remission of rituximab and conventional treatment for acquired thrombotic thrombocytopenic purpura: a systematic review and meta-analysis. Clin Appl Thromb Hemost 2019;25:1076029618825309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lammle B Thrombotic microangiopathy: caplacizumab accelerates resolution of acute acquired TTP. Nat Rev Nephrol. 2016;12:259–260. [DOI] [PubMed] [Google Scholar]
- 20.Peyvandi F, Scully M, Kremer Hovinga JA, et al. Caplacizumab reduces the frequency of major thromboembolic events, exacerbations and death in patients with acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2017;15:1448–1452. [DOI] [PubMed] [Google Scholar]
- 21.Coppo P, Cuker A, George JN. Thrombotic thrombocytopenic purpura: toward targeted therapy and precision medicine. Res Pract Thromb Haemost. 2019;3:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estcourt LJ. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura (HERCULES trial). Transfus Med. 2019;29:146–148. [DOI] [PubMed] [Google Scholar]
- 23.Khan S, Landry K, Umyarova E. Caplacizumab treatment for acquired refractory thrombotic thrombocytopenic purpura. Br J Haematol. 2020;191:e44–e46. [DOI] [PubMed] [Google Scholar]
- 24.Schelpe AS, Petri A, Roose E, et al. Antibodies that conformationally activate ADAMTS13 allosterically enhance metalloprotease domain function. Blood Adv. 2020;4:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roose E, Schelpe AS, Tellier E, et al. Open ADAMTS13, induced by antibodies, is a biomarker for subclinical immune-mediated thrombotic thrombocytopenic purpura. Blood. 2020;136:353–361. [DOI] [PubMed] [Google Scholar]
- 26.Graca NAG, Ercig B, Carolina Velasquez Pereira L, et al. Modifying ADAMTS13 to modulate binding of pathogenic autoantibodies of patients with acquired thrombotic thrombocytopenic purpura. Haematologica. 2020;105:2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petri A, Kim HJ, Xu Y, et al. Hrombotic thrombocytopenic purpura-hemolytic uremic syndrome: diagnosis and treatment. Nat Commun. 2019;10:3781.31439947 [Google Scholar]
- 28.Zhu J, Muia J, Gupta G, et al. Exploring the “minimal” structure of a functional ADAMTS13 by mutagenesis and small-angle X-ray scattering. Blood. 2019;133:1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muia J, Zhu J, Greco SC, et al. Phylogenetic and functional analysis of ADAMTS13 identifies highly conserved domains essential for allosteric regulation. Blood. 2019;133:1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halkidis K, Siegel DL, Zheng XL. A human monoclonal antibody against the distal carboxyl terminus of ADAMTS-13 modulates its susceptibility to an inhibitor in thrombotic thrombocytopenic purpura. J Thromb Haemost. 2021;19:1888–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deforche L, Roose E, Vandenbulcke A, et al. Linker regions and flexibility around the metalloprotease domain account for conformational activation of ADAMTS-13. J Thromb Haemost. 2015;13:2063–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muia J, Zhu J, Gupta G, et al. Allosteric activation of ADAMTS13 by von Willebrand factor. Proc Natl Acad Sci USA. 2014;111:18584–18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casina VC, Hu W, Mao JH, et al. High-resolution epitope mapping by HX MS reveals the pathogenic mechanism and a possible therapy for autoimmune TTP syndrome. Proc Natl Acad Sci USA. 2015;112:9620–9625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.South K, Freitas MO, Lane DA. Conformational quiescence of ADAMTS13 prevents proteolytic promiscuity. J Thromb Haemost. 2016;14:2011–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch CJ, Lane DA, Luken BM. Control of VWF A2 domain stability and ADAMTS13 access to the scissile bond of full-length VWF. Blood. 2014;123:2585–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.South K, Luken BM, Crawley JT, et al. Conformational activation of ADAMTS13. Proc Natl Acad Sci U S A. 2014;111:18578–18583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Groot R, Lane DA, Crawley JT. The ADAMTS13 metalloprotease domain: roles of subsites in enzyme activity and specificity. Blood. 2010;116:3064–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–494. [DOI] [PubMed] [Google Scholar]
- 39.Gerritsen HE, Robles R, Lammle B, Furlan M. Partial amino acid sequence of purified von Willebrand factor-cleaving protease. Blood. 2001;98:1654–1661. [DOI] [PubMed] [Google Scholar]
- 40.Fujikawa K, Suzuki H, McMullen B, Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98:1662–1666. [DOI] [PubMed] [Google Scholar]
- 41.Soejima K, Mimura N, Hirashima M, et al. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J Biochem. 2001;130:475–480. [DOI] [PubMed] [Google Scholar]
- 42.Zheng XL. Structure-function and regulation of ADAMTS-13 protease. J Thromb Haemost. 2013;11(Suppl 1):11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Amo-Maestro L, Sagar A, Pompach P, et al. An integrative structural biology analysis of von Willebrand factor binding and processing by ADAMTS-13 in solution. J Mol Biol. 2021;433:166954. [DOI] [PubMed] [Google Scholar]
- 44.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci USA. 2009;106:19274–19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ai J, Smith P, Wang S, Zhang P, Zheng XL. The proximal carboxyl-terminal domains of ADAMTS13 determine substrate specificity and are all required for cleavage of von Willebrand factor. J Biol Chem. 2005;280:29428–29434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X, Nishio K, Majerus EM, Sadler JE. Cleavage of von Willebrand factor requires the spacer domain of the metalloprotease ADAMTS13. J Biol Chem. 2003;278:30136–30141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao J, Jin SY, Xue J, Sorvillo N, Voorberg J, Zheng XL. Essential domains of a disintegrin and metalloprotease with thrombospondin type 1 repeats-13 metalloprotease required for modulation of arterial thrombosis. Arterioscler Thromb Vasc Biol. 2011;31:2261–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin SY, Xiao J, Bao J, Zhou S, Wright JF, Zheng XL. AAV-mediated expression of an ADAMTS13 variant prevents shigatoxin-induced thrombotic thrombocytopenic purpura. Blood. 2013;121(3825–9):S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.South K, Freitas MO, Lane DA. A model for the conformational activation of the structurally quiescent metalloprotease ADAMTS13 by von Willebrand factor. J Biol Chem. 2017;292:5760–5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HJ, Xu Y, Petri A, Vanhoorelbeke K, Crawley JTB, Emsley J. Crystal structure of ADAMTS13 CUB domains reveals their role in global latency. Sci Adv. 2021;7:eabg4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, Wu Z, Xie X, et al. Characterization of the interactions of ADAMTS13 CUB1 domain to WT- and GOF-Spacer domain by molecular dynamics simulation. J Mol Graph Model. 2021;109:108029. [DOI] [PubMed] [Google Scholar]
- 52.Jian C, Xiao J, Gong L, et al. Gain-of-function ADAMTS13 variants that are resistant to autoantibodies against ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Blood. 2012;119:3836–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rottensteiner H, Seyfried BK, Kaufmann S, et al. Identification of cysteine thiol-based linkages in ADAMTS13 in support of a non-proteolytic regulation of von Willebrand factor. J Thromb Haemost. 2019;17:2099–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roose E, Veyradier A, Vanhoorelbeke K. Insights into ADAMTS13 structure: impact on thrombotic thrombocytopenic purpura diagnosis and management. Curr Opin Hematol. 2020;27:320–326. [DOI] [PubMed] [Google Scholar]
- 55.Velasquez Pereira LC, Roose E, Graca NAG, et al. Immunogenic hotspots in the spacer domain of ADAMTS13 in immune-mediated thrombotic thrombocytopenic purpura. J Thromb Haemost. 2021;19:478–488. [DOI] [PubMed] [Google Scholar]
- 56.Ostertag EM, Kacir S, Thiboutot M, et al. ADAMTS13 autoantibodies cloned from patients with acquired thrombotic thrombocytopenic purpura: 1. Structural and functional characterization in vitro. Transfusion. 2016;56:1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas MR, de Groot R, Scully MA, Crawley JT. Pathogenicity of anti-ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. EBioMedicine. 2015;2:942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Igari A, Nakagawa T, Moriki T, et al. Identification of epitopes on ADAMTS13 recognized by a panel of monoclonal antibodies with functional or non-functional effects on catalytic activity. Thromb Res. 2012;130:e79–e83. [DOI] [PubMed] [Google Scholar]
- 59.Kremer Hovinga JA, Lammle B. Role of ADAMTS13 in the pathogenesis, diagnosis, and treatment of thrombotic thrombocytopenic purpura. Hematol Am Soc Hematol Educ Program. 2012;2012:610–616. [DOI] [PubMed] [Google Scholar]
- 60.Pos W, Sorvillo N, Fijnheer R, et al. Residues Arg568 and Phe592 contribute to an antigenic surface for anti-ADAMTS13 antibodies in the spacer domain. Haematologica. 2011;96:1670–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi Y, Moriki T, Igari A, et al. Epitope analysis of autoantibodies to ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Thromb Res. 2011;128:169–173. [DOI] [PubMed] [Google Scholar]
- 62.Zheng XL, Wu HM, Shang D, et al. Multiple domains of ADAMTS13 are targeted by autoantibodies against ADAMTS13 in patients with acquired idiopathic thrombotic thrombocytopenic purpura. Haematologica. 2010;95:1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luken BM, Turenhout EA, Hulstein JJ, Van Mourik JA, Fijnheer R, Voorberg J. The spacer domain of ADAMTS13 contains a major binding site for antibodies in patients with thrombotic thrombocytopenic purpura. Thromb Haemost. 2005;93:267–274. [DOI] [PubMed] [Google Scholar]
- 64.Klaus C, Plaimauer B, Studt JD, et al. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103:4514–4519. [DOI] [PubMed] [Google Scholar]
- 65.Ostertag EM, Bdeir K, Kacir S, et al. ADAMTS13 autoantibodies cloned from patients with acquired thrombotic thrombocytopenic purpura: 2. Pathogenicity in an animal model. Transfusion. 2016;56:1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roose E, Schelpe AS, Joly BS, et al. An open conformation of ADAMTS-13 is a hallmark of acute acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2018;16:378–388. [DOI] [PubMed] [Google Scholar]
- 67.Deforche L, Tersteeg C, Roose E, et al. Generation of anti-Murine ADAMTS13 antibodies and their application in a mouse model for acquired thrombotic thrombocytopenic purpura. PLoS One. 2016;11:e0160388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129:93–100. [DOI] [PubMed] [Google Scholar]
- 69.Di Stasio E, Lancellotti S, Peyvandi F, Palla R, Mannucci PM, De Cristofaro R. Mechanistic studies on ADAMTS13 catalysis. Biophys J. 2008;95:2450–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roose E, Vidarsson G, Kangro K, et al. Anti-ADAMTS13 autoantibodies against cryptic epitopes in immune-mediated thrombotic thrombocytopenic purpura. Thromb Haemost. 2018;118:1729–1742. [DOI] [PubMed] [Google Scholar]
- 71.Jin SY, Skipwith CG, Zheng XL. Amino acid residues Arg(659), Arg(660), and Tyr(661) in the spacer domain of ADAMTS13 are critical for cleavage of von Willebrand factor. Blood. 2010;115:2300–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zanardelli S, Crawley JT, Chion CK, Lam JK, Preston RJ, Lane DA. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281:1555–1563. [DOI] [PubMed] [Google Scholar]
- 73.Henry ER, Jones CM, Hofrichter J, Eaton WA. Can a two-state MWC allosteric model explain hemoglobin kinetics? Biochemistry. 1997;36:6511–6528. [DOI] [PubMed] [Google Scholar]
- 74.Senozan NM, DeVore JA, Lesniewski EK. Hemoglobin-oxygen-carbon monoxide equilibria with the MWC model. Biophys Chem. 1998;75:141–150. [DOI] [PubMed] [Google Scholar]
- 75.Henry ER, Harper J, Glass KE, Metaferia B, Louis JM, Eaton WA. MWC allosteric model explains unusual hemoglobin-oxygen binding curves from sickle cell drug binding. Biophys J. 2021;120:2543–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cornish-Bowden A, Cardenas ML. Specificity of non-Michaelis-Menten enzymes: necessary information for analyzing metabolic pathways. J Phys Chem B. 2010;114:16209–16213. [DOI] [PubMed] [Google Scholar]


