Abstract
A single-platform technology that uses an internal bead standard and three-color flow cytometry to determine CD4 and CD8 absolute counts was evaluated for reproducibility and agreement. Values obtained using TruCount absolute-count tubes were compared to those obtained using a two-color predicate methodology. Sixty specimens from human immunodeficiency virus type 1-infected donors were shipped to five laboratories. Each site also analyzed replicates of 14 human immunodeficiency virus type 1-infected local specimens at 6 h and again at 24 h. The interlaboratory variability was significantly less with TruCount (median difference in percent coefficient of variation [%CV] between the two methods was −8% and −3% for CD4 and CD8, respectively) than with the predicate method. Intralaboratory variability was smaller, with a median difference in %CV of −1% for both CD4 and CD8 with 6-h samples and −2% and −3% for CD4 and CD8, respectively, with 24-h samples. Use of TruCount for shipped samples resulted in a median CD4 count change of 7 cells (50th estimated percentile) when all laboratories and CD4 strata were combined. For on-site samples, the median CD4 count change was 10 CD4 cells for 6-h samples and 2 CD4 cells for 24-h samples. Individual site biases occurred in both directions and cancelled each other when the data were combined for all laboratories. Thus, the combined data showed a smaller change in median CD4 count than what may have occurred at an individual site. In summary, the use of TruCount decreased both the inter- and intralaboratory variability in determining absolute CD4 and CD8 counts.
Human immunodeficiency virus type 1 (HIV-1) infects cells that express the CD4 receptor (8) and, as a result, depletes its host of CD4 lymphocytes (11). This depletion of CD4 T lymphocytes has been linked to the immunopathogenesis of HIV infection and progression of the disease (9, 13). A CD4 count of ≤200 cells/μl has been included as an AIDS-defining event (5), as these measurements are useful predictors for the onset of opportunistic diseases such as Pneumocystis carinii pneumonia (4). With the advent of highly active antiretroviral therapy, CD4 T-lymphocyte measurements have been used to monitor immune reconstitution (1).
The current predicate methodology for determining absolute CD4 T-lymphocyte counts is dependent upon immunophenotypic identification of cells with fluorescently labeled monoclonal antibodies directed against the CD4 antigen. Relative percentages of CD4 T cells are determined with a flow cytometer. An absolute CD4 count is derived by multiplying the percentage of lymphocytes that are CD3+ CD4+ by the absolute lymphocyte count determined with a hematology instrument. However, the overnight shipment of blood may result in increased intrinsic variability in the absolute lymphocyte count depending on the hematology instrument that is used (10, 16). Therefore, the absolute CD4 count in overnight samples may have increased variability due solely to the hematological determinants.
The need for precise and reproducible monitoring of CD4 T-lymphocyte levels in HIV-infected patients has led several companies to develop simpler methods for measuring absolute CD4 and CD8 T-lymphocyte counts (2, 7, 14, 15, 17). The new single-platform system developed by BD Biosciences (San Jose, Calif.) eliminates the need for multiple technologies (i.e., flow cytometry and hematology) and should be less expensive than predicate methods when labor, cost and inconvenience of repeat samples, and hematology costs are considered. TruCount absolute-count tubes contain a lyophilized pellet that dissolves during sample preparation, releasing a known number of fluorescent beads. By gating the bead population during analysis, absolute cell counts can be readily determined by a simple calculation.
The purpose of this study was to evaluate the single-platform methodology of TruCount tubes as an alternative method for determining CD4 and CD8 absolute counts and to compare these values with those obtained by predicate methodology. Both reproducibility and agreement with the predicate method were measured on centrally shipped specimens as well as on replicate samples of specimens obtained at individual sites. In addition, the sample stability of on-site replicate 6-h and 24-h paired specimens was investigated.
MATERIALS AND METHODS
Evaluation sites and instrumentation.
The five participating laboratories, certified by the National Institute of Allergy and Infectious Disease (NIAID) flow cytometry proficiency program, were chosen to represent different geographical locations from within the United States: two on the east coast, one in the Midwest, and two on the west coast. All laboratories used a FACScan flow cytometer equipped with either a Hewlett Packard or Macintosh Quadra computer. For complete blood count and lymphocyte differential determinations, three laboratories used Coulter STKS instruments, a fourth laboratory used a Coulter MD 16 instrument, and the fifth laboratory used a Roche Helios instrument.
Sample collection.
Peripheral blood samples were obtained only from HIV-1-infected persons. Approval and informed consent were obtained from all participants. A central contractor, FAST Systems Inc., Gaithersburg, Md., shipped aliquots of EDTA-anticoagulated blood from HIV-1-infected donors to individual sites until a total of 60 common specimens were analyzed at all sites for flow cytometry and hematology. The mailings were received at the sites the day following the blood drawing and were analyzed on the day of receipt. The specimens were stratified with regard to CD4 absolute count determined by the predicate methodology, so that approximately one-third of the shipped samples were in each stratum: <200, between 200 and 500, and >500 cells/μl.
In addition, each laboratory obtained EDTA-anticoagulated blood from 14 local HIV-1-infected donors in a single blood drawing consisting of two 7-ml and sixteen 2.5-ml tubes. Eight 2.5-ml EDTA tubes were sent to the hematology laboratory immediately for replicate determinations. One 7-ml EDTA tube was used for eight replicate determinations of CD3+ CD4+ and CD3+ CD8+ using the two different flow cytometric methods. The remaining EDTA tubes were kept overnight at room temperature for 24-h measurements. Donors were prescreened for CD4 count to ensure that seven donors had CD4 counts of <200 cells/μl and the other seven donors had CD4 counts of ≥200 cells/μl. Sites continued to process donor specimens until 14 acceptable 6-h and 24-h paired data sets were obtained.
Antibody staining procedure and data collection.
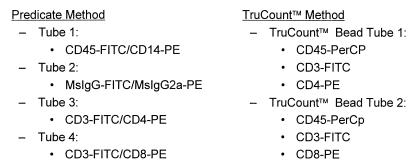
The immunophenotyping panels performed on all shipped and on-site specimens are listed in Fig. 1. All reagents and computer software used for data acquisition and analysis were supplied by BD Biosciences. For the predicate method, Simultest two-color antibodies were used for staining and SimulSET software was used for automated data collection. The 1997 Centers for Disease Control revised guidelines for the performance of CD4 determinations were followed for samples processed by the two-color, stain, wash, and fix predicate method (6). For the single-platform method, 20 μl of TriTEST three-color antibodies and 50 μl of whole blood were added to bead-containing TruCount tubes. Tubes were incubated for 20 min at room temperature before 450 μl of FACS Lysing Solution was added. Tubes were analyzed the same day with CELLQUEST software. The bead population and the CD45 lymphocyte versus side scatter population were manually gated. The absolute count using TruCount tubes was calculated from the appropriate dot plot values entered into a spreadsheet that was formatted to use the formula [(no. of events in quadrant containing cell population)/(no. of events in absolute-count bead region [R2])] × [(total no. of absolute-count beads)/(test volume [50 μl])].
FIG. 1.
Antibody staining profiles. FITC, fluorescein isothiocyanate; PE, phycoerythrin; PerCP, peridinin chlorophyll protein; MsIgG, mouse immunoglobulin G.
Hematology measurements.
The five hematology laboratories that participated in this study maintained performance that conformed to accepted standards of practice (e.g., College of American Pathologists and National Committee for Clinical Laboratory Standards). All hematology samples were drawn at the same time as the specimens for flow cytometry. For shipped specimens, the hematology measurements were performed within 33 h of specimen draw. For on-site specimens, the measurements were performed within 7 h of draw for same-day specimens and within 33 h of draw for the paired 24-h specimen data. White blood cell (WBC) and leukocyte differential (including percent lymphocyte) counts were performed on an automated instrument. If the specimen was rejected or flagged in the lymph region by the machine, the value was flagged in the database spreadsheet and subsequently eliminated from the study analyses. A cell designated as an atypical lymph or large unstained cell was included in the total lymphocyte number.
Analyses.
Criteria for accepting data obtained with SimulSET software included the following: (i) gated lymphocyte purity >85%, (ii) lymphocyte recovery within the gate >90%, and (iii) differences in the CD3 percentages between the CD3+ CD4+ and CD3+ CD8+ tubes ≤7%. Data from individual sites were entered into a spreadsheet designed by the Statistical Data Analysis Center, Harvard School of Public Health, Boston, Mass., and imported into a central database for analyses. Comparisons of the variability of the TruCount method versus the predicate method were based on the Wilcoxon signed-rank test (12) applied to the differences (TruCount method minus predicate method) in the percent coefficient of variation (%CV) of reported CD4 and CD8 counts for each specimen (between-laboratory %CV in the case of the centrally shipped specimens and within-replicate %CV in the case of local donor specimens). The accuracy of the CD4 and CD8 counts determined by the TruCount method versus the predicate method was tested by the Wilcoxon signed-rank test applied to the differences (TruCount method minus predicate method) in reported CD4 and CD8 counts for each centrally shipped specimen at each laboratory. In the case of CD4 and CD8 counts on replicates from local donors, a log rank test stratified by donor was used. For the primary endpoints, statistical significance is defined as P < 0.05. The primary endpoints for both CD4 and CD8 counts, and combining all CD4 strata, are (i) intralaboratory variability in all laboratories combined using local specimens 24 h old, (ii) interlaboratory variability using centrally shipped specimens, (iii) intralaboratory agreement using local specimens 24 h old, and (iv) intralaboratory agreement using centrally shipped specimens. The P values for the secondary endpoints are exploratory only. Tertiary analyses on the CD4 and CD8 subset percentages were carried out similarly.
RESULTS
Interlaboratory variability for shipped specimens.
CD4 and CD8 absolute counts were obtained for 60 samples shipped to five different laboratories. Statistical analyses of the %CVs of the TruCount method minus the %CVs of the predicate method were performed on the database as a whole, on the different CD4 strata, and on individual site data. Table 1 shows that the median %CV for the TruCount method was 8% and 3% less than the predicate method for CD4 and CD8, respectively. When analyzed with regard to CD4 stratum, the <200 cells/μl group showed a significant 23% (CD4) and 9% (CD8) median difference in %CVs for the two methods. These large differences were due to the large median %CVs for the predicate method for samples with a CD4 count of <200 cells/μl. For the remaining two CD4 strata, the median differences between methods were again significantly lower (3 to 7%), favoring the TruCount method.
TABLE 1.
Interlaboratory variability for CD4 and CD8 counts for shipped samples
| CD4 stratum | n | Median CD4 count (cells/μl) | Median %CV absolute CD4 count
|
Median CD8 count (cells/μl) | Median %CV absolute CD8 count
|
Median difference in %CVs (TruCount − predicate)a
|
|||
|---|---|---|---|---|---|---|---|---|---|
| TruCount | Predicate | TruCount | Predicate | CD4 | CD8 | ||||
| All | 60 | 315 | 9 | 16 | 988 | 7 | 11 | −8* | −3* |
| <200 | 21 | 130 | 11 | 28 | 620 | 6 | 22 | −23* | −9* |
| 200–500 | 24 | 336 | 8 | 15 | 989 | 7 | 8 | −7* | −2 |
| >500 | 15 | 714 | 9 | 10 | 1,256 | 7 | 10 | −3* | −3* |
*, P < 0.05, Wilcoxon signed-rank test.
Intralaboratory variability for 6-h and 24-h replicate samples.
Each of the five sites solicited 14 donors whose samples were analyzed as eight replicates at 6 h and eight replicates at 24 h. Therefore, a total of 70 paired samples (35 with a CD4 count of <200 cells/μl and 35 with a CD4 count of ≥200 cells/μl) constituted the database. Table 2 shows that the median difference in %CVs for 6-h replicate samples was −1% (%CV for TruCount − %CV for predicate method) for both CD4 and CD8 counts by both methods. When analyzed with regard to CD4 strata, specimens with CD4 counts of <200 cells/μl showed significant differences in the %CVs for CD4 and CD8 counts (−3 and −1%, respectively). For samples with ≥200 CD4 cells/μl, no differences in median %CVs between the two methods were seen. Individual site performance reflected what was observed for the database as a whole.
TABLE 2.
Intralaboratory variability for CD4 and CD8 counts for 6-h replicate samples
| CD4 stratum or laboratory | n | Median %CV absolute CD4 count
|
Median %CV absolute CD8 count
|
Median difference in %CVs (TruCount − predicate)a
|
|||
|---|---|---|---|---|---|---|---|
| TruCount | Predicate | TruCount | Predicate | CD4 | CD8 | ||
| Stratum | |||||||
| All | 70 | 7 | 7 | 5 | 5 | −1* | −1* |
| <200 | 35 | 8 | 11 | 5 | 6 | −3* | −1* |
| ≥200 | 35 | 6 | 6 | 5 | 5 | 0 | 0 |
| Laboratory | |||||||
| A | 14 | 6 | 7 | 5 | 5 | −1* | 0 |
| B | 14 | 7 | 7 | 5 | 5 | −1 | −1 |
| C | 14 | 8 | 8 | 4 | 5 | −3 | −1 |
| D | 14 | 7 | 6 | 5 | 6 | 0 | −1 |
| E | 14 | 8 | 8 | 5 | 6 | −3 | −1 |
*, P <0.05, Wilcoxon signed-rank test.
For the on-site replicate samples held overnight, the TruCount tubes generated overall median %CV differences that were less than the predicate method values for both CD4 and CD8 counts (2 and 3%, respectively) (Table 3). When the samples with a CD4 count of <200 cells/μl were analyzed, the median differences in %CVs were 3 and 4%, TruCount values being less than predicate method values for CD4 and CD8 counts, respectively. Samples with a CD4 count of ≥200 cells/μl showed significant median differences with %CVs of −2% for CD4 counts but an insignificant −1% for CD8 counts. In general, individual site performance reflected what was observed for the database as a whole.
TABLE 3.
Intralaboratory variability for CD4 and CD8 counts for 24-h replicate samples
| CD4 stratum or laboratory | n | Median %CV absolute CD4 count
|
Median %CV absolute CD8 count
|
Median difference in %CVs (TruCount − predicate)
|
|||
|---|---|---|---|---|---|---|---|
| TruCount | Predicate | TruCount | Predicate | CD4 | CD8 | ||
| Stratum | |||||||
| All | 70 | 7 | 9 | 4 | 7 | −2* | −3* |
| <200 | 35 | 8 | 10 | 4 | 8 | −3* | −4* |
| ≥200 | 35 | 6 | 8 | 5 | 6 | −2* | −1 |
| Laboratory | |||||||
| A | 14 | 5 | 8 | 4 | 6 | −3* | 2* |
| B | 14 | 8 | 10 | 6 | 6 | −1 | 0 |
| C | 14 | 9 | 9 | 4 | 8 | −3 | −5* |
| D | 14 | 6 | 6 | 4 | 4 | −1 | 0 |
| E | 14 | 7 | 10 | 4 | 9 | −4* | −4* |
*, P < 0.05 using the Wilcoxon Signed Rank test.
Agreement for CD4 and CD8 absolute counts with the predicate method for shipped samples.
Agreement between methods was assessed by subtracting the absolute count obtained by the predicate method from the absolute count obtained by the TruCount method for the same sample, and the 10th, 50th, and 90th percentiles of the differences were estimated. For shipped samples, with all CD4 strata and laboratories combined, the median of the differences for CD4 and CD8 counts were 7 and −51 cells, respectively (Table 4). With all CD4 strata combined, the median of the differences for CD4 counts from individual laboratories ranged from −34 to 25 cells. Individual site biases were noted for both CD4 and CD8 counts between the two methods. Laboratories A and B obtained lower values for CD4 counts using TruCount, while laboratories C, D, and E obtained higher values with the tubes.
TABLE 4.
Agreement between shipped-sample CD4 and CD8 countsa
| CD4 stratum and laboratory | n | Predicate CD4 count | Agreement CD4 count
|
Pb | Predicate CD8 count | Agreement CD8 count
|
P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10th | 50th | 90th | 10th | 50th | 90th | ||||||
| All strata | |||||||||||
| All labs | 411 | 272 | −67 | 7 | 79 | <0.01 | 992 | −238 | −51 | 100 | <0.01 |
| A | 84 | 311 | −103 | −17 | 61 | <0.01 | 1,068 | −310 | −136 | 7 | <0.01 |
| B | 74 | 378 | −135 | −34 | 55 | <0.01 | 1,129 | −437 | −129 | 35 | <0.01 |
| C | 79 | 237 | −16 | 13 | 73 | <0.01 | 845 | −86 | 11 | 136 | |
| D | 86 | 232 | −24 | 12 | 70 | <0.01 | 984 | −179 | −44 | 64 | <0.01 |
| E | 88 | 200 | −25 | 25 | 181 | <0.01 | 927 | −190 | 3 | 220 | |
| <200 | |||||||||||
| All labs | 168 | 115 | −49 | 2 | 40 | 721 | −274 | −34 | 104 | <0.01 | |
| A | 33 | 168 | −83 | −16 | 9 | <0.01 | 875 | −585 | −157 | −29 | <0.01 |
| B | 25 | 178 | −94 | −14 | 35 | <0.03 | 964 | −491 | −144 | 108 | <0.01 |
| C | 36 | 106 | −13 | 10 | 42 | <0.02 | 584 | −68 | 19 | 159 | |
| D | 36 | 105 | −10 | 7 | 48 | <0.01 | 625 | −114 | −16 | 61 | |
| E | 38 | 103 | −24 | 11 | 55 | <0.01 | 680 | −191 | −2 | 162 | |
| 200–500 | |||||||||||
| All labs | 156 | 337 | −77 | 17 | 101 | <0.01 | 1,080 | −230 | −65 | 99 | <0.01 |
| A | 33 | 380 | −115 | 9 | 76 | 1,202 | −247 | −108 | 77 | <0.01 | |
| B | 31 | 396 | −169 | −46 | 62 | <0.01 | 1,159 | −526 | −129 | 29 | <0.01 |
| C | 27 | 318 | −28 | 14 | 83 | <0.01 | 1,003 | −86 | 1 | 114 | |
| D | 33 | 319 | −21 | 21 | 83 | <0.01 | 1,066 | −201 | −93 | 53 | <0.01 |
| E | 32 | 274 | −239 | 68 | 225 | <0.01 | 1,055 | −135 | 9 | 852 | |
| >500 | |||||||||||
| All labs | 87 | 680 | −102 | 12 | 144 | 1,191 | −237 | −63 | 108 | <0.01 | |
| A | 18 | 742 | −145 | −29 | 81 | 1,198 | −202 | −96 | 73 | <0.01 | |
| B | 18 | 752 | −182 | −49 | 155 | 1,297 | −373 | −119 | 59 | <0.01 | |
| C | 16 | 677 | −26 | 47 | 166 | <0.01 | 1,133 | −130 | 3 | 142 | |
| D | 17 | 622 | −49 | 12 | 141 | 1,234 | −237 | −19 | 153 | ||
| E | 18 | 602 | −14 | 74 | 262 | <0.01 | 1,138 | −307 | −7 | 184 | |
Agreement was determined by subtracting the absolute count obtained by the predicate method from the absolute count obtained by the TruCount method for the same sample. The estimated 10th, 50th, and 90th percentiles of the differences in absolute counts (cells per microliter) are given.
P value for the estimated 50th percentile by the Wilcoxon signed-rank test.
Agreement of CD4 and CD8 absolute counts for on-site replicate samples.
For 6-h replicate samples, the median of the differences for CD4 and CD8 counts between TruCount and the predicate method for all CD4 strata and laboratories were 10 and 23 cells, respectively (Table 5). The median of the differences for CD4 and CD8 counts for the 24-h replicate samples were 2 and −21 cells, respectively; these differences were similar in direction and magnitude to those obtained for the shipped samples (CD4 results similar to and CD8 TruCount results lower than those with the predicate method). Individual site biases were noted for both CD4 and CD8 counts between the two methods for both the 6-h and the 24-h replicate samples. Laboratory B obtained lower values for CD4 counts using TruCount, while laboratories A, C, D, and E obtained higher values with this method.
TABLE 5.
Agreement between on-site replicate sample CD4 and CD8 countsa
| CD4 stratum and laboratory | CD4 count (cells/μl)
|
CD8 count (cells/μl)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 h
|
24 h
|
6 h
|
24 h
|
|||||||||
| 10th | 50th | 90th | 10th | 50th | 90th | 10th | 50th | 90th | 10th | 50th | 90th | |
| All strata | ||||||||||||
| All labs | −29 | 10b | 72 | −62 | 2 | 65 | −120 | 23* | 194 | −206 | −21* | 140 |
| A | −22 | 18* | 72 | −93 | 7 | 55 | −75 | 59* | 207 | −379 | −2 | 212 |
| B | −59 | −7* | 69 | −132 | −33* | 14 | −304 | −42* | 193 | −396 | −140* | 9 |
| C | −10 | 9* | 66 | −41 | 2 | 111 | −52 | 39* | 257 | −169 | 4 | 234 |
| D | −25 | 14* | 75 | −5 | 20* | 89 | −142 | −1 | 158 | −106 | 11 | 103 |
| E | −5 | 21* | 74 | −29 | 4 | 43 | −49 | 32* | 183 | −107 | −10* | 110 |
| <200 | ||||||||||||
| All labs | −15 | 5* | 38 | −35 | 1 | 38 | −102 | 21* | 202 | −185 | −4* | 154 |
| A | −14 | 11* | 41 | −156 | 10* | 31 | −66 | 63* | 227 | −972 | 44 | 246 |
| B | −32 | −1* | 58 | −48 | −25* | 3 | −233 | −33 | 205 | −493 | −142* | −49 |
| C | −6 | 0 | 23 | −20 | −1 | 54 | −20 | 39* | 208 | −159 | 20 | 208 |
| D | −4 | 6* | 36 | −3 | 12* | 52 | −99 | −4 | 183 | −72 | 14 | 115 |
| E | −5 | 9* | 43 | −14 | 6* | 39 | −57 | 23 | 189 | −67 | 10 | 132 |
| ≥200 | ||||||||||||
| All labs | −50 | 24* | 105 | −92 | 6 | 101 | −155 | 25* | 182 | −223 | −38* | 124 |
| A | −27 | 24* | 87 | −71 | 2 | 73 | −77 | 57* | 201 | −288 | −23 | 172 |
| B | −84 | −15 | 112 | −165 | −58* | 42 | −325 | −50* | 156 | −362 | −136* | 36 |
| C | −31 | 26* | 95 | −64 | 34 | 149 | −73 | 38* | 240 | −181 | −26 | 312 |
| D | −90 | 36* | 100 | −21 | 51* | 132 | −358 | 15 | 108 | −158 | −10 | 89 |
| E | −7 | 32* | 128 | −46 | 2 | 48 | −31 | 51* | 170 | −148 | −45* | 82 |
See Table 4, footnote a.
*, P < 0.05 for the estimated 50th percentile by the Wilcoxon signed-rank test.
Interlaboratory variability of subset percentages.
In an attempt to estimate the contribution of the intrinsic variability of the WBC and leukocyte differential counts to the combined CD4 absolute-count variability, the subset percentages (CD3+ CD4+ and CD3+ CD8+) obtained by both methods were evaluated. For both shipped and 24-h on-site replicate samples, the median %CVs obtained for CD4 and CD8 subset percentages were significantly less using the TruCount method than the predicate method (Table 6). The median difference in %CVs for CD4 subset percentage was almost half for shipped samples and about one third less for 24-h on-site replicate samples using the TruCount tubes. For CD8 subset percentages, these differences were of a lower magnitude: the median difference in %CVs was about 1% lower using the TruCount tubes for both shipped and 24-h on-site replicate samples. Similar differences were seen when the data were analyzed with regard to individual CD4 count stratum.
TABLE 6.
Variability of subset percentages for shipped and 24-h on-site replicate samples
| Sample group and CD4 stratum | n | Median %CV for subset
|
Median difference in %CVsa
|
||||
|---|---|---|---|---|---|---|---|
| CD4
|
CD8
|
||||||
| TruCount | Predicate | TruCount | Predicate | CD4 | CD8 | ||
| Shipped | |||||||
| All | 60 | 5 | 9 | 2 | 3 | −4* | −1* |
| <200 | 21 | 8 | 12 | 3 | 3 | −2* | −1* |
| 200–500 | 24 | 4 | 9 | 2 | 3 | −5* | −1 |
| >500 | 15 | 4 | 6 | 2 | 3 | −2* | 0 |
| Replicates | |||||||
| All | 70 | 4 | 6 | 2 | 2 | −1* | −1* |
| <200 | 35 | 7 | 9 | 1 | 2 | −2* | −1* |
| ≥200 | 35 | 3 | 4 | 2 | 2 | −1 | 0* |
Calculated as TruCount %CV − predicate %CV. *, P < 0.05, Wilcoxon signed-rank test.
Sample stability of paired specimens analyzed at 6 and 24 h.
An indicator of sample stability was determined by subtracting the 6-h count from the 24-h count for the same paired samples analyzed by the same method. Therefore, differences in sample stability were determined separately for the predicate and TruCount methods. For CD4 counts, the median of the difference in fresh and day-old samples using TruCount tubes was −2 cells, compared with 4 cells using the predicate method when all laboratories and CD4 strata were combined (Table 7). In other words, TruCount tubes gave slightly higher CD4 values at 6 h than at 24 h, while the predicate method gave slightly lower values at 6 h than at 24 h. Similar differences were observed when the CD4 strata were evaluated. Each laboratory had its individual bias for paired samples at 6 and 24 h. Table 8 shows a similar data profile for 6-h and 24-h paired CD8 counts. When all strata and laboratories were combined, the median of the differences in CD8 counts using TruCount was higher for fresh specimens (15 cells) but lower for day-old specimens (35 cells). Similar individual site biases were observed for CD8 counts for the paired samples at both time points. In general, the stability data showed that use of TruCount tubes resulted in higher CD4 and CD8 counts at 6 h than at 24 h.
TABLE 7.
Agreement between 24-h and 6-h CD4 counts by methoda
| CD4 stratum and laboratory | n | Median CD4 count
|
Agreement, TruCount
|
Pb | Median CD4 count
|
Agreement, predicate
|
P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24-h | 6-h | 10th | 50th | 90th | 24-h | 6-h | 10th | 50th | 90th | ||||
| All strata | |||||||||||||
| All labs | 1,120 | 211 | 219 | −39 | −2 | 39 | <0.01 | 209 | 200 | −38 | 4 | 81 | <0.01 |
| A | 224 | 269 | 279 | −26 | 6 | 56 | <0.01 | 289 | 254 | −12 | 25 | 115 | <0.01 |
| B | 224 | 173 | 183 | −47 | −8 | 42 | <0.01 | 200 | 210 | −22 | 20 | 114 | <0.01 |
| C | 224 | 237 | 224 | −18 | 1 | 56 | 190 | 194 | −45 | 0 | 65 | ||
| D | 224 | 213 | 217 | −42 | −3 | 26 | <0.01 | 179 | 191 | −80 | −12 | 13 | <0.01 |
| E | 224 | 186 | 192 | −56 | −9 | 10 | <0.01 | 158 | 177 | −34 | 0 | 47 | |
| <200 | |||||||||||||
| All labs | 560 | 104 | 108 | −23 | −3 | 15 | <0.01 | 109 | 103 | −26 | 0 | 33 | |
| A | 80 | 171 | 173 | −16 | 4 | 26 | <0.05 | 152 | 144 | −12 | 10 | 160 | <0.01 |
| B | 112 | 115 | 119 | −29 | −7 | 18 | <0.01 | 136 | 113 | −19 | 13 | 74 | <0.01 |
| C | 112 | 60 | 58 | −13 | 0 | 14 | 39 | 49 | −34 | −1 | 28 | ||
| D | 128 | 109 | 115 | −22 | −2 | 10 | <0.01 | 90 | 104 | −30 | −5 | 7 | <0.01 |
| E | 128 | 104 | 115 | −26 | −6 | 2 | <0.01 | 87 | 96 | −34 | −2 | 19 | |
| ≥200 | |||||||||||||
| All labs | 560 | 400 | 387 | −59 | −1 | 62 | 401 | 370 | −57 | 18 | 103 | <0.01 | |
| A | 144 | 320 | 312 | −35 | 8 | 66 | 361 | 305 | −15 | 37 | 104 | <0.01 | |
| B | 112 | 401 | 401 | −72 | −11 | 67 | 498 | 443 | −25 | 52 | 142 | <0.01 | |
| C | 112 | 418 | 389 | −31 | 18 | 91 | <0.01 | 402 | 368 | −66 | 12 | 98 | |
| D | 96 | 473 | 492 | −66 | −8 | 39 | 406 | 451 | −116 | −33 | 31 | <0.01 | |
| E | 96 | 269 | 279 | −82 | −23 | 26 | <0.01 | 262 | 252 | −34 | 9 | 91 | <0.02 |
Agreement was determined by subtracting the 6-h absolute count from the 24-h absolute count (cells per microliter) obtained for the same sample by the same method. The estimated 10th, 50th, and 90th percentiles of the differences in absolute counts were determined.
P value for the estimated 50th percentile, the Wilcoxon signed-rank test.
TABLE 8.
Agreement between 24-h and 6-h CD8 counts by methoda
| CD4 stratum and laboratory | n | Median CD8 count
|
Agreement, TruCount
|
P | Median CD8 count
|
Agreement, predicate
|
P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24-h | 6-h | 10th | 50th | 90th | 24-h | 6-h | 10th | 50th | 90th | ||||
| All strata | |||||||||||||
| All labs | 1,120 | 795 | 828 | −132 | −15 | 133 | <0.01 | 816 | 820 | −109 | 35 | 221 | <0.01 |
| A | 224 | 908 | 945 | −80 | 65 | 211 | <0.01 | 1,051 | 896 | −10 | 119 | 401 | <0.01 |
| B | 224 | 970 | 995 | −154 | −43 | 195 | <0.01 | 1,189 | 1,065 | −73 | 99 | 287 | <0.01 |
| C | 224 | 848 | 842 | −132 | −10 | 104 | <0.01 | 851 | 827 | −158 | 42 | 214 | <0.01 |
| D | 224 | 756 | 774 | −99 | −16 | 47 | <0.01 | 751 | 810 | −149 | −22 | 68 | <0.01 |
| E | 224 | 675 | 765 | −158 | −51 | 5 | <0.01 | 706 | 674 | −148 | 0 | 92 | |
| <200 | |||||||||||||
| All labs | 560 | 695 | 716 | −116 | −19 | 84 | <0.01 | 702 | 684 | −117 | 18 | 213 | <0.01 |
| A | 80 | 786 | 754 | −47 | 74 | 202 | <0.01 | 731 | 604 | −11 | 113 | 973 | <0.01 |
| B | 112 | 684 | 752 | −138 | −49 | 106 | <0.01 | 1,136 | 985 | −78 | 122 | 316 | <0.01 |
| C | 112 | 628 | 634 | −173 | −21 | 24 | <0.01 | 764 | 594 | −183 | 5 | 199 | |
| D | 128 | 624 | 640 | −96 | −11 | 24 | <0.01 | 650 | 694 | −116 | −22 | 55 | <0.01 |
| E | 128 | 691 | 765 | −107 | −29 | 13 | <0.01 | 668 | 694 | −124 | −8 | 78 | |
| ≥200 | |||||||||||||
| All labs | 560 | 926 | 945 | −159 | −8 | 180 | 928 | 913 | −96 | 54 | 224 | <0.01 | |
| A | 144 | 1,048 | 1,132 | −139 | 62 | 216 | <0.01 | 1,226 | 1,065 | −10 | 123 | 320 | <0.01 |
| B | 112 | 1,076 | 1,050 | −204 | −31 | 274 | 1,233 | 1,104 | −66 | 73 | 219 | <0.01 | |
| C | 112 | 1,012 | 937 | −87 | 28 | 181 | <0.05 | 926 | 887 | −110 | 68 | 281 | <0.01 |
| D | 96 | 889 | 894 | −109 | −21 | 84 | <0.05 | 867 | 893 | −249 | −23 | 80 | |
| E | 96 | 664 | 754 | −293 | −79 | −10 | <0.01 | 719 | 661 | −166 | 12 | 108 | |
See Table 7, footnotes a and b.
DISCUSSION
The current method for the measurement of CD4 absolute counts is expensive and time-consuming and requires multiple manipulations. In addition, most laboratories require separate tubes of blood for the flow cytometry and hematology measurements. Since the hematology measurement is often sensitive to small changes in the blood components, the intrinsic variability of this measurement is especially difficult to minimize. Since the interlaboratory variability in CD4 absolute count for shipped specimens has been unacceptably large, the values for overnight samples may not be reliable. Another confounding factor is that different hematology instruments may have biases toward higher or lower lymphocyte counts (3, 18). This poses a serious problem for patients on HIV-1 intervention protocols if they change where their laboratory CD4 determinations are made. The high variability between any two laboratories may make longitudinal comparisons of CD4 counts inaccurate. The availability of single-platform technologies, which determine CD4 or CD8 absolute counts using only flow cytometry, would decrease this problem and make absolute counts between institutions less variable.
In the current study, which compared single-platform TruCount tubes with a multiplatform predicate method, both the interlaboratory and intralaboratory variability was significantly less using TruCount tubes for both shipped and on-site replicate samples. At the time that the schema was developed, the predicate method in general use did not use CD45 gating. The purpose of this study was to determine the differences in reproducibility and agreement that would occur if a laboratory switched from their current two-color predicate method to a single-platform method. For shipped samples, the variability between CD4 absolute counts using TruCount tubes was about half that using the predicate method. One could argue that this decrease in variability was largely due to the CD45 gating strategy used in the TruCount method and that this decrease in variability could also be achieved with any multiplatform method that used three-color and CD45 gating. If this were true, the difference in variability observed in the subset percentage data for the two methods should account for the majority of the decreased variability in the absolute-count data. However, an analysis of the CD4 subset percentages showed that only a small portion of the decreased variability could be attributed to the use of CD45 gating. This suggests that the TruCount method, in addition to improving the precision of determining lymphocyte subsets over the predicate method, also eliminated the intrinsic variability contributed by the hematology measurements.
When the difference in absolute counts for the same sample by the two methods was calculated, small differences were detected for the database as a whole. For example, the agreement for the 60 shipped samples was a median CD4 count change of 7 cells. Likewise, for on-site replicate samples, the agreement was 10 CD4 cells for 6-h samples and 2 cells for 24-h replicate samples. However, these values were misleading because individual sites had biases in the absolute counts that varied in both magnitude and direction. Individual site evaluations showed significant changes in absolute-count values between the two methods, but since site subset percentages did not show these directional biases (data not shown), one must conclude that the site's bias in the absolute-count determinations arose from the site's hematology instrument. For example, laboratory B used a Roche Helios hematology instrument, and that site's predicate-method absolute counts were consistently higher than those obtained by the TruCount method determinations compared with the other sites, which used Coulter hematology instruments. Thus, it is important that a site perform a comparative study to determine whether a bias in their reported CD4 and CD8 absolute counts will occur if they switch to a single-platform methodology.
Single-platform technology can be more cost-effective than the predicate method when the cost of the WBC and lymphocyte differential counts, the time saved with reduced sample manipulation, and the cost and inconvenience of redrawing blood are considered. For example, the 1999 BD Biosciences list price for the antibody reagents used in the predicate method Simultest panel cost $32.30 per patient test. In comparison, the TriTEST antibody reagents cost $25.40 per patient test. TruCount tubes add $9.60 to the cost of the TriTEST reagents, to bring the single-platform cost to $35.00 for a patient test. Considering that a WBC and lymphocyte differential determination costs more than $2.70 (TruCount minus Simultest costs) and often more than $9.60 (TruCount minus TriTEST costs), the single-platform determination is more cost-effective than the multiplatform methods for reagents alone. In addition, many flow cytometry laboratories are not able to receive hematology laboratory reports by direct data transmission. Since the hematology results must be received by the flow cytometry laboratory before CD4 and CD8 absolute counts can be calculated, misplaced reports can result in absolute-count reporting delays. Occasionally, the tube of blood for the hematology laboratory is not drawn and the CD4 and CD8 absolute counts cannot be reported for the patient sample. In other circumstances, sample quality is so poor that the hematology instrument cannot perform a reliable lymphocyte differential, and again absolute counts cannot be calculated. When hematology values are unattainable for these various reasons, bead-based, single-platform technology can provide meaningful absolute-count data.
Some single-platform methods do not directly determine lymphocyte percentages and only give CD4 and CD8 absolute counts. Examples include the FACSCount system, the Ortho Cytoron Absolute system, the Zymmune CD4/CD8 cell monitoring kit from Zynaxis, Inc. (7), volumetric capillary cytometry from Biometric Imagining, Inc. (15), and the TRAx CD4 test kit from T Cell Diagnostics, Inc. (17). However, TruCount tubes and Flow-Count fluorospheres (Beckman Coulter) provide lymphocyte subset percentages as well as absolute counts. The determination of both of these clinical parameters is important because the lymphocyte subset values are often required for monitoring pediatric HIV-positive populations.
In the present study, the predicate method used forward light scatter versus side scatter in comparison to the TruCount method, which used CD45 lymphocyte gating for sample acquisition and analysis. CD45 lymphocyte gating is also routinely used today by flow cytometric laboratories performing three-color, multiplatform analyses and is not unique to the bead-based, single-platform technology used in this study. Degenerative samples that must be analyzed with a gating strategy based on forward light scatter versus side scatter often fail to meet acceptable gating criteria and result in blood specimens having to be retested or redrawn. For example, in order for the 60 shipped samples to be analyzable at all five sites, a total of 90 specimens were actually sent to the laboratories. The majority of extra specimens were needed because of unacceptable gating criteria for the shipped samples by the predicate method. However, for those samples that could not be analyzed with the predicate method, CD4 and CD8 absolute counts were almost always obtained by using CD45 lymphocyte gating. The TruCount method had an additional advantage over a three-color multiplatform method in that it could “rescue” samples for which an absolute count could not be calculated because of invalid or incomplete hematology values.
In summary, the TruCount method gave better reproducibility and agreement for both shipped and on-site replicate samples than a multiplatform predicate method. In addition to being more cost-efficient, valid absolute cell counts for degenerative samples could be consistently obtained by the single-platform method. Therefore, the results of this multisite study support the use of bead-based, single-platform technology for routine clinical assessment of CD4 and CD8 absolute cell counts.
ACKNOWLEDGMENTS
This work was funded in part by the NIAID Immunophenotyping Quality Assessment Program, contract NO-AI-45175.
We thank BD Biosciences (Ann Shiba and Jim Lowder) for additional financial support and contribution of reagents and training of site personnel. Special appreciation is also extended to the site technologists Eileen Bessent, Gina Bonifacio, Todd Christian, David Devernoe, Fred Menendez, and Kristen Stank, who prepared and analyzed the samples.
REFERENCES
- 1.Autran B, Carcelain G, Li T S, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 2.Bene M C, Kolopp Sarda M N, El Kaissouni J, De March Kennel A, Mole C, Kohler C, Faure G C. Automated cell count in flow cytometry: a valuable tool to assess CD4 absolute levels in peripheral blood. Am J Clin Pathol. 1998;110:321–326. doi: 10.1093/ajcp/110.3.321. [DOI] [PubMed] [Google Scholar]
- 3.Bentley S A, Johnson A A, Bishop C A. A parallel evaluation of four automated hematology analyzers. Am J Clin Pathol. 1993;100:626–632. doi: 10.1093/ajcp/100.6.626. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control. Recommendations for prophylaxis against Pneumocystic carinii pneumonia for adults and adolescents infected with human immunodeficiency virus. Morbid Mortal Weekly Rep. 1992;41(RR-4):1–11. [PubMed] [Google Scholar]
- 5.Centers for Disease Control. Revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morbid Mortal Weekly Rep. 1992;41(RR-17):1–35. [PubMed] [Google Scholar]
- 6.Centers for Disease Control. 1997 revised guidelines for performing CD4+ T-cell determinations in persons infected with human immunodeficiency virus (HIV) Morbid Mortal Weekly Rep. 1997;46(RR-2):1–29. [PubMed] [Google Scholar]
- 7.Denny T N, Jensen B D, Gavin E I, Louzao A G, Vella F A, Oleske J M, Wong W. Determination of CD4 and CD8 lymphocyte subsets by a new alternative fluorescence immunoassay. Clin Diagn Lab Immunol. 1995;2:330–336. doi: 10.1128/cdli.2.3.330-336.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Wolf F, Roos M, Lange J M A, Houweling J T M, Coutinho R A, van der Noordaa J, Schellekens P T, Goudsmit J. Decline in CD4+ cell numbers reflects increase in HIV-1 replication. AIDS Res Hum Retroviruses. 1988;4:433–440. doi: 10.1089/aid.1988.4.433. [DOI] [PubMed] [Google Scholar]
- 9.Fahey J L, Taylor J M G, Detels R, Hoffman B, Melmed R, Nishanian P, Giorgi J V. The prognostic value of cellular and serological markers in infection with human immunodeficiency virus type 1. N Engl J Med. 1990;322:166–172. doi: 10.1056/NEJM199001183220305. [DOI] [PubMed] [Google Scholar]
- 10.Koepke J A, Landay A L. Precision and accuracy of absolute lymphocyte counts. Clin Immunol Immunopathol. 1989;52:19–27. doi: 10.1016/0090-1229(89)90189-x. [DOI] [PubMed] [Google Scholar]
- 11.Lang W, Perkins H, Anderson R E, Royce R, Jewell N, Winkelstein W., Jr Patterns of T-lymphocyte changes with human immunodeficiency virus infection: from seroconversion to the development of AIDS. J Acquir Immune Defic Syndr. 1989;2:63–69. [PubMed] [Google Scholar]
- 12.Lehman E L, D'Abrera H J M. Non-parametrics: statistical methods based on ranks. Oakland, Calif: Holden Day, Inc.; 1975. [Google Scholar]
- 13.Masur H, Ognibene F P, Yarchoan R, Shelhamer J H, Baird B F, Travis W, Suffredini A F, Deyton L, Kovacs J A, Falloon J, Davey R, Polis M, Metcalf J, Baseler M, Wesley R, Gill V J, Fauci A S, Lane H C. CD4 counts as predictors of opportunistic pneumonias in human immunodeficiency virus (HIV) infection. Ann Intern Med. 1989;111:223–231. doi: 10.7326/0003-4819-111-3-223. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson J K A, Velleca W M, Jubert S, Green T A, Bryan L. Evaluation of alternative CD4 technologies for the enumeration of CD4 lymphocytes. J Immunol Methods. 1994;177:43–54. doi: 10.1016/0022-1759(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 15.O'Gorman M R G, Gelman R, et al. Site Investigators. Inter- and intrainstitutional evaluation of automated volumetric capillary cytometry for the quantitation of CD4- and CD8-positive T lymphocytes in the peripheral blood of persons infected with human immunodeficiency virus. Clin Diagn Lab Immunol. 1997;4:173–179. doi: 10.1128/cdli.4.2.173-179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paxton H, Bendele T. Effect of time, temperature, and anticoagulant on flow cytometry and hematological values. Ann N Y Acad Sci. 1993;677:440–443. doi: 10.1111/j.1749-6632.1993.tb38811.x. [DOI] [PubMed] [Google Scholar]
- 17.Paxton H, Pins M, Denton G, McGonigle A D, Meisner R S, Phair J P. Comparison of CD4 cell count by a simple enzyme-linked immunosorbent assay using a TRAx CD4 test kit and by flow cytometry and hematology. Clin Diagn Lab Immunol. 1995;2:104–114. doi: 10.1128/cdli.2.1.104-114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson G, Morgan L, Evans M, McDermott S, Pereira S, Wansbrough-Jones M, Griffin G. Effect of type of haematology analyser on CD4 count. Lancet. 1992;340:485. doi: 10.1016/0140-6736(92)91807-k. [DOI] [PubMed] [Google Scholar]