Abstract
Significance:
Mitochondria produce most of the cellular ATP through the process of oxidative phosphorylation. Energy metabolism in the mitochondria is associated with the production of reactive oxygen species (ROS). Excessive ROS production leads to oxidative stress and compromises cellular physiology. Energy metabolism in the mitochondria depends on nutrient flux and cellular metabolic needs, which are in turn connected with the feeding/fasting cycle. In animals, the feeding/fasting cycle is controlled by the circadian clock that generates 24-h rhythms in behavior, metabolism, and signaling.
Recent Advances:
Here, we discuss the role of the circadian clock and rhythms in mitochondria on ROS homeostasis. The circadian clock is involved in mitochondrial ROS production and detoxification through the control of nutrient flux and oxidation, uncoupling, antioxidant defense, and mitochondrial dynamics.
Critical Issues:
Little is known on the molecular mechanisms of circadian control of mitochondrial functions. The circadian clock regulates the expression and activity of mitochondrial metabolic and antioxidant enzymes. The regulation involves a direct transcriptional control by Circadian Locomotor Output Cycles Kaput/brain and muscle ARNT-like 1(CLOCK/BMAL1), nuclear factor erythroid-2-related factor 2 (NRF2) transcriptional network, and sirtuin-dependent posttranslational protein modifications.
Future Perspectives:
We hypothesize that the circadian clock orchestrates mitochondrial physiology to synchronize it with the feeding/fasting cycle. Circadian coordination of mitochondrial function couples energy metabolism with diets and contributes to antioxidant defense to prevent metabolic diseases and delay aging. Antioxid. Redox Signal. 37, 647–663.
Keywords: metabolism, oxidative stress, antioxidant defense, circadian rhythms, gene expression, caloric restriction, fasting, longevity
Introduction
Energy metabolism is central for cellular homeostasis. Cells generate energy by oxidizing nutrients: carbohydrates, amino acids, and lipids. Nutrient oxidation occurs within different cellular compartments, and the mitochondria play a primary role in the generation of energy-rich ATP molecules through the process of oxidative phosphorylation. Side products of oxidative phosphorylation are reactive oxygen species (ROS) such as superoxide anion, generated when high-energy electrons are transferred to molecular oxygen.
Peroxisome is another cellular organelle that oxidizes long-chain, branch-chain, and dicarboxylic fatty acids. Oxidation in the peroxisome is coupled with generation of hydrogen peroxide (H2O2), another major cellular ROS. Thus, energy production in cells is associated with the generation of ROS in different cellular compartments. ROS and reactive nitrogen species (RNS) are important signaling molecules (29). They regulate multiple cellular processes such as transcription, secretion, and proliferation. However, due to their high chemical reactivity, uncontrolled ROS production causes oxidative damage to DNA, lipids, and proteins. This oxidative stress compromises the functions of biological macromolecules, which in turn affects cellular physiology, leads to cellular damage, and ultimately induces cell death (29).
Mounting evidence connects oxidative stress with cardiovascular diseases, cancer, diabetes, neurodegeneration, and accelerated aging (12, 29, 42, 153). To prevent these unwanted events, cells developed a highly sophisticated network of proteins and cofactors, together known as the cellular antioxidant system (29, 39). ROS are detoxified through a chain of reactions catalyzed by different enzymes. The efficient detoxification of ROS would require coordinated expression and/or activity of these enzymes and their cofactors. It was proposed that one of the functions of the circadian system is the orchestration of cellular oxidative stress response (74, 98, 114).
The circadian system is a network of circadian clocks that are present in every tissue and cell in most organisms (21, 25, 67). Cellular molecular oscillators, formed as transcriptional/translational feedback loops, generate 24-h rhythms in gene expression and signaling (67, 101). Cellular rhythms integrate with organism rhythms in metabolism, physiology, and behavior. Circadian disruption in humans is associated with the development of cardiometabolic diseases, cancer, and neurodegeneration (45, 98, 124).
Interestingly, oxidative stress is a contributing factor for the same diseases. Several animal models of circadian clock deficiency display chronic oxidative stress and defective antioxidant defense (8, 72, 99, 117, 158). Reduced mitochondrial volume, diminished respiration rate, and increased oxidative damage were found in circadian clock mutants (4, 95, 99, 131, 162), which impacted their physiology. The circadian clock is implicated in coordinating the antioxidant defense through both transcriptional-dependent and transcriptional-independent mechanisms (48). The circadian clock regulates rhythms in the expression and activity of nuclear factor erythroid-2-related factor 2 (NRF2), which is a leucine zipper transcription factor and master regulator of antioxidant defense that drives the transcription of major antioxidant enzymes.
The circadian clock also regulates rhythmic production of melatonin, a recognized ROS scavenger (48). Transcription-independent circadian mechanisms of redox control also exist in red blood cells, and they are linked with rhythmic oxidation/reduction of peroxiredoxin (PRDX) proteins (30).
The role of the circadian clock in ROS detoxification is confirmed by multiple studies. Whether the circadian clock is involved in the control of ROS production is not fully resolved. Energy metabolism is linked with the feeding/fasting cycle and is under the control of the circadian clock (1, 49, 154, 155). Mitochondria explore several strategies to maintain ROS homeostasis. Here, we discuss the circadian clock as a master regulator of ROS and mitochondria physiology in the context of ROS.
Circadian Control of Mitochondrial ROS Production
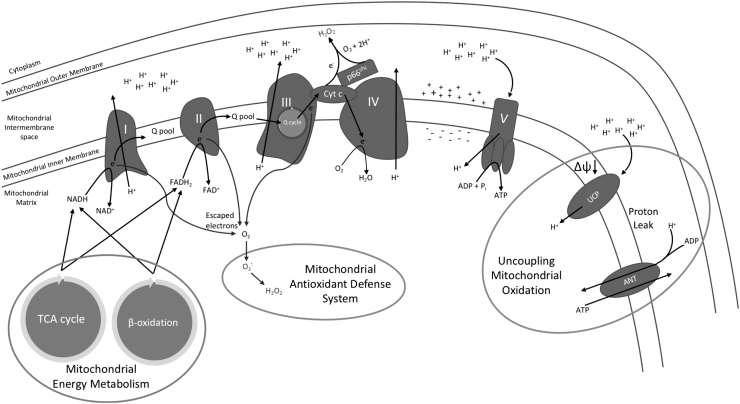
ROS are generated in several cellular compartments: in peroxisomes during oxidation of fatty acids and degradation of xenobiotics, at cell plasmatic membrane by associated dehydrogenases, but the main site of cellular ROS production is the mitochondrion. Mitochondrial energy metabolism is tightly linked with ROS production (Fig. 1). Mitochondria are capable of oxidizing different classes of nutrients: pyruvate, amino acids, and fatty acids, which generate most of the cellular ATP. Nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2) molecules generated through glycolysis, β-oxidation, and TCA cycle will donate high-energy electrons to the mitochondrial electron transport chain (ETC).
FIG. 1.
Mitochondrial energy production and management linked with ROS. In the mitochondrial matrix, high-energy electron carrier molecules, including NAD+ and FAD+, are reduced during processes of energy metabolism, including the TCA cycle and β-oxidation. These high-energy molecules travel to complex I and II to be oxidized, which results in the reduction of ubiquinone (Q). Reduced Q, QH2, will diffuse through the mitochondrial inner membrane to complex III for participation in the Q cycle, which oxidizes QH2 and reduces Cyt C. Reduced cytochrome C will transfer electrons to molecular oxygen in complex IV to produce water. Some of the reduced cytochrome C will be oxidized by p66 to produce H2O2. During the process of electron transfer from NADH and FADH2 through the electron transport chain to molecular oxygen, some electrons may be lost from the electron transport chain, which interact with molecular oxygen and produce superoxide anion, which is processed into H2O2 and then managed by the mitochondrial antioxidant defense system. Throughout the electron transfer chain, complexes I, III, and IV move protons from the matrix into the mitochondrial intermembrane space, which induces a chemical and electrical gradient that drives ATP synthase, complex V, and leads to the production of ATP while transferring protons down the gradient. In times of excess electrons being processed through the electron transfer chain, uncoupling of mitochondrial oxidation is necessary to alleviate production of ROS. This uncoupling process takes place through UCP along with ANT. Both proteins allow protons to leak back into the mitochondrial matrix, which reduces the magnitude of the chemical and electrical gradient and allows for electrons to move through the complexes with a lower chance of ROS production. ANT, adenine nucleotide translocator; Cyt C, cytochrome C; FADH2, flavin adenine dinucleotide; H2O2, hydrogen peroxide; NADH, nicotinamide adenine dinucleotide; ROS, reactive oxygen species; UCP, uncoupling proteins.
The ETC is composed of four multisubunit transmembrane protein complexes (I–IV) and two diffusive carriers, ubiquinone and cytochrome C. Electron transport through the ETC and the free energy released from this process drives proton pumping from the mitochondrial matrix to the intermembrane space that creates an electrical gradient (ΔΨm) as well as a chemical gradient (ΔpH). The proton gradient is used to produce ATP molecules through a transmembrane ATP synthase. The oxidative phosphorylation mechanism is not 100% efficient, and naturally, some electrons leak out of the ETC system.
The major sites for electron escape are Complexes I and III. Complex II can generate ROS in the presence of FADH2 (121). Complex IV is an endpoint of electron transfer, and whether complex IV is a direct source of ROS is unresolved. Escaped electrons interact with molecular oxygen and generate superoxide anion, from which other types of ROS such as H2O2 can be generated. High rates of electron transport create a high-proton motive force that causes the ETC to slow down because the protons are pumped against a stronger force. Slowing down the ETC results in electrons spending more time on complexes of the ETC, which increases their potential to escape. Mitochondria use different strategies to reduce electron escape from the ETC.
The transfer of electrons to ETC depends on NADH and FADH2 supply (Fig. 1). NADH and FADH2 are generated as a result of nutrient oxidation. The choice of the substrate for oxidation depends on the stage of the feeding/fasting cycle, with preferential carbohydrate oxidation during feeding and fatty acids during fasting (11). There are several recent reviews on circadian clock control of nutrient digestion and transport to the cell (45). The expression of enzymes involved in glycolysis, mitochondrial β-oxidation of fatty acids, and TCA cycle is highly rhythmic across the day and responds to the diets in a clock-dependent manner (88, 112, 115).
One of the potential mechanistic connections is the peroxisome proliferator-activated receptor (PPAR) network, which is a master regulator of fat oxidation and energy production. PPARs make up a family of transcription factors that include three isoforms, α, β/δ, and γ (17). These transcriptional factors belong to a larger superfamily of nuclear receptors that are activated upon binding their endogenous fatty acid ligands along with various industrial, pharmaceutical, and phytological chemicals (17). Activated PPARs heterodimerize with retinoid X receptor and bind PPAR responsive elements to regulate transcription of target genes (17).
The three highly homologous isoforms are differentially expressed among tissues and elicit various cellular functions. All PPAR isoforms across tissues have demonstrated circadian rhythmicity in their expression, while the alpha and gamma isoforms display direct interaction with core clock genes (13, 58, 166). It is hypothesized that PPARα and PPARγ bind a PPAR response element (PPRE) in the promoter of Bmal1 and Nr1d2 genes to regulate their transcription (17). Plus, the expression of components of the ETC might also be under clock control (107).
In agreement with this, diurnal regulation of mitochondrial respiration was blunted in mice lacking PER1/2 (106). Thus, the circadian clock tightly controls the rate of nutrient oxidation to guarantee that it is in balance with ATP production to minimize ROS generation.
Transfer of electrons through the ETC and the movement of protons are coupled with ATP synthesis, and most protons are transferred back to the mitochondrial matrix through ATP synthase (Fig. 1). Some protons can leak through the inner mitochondrial membrane without ATP production, which results in uncoupling and energy dissipated as heat. Mild uncoupling lowers the electrochemical potential, minimizes electron leak, and prevents ROS production (12, 82). Basal proton leakage occurs by diffusion of protons through the inner membrane (62a) and through adenine nucleotide translocase.
Facilitated process of uncoupling is mostly supported by uncoupling proteins (UCP). The mammalian UCP family consists of five proteins (UCP1–5). Members of the family have similar activities but different tissue distribution. UCPs are localized in the inner mitochondrial membrane and they catalyze the transport of protons across the membrane. UCP1 is expressed in brown adipose tissues, UCP2 is ubiquitously expressed in many tissues, and UCP3 is predominantly expressed in skeletal muscle. UCP4 and UCP5 are expressed predominantly in nervous tissues.
In addition, UCPs are linked with many diseases such as cancer, neurodegeneration, and chronic inflammation. UCP1 plays an important role in thermogenesis, but the data on UCP1 in ROS homeostasis are conflicting (27, 108, 132). The role of UCP2 in the regulation of mitochondrial ROS level is well documented. Increased expression of UCP2 is associated with reduced ROS production (78, 105, 145). UCP3 plays a similar role by inhibiting ROS production and oxidative stress in skeletal muscle (10) and in isolated mitochondria (147). UCP4 and UCP5 reduce oxidative stress in the neural system and might play some other roles.
The circadian clock regulates the expression and activity of UCPs. UCP1 is rhythmically expressed in brown adipose tissue and is directly regulated by the circadian transcriptional repressor Nr1D1 (also known as Rev-erb-α) (36). PER2 acts as a coactivator of PPARα transcriptional factor in FABP3/fatty acid-dependent activation of UCP1 (15). Brain and muscle ARNT-like 1 (BMAL1) regulates UCP2 expression and uncoupling in β cells (76) and the heart (69). UCP3 expression oscillates in the skeletal muscle in a circadian manner (87).
The expression of UCPs is impaired in circadian clock mutants, which further supports the role of the clock in their regulation. Clock-dependent rhythmic expression of UCPs contributes to the circadian control of thermogenesis and other metabolic functions. Whether rhythms in UCP expression contribute to circadian ROS production in mitochondria is unknown. Such regulatory mechanisms are possible, but the effect might be tissue specific and needs to be investigated.
ETC-associated ROS are side products of oxidative phosphorylation. Mitochondria also have active and highly controlled mechanisms of ROS generation, probably for regulatory purposes. Recent findings have shown that activated p66shc is responsible for ∼30% of ROS produced in the mitochondria (39). Activated p66shc directs H2O2 production by oxidizing reduced cytochrome C and catalyzing the reduction of O2 to H2O2 with electrons from the electron transfer chain (38, 39). ETC-generated ROS are in the mitochondrial matrix; in contrast to that, p66 generates ROS in the intermembrane space, which allows ROS to leak into the cytoplasm (39). Increased p66shc activity leads to mitochondrial H2O2 leakage to the cytoplasm, which induces apoptosis and may contribute to mitochondrial fusion or mitophagy (38, 39, 116).
Deficiency in p66shc reduces mitochondrial H2O2 production, stabilizes mitochondrial dynamics, and increases longevity (39, 90, 116) Furthermore, mice lacking p66shc display alterations in the hepatic circadian transcriptome, along with reduced levels of NAD+ and ratio of oxidized to reduced NAD, which may be correlated to decreased expression of nicotinamide phosphoribosyltransferase (NAMPT) (116). p66shc gene contains an E-box element in its promoter, indicating potential transcriptional control by the core clock machinery (116).
p66shc expression is rhythmically expressed in the suprachiasmatic nucleus and liver and is critical for maintaining redox control of Circadian Locomotor Output Cycles Kaput (CLOCK) cysteinyl thiols (116). It has also been shown to be a critical component for maintaining normal circadian rhythms, which implicates it as a significant enzyme for managing mitochondrial H2O2 in regulating gene expression, metabolic homeostasis, and behavior.
Circadian Control of Mitochondrial Antioxidant Network
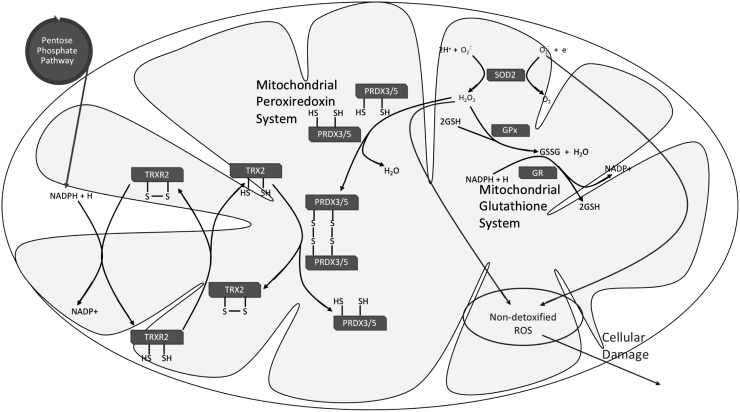
Oxidative stress occurs when the antioxidant system is not sufficient to match ROS production (118, 134). Cells keep their oxidative stress status in check by controlling the rate of ROS production and accumulation, along with the scavenging activity of antioxidants. The mitochondrion being a major site for ROS production also has its antioxidant system, which comprises superoxide dismutase (SOD), glutathione (GSH), and the PRDX-thioredoxin system (Fig. 2). The SOD proteins are the first line of defense against superoxide radicals (127).
FIG. 2.
Mitochondrial antioxidant system. The mitochondrial antioxidant defense comprises SOD, glutathione, and the PRDX system. SOD2 functions as the first line of defense against mitochondrial ROS by catalyzing the dismutation of superoxide (O2−) to H2O2 or molecular oxygen. The reduced form of GSH is used for the reduction of H2O2 to H2O, and in turn glutathione is oxidized to GSSG and needs to be converted back to its reduced state to maintain the cycle. GPx catalyzes the GSH-dependent reduction of H2O2, while GR catalyzes the reduction of GSSG to GSH in a reaction that requires NADPH. Another route for H2O2 reduction is the PRDX system. PRDX3/5 in their reduced state is used for the reduction of H2O2 and this is followed by the subsequent oxidation of PRDX3/5. Oxidized PRDX3/5 can be recycled back to its reduced state by accepting hydrogen from reduced thioredoxin TRX2. Mitochondrial TRXR2 is needed for maintaining reduced levels of TRX2 and this reaction requires NADPH. GPx, glutathione peroxidase; GR, glutathione reductase; GSH, glutathione; GSSG, oxidized glutathione; PRDX, peroxiredoxin; SOD, superoxide dismutase; TRX2, thioredoxin-2; TRXR2, thioredoxin reductase 2.
There are three different SODs, SOD1, SOD2, and SOD3, located in different cellular compartments (33, 169). SOD2 is also referred to as the manganese-dependent SOD, and it is predominantly localized in the mitochondria. The mitochondrial pool of superoxide is mostly generated during the process of electron transfer during oxidative phosphorylation. This radical is very reactive and high concentrations have deleterious effects on the cell. SODs catalyze the dismutation of superoxide (O2−) to H2O2 (33, 114, 127, 169). H2O2 produced is further detoxified either via the glutathione or PRDX system.
Glutathione is synthesized in the cytosol and can be transported across the mitochondrial inner membrane. It is a tripeptide that is highly studied for its antioxidant attributes. Various cellular compartments maintain varying levels of glutathione, and the mitochondria comprise about 10%–15% of cellular glutathione content (84, 127). Most of the mitochondrial glutathione is maintained in its reduced form as GSH, which is needed for the detoxification of H2O2 (84, 97).
The glutathione system consists of two enzymes, glutathione peroxidase (GPx) and glutathione reductase (GR). GPx is a selenium containing enzyme that catalyzes the reduction of H2O2 to water (H2O), and this process utilizes GSH. Following the reaction, GSH is oxidized to GSSG. In turn, GSSG can be reduced back to GSH via the GR enzyme. The reduction of GSSG by GR requires NADPH (84, 97, 127) This is important for recycling glutathione back to its reduced state, making it available for another round of H2O2 reduction.
While this is out of the scope of the current review, we want to mention that glutathione-S-transferase (GST), the enzyme that conjugates glutathione to various electrophilic compounds, shows significant circadian oscillation in mammals (24, 59, 150), plants (34, 35), and flies (75). Rhythms in GST activity contribute to circadian variability in detoxification of xenobiotics, toxins, and pharmacological drugs.
PRDX is a thiol-dependent peroxidase, which confers another route for the detoxification of H2O2 to H2O. In this process, PRDX becomes oxidized and needs to be recycled back to its reduced state for another cycle of H2O2 reduction. To do this, the system is coupled to thioredoxin. Thioredoxin in its reduced state serves as a hydrogen donor used for the reduction of oxidized PRDX, and this is followed by the subsequent oxidation of thioredoxin (127). Conversion of thioredoxin back to its reduced state is catalyzed by the NADPH-dependent thioredoxin reductase, and this process is important for the continuation of the PRDX scavenging cycle (127).
Excessive ROS production may have detrimental effects on cell physiology by damaging biological macromolecules, which compromises cellular functions and could lead to cellular death.
The cycling of fuel substrates and the level of oxidative phosphorylation occur across the day. Therefore, it is surprising that the antioxidant defense system is highly rhythmic. Daily oscillations of antioxidant enzymes and the redox ratio of scavenging molecules occur in both the cytoplasm and mitochondria (117, 119). These rhythms are disrupted in circadian clock mutants, and clock deficiency is associated with oxidative stress.
Circadian transcriptional factor BMAL1 is central for circadian antioxidant defense, and mice deficient for BMAL1 represent the most striking example of oxidative stress upon circadian disruption. Total Bmal1−/− mice have increased ROS level in the liver (72). Total and neuron-specific Bmal1−/− mice demonstrate synaptic degeneration and neuropathology most likely through increased oxidative stress (99). Bmal1−/− fibroblasts also have an increased ROS level, which is associated with senescence (65).
β cell-specific BMAL1 deficiency leads to accumulation of ROS and mitochondrial uncoupling (76). Circadian regulation of redox control is conserved in Drosophila (74), but not every knockout of the circadian clock is associated with increased ROS. Per and Tim deficiency in Drosophila males extends the life span, increases uncoupling of respiration, and lowers intestinal ROS level (152).
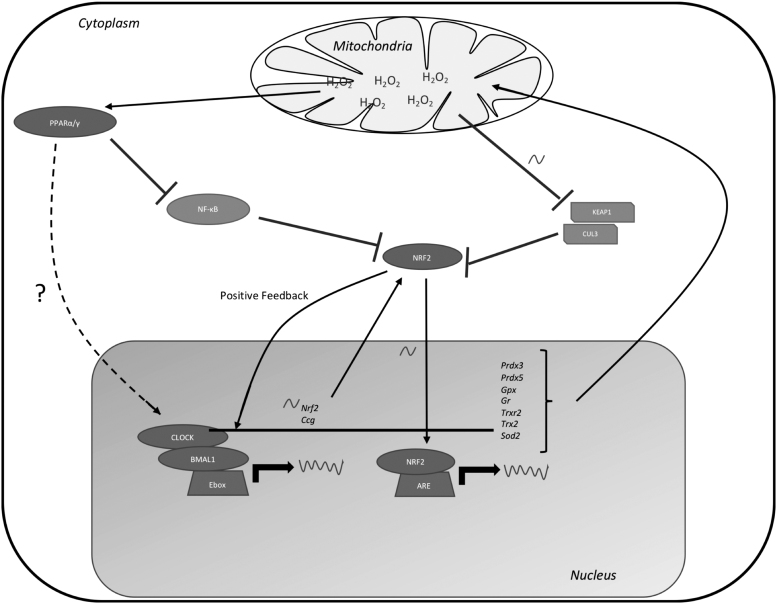
The expression of many antioxidant enzymes is regulated by the clock directly through clock responsive elements or indirectly through antioxidant response elements (ARE) and retinoid response elements in their promoters (Fig. 3). NRF2 plays one of the central roles in the circadian antioxidant defense. NRF2 regulates the transcription of genes that manage mitochondrial oxidative stress and redox status (128, 159) by binding to ARE in their promoters (23, 159).
FIG. 3.
Circadian interaction with transcriptional regulation of antioxidant defense system. Core clock components drive the rhythmic expression of Nrf2. Mitochondrial H2O2 that escapes the mitochondria rhythmically inhibits KEAP1, which allows NRF2 to translocate into the nucleus and regulate expression of mitochondrial antioxidant defense enzymes and pentose phosphate pathway enzymes. NRF2 interacts with the core clock through positive feedback to regulate its own expression. Mitochondrial H2O2 drives PPAR nuclear receptors to inhibit activity of NF-κB and is speculated to promote activity of core clock transcriptional regulation for management of mitochondrial oxidative stress enzymes. KEAP1, Kelch-like ECH-associated protein 1; NF-κB, nuclear factor-κB; NRF2, nuclear factor erythroid-2-related factor 2; PPAR, peroxisome proliferator-activated receptor.
There are over 250 genes with ARE in their promoters (23), including antioxidant enzymes and enzymes involved in the homeostasis of ROS scavengers such as glutathione, NADH, and NADPH (28, 44, 54, 94, 117, 119, 126, 140, 163). NRF2 activity, localization, and expression are directly impacted by the oxidative state of the cell through interactions with Kelch-like ECH-associated protein 1 (KEAP1) (60, 68, 142). KEAP1 is a negative regulator of NRF2 nuclear translocation by promoting its ubiquitination through CULLIN E3 ligase, which leads to the subsequent proteasomal degradation of NRF2 (60, 142).
Daily oscillations in ROS, RNS, and electrophiles create oscillatory interactions with reactive cysteine residues of KEAP1, allowing for circadian timekeeping of NRF2 activity and degradation (116, 159, 165). Plus, the positive arm of the circadian system holds transcriptional control over NRF2 through an E-box element in its promoter (141, 143). NRF2 expression and activity are regulated by the clock in several mouse models such as lung fibrosis, neurodegeneration, and human lens epithelial cells, which suggest that the NRF2/clock interaction exists in different tissues and warrants further study (19, 99, 117).
Above in the Circadian Control of Mitochondrial ROS Production section, we have already discussed the PPAR-clock cross talk. PPAR transcriptional factor family is also implicated in antioxidant defense. Several antioxidant enzymes such are SODs, catalase, and Gpx-3 are under PPAR transcriptional control through PPREs in their promoters (57, 149). Treatment of cells with the PPARγ agonist increases the expression some antioxidant genes such as Sod2 and Gpx-3, while the PPARα agonist had an antioxidant and antifibrotic effect in mice (26, 40). PPARγ exerts antioxidant effects through suppressing nuclear factor-κB and allowing ROS to be depleted and antioxidant enzymes to be promoted (3, 113).
Interestingly, several reports have associated PPAR with NRF2 in the management of mitochondrial oxidative stress through the context of metabolic disorders and drug-induced injury (81).
Circadian Clock and Mitochondrial Sirtuins
Sirtuins (SIRTs) are members of class III histone deacetylases (100). They play an important role in the regulation of protein function by regulating their posttranslational modifications such as acetylation, malonylation, succinylation, and glutarylation. There are currently seven known SIRTs (SIRT1–7) that have been identified in mammals, and they are distributed across various subcellular compartments (89). Localized in the nucleus are SIRT1, SIRT6, and SIRT7. SIRT2 is situated mostly in the cytoplasm, but can also be found in the nucleus. While SIRT3, SIRT4, and SIRT5 are in the mitochondria (100).
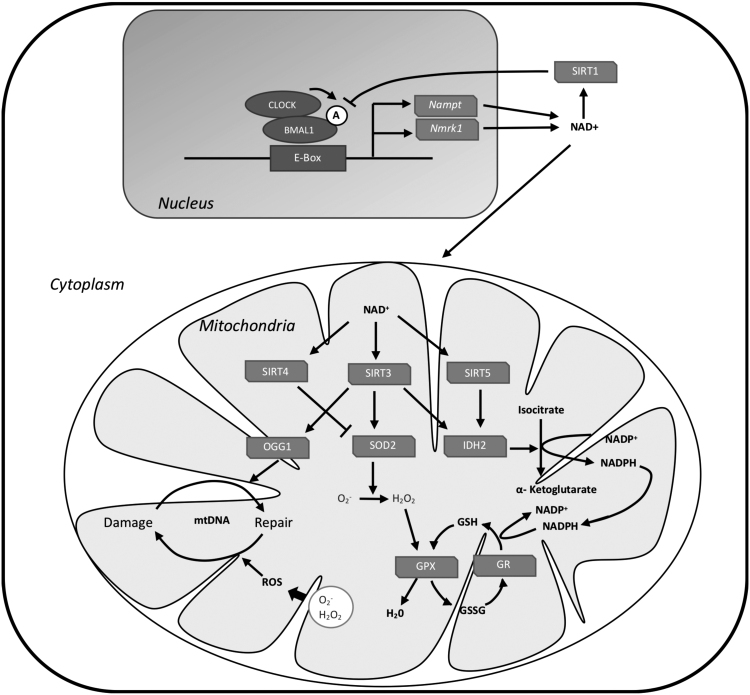
Activity of SIRTs relies on the availability of NAD+ for their function. NAD+ is synthesized via different routes (Fig. 4); de novo biosynthesis from tryptophan, the deamidated pathway from nicotinic acid, and through the amidated route (96, 138). The amidated route occurs either through nicotinamide riboside kinase 1 (NRK1), which synthesizes NAD+ from dietary supplies of nicotinamide ribose, or through NAMPT, which synthesizes NAD+ by recycling nicotinamide (86). Most of the mammalian NAD+ is synthesized via the amidated route (96). BMAL1 binds to the Nmrk1 gene (which codes for NRK1 protein) and promotes its expression and rhythmicity. The rhythms in the expression of Nmrk1 are absent in Bmal1−/− and Cry1,2−/− mice (73, 86). CLOCK and BMAL1 dimerize and bind to E-box elements in the Nampt gene and promote its transcription (Fig. 4).
FIG. 4.
Regulation of mitochondrial SIRTs by the circadian clock. CLOCK and BMAL1 heterodimerize and bind to E-box elements to promote the transcription of Nampt gene. This transcriptional process is favored by CLOCK-dependent acetylation of BMAL1. NAMPT is an important enzyme in the salvage route in NAD+ biosynthesis. Another route for NAD+ biosynthesis is via NRK1 enzyme, which produces NAD+ from dietary supplies of nicotinamide ribose. This process is also under circadian control. BMAL1 binds to Nmrk1 gene and promotes its expression, but it is not clear if it has an E-box element. Circadian rhythms in NAD+ are responsible for circadian rhythms in the activity of SIRTs. SIRT3, SIRT4, and SIRT5 are localized in mitochondria and they contribute to mitochondrial antioxidant defense. SIRT3 deacetylates SOD2, IDH2, and OGG1. IDH2 produces NADPH used to reduce oxidized glutathione, and OGG1 is a DNA repair enzyme that protects mitochondrial DNA from ROS damage. SIRT5 contributes to antioxidant defense via IDH2 desuccinylation. SIRT4 may have an opposing effect on antioxidant defense; it prevents the deacetylation of SOD2 by SIRT3. Feedback regulation occurs between SIRTs and the circadian clock via SIRT1-dependent deacetylation of BMAL1. BMAL1, brain and muscle ARNT-like 1; CLOCK, Circadian Locomotor Output Cycles Kaput; IDH2, isocitrate dehydrogenase 2; NAMPT, nicotinamide phosphoribosyltransferase; NRK1, nicotinamide riboside kinase 1; OGG1, 8-oxoguaninine-DNA glycosylase 1; SIRT, sirtuin.
Both the mRNA and protein expressions of the NAD+ salvage enzyme NAMPT are rhythmic and the rhythms in the expression are compromised in circadian clock mutants (103, 122). In agreement with changes in NAMPT expression in circadian clock mutant mice, total and mitochondrial levels of NAD+ are affected (86, 115, 122). There is feedback interaction between SIRTs and the circadian clock. CLOCK acts as a histone acetyltransferase and directly acetylates BMAL1, and this is important for promoting the transcriptional activity (51). SIRT1, which is mainly localized in the nucleus, regulates this process by deacetylating BMAL1 and PER2 (5, 102).
SIRT3 is the major mitochondrial protein deacetylase (Fig. 4). Acetylation is receiving more attention as a key posttranslational modification that can regulate various mitochondrial processes, including fatty acid oxidation and the TCA cycle (50, 52, 123). Studies on SIRT3 knockout mice reveal increased global liver mitochondrial protein acetylation, decreased β-oxidation, decreased ketogenesis, and increased oxidative stress (50, 118, 133). In addition, SIRT3 has been shown to confer cell-protective properties against ROS toxicity by boosting the antioxidant defense and by protecting against mitochondrial damage (80).
A notable mitochondrial protein that is regulated by SIRT3 through deacetylation is SOD2. Decreased activity of SIRT3 leads to increased acetylation and decreased activity of SOD2 (120). Some other key acetylated mitochondrial proteins are ornithine-transcarbamoylase (OTC) and isocitrate dehydrogenase 2 (IDH2) (47). IDH2 contributes to the production of NADPH by catalyzing the oxidation of isocitrate to α-ketoglutarate. The production of NADPH is required for the reduction of oxidized glutathione and is thereby needed for antioxidant defense (171). SIRT3 deacetylates and promotes IDH2 activity (136, 168). In addition, SIRT3 also deacetylates 8-oxoguaninine-DNA glycosylase 1 (OGG1), which catalyzes the base excision repair of mitochondrial DNA and protects it from ROS toxicity (18, 66).
The circadian clock regulates SIRT3 activity via the availability of NAD+ (115) and is in agreement with rhythmic acetylation of SOD2 (86). SIRT3 activity is decreased in Bmal1−/− mice, and this leads to an increase in mitochondrial protein acetylation, including SOD2, OTC, and IDH2 (115). SIRT4 and SIRT5 are two other members of the SIRT family that are localized in the mitochondria and play an important role in mitochondrial physiology. While it is not clear if the circadian clock interacts with SIRT5 or SIRT4, it remains plausible considering that the activities of SIRT deacetylases are regulated by NAD+ availability, which is under the control of the circadian clock.
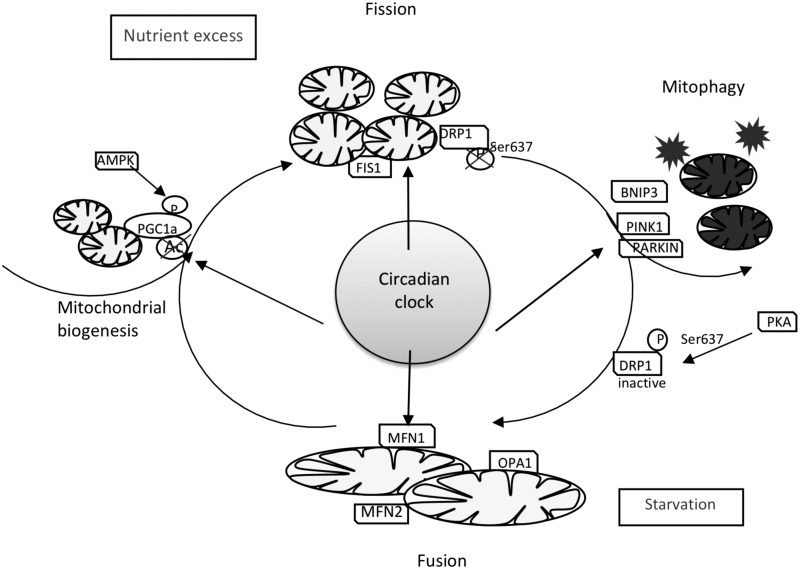
Circadian Regulation of Mitochondrial Dynamics
Mitochondria are dynamic organelles that respond to various challenges by changing their shape through fusion/fission processes and their numbers through mitophagy and mitogenesis. Balance in these processes has a dramatic effect on cellular homeostasis (Fig. 5). Under stress, fusion of mitochondria occurs, which increases their oxidative capacity. Fusion of damaged and undamaged mitochondria compensates for loss of function (167) and induces a prosurvival response to stress (148). Mitochondrial fission is essential for cell growth as well as elimination and replacement of damaged organelles and components. Fragmentation of mitochondria networks is associated with apoptosis (53, 139).
FIG. 5.
Circadian clock control of mitochondrial dynamics. Feeding/fasting cycle creates temporal oscillation of nutrient flux to mitochondria. Both nutrient excess and nutrient starvation are a challenge, and to respond to it, mitochondria engage mitogenesis/mitophagy and fission/fusion processes. The circadian clock contributes to these processes to maintain healthy mitochondria. The circadian clock regulates the expression of several key proteins in mitochondrial fission/fusion such as DRP1, FIS1, and MFN1. Circadian control of mitophagy occurs through regulation of PINK1 and BNIP3 expression. Cross talk between the circadian clock and PGC1a pathways is involved in mitochondrial biogenesis. BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; DRP1, dynamin-related protein 1; FIS1, fission 1; MFN1, mitofusin 1; PINK1, PTEN-induced kinase 1.
Nutrient challenges can affect mitochondrial dynamics: nutrient excess causes mitochondria fission and creates fragmented mitochondria. Organismal fasting or nutrient depletion in cell culture induces mitochondria fusion, which is associated with increased capacity for ATP synthesis (79, 157). Mitochondrial fission is controlled by dynamin-related protein 1 (DRP1), a cytosolic GTPase that is recruited to the mitochondria by several proteins, including fission 1 (FIS1) (93). Mff1, Mief1, and MiD49/51 also were proposed to regulate DRP1 recruitment and function (110, 111, 170).
The key players in fusion machinery are mitofusins 1 and 2, located on the outer mitochondrial membrane, and optic atrophy 1 (OPA1), which resides in the inner membrane or intermembrane space (137). OPA1 was also proposed to regulate mitochondria cristae structure (2, 31).
Circadian oscillations of mitochondrial morphology were noticed about 40 years ago (151). It was hypothesized that the circadian clock regulates mitochondrial dynamics and helps prepare cells for day/night feeding/fasting cycles, while clock disruption is associated with an inability to adapt to different nutrient conditions (Fig. 5).
The expression of several fission/mitophagy genes in the liver is regulated in response to feeding, and this response was disrupted in the liver of liver-specific Bmal1−/− mice. In agreement with that, the mitochondrial dynamics was significantly impaired in this circadian mutant, which led to dysfunctional mitochondria (61). Schmitt et al. demonstrated that clock regulates mitochondrial morphology through DRP1 (130). DRP1 is a key mediator of mitochondria fission, and its activity is regulated by phosphorylation. DRP1 phosphorylation by PKA inhibits its activity and leads to the formation of an elongated mitochondria (43, 109). Calcineurin dephosphorylates DRP1 and activates mitochondrial fission (14). The activity of calcineurin is under circadian control (55).
Pharmacological inhibition of calcineurin in cells blocks circadian oscillations in DRP1 phosphorylation and mitochondrial rhythms (130). CLOCK can bind to Drp1 mRNA, which affects its stability and regulates mitochondrial dynamics and function (164). Circadian control of mitochondrial dynamics through expression of fission protein FIS1 was abolished in Bmal1−/− mice, and liver overexpression of Fis1 normalized mitochondria function and reduced oxidative damage (61).
PGC1α and PGC1β are master regulators of mitochondrial biogenesis and energy production through activation of several transcriptional factors, such as NRF1 and 2, PPARs, and ERRs (64, 156), while PINK1, PARKIN, and BNIP3 are involved in removing damaged mitochondria through mitophagy (41). PGC1α activity is regulated by many proteins that are connected to the energy status of the cell such as AMPK, SIRT1, MAPK, CaMKIV, and PKC.
In agreement with that, PGC1α phosphorylation and acetylation are affected by the feeding/fasting cycle (37, 41, 62). PGC1α expression is also regulated by diet such as caloric restriction (CR) (20). Mitochondrial pathology observed in circadian clock mutants was linked with disrupted expression of PGC1α. Interestingly, PGC1α regulates circadian clock gene expression, thus providing a feedback to the clock from mitochondria. Several pieces of evidence connect the circadian clock and mitophagy. BMAL1 is involved in quality control of mitochondria through mitophagy regulation. Expression of PINK1 is rhythmic, and it is the direct target of BMAL1 (71, 125). In addition, BMAL1 promotes the expression of Bnip3 gene by binding to an E-box element in its promoter (77).
ROS Signals to Clock in Feedback Regulation
ROS, RNS, and electrophilic molecules are recognized as signaling molecules that lead to changes in cellular redox status (29). Mitochondria-produced ROS directly oxidize numerous cellular proteins such as receptors, kinases, and phosphatases, which impacts their biological functions, and, ultimately, modulates signal transduction pathways, gene expression, and metabolism (94, 160, 161). Cellular circadian oscillators are also a target for ROS (Fig. 6). Cellular redox status provides a rhythmic signal for maintaining circadian regulation of metabolic homeostasis (56, 91, 104, 116). Increased ROS lead to a decrease in the ratio of NADP+/NADPH to manage the oxidative environment, and in turn provides a regulatory component for the circadian clock (126, 163).
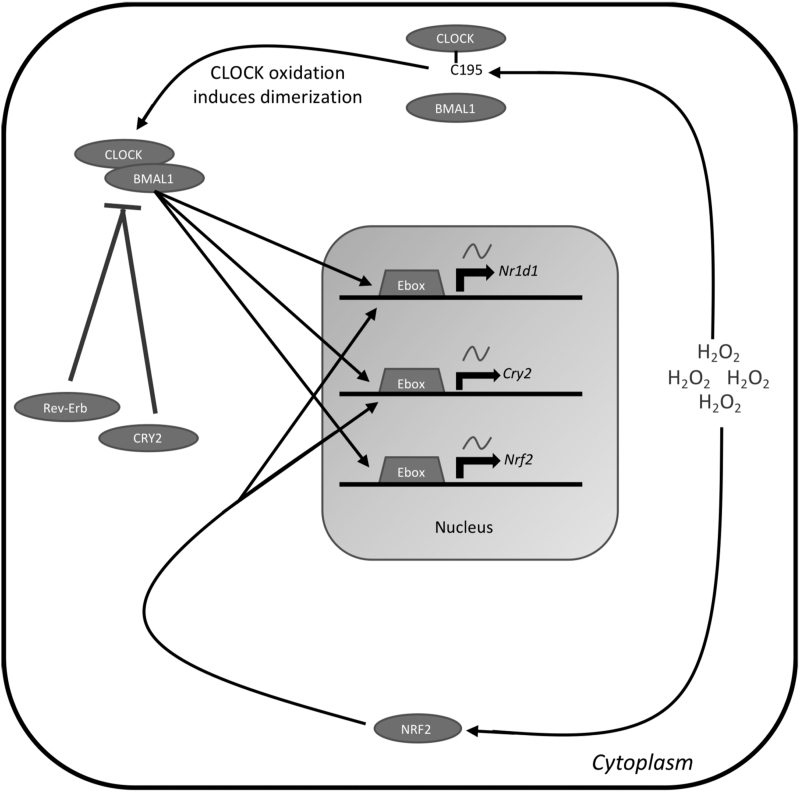
FIG. 6.
ROS feedback to the clock. Cytoplasmic ROS act as a posttranslational modifier and signaling molecule, directing the cell to increase antioxidant defense. H2O2 in the cytoplasm will interact directly with CLOCK and oxidize cysteine195. This oxidation event promotes dimerization between CLOCK and BMAL1 and their subsequent nuclear translocation. Cytoplasmic H2O2 will also stimulate NRF2 nuclear translocation and activity. BMAL1 targets NRF2, which will promote further antioxidant defense. NRF2 targets Cry2 and Nr1d1, which will act as a negative feedback mechanism to inhibit BMAL1 transcriptional activity and reduce the antioxidant response.
Overwhelming the mitochondrial antioxidant system with increased oxidation causes the clock to be shifted and reset, and may even be attenuated depending on how long the oxidative stress persists (143, 144). This indicates a threshold for which oxidative stress uncouples the circadian system from redox homeostasis and coordinates prosurvival signals and gene expression through stress-resistant and stress-responsive pathways, including heat shock factors and proteins for maintaining protein homeostasis (143, 144). Organismal oxidative stress is critical for maintaining circadian function and timing, and CLOCK and BMAL1 are heavily impacted by the levels of ROS and electrophiles (116).
Protein interaction with H2O2 is an important rhythmic modification that regulates signal transduction and enzymatic activity. Specifically, oxidation of cysteine195 by H2O2 in the PAS domain of CLOCK promotes CLOCK interaction with BMAL1, DNA binding, and transcriptional control of downstream targets. H2O2 targeting of CLOCK provides a direct coupling between redox signaling and the circadian clock (116). The production of H2O2 regulates core clock genes, including retinoic acid-related orphan receptor (ROR) and NR1D1/2, through a PRDX/STAT3 pathway (63). Interestingly, it is well documented that a robust circadian clock increases longevity, however, it may be speculated that increased longevity can be attained with a disrupted circadian clock if oxidative stress is reduced (9, 116).
Transcriptional factor NRF2 mediates the clock effects on antioxidant defense as discussed above in the Circadian Control of Mitochondrial Antioxidant Network section. Recent data suggest that ROS can signal to the clock through the NRF2-dependent pathway. Indeed, NRF2 interacts with the clock by binding the promoters of Cry2 and Nr1d1 genes, which induces their expression (159).
Increased expression of Cry2 drives negative feedback that inhibits its own gene expression along with the expression of Nrf2 and other core clock genes (159). Nr1d1 induction by NRF2 will interact with ROR elements in the Bmal1 promoter to inhibit the production of Bmal1 and halt the transcription of Nrf2 and its target genes. This interaction between the circadian clock and NRF2 creates a direct link between regulation of the circadian system and redox homeostasis (7).
Diets, Feeding/Fasting Cycle, and Circadian Mitochondria
Diets have a strong impact on metabolism and physiology. Some diets such as high-glucose or high-fat diets disrupt metabolism and provoke the development of pathologies such as cardiovascular diseases, metabolic syndrome, and diabetes (7). Other diets such as CR or diets that explore periodic fasting might improve metabolism, reduce the rate of diseases, and increase longevity (22, 92). Oxidative stress contributes to development of the above diseases, and diets are known to impact ROS homeostasis, for example, a high-fat diet is associated with increased oxidative stress, while CR is associated with reduced oxidative stress. Diets also significantly impact circadian clock and rhythms in the liver and other tissues.
Indeed, there is a large body of evidence that circadian rhythms are significantly reprogrammed in response to various diets (46, 83, 129, 135, 146). Some genes become rhythmic, while others lose rhythmicity. Interestingly, about the same fraction of genes, 10%–20%, oscillates on different diets; the overlap of rhythmic transcripts between different diets is less than 25%. The expression of clock genes is also affected by diet. Diets that have a negative impact on health dampen the rhythms (70), and diets that improve metabolism enhance the rhythms (6, 16). This leads to the hypothesis that positive or negative metabolic effects of diets are linked with the effect of diet on circadian clock and rhythms. Diets frequently impact the pattern of food consumption, and therefore, feeding/fasting rhythms, which are linked with rhythms in energy metabolism (7, 32).
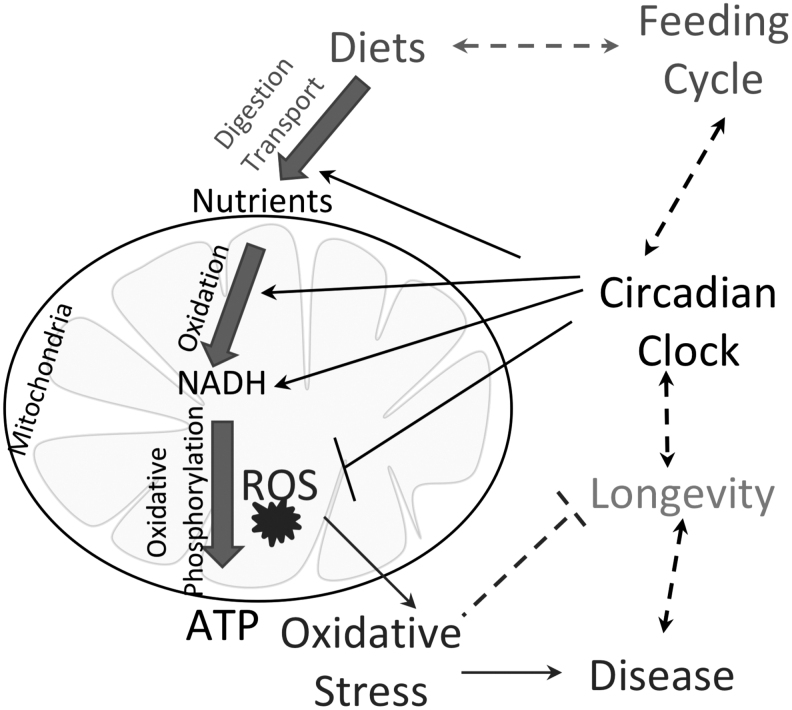
As discussed above, mitochondria adjust their physiology responding to nutrient flux and metabolic needs to balance ATP and ROS production. We hypothesize that reprogramming of circadian rhythms in response to various diets plays an important role in mitochondrial antioxidant defense by orchestrating the expression of various proteins in mitochondria to synchronize their activities with rhythms in ROS generation (Fig. 7). Periodicity of the feeding/fasting cycle will entrain rhythms in antioxidant defense in a manner similar with other circadian metabolic entrainments. Some of the metabolic benefits of periodic fasting based diets might be due to this entrainment.
FIG. 7.
Circadian coupling of mitochondria and ROS homeostasis. Daily feeding/fasting cycle dictates nutrient flux to mitochondria, where the nutrient oxidation is coupled with production of energy-rich ATP molecules in the process of oxidative phosphorylation. Due to electron leakage, mitochondrial energy production is associated with ROS generation. Excessive ROS lead to oxidative damage and contribute to diseases, and impact longevity. Almost every stage of the nutrient oxidation/energy production chain and mitochondrial antioxidant defense is under circadian control. The circadian clock is intertwined with daily rhythms, including the feeding cycle. Due to that, the circadian clock is capable of coordinating ROS homeostasis with organism daily activity and optimizes metabolism to reduce the risk of disease and delay aging.
Thus, a robust circadian clock helps to prevent oxidative stress and damage to cellular structures and ultimately contributes to health and longevity. Feeding during the wrong time, for example, during the time of rest or expected fasting will result in ROS production when clock-controlled antioxidant defense is not ready to manage it. Consequently, the antioxidant defense will be compromised resulting in oxidative damage. Circadian disruption is associated with disrupted feeding rhythms, and increased oxidative stress in circadian clock mutants is in line with the model. However, it is important to keep in mind that some of the effects of the circadian mutations might be rhythm independent.
Conclusions
Mitochondria have a broad range of functions that include energy production, lipid metabolism, calcium homeostasis, generation and detoxification of ROS, and the initiation of apoptosis. Maintenance of proper mitochondrial physiology is essential for cellular metabolism and organismal survival. ROS are produced as side products of mitochondrial energy metabolism, mostly in the electron transfer chain. Excessive ROS production might have detrimental effects on cell physiology by damaging biological macromolecules, which compromise cellular function and may result in cell death. Therefore, the mitochondrial antioxidant defense system is critically important for cell life cycle and survival. The circadian clock is integrated with mitochondrial physiology and contributes to ROS homeostasis by orchestrating mitochondrial ROS production and antioxidant defense.
Abbreviations Used
- AMPK
AMP-activated protein kinase
- ANT
adenine nucleotide translocator
- ARE
antioxidant response elements
- BMAL1
brain and muscle ARNT-like 1
- BNIP3
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3
- CaMKIV
calcium/calmodulin-dependent protein kinase IV
- CLOCK
Circadian Locomotor Output Cycles Kaput
- CR
caloric restriction
- CRY1
cryptochrome circadian regulator 1
- CRY2
cryptochrome circadian regulator 2
- Cyt C
cytochrome C
- DRP1
dynamin-related protein 1
- ETC
electron transport chain
- FABP3
fatty acid binding protein 3
- FADH2
flavin adenine dinucleotide
- FIS1
mitochondrial fission 1 protein
- GPx
glutathione peroxidase
- GR
glutathione reductase
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- GST
glutathione-S-transferase
- H2O2
hydrogen peroxide
- IDH2
isocitrate dehydrogenase 2
- KEAP1
Kelch-like ECH-associated protein 1
- MAPK
mitogen-activated protein kinase
- MFF1
mitochondrial fission factor 1
- MFN
mitofusin
- NADH
nicotinamide adenine dinucleotide
- NAMPT
nicotinamide phosphoribosyltransferase
- NF-κB
nuclear factor-κB
- Nr1d1
nuclear receptor subfamily 1 group D member 1
- Nr1d2
nuclear receptor subfamily 1 group D member 2
- NRF1
nuclear factor erythroid 2-related factor 1
- NRF2
nuclear factor erythroid-2-related factor 2
- NRK1
nicotinamide riboside kinase 1
- OGG1
8-oxoguaninine-DNA glycosylase 1
- OPA1
optic atrophy 1; OPA1 mitochondrial dynamin-like GTPase
- OTC
ornithine-transcarbamylase
- Per1
period circadian regulator 1
- Per2
period circadian regulator 2
- PGC1a
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PINK1
PTEN-induced kinase 1
- PKA
protein kinase A
- PKC
protein kinase C
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response element
- PRDX
peroxiredoxin
- RNS
reactive nitrogen species
- ROR
retinoic acid-related orphan receptor
- ROS
reactive oxygen species
- SIRT
sirtuin
- SOD
superoxide dismutase
- TRX2
thioredoxin-2
- TRXR
thioredoxin reductase
- UCP
uncoupling proteins
Authors' Contributions
All authors conceived and designed the study. All authors wrote the article. All authors contributed to reading, revision, and approval of the article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the U.S. National Institutes of Health/National Institute on Aging Grant R01AG039547 and funds from the Center for Gene Regulation in Health and Disease (Cleveland State University) to Roman V. Kondratov.
References
- 1. Acosta-Rodríguez VA, de Groot MHM, Rijo-Ferreira F, Green CB, and Takahashi JS. Mice under caloric restriction self-impose a temporal restriction of food intake as revealed by an automated feeder system. Cell Metab 26: 267–277.e2, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alavi MV and Fuhrmann N. Dominant optic atrophy, OPA1, and mitochondrial quality control: understanding mitochondrial network dynamics. Mol Neurodegener 8: 1–11, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alqahtani S and Mahmoud AM. Gamma-glutamylcysteine ethyl ester protects against cyclophosphamide-induced liver injury and hematologic alterations via upregulation of PPARγ and attenuation of oxidative stress, inflammation, and apoptosis. Oxid Med Cell Longev 2016: 401609, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, and Esser KA. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A 107: 19090–19095, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, and Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328, 2008. [DOI] [PubMed] [Google Scholar]
- 6. Astafev AA, Patel SA, and Kondratov RV. Calorie restriction effects on circadian rhythms in gene expression are sex dependent. Sci Rep 7: 9716, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bass J. Circadian topology of metabolism. Nature 491: 348–356, 2012. [DOI] [PubMed] [Google Scholar]
- 8. Beaver LM, Klichko VI, Chow ES, Kotwica-Rolinska J, Williamson M, Orr WC, Radyuk SN, and Giebultowicz JM. Circadian regulation of glutathione levels and biosynthesis in Drosophila melanogaster. PLoS One 7: e50454, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Betts DH, Bain NT, and Madan P. The p66Shc adaptor protein controls oxidative stress response in early bovine embryos. PLoS One 9: e86978, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brand MD and Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab 2: 85–93, 2005. [DOI] [PubMed] [Google Scholar]
- 11. Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, and Hellerstein MK. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am J Physiol Endocrinol Metab 298: E108–E116, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cadenas S. Mitochondrial uncoupling, ROS generation and cardioprotection. Biochim Biophys Acta Bioenerg 1859: 940–950, 2018. [DOI] [PubMed] [Google Scholar]
- 13. Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, and Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20: 1715–1727, 2006. [DOI] [PubMed] [Google Scholar]
- 14. Cereghetti GM, Stangherlin A, Martins De Brito O, Chang CR, Blackstone C, Bernardi P, and Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A 105: 15803–15808, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chappuis S, Ripperger JA, Schnell A, Rando G, Jud C, Wahli W, and Albrecht U. Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol Metab 2: 184–193, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaudhari A, Gupta R, Patel S, Velingkaar N, and Kondratov R. Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol Biol Cell 28: 834–842, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L and Yang G. PPARs integrate the mammalian clock and energy metabolism. PPAR Res 5: 653017, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng Y, Ren X, Gowda ASP, Shan Y, Zhang L, Yuan YS, Patel R, Wu H, Huber-Keener K, Yang JW, Liu D, Spratt TE, and Yang JM. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis 4: e731, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chhunchha B, Kubo E, and Singh DP. Clock protein Bmal1 and Nrf2 cooperatively control aging or oxidative response and redox homeostasis by regulating rhythmic expression of Prdx6. Cells 9: 1861, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, and Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med 4: e76, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collins B, Mazzoni EO, Stanewsky R, and Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr Biol 16: 441–449, 2006. [DOI] [PubMed] [Google Scholar]
- 22. Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons A, Kemnitz JW, and Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325: 201–204, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuadrado A, Manda G, Hassan A, Alcaraz MJ, Barbas C, Daiber A, Ghezzi P, León R, López MG, Oliva B, Pajares M, Rojo AI, Robledinos-Antón N, Valverde AM, Guney E, and Schmidt HHHW. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol Rev 70: 348–383, 2018. [DOI] [PubMed] [Google Scholar]
- 24. Davies MH, Bozigian HP, Merrick BA, Birt DF, and Schnell RC. Circadian variations in glutathione-S-transferase and glutathione peroxidase activities in the mouse. Toxicol Lett 19: 23–27, 1983. [DOI] [PubMed] [Google Scholar]
- 25. Dibner C, Schibler U, and Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549, 2010. [DOI] [PubMed] [Google Scholar]
- 26. Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, Bacanamwo M, Chen YE, Schneider MD, Mangelsdorf DJ, Evans RM, and Yang Q. Cardiac peroxisome proliferator-activated receptor γ is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res 76: 269–279, 2007. [DOI] [PubMed] [Google Scholar]
- 27. Dlasková A, Clarke KJ, and Porter RK. The role of UCP 1 in production of reactive oxygen species by mitochondria isolated from brown adipose tissue. Biochim Biophys Acta Bioenerg 1797: 1470–1476, 2010. [DOI] [PubMed] [Google Scholar]
- 28. Dong J, Sulik KK, and Chen SY. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: implications for the prevention of fetal alcohol spectrum disorders. Antioxidants Redox Signal 10: 2023–2033, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Droge W. Free radicals in the physiological control of cell function. Physiol Rev 82: 47–95, 2002. [DOI] [PubMed] [Google Scholar]
- 30. Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, and Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, and Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189, 2006. [DOI] [PubMed] [Google Scholar]
- 32. Froy O. Circadian aspects of energy metabolism and aging. Ageing Res Rev 12: 931–940, 2013. [DOI] [PubMed] [Google Scholar]
- 33. Fukai T and Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal 15: 1583–1606, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gallé Á, Czékus Z, Bela K, Horváth E, Csiszár J, and Poór P. Diurnal changes in tomato glutathione transferase activity and expression. Acta Biol Hung 69: 505–509, 2018. [DOI] [PubMed] [Google Scholar]
- 35. Gallé Á, Czékus Z, Bela K, Horváth E, Ördög A, Csiszár J, and Poór P. Plant glutathione transferases and light. Front Plant Sci 9: 1944, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, Pryma DA, Khurana TS, and Lazar MA. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature 503: 410–413, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, and Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26: 1913–1923, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M, Pinton P, Rizzuto R, Bernardi P, Paolucci F, and Pelicci PG. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell 122: 221–233, 2005. [DOI] [PubMed] [Google Scholar]
- 39. Giorgio M, Trinei M, Migliaccio E, and Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol 8: 722–728, 2007. [DOI] [PubMed] [Google Scholar]
- 40. Girnun GD, Domann FE, Moore SA, and Robbins ME. Identification of a functional peroxisome proliferator-activated receptor response element in the rat catalase promoter. Mol Endocrinol 16: 2793–2801, 2002. [DOI] [PubMed] [Google Scholar]
- 41. Goede Pde, Wefers J, Brombacher EC, Schrauwen P, and Kalsbeek A. Circadian rhythms in mitochondrial respiration. J Mol Endocrinol 60: 115–130, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goljanek-Whysall K, Iwanejko LA, Vasilaki A, Pekovic-Vaughan V, and McDonagh B. Ageing in relation to skeletal muscle dysfunction: redox homoeostasis to regulation of gene expression. Mamm Genome 27: 341–357, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gomes LC, Di Benedetto G, and Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13: 589–598, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Greco T, Shafer J, and Fiskum G. Sulforaphane inhibits mitochondrial permeability transition and oxidative stress. Free Radic Biol Med 51: 2164–2171, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Green CB, Takahashi JS, and Bass J. The meter of metabolism. Cell 134: 728–742, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greenwell BJ, Trott AJ, Beytebiere JR, Pao S, Bosley A, Beach E, Finegan P, Hernandez C, and Menet JS. Rhythmic food intake drives rhythmic gene expression more potently than the hepatic circadian clock in mice. Cell Rep 27: 649–657, 2019. [DOI] [PubMed] [Google Scholar]
- 47. Hallows WC, Yu W, Smith BC, Devires MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, and Denu JM. Sirt3 Promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 41: 139–149, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hardeland R, Coto-Montes A, and Poeggeler B. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int 20: 921–962, 2003. [DOI] [PubMed] [Google Scholar]
- 49. Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, and Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15: 848–860, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, and Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49: 186–199, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, and Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450: 1086–1090, 2007. [DOI] [PubMed] [Google Scholar]
- 52. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Alt FW, Kahn CR, and Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoppins S and Nunnari J. Mitochondrial dynamics and apoptosis—the ER connection. Science 337: 1052–1054, 2012. [DOI] [PubMed] [Google Scholar]
- 54. Hu B, Wu Y, Liu J, Shen X, Tong F, Xu G, and Shen R. GSK-3beta inhibitor induces expression of Nrf2/TrxR2 signaling pathway to protect against renal ischemia/reperfusion injury in diabetic rats. Kidney Blood Press Res 41: 937–946, 2016. [DOI] [PubMed] [Google Scholar]
- 55. Huang CCY, Ko ML, Vernikovskaya DI, and Ko GYP. Calcineurin serves in the circadian output pathway to regulate the daily rhythm of l-type voltage-gated calcium channels in the retina. J Cell Biochem 113: 911–922, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Imai S. “Clocks” in the NAD World: NAD as a metabolic oscillator for the regulation of metabolism and aging. Biochim Biophys Acta 1804: 1584–1590, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Inoue I, Noji S, Awata T, Takahashi K, Nakajima T, Sonoda M, Komoda T, and Katayama S. Bezafibrate has an antioxidant effect: peroxisome proliferator-activated receptor α is associated with Cu2+, Zn2+-superoxide dismutase in the liver. Life Sci 63: 135–144, 1998. [DOI] [PubMed] [Google Scholar]
- 58. Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, Komoda T, and Katayama S. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb 12: 169–174, 2005. [DOI] [PubMed] [Google Scholar]
- 59. Inoue N, Imai K, and Aimoto T. Circadian variation of hepatic glutathione S-transferase activities in the mouse. Xenobiotica 29: 43–51, 1999. [DOI] [PubMed] [Google Scholar]
- 60. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, and Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen NH, Alexander RK, Unluturk U, Li X, Kong X, Hyde AL, Gangl MR, Mair WB, and Lee CH. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab 22: 709–720, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jäer S, Handschin C, St-Pierre J, and Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci U S A 104: 12017–12022, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62a. Jastroch M, Divakaruni AS, Mookerjee S, Treberg JR, and Brand MD. Mitochondrial proton and electron leaks oxygen consumption and pH data. Essays Biochem 47: 53–67, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ji G, Lv K, Chen H, Wang Y, Zhang Y, Li Y, and Qu L. Hydrogen peroxide modulates clock gene expression via PRX2-STAT3-REV-ERBα/β pathway. Free Radic Biol Med 145: 312–320, 2019. [DOI] [PubMed] [Google Scholar]
- 64. Jornayvaz FR and Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem 47: 69–84, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khapre RV, Kondratova AA, Susova O, and Kondratov RV. Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle 10: 4162–4169, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kincaid B and Bossy-Wetzel E. Forever young: SIRT3 a shield against mitochondrial meltdown, aging, and neurodegeneration. Front Aging Neurosci 5: 48, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ko CH and Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 15: R271–R277, 2006. [DOI] [PubMed] [Google Scholar]
- 68. Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, and Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kohsaka A, Das P, Hashimoto I, Nakao T, Deguchi Y, Gouraud SS, Waki H, Muragaki Y, and Maeda M. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One 9: e112811, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, and Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6: 414–421, 2007. [DOI] [PubMed] [Google Scholar]
- 71. Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, and Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 20: 1868–1873, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Koronowski KB, Kinouchi K, Welz PS, Smith JG, Zinna VM, Shi J, Samad M, Chen S, Magnan CN, Kinchen JM, Li W, Baldi P, Benitah SA, and Sassone-Corsi P. Defining the independence of the liver circadian clock. Cell 177: 1448–1462.e14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Krishnan N, Davis AJ, and Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem Biophys Res Commun 374: 299–303, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Krishnan N, Kretzschmar D, Rakshit K, Chow E, and Giebultowicz JM. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging (Albany NY) 1: 937–948, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee J, Kim MS, Li R, Liu VY, Fu L, Moore DD, Ma K, and Yechoor VK. Loss of Bmal1 leads to uncoupling and impaired glucose-stimulated insulin secretion in β-cells. Islets 3: 381–388, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li E, Li X, Huang J, Xu C, Liang Q, Ren K, Bai A, Lu C, Qian R, and Sun N. BMAL1 regulates mitochondrial fission and mitophagy through mitochondrial protein BNIP3 and is critical in the development of dilated cardiomyopathy. Protein Cell 11: 1–19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li L and Zou L. Sensing, signaling, and responding to DNA damage: organization of the checkpoint pathways in mammalian cells. J Cell Biochem 94: 298–306, 2005. [DOI] [PubMed] [Google Scholar]
- 79. Liesa M and Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab 17: 491–506, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu J, Li D, Zhang T, Tong Q, Ye RD, and Lin L. SIRT3 protects hepatocytes from oxidative injury by enhancing ROS scavenging and mitochondrial integrity. Cell Death Dis 8: e3158, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mahmoud AM, Alexander MY, Tutar Y, Wilkinson FL, and Venditti A. Oxidative stress in metabolic disorders and drug-induced injury: the potential role of Nrf2 and PPARs activators. Oxid Med Cell Longev 2017: 2017, 2508909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mailloux RJ and Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med 51: 1106–1115, 2011. [DOI] [PubMed] [Google Scholar]
- 83. Makwana K, Gosai N, Poe A, and Kondratov RV. Calorie restriction reprograms diurnal rhythms in protein translation to regulate metabolism. FASEB J 33: 4473–4489, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Marí M, Morales A, Colell A, García-Ruiz C, and Fernández-Checa JC. Mitochondrial glutathione, a key survival antioxidant. Antioxidants Redox Signal 11: 2685–2700, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. This reference has been deleted.
- 86. Mauvoisin D, Atger F, Dayon L, Núñez Galindo A, Wang J, Martin E, Da Silva L, Montoliu I, Collino S, Martin FP, Ratajczak J, Cantó C, Kussmann M, Naef F, and Gachon F. Circadian and feeding rhythms orchestrate the diurnal liver acetylome. Cell Rep 20: 1729–1743, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, and Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics 31: 86–95, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mezhnina V, Pearce R, Poe A, Velingkaar N, Astafev A, Ebeigbe OP, Makwana K, Sandlers Y, and Kondratov RV. CR reprograms acetyl-CoA metabolism and induces long-chain acyl-CoA dehydrogenase and CrAT expression. Aging Cell 19: e13266, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Michan S and Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J 404: 1–13, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Migliaccio E, Giogio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, and Pelicci PG. The p66(shc) adaptor protein controls oxidative stress response and life span in mammals. Nature 402: 309–313, 1999. [DOI] [PubMed] [Google Scholar]
- 91. Milev NB, Rhee S, and Reddy AB. Cellular timekeeping: it's redox o'clock. Cold Spring Harb Perspect Biol 10: a027698, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mitchell SJ, Bernier M, Mattison JA, Aon MA, Kaiser TA, Anson RM, Ikeno Y, Anderson RM, Ingram DK, and de Cabo R. Daily fasting improves health and survival in male mice independent of diet composition and calories. Cell Metab 29: 221–228, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mitra K. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. Bioessays 35: 955–964, 2013. [DOI] [PubMed] [Google Scholar]
- 94. Moldogazieva NT, Mokhosoev IM, Feldman NB, and Lutsenko SV. ROS and RNS signalling: adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic Res 52: 507–543, 2018. [DOI] [PubMed] [Google Scholar]
- 95. Moorsel DV, Hansen J, Havekes B, Scheer FAJL, Jörgensen JA, Hoeks J, Schrauwen-Hinderling VB, Duez H, Lefebvre P, Schaper NC, Hesselink MKC, Staels B, and Schrauwen P. Demonstration of a day-night rhythm in human skeletal muscle oxidative capacity. Mol Metab 5: 635–645, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mori V, Amici A, Mazzola F, Di Stefano M, Conforti L, Magni G, Ruggieri S, Raffaelli N, and Orsomando G. Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS One 9: e113939, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Murphy MP. Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxid Redox Signal 16: 476–495, 2012. [DOI] [PubMed] [Google Scholar]
- 98. Musiek ES and Holtzman DM. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 354: 1004–1008, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, Roh JH, Ortiz-Gonzalez X, Dearborn JT, Culver JP, Herzog ED, Hogenesch JB, Wozniak DF, Dikranian K, Giasson BI, Weaver DR, Holtzman DM, and FitzGerald GA. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest 123: 5389–5400, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nakagawa T and Guarente L. Sirtuins at a glance. J Cell Sci 124: 833–838, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nakahata Y, Grimaldi B, Sahar S, Hirayama J, and Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol 19: 230–237, 2007. [DOI] [PubMed] [Google Scholar]
- 102. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, and Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nakahata Y, Sahar S, Astarita G, Kaluzova M, and Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ndiaye MA, Nihal M, Wood GS, and Ahmad N. Skin, reactive oxygen species, and circadian clocks. Antioxid Redox Signal 2: 2982–2996, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nègre-Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, Salvayre R, Pénicaud L, and Casteilla L. A role for uncoupling protein-2 as a regulator of mitochondrial hydrogen peroxide generation. FASEB J 11: 809–815, 1997. [PubMed] [Google Scholar]
- 106. Neufeld-Cohen A, Robles MS, Aviram R, Manella G, Adamovich Y, Ladeuix B, Nir D, Rousso-Noori L, Kuperman Y, Golik M, Mann M, and Asher G. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc Natl Acad Sci U S A 113: E1673–E1682, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nohara K, Mallampalli V, Nemkov T, Wirianto M, Yang J, Ye Y, Sun Y, Han L, Esser KA, Mileykovskaya E, D'Alessandro A, Green CB, Takahashi JS, Dowhan W, Yoo SH, and Chen Z. Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat Commun 10: 3923, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Oelkrug R, Kutschke M, Meyer CW, Heldmaier G, and Jastroch M. Uncoupling protein 1 decreases superoxide production in brown adipose tissue mitochondria. J Biol Chem 285: 21961–21968, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Otera H, Ishihara N, and Mihara K. New insights into the function and regulation of mitochondrial fission. Biochim Biophys Acta Mol Cell Res 1833: 1256–1268, 2013. [DOI] [PubMed] [Google Scholar]
- 110. Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, and Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, and Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12: 565–573, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, and Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002. [DOI] [PubMed] [Google Scholar]
- 113. Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, and Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437: 759–763, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Patel SA, Velingkaar NS, and Kondratov RV. Transcriptional control of antioxidant defense by the circadian clock. Antioxid Redox Signal 20: 2997–3006, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, Levine DC, Bacsik DJ, Gius D, Newgard CB, Goetzman E, Chandel NS, Denu JM, Mrksich M, and Bass J. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342: 1243417, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pei JF, Li XK, Li WQ, Gao Q, Zhang Y, Wang XM, Fu JQ, Cui SS, Qu JH, Zhao X, Hao DL, Ju D, Liu N, Carroll KS, Yang J, Zhang EE, Cao JM, Chen HZ, and Liu DP. Diurnal oscillations of endogenous H2O2 sustained by p66Shc regulate circadian clocks. Nat Cell Biol 21: 1553–1564, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, Guo B, Sagami A, Taguchi K, Bechtold D, Loudon A, Yamamoto M, Chan J, van der Horst GTJ, Fukada Y, and Meng QJ. The circadian clock regulates rhythmic activation of the NRF2/glutathionemediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev 28: 548–560, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pizzino G, Irrera N, Cucinotta M, Pallio G, Mannino F, Arcoraci V, Squadrito F, Altavilla D, and Bitto A. Oxidative stress: harms and benefits for human health. Oxid Med Cell Longev 2017: 8416763, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ponce IT, Rezza IG, Delgado SM, Navigatore LS, Bonomi MR, Golini RL, Gimenez MS, and Anzulovich AC. Daily oscillation of glutathione redox cycle is dampened in the nutritional vitamin A deficiency. Biol Rhythm Res 43: 351–372, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Qiu X, Brown K, Hirschey MD, Verdin E, and Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667, 2010. [DOI] [PubMed] [Google Scholar]
- 121. Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, and Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem 287: 27255–27264, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai SI, and Bass J. Circadian clock feedback cycle through NAMPT-Mediated NAD+ biosynthesis. Science 324: 651–654, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, and Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A 110: 6601–6606, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Reinke H and Asher G. Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol 20: 227–241, 2019. [DOI] [PubMed] [Google Scholar]
- 125. Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, and Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rey G, Valekunja UK, Feeney KA, Wulund L, Milev NB, Stangherlin A, Ansel-Bollepalli L, Velagapudi V, O'Neill JS, and Reddy AB. The pentose phosphate pathway regulates the circadian clock. Cell Metab 24: 462–473, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ribas V, García-Ruiz C, and Fernández-Checa JC. Glutathione and mitochondria. Front Pharmacol 5: 151, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ryoo I-G and Kwak MK. Regulatory crosstalk between the oxidative stress-related transcription factor Nfe2l2/Nrf2 and mitochondria. Toxicol Appl Pharmacol 359: 24–33, 2018. [DOI] [PubMed] [Google Scholar]
- 129. Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, and Sassone-Corsi P. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170: 664–677.e11, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Schmitt K, Grimm A, Dallmann R, Oettinghaus B, Restelli LM, Witzig M, Ishihara N, Mihara K, Ripperger JA, Albrecht U, Frank S, Brown SA, and Eckert A. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab 27: 657–666.e5, 2018. [DOI] [PubMed] [Google Scholar]
- 131. Scrima R, Cela O, Merla G, Augello B, Rubino R, Quarato G, Fugetto S, Menga M, Fuhr L, Relógio A, Piccoli C, Mazzoccoli G, and Capitanio N. Clock-genes and mitochondrial respiratory activity: evidence of a reciprocal interplay. Biochim Biophys Acta Bioenerg 1857: 1344–1351, 2016. [DOI] [PubMed] [Google Scholar]
- 132. Shabalina IG, Petrovic N, deJong JMA, Kalinovich AV, Cannon B, and Nedergaard J. UCP1 in Brite/Beige adipose tissue mitochondria is functionally thermogenic. Cell Rep 5: 1196–1203, 2013. [DOI] [PubMed] [Google Scholar]
- 133. Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, and Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12: 654–661, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Singh CK, Chhabra G, Ndiaye MA, Garcia-Peterson LM, MacK NJ, and Ahmad N. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal 28: 643–661, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Solanas G, Peixoto FO, Perdiguero E, Jardí M, Ruiz-Bonilla V, Datta D, Symeonidi A, Castellanos A, Welz PS, Caballero JM, Sassone-Corsi P, Muñoz-Cánoves P, and Benitah SA. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170: 678–692.e20, 2017. [DOI] [PubMed] [Google Scholar]
- 136. Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, and Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under Caloric Restriction. Cell 143: 802–812, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Song Z, Chen H, Fiket M, Alexander C, and Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178: 749–755, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Stein LR and Imai SI. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 23: 420–428, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Suen DF, Norris KL, and Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev 22: 1577–1590, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sun J, Ren X, and Simpkins JW. Sequential upregulation of superoxide dismutase 2 and heme oxygenase 1 by tert-butylhydroquinone protects mitochondria during oxidative stress. Mol Pharmacol 88: 437–449, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sun Q, Zeng C, Du L, and Dong C. Mechanism of circadian regulation of the NRF2/ARE pathway in renal ischemia‑reperfusion. Exp Ther Med 21: 1–9, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Suzuki T and Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic Biol Med 88: 93–100, 2015. [DOI] [PubMed] [Google Scholar]
- 143. Tamaru T, Hattori M, Ninomiya Y, Kawamura G, Varès G, Honda K, Mishra DP, Wang B, Benjamin I, Sassone-Corsi P, Ozawa T, and Takamatsu K. ROS stress resets circadian clocks to coordinate pro-survival signals. PLoS One 8: e82006, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tchouagué M, Grondin M, Glory A, and Averill-Bates D. Heat shock induces the cellular antioxidant defenses peroxiredoxin, glutathione and glucose 6-phosphate dehydrogenase through Nrf2. Chem Biol Interact 310: 108717, 2019. [DOI] [PubMed] [Google Scholar]
- 145. Teshima Y, Akao M, Jones SP, and Marbán E. Uncoupling protein-2 overexpression inhibits mitochondrial death pathway in cardiomyocytes. Circ Res 93: 192–200, 2003. [DOI] [PubMed] [Google Scholar]
- 146. Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, Baldi P, and Sassone-Corsi P. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab 26: 523–538, 2017. [DOI] [PubMed] [Google Scholar]
- 147. Toime LJ and Brand MD. Uncoupling protein-3 lowers reactive oxygen species production in isolated mitochondria. Free Radic Biol Med 49: 606–611, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, and Martinou JC. SlP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28: 1589–1600, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Toyama T, Nakamura H, Harano Y, Yamauchi N, Morita A, Kirishima T, Minami M, Itoh Y, and Okanoue T. PPARα ligands activate antioxidant enzymes and suppress hepatic fibrosis in rats. Biochem Biophys Res Commun 324: 697–704, 2004. [DOI] [PubMed] [Google Scholar]
- 150. Tuñoacuten MJ, González P, López P, Salido GM, and Madrid JA. Circadian rhythms in glutathione and glutathione-s transferase activity of rat liver. Arch Physiol Biochem 100: 83–87, 1992. [DOI] [PubMed] [Google Scholar]
- 151. Uchiyama Y. Circadian alterations in tubular structures on the outer mitochondrial membrane of rat hepatocytes. Cell Tissue Res 214: 519–527, 1981. [DOI] [PubMed] [Google Scholar]
- 152. Ulgherait M, Chen A, McAllister SF, Kim HX, Delventhal R, Wayne CR, Garcia CJ, Recinos Y, Oliva M, Canman JC, Picard M, Owusu-Ansah E, and Shirasu-Hiza M. Circadian regulation of mitochondrial uncoupling and lifespan. Nat Commun 11: 1927, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Valko M, Rhodes CJ, Moncol J, Izakovic M, and Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact 160: 1–40, 2006. [DOI] [PubMed] [Google Scholar]
- 154. Velingkaar N, Mezhnina V, Poe A, and Kondratov RV. Two-meal caloric restriction induces 12-hour rhythms and improves glucose homeostasis. FASEB J 35: e21342, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Velingkaar N, Mezhnina V, Poe A, Makwana K, Tulsian R, and Kondratov RV. Reduced caloric intake and periodic fasting independently contribute to metabolic effects of caloric restriction. Aging Cell 19: e13138, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ventura-Clapier R, Garnier A, and Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1α. Cardiovasc Res 79: 208–217, 2008. [DOI] [PubMed] [Google Scholar]
- 157. Wai T and Langer T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol Metab 27: 105–117, 2016. [DOI] [PubMed] [Google Scholar]
- 158. Warburg O. On respiratory impairment in cancer cells. Science 124: 269–270, 1956. [PubMed] [Google Scholar]
- 159. Wible RS, Ramanathan C, Sutter CH, Olesen KM, Kensler TW, Liu AC, and Sutter TR. NRF2 regulates core and stabilizing circadian clock loops, coupling redox and timekeeping in mus musculus. Elife 7: 1–27, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wible RS and Sutter TR. Soft cysteine signaling network: the functional significance of cysteine in protein function and the soft acids/bases thiol chemistry that facilitates cysteine modification. Chem Res Toxicol 30: 729–762, 2017. [DOI] [PubMed] [Google Scholar]
- 161. Wible RS, Tran QT, Fathima S, Sutter CH, Kensler TW, and Sutter TR. Pharmacogenomics of chemically distinct classes of keap1-Nrf2 activators identify common and unique gene, protein, and pathway responses in vivo. Mol Pharmacol 93: 297–308, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MKC, Paquet C, Delhaye S, Shin Y, Kamenecka TM, Schaart G, Lefebvre P, Nevière R, Burris TP, Schrauwen P, Staels B, and Duez H. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 19: 1039–1046, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wu KC, Cui JY, and Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci 123: 590–600, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Xu L, Cheng Q, Hua B, Cai T, Lin J, Yuan G, Yan Z, Li X, Sun N, Lu C, and Qian R.. Circadian gene Clock regulates mitochondrial morphology and functions by posttranscriptional way. bioRxiv 2018. DOI: 10.1101/365452. [DOI] [Google Scholar]
- 165. Yamamoto M, Kensler TW, and Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol Rev 98: 1169–1203, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, and Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell 126: 801–810, 2006. [DOI] [PubMed] [Google Scholar]
- 167. Youle RJ and Van Der Bliek AM. Good for discussion: mitochondrial fission, fusion, and stress. Science 337: 1062–1065, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yu W, Dittenhafer-Reed KE, and Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem 287: 14078–14086, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Zelko IN, Mariani TJ, and Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med 33: 337–349, 2002. [DOI] [PubMed] [Google Scholar]
- 170. Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlén P, Tomilin N, Shupliakov O, Lendahl U, and Nistér M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J 30: 2762–2778, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zhou L, Wang F, Sun R, Chen X, Zhang M, Xu Q, Wang Y, Wang S, Xiong Y, Guan K, Yang P, Yu H, and Ye D. SIRT 5 promotes IDH 2 desuccinylation and G6 PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep 17: 811–822, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]