ABSTRACT
Hippo signaling, an evolutionarily conserved kinase cascade involved in organ size control, plays key roles in various tissue developmental processes, but its role in craniofacial development remains poorly understood. Using the transgenic Wnt1-Cre2 driver, we inactivated the Hippo signaling components Lats1 and Lats2 in the cranial neuroepithelium of mouse embryos and found that the double conditional knockout (DCKO) of Lats1/2 resulted in neural tube and craniofacial defects. Lats1/2 DCKO mutant embryos had microcephaly with delayed and defective neural tube closure. Furthermore, neuroepithelial cell shape and architecture were disrupted within the cranial neural tube in Lats1/2 DCKO mutants. RNA sequencing of embryonic neural tubes revealed increased TGFB signaling in Lats1/2 DCKO mutants. Moreover, markers of epithelial-to-mesenchymal transition (EMT) were upregulated in the cranial neural tube. Inactivation of Hippo signaling downstream effectors, Yap and Taz, suppressed neuroepithelial defects, aberrant EMT and TGFB upregulation in Lats1/2 DCKO embryos, indicating that LATS1/2 function via YAP and TAZ. Our findings reveal important roles for Hippo signaling in modulating TGFB signaling during neural crest EMT.
Keywords: Lats1, Lats2, Hippo signaling, Cranial neural tube, Cranial neural crest, Craniofacial development
Summary: Hippo signaling kinases LATS1 and LATS2 phosphorylate downstream effectors YAP/TAZ to regulate the TGFB-induced epithelial-to-mesenchymal transition in the migratory neural crest on the dorsal cranial neural tube.
INTRODUCTION
Tissues of the craniofacial complex are derived primarily from the neural crest (NC), which is a transient, migratory, multipotent cell population originating in the embryonic dorsal neural tube (NT) (Santagati and Rijli, 2003). NC cells arise at the border between the dorsal NT and the non-neural ectoderm. While in the NT, premigratory NC cells have characteristics of epithelial cells. NC cells lose apicobasal polarity, undergo epithelial-to-mesenchymal transition (EMT), and delaminate from the neuroepithelium to migrate towards specific regions in the embryo and subsequently differentiate (Bronner and Simões-Costa, 2016; Gandhi and Bronner, 2021; Theveneau and Mayor, 2012).
Developmental defects in NT and NC formation result in human congenital anomalies collectively known as NT defects (NTDs) and neurocristopathies, respectively. NTDs are among the most common birth defects of the central nervous system worldwide, with a prevalence that varies according to ethnic and racial background, geographic location and surveillance program accessibility (Blencowe et al., 2018; Wallingford et al., 2013; Williams et al., 2016; Zaganjor et al., 2016). Although surgery can be used to correct structural defects in some cases, numerous secondary conditions are associated with NTDs, such as neurological impairment, movement problems or paralysis, and sensorial problems (Adzick et al., 2011; Botto et al., 1999; Greene and Copp, 2014; Wallingford et al., 2013). These complications add significant disease burden (Grosse et al., 2016; Rofail et al., 2013; Yi et al., 2011). Better understanding of the cellular and molecular mechanisms required for proper NT closure are essential for developing methods for predicting and preventing NTDs.
Hippo signaling, an evolutionarily conserved organ size control pathway, has been implicated in the development of multiple tissues (Misra and Irvine, 2018; Wang et al., 2018). In mammals, the core Hippo signaling pathway consists of a kinase cascade in which MST1/2 (STK4/3) kinases phosphorylate and activate LATS1/2. In turn, LATS1/2 phosphorylate the transcriptional effectors YAP (YAP1) and TAZ (WWTR1), promoting their cytoplasmic retention. In the absence of activated Hippo pathway kinases, YAP/TAZ remain unphosphorylated and translocate to the nucleus to activate transcription. Furthermore, YAP/TAZ mediate cellular responses to mechanical cues, such as changes in tissue stiffness and architecture (Chakraborty et al., 2017; Du et al., 2021; Dupont et al., 2011; Meng et al., 2018). Previous studies have confirmed a role for YAP/TAZ in NC migration and lineage diversification (Hindley et al., 2016; Kumar et al., 2019; Wang et al., 2016). Here, we elucidate the role of the Hippo core kinases LATS1 and LATS2 (LATS1/2) during early craniofacial development and neurulation. Using a Wnt1-Cre2 driver, we show that Lats1/2 deficiency results in neuroepithelial disorganization and uncover important roles of LATS1/2 in TGFB-induced EMT on the dorsal cranial NT.
RESULTS
Lats1/2 double mutant embryos exhibit craniofacial defects
Hippo kinases LATS1/2 are necessary for murine development; their disruption results in perinatal and embryonic lethality (McPherson et al., 2004; St John et al., 1999). Nevertheless, the role of these kinases has not been explored in tissues involved in craniofacial development. Lats1 and Lats2 are found in tissues of ectodermal and mesodermal origin in the developing head (McPherson et al., 2004; Yu et al., 2017). At embryonic day (E)10.5, we observed both Lats1 and Lats2 transcripts in the cranial neuroepithelium indicated by RNAscope, with Lats1 more enriched than Lats2 (Fig. S1A). Moreover, we analyzed previous transcriptomic data from mouse NTs at E8.5, E9.5 and E10.5, and found that Lats1/Lats2 expression remains relatively constant during neurulation (Fig. S1B). Consistently, the expression of Lats1 in the NT was relatively higher than that of Lats2.
We inactivated the Hippo pathway components Lats1/2 in the NC and neuroepithelium using a transgenic Wnt1-Cre2 driver (Dinsmore et al., 2022; Lewis et al., 2013). We collected embryos at different time points and examined morphologic phenotypes. Among the different mutant genotypes, Wnt1-Cre2, Lats1F/Δ, Lats2F/Δ double conditional knockout (DCKO) mutant embryos were embryonic lethal after E10.5 (Table S2). No Lats1/2 DCKOs were found alive by stage E11.0, but genotyping of dead or resorbed embryos could be performed. Lats1/2 compound mutants (Wnt1-Cre2, Lats1F/+, Lats2F/Δ and Wnt1-Cre2, Lats1F/Δ, Lats2F/+) and heterozygous embryos (Wnt1-Cre2, Lats1F/+, Lats2F/+) had no obvious morphologic defects compared with Wnt1-Cre2-negative littermate controls. Lats1 or Lats2 haploinsufficiency was sufficient to circumvent embryonic lethality caused by deletion of both Lats1 and Lats2, indicating redundant roles in the NC.
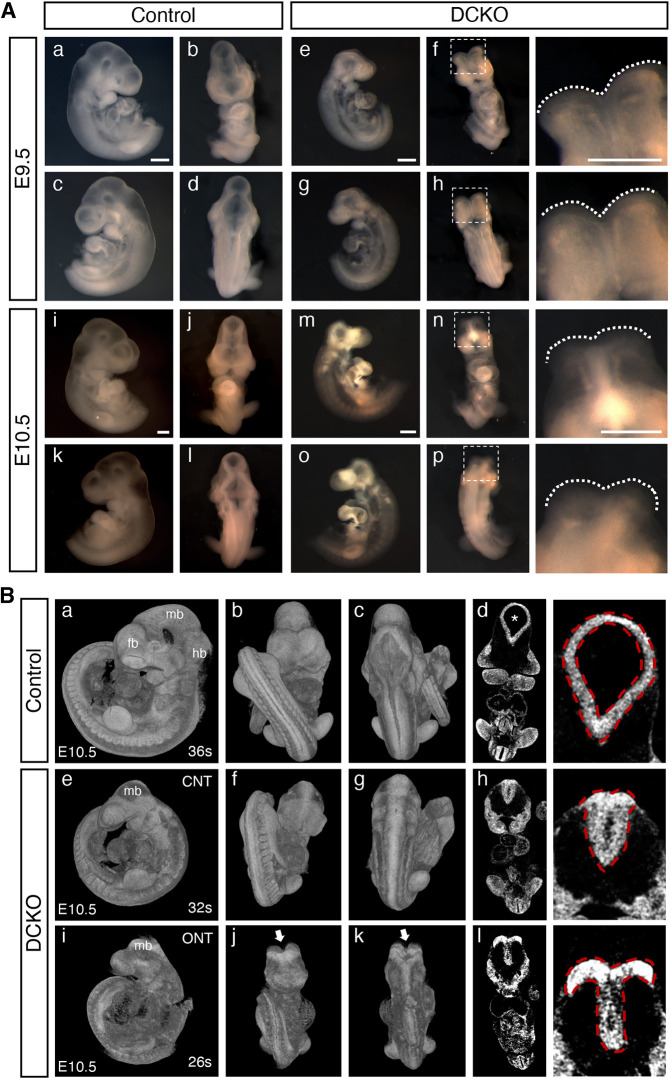
Lats1/2 DCKOs had distinctive craniofacial and NT phenotypes compared with littermate controls (Fig. 1A). In mouse embryonic development, closure of the cranial NT is completed by E9.0 (16-20 somites) before counteracting forces can interfere with neural fold fusion (Yamaguchi and Miura, 2013). All control embryos had a fully closed NT at both E9.5 and E10.5, whereas all Lats1/2 DCKOs had an open NT at E9.5 and about half had an open NT at E10.5 (Fig. 1A, Table S3). Closer examination by 3D reconstruction of microcomputed tomographic (µCT) imaging of E10.5 embryos showed abnormal NT closure in the absence of Lats1/2 (Fig. 1B). Compared with control embryos (Fig. 1Bd), Lats1/2 DCKO embryos lacked ventricular space within the NT (Fig. 1Bh,l). Parts of the midbrain, hindbrain and forebrain regions were missing in all Lats1/2 DCKOs compared with controls (Fig. 1A,B). These findings suggest that Lats1 and Lats2 are required for proper NT fusion during cranial development.
Fig. 1.
Lats1/2 double conditional knockout (DCKO) mutant embryos have craniofacial phenotypes with severe NTDs. (A) Bright-field images of E9.5 and E10.5 embryos. Compared with controls (a-d,i-l), DCKO embryos have smaller heads and parts of the forebrain, midbrain and hindbrain missing (e-h,m-p). DCKO embryos have open cranial neural tubes (NTs) (f,h,n,p, outlined). Boxed areas are shown at higher magnification (right). Scale bars: 500 µm. (B) 3D reconstruction of microcomputed tomographic (µCT) imaging of E10.5 embryos. Compared with control embryos (a-c), DCKO embryos have smaller heads and abnormal NTs (e-g,i-k, outlined; arrows indicate open NT). Coronal µCT slices (higher magnification on right) illustrate the NT ventricular space in control embryos (asterisk, d), which is absent in the DCKO embryos (h,l). CNT, closed neural tube; fb, forebrain; hb, hindbrain; mb, midbrain; ONT, open neural tube.
Lats1/2 are required for neuroepithelial cell attachment
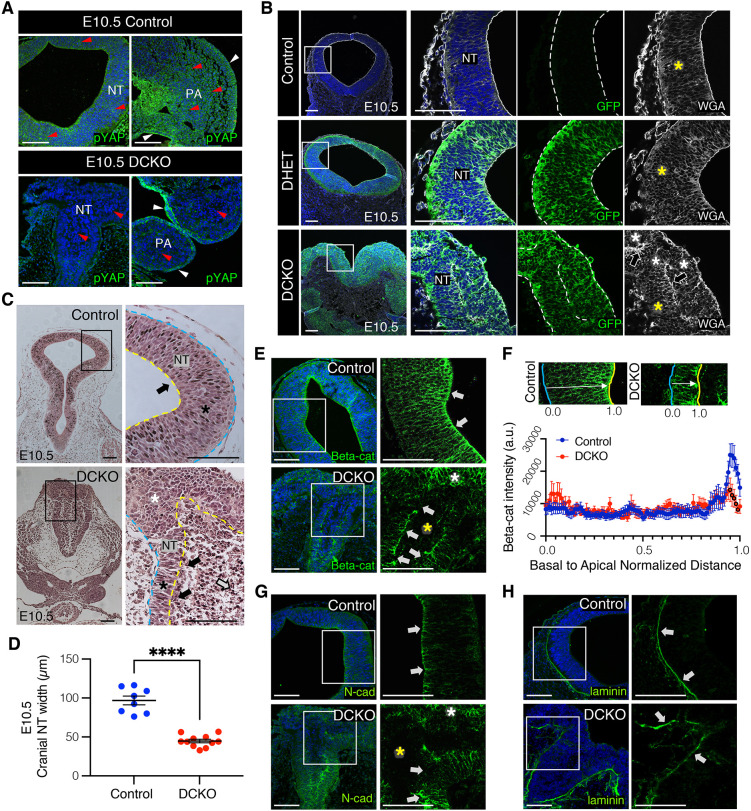
LATS kinase activity was reduced in the neuroepithelium and NC-derived cells of E10.5 Lats1/2 DCKO mutants, indicated by decreased phosphorylated (p)YAP in the NT and pharyngeal arches relative to control embryos (Fig. 2A). pYAP remained unaltered in non-NC-derived tissues. The R26mTmG (mTmG) reporter, which expresses eGFP upon Cre activation, was included to track recombination in the neuroepithelium (Muzumdar et al., 2007). At E10.5, we detected recombination in the dorsal NT- and NC-derived cells in Lats1/2 double heterozygous (DHET) and Lats1/2 DCKO embryos (Fig. 2B). Interestingly, GFP+ neuroepithelial cells in the Lats1/2 DCKOs no longer had an elongated morphology across the apicobasal axis. Wheat germ agglutinin (WGA) staining of the cellular plasma membrane revealed neuroepithelial cells in Lats1/2 DCKO NTs without the tight pseudostratified architecture seen in the controls (Fig. 2B, Fig. S2A). Furthermore, the ventricular space in the Lats1/2 DCKO NT was occupied by neuroepithelium-derived GFP+ cells not attached to the basement membrane (Fig. S2A).
Fig. 2.
Lats1/2 deficiency disrupts neuroepithelial cell behavior and polarization. (A) Phosphorylated YAP (pYAP) is not detected by immunostaining in the neuroepithelium or neural crest (NC) of E10.5 DCKOs (red arrowheads), but is detected in non-neuroepithelium-derived tissues (white arrowheads). (B) Immunostaining of membrane-bound GFP. A mTmG reporter was crossed into the genetic model to track recombination. Polarized and pseudostratified neuroepithelium were found in control and Lats1/2 double heterozygous (DHET) NTs (yellow asterisks). GFP+ neuroepithelial cells lose attachment in the DCKO NT (white asterisks, arrows). Wheat germ agglutinin (WGA) visualizes the cell membrane. (C) H&E staining of coronal sections at E10.5. Neuroepithelium extends from the basal edge to the apical edge (controls). The apical surface of the DCKO NT was interrupted, with infiltrating cells detected inside the ventricle (white arrow). Polarized neuroepithelium was found in control and DCKO NTs (black asterisks), but unpolarized cells were detected in the apical edge of the latter (black arrows). Loss of organization was observed in the dorsal folds of DCKO NTs (white asterisk). (D) The neuroepithelium width was measured from basal to apical edge at E10.5 (control, n=8; DCKO, n=11). Data compared using a two-tailed unpaired t-test, ****P<0.0001. (E) Immunostaining of beta-catenin (Beta-cat) at E10.5. The apical bias is lost in DCKO NTs (white arrows), with ectopic expression in the dorsal folds (white asterisk) but not the infiltrating cells (yellow asterisk). (F) Top: Beta-cat staining intensity across the apicobasal axis was recorded and normalized by NT width. Bottom: Average intensity line profiles binned into 100 intervals (see Materials and Methods) from control and DCKO NTs (control NTs, n=3; DCKO NTs, n=3). Data compared using a two-tailed unpaired t-test; black outlines indicate significance (bins 95-99: 95, P<0.05; 96, P<0.05; 97, P<0.05; 98, P<0.01; 99, P<0.05). a.u., arbitrary units. (G) Immunostaining of N-cadherin (N-cad). Apical bias was changed in DCKO NTs (white arrows), and ectopically expressed in the dorsal folds (white asterisk) but not in the infiltrating cells (yellow asterisk). (H) Immunostaining of the basement membrane marker laminin (basal edge marked by white arrows). Boxed areas are shown at higher magnification (right). Scale bars: 100 μm. NT, neural tube; PA, pharyngeal arches (NC-derived). Outline delineates the neuroepithelium. Yellow lines indicate apical edge; blue lines indicate basal edge.
Histological analysis further revealed that, compared with controls, Lats1/2 DCKOs had infiltrating neuroepithelium-derived cells within the NT ventricle and an interrupted non-neural ectoderm layer at E10.5 (Fig. 2C). Ventricular cellular infiltration could also be detected at E9.5, before Lats1/2 DCKO cranial neural fold convergence (Fig. S2B). Additionally, Lats1/2 DCKO NTs had a narrower pseudostratified neuroepithelium and thicker dorsal folds (Fig. 2D), suggesting dorsal NC expansion and migration into the ventricle. Interestingly, trunk and caudal NT closures were not affected in the Lats1/2 DCKOs, suggesting a specific role for Lats1/2 during cranial neurulation (Fig. S2B,C). To assess changes in neuroepithelial proliferation, we performed immunofluorescence for the proliferation marker phospho-histone H3 (pHH3). Although no significant difference was observed in the number of proliferating cells at E9.5, the organization of pHH3+ mitotic cells appeared slightly disrupted in the NTs of Lats1/2 DCKOs compared with controls (Fig. S2D,E). Proliferative cells were also detected in the ventricle-infiltrating, neuroepithelium-derived cell population of Lats1/2 DCKO NTs (Fig. S2D, red arrowheads).
Polarized neuroepithelial cells have a pseudostratified configuration with nuclei distributed along the apicobasal axis and undergo interkinetic nuclear migration (INM) (Norden, 2017). INM is a common feature of developing neuroepithelia, characterized by nuclear migration along the apicobasal axis of the tissue in phase with the cell cycle (Cearns et al., 2016; Kosodo et al., 2011; Norden et al., 2009; Sauer, 1935). In INM, mitosis occurs exclusively on the apical edge of the NT. In control embryos, mitotic cells in the NT are found apically, surrounding the ventricle (Fig. S2D, see also Fig. 6C). However, at E10.5, mitotic cells in Lats1/2-deficient NTs were found scattered along the apicobasal axis of the neuroepithelium (see Fig. 6C). Cell polarity is essential for nuclear position and movement within the apicobasal axis of the NT (Baye and Link, 2007; Chenn et al., 1998; Imai et al., 2006; Spear and Erickson, 2012). In recent reports, Lats1/2 have been associated with apicobasal polarity during epithelial branching in the developing lung (Nantie et al., 2018), pancreas (Braitsch et al., 2019) and kidney (Reginensi et al., 2016). To evaluate apicobasal attachment, we analyzed the localization of polarized proteins in the NT. At E9.5 and E10.5, beta-catenin and N-cadherin, both part of the adherens junction complex found on the apical side of the neuroepithelium (Herrera et al., 2014; Miyamoto et al., 2015), were expressed with an apical bias in control NTs (Fig. 2E-G, Fig. S2F). At E9.5, beta-catenin and N-cadherin expression was also detected along the apical edge of the Lats1/2 DCKO NT and in ventricular infiltrating cells (Fig. S2F). By E10.5, beta-catenin and N-cadherin expression lost their apical bias in neuroepithelia and were not detected in the neuroepithelial-derived cell population inside the Lats1/2 DCKO NT (Fig. 2E-G). Interestingly, N-cadherin downregulation has been associated with retinal and NT neuronal progenitor detachment from the apical region of the neuroepithelium (Das and Storey, 2014; Rousso et al., 2012; Wong et al., 2012). Moreover, neuroepithelia in the dorsal region of the Lats1/2 DCKO NT ectopically expressed beta-catenin and N-cadherin, suggesting abnormal attachment in that region (Fig. 2E,G, white asterisks). Instead, the basement membrane marker laminin was detected in the basal edge of the NT in both controls and Lats1/2 DCKOs (Fig. 2H). These observations suggest that Lats1 and Lats2 are required for proper neuroepithelia behavior, organization and apical attachment.
Fig. 6.
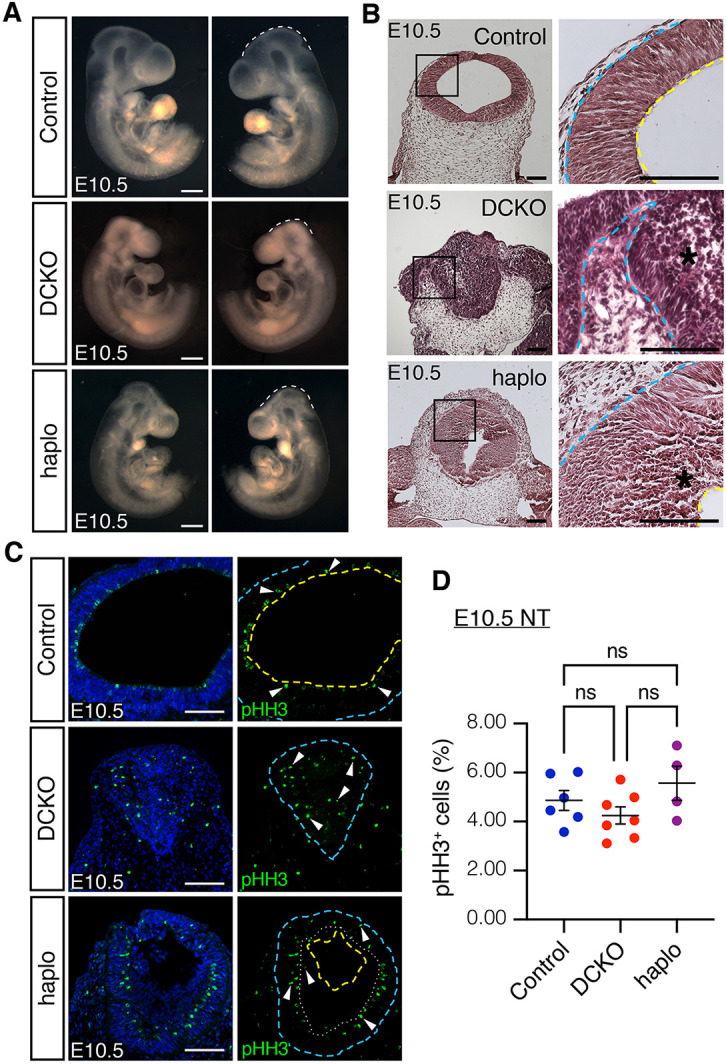
Yap/Taz haploinsufficiency reduces cell infiltration and partially recovers neuroepithelial cell architecture in Lats1/2-deficient neural tubes. (A) Bright-field images of Lats1/2 DCKO embryos with Yap/Taz heterozygosity (haplo embryos), which are phenotypically like Lats1/2 DCKO embryos. Scale bars: 500 μm. (B) H&E staining of coronal sections at E10.5. Neuroepithelial cell organization is partially recovered in haplo NTs, while cell infiltration into the ventricle is still detected (black asterisk). Boxed areas are shown at higher magnification (right). (C) Immunostaining of proliferation marker pHH3 show mitotic cells on the apical edge of the polarized neuroepithelium for control and haplo NTs (dotted line), in contrast to DCKO with mitotic cells along the apicobasal axis. Nuclei stained with DAPI (blue). Scale bars: 100 μm. Outline delineates neuroepithelium, with yellow lines indicating apical edge and blue lines indicating basal edge. (D) Quantification of pHH3+ cells in the NT (control, n=6; DCKO, n=7; haplo, n=4). Data compared using one-way ANOVA followed by Tukey's multiple comparisons test. NT, neural tube; ns, not significant. All error bars represent s.e.m.
Transcriptional profiling suggests increased TGFB1-dependent signaling in the neural tube of Lats1/2 DCKO mutants
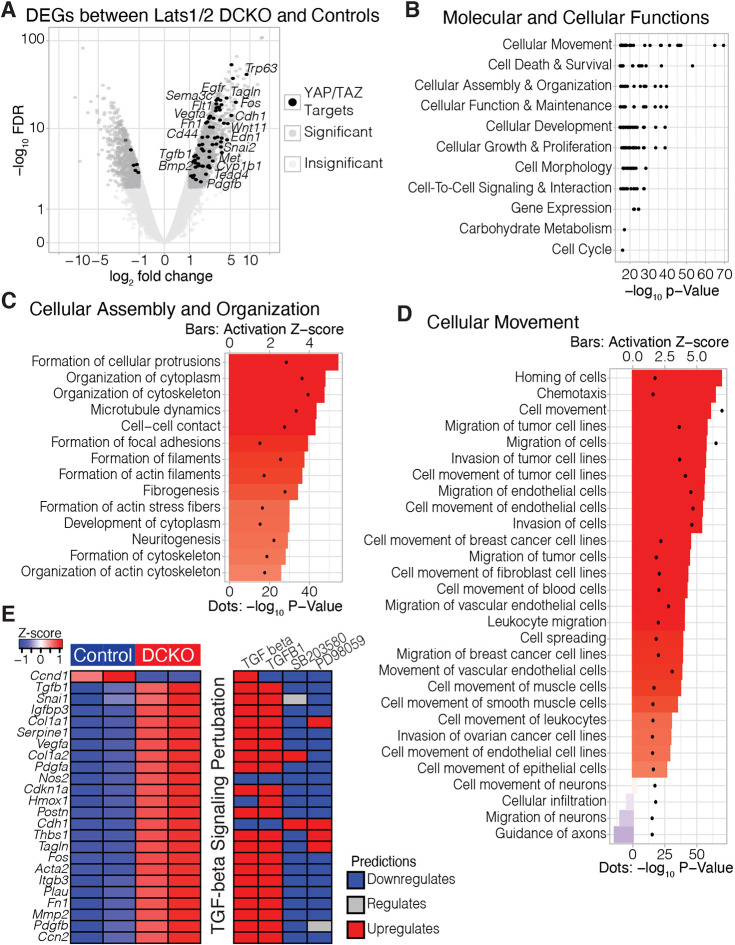
Based on the observed phenotypes, we hypothesized that the loss of Lats1/2 in neuroepithelial cells was disrupting normal morphogenetic pathways of NT formation. To identify the molecular mechanisms underlying the Lats1/2 DCKO mutant phenotype, we performed transcriptional profiling of E10.5 microdissected cranial NTs from Lats1/2 DCKO and control embryos (Fig. S3A). Quantification and recombination efficiency of Lats1 and Lats2 floxed alleles in microdissected NT tissue confirmed that the expression of Lats1/2 was significantly reduced in the neuroepithelium of Lats1/2 DCKO compared with controls (Fig. S3B,C). We identified 2134 differentially expressed genes (DEGs) [false discovery rate (FDR)<0.01; |log2 fold change (log2FC)|>1], with 1265 upregulated and 869 downregulated genes, in Lats1/2 DCKOs compared with controls (Fig. 3A, Fig. S3D,E). Some variability was detected between DCKO samples, possibly due to variation in phenotype severity and low sample number (Fig. S3E). Among the DEGs, we identified 75 genes regulated by either YAP or TAZ (Fig. 3A). The significant enrichment of YAP/TAZ targets among the upregulated genes suggested YAP/TAZ hyperactivation in the neuroepithelium.
Fig. 3.
Transcriptional profiling of Lats1/2 DCKO neural tubes shows disruption to pathways associated with cell migration, cytoskeleton and neurogenesis. (A) Volcano plot of differential gene expression between DCKO and control NTs at E10.5. Significant genes (FDR<0.01; |log2FC|>1) are in dark gray. Significant genes predicted to be upregulated by TAZ and/or YAP are highlighted in black. (B) Ranked order of the most significant molecular and cellular function terms identified by Ingenuity Pathway Analysis (IPA). Labels indicate parent terms, and black dots represent significant child terms. (C,D) Child terms under ‘cellular assembly and organization’ (C) and ‘cellular movement’ (D) ranked by activation Z-score, i.e. the consistency of gene expression changes associated with each term. Activation Z-scores are indicated by bars; P-values are represented by dots. (E) Per replicate heatmap (left) displaying the intersection of TGFB target genes identified by TGFB, TGFB1, SB203580 and PD98059, and how those genes are predicted to react to the application of any of those compounds (right).
To investigate the pathways associated with the 2134 DEGs, we utilized Ingenuity Pathway Analysis (IPA; Krämer et al., 2014). We subset the top 500 most significant disease and biofunction terms (by P-value) to only those associated with ‘molecular and cellular functions’ (Fig. 3B). Among the top parent terms, we found ‘cellular assembly and organization’ (P-value range: 4.00×10−40 to 6.79×10−16; Fig. 3C) and ‘cellular movement’ (P-value range: 2.69×10−70 to 8.34×10−16; Fig. 3D). In the absence of Lats1/2, these processes were predicted to be strongly activated in the NT. For example, ‘cell movement’ had an activation Z-score of 6.03 (P=2.69×10−70) and ‘organization of cytoskeleton’ had an activation Z-score of 4.72 (P=4.00×10−40). These findings are consistent with known Hippo signaling functions in the NC (Hindley et al., 2016; Wang et al., 2016) and other contexts (Ma et al., 2017; Sun and Irvine, 2016).
To further uncover molecular mechanisms underlying the Lats1/2 DCKO mutant phenotype, we used IPA to examine the predicted upstream regulators of the Lats1/2 DCKO DEGs. Using the more significant upstream regulators with clear directional outputs (P<1×10−10; |activation Z-score|>5), we clustered predicted upstream regulators on the basis of their downstream targets. In particular, we focused on a cluster of TGFB signaling regulators. TGFB1 was the most significant regulator (P=1.21×10−53; 364 target molecules in dataset) with the most polarized activity (activation Z-score=9.61), predicting a strong activation of TGFB1-dependent signaling. Consistent with this, Tgfb1 was significantly upregulated (log2fc=1.49; FDR=1.25×10−4) in Lats1/2 DCKO NTs. Furthermore, when we examined the intersection of targets of TGFB, TGFB1 and the inhibitors of TGFB signaling SB203580 and PD98059, we identified the consistent activation of targets associated with cell migration and cytoskeletal regulation (Fig. 3E), such as Snai1 (log2fc=1.34; FDR=2.45×10−4), Cdh1 (log2fc=3.65; FDR=2.24×10−19) and Tagln (log2fc=4.37; FDR=6.00×10−23). Together, our data suggest that, in the absence of Lats1 and Lats2 from the neuroepithelium, TGFB signaling and genes associated with cell migration are increased, consistent with reported roles of TGFB in NC development (Xu et al., 2018).
Lats1/2 deficiency promotes aberrant EMT
As indicated by our RNA-sequencing (RNA-seq) data and DEG analysis, genes involved in cell migration and movement were highly upregulated in the Lats1/2 DCKO NT compared with controls (Fig. 3D,E). After the premigratory NC population undergoes EMT, NC cells start delaminating and migrating upon EMT gene activation (Soldatov et al., 2019). We hypothesized that Lats1/2 deletion in the dorsal NT, which contains NC progenitors, may promote EMT in the NT. Hippo/YAP/TAZ signaling has been implicated in EMT regulation in other developmental contexts (Bhattacharya et al., 2020; Diepenbruck et al., 2014; Lei et al., 2008; Ling et al., 2017; Wang et al., 2016; Xiao et al., 2018). Moreover, EMT requires the loss of apicobasal polarity for epithelial cells to acquire mesenchymal characteristics (Jung et al., 2019). Furthermore, the loss of cellular attachments in the neuroepithelium could promote NC progenitors in the NT to transition into nonpolarized mesenchymal-like cells.
During development, EMT can be triggered by several transcription factors, including SNAI1, SNAI2, TWIST1, ZEB1 and ZEB2 (Gonzalez and Medici, 2014). Twist1 (log2fc=1.72; FDR=3.55×10−8), Snai2 (log2fc=2.33; FDR=3.66×10−6) and Snai1 (log2fc=1.34; FDR=2.45×10−4) transcripts were significantly upregulated in Lats1/2 DCKO NTs compared with controls (Fig. 4A). To study the role of Lats1/2 in NC EMT, we used the O9-1 cell line, which is a stable, multipotent, mesenchymal cranial NC cell line derived from Wnt1-Cre2; R26R-GFP-expressing cells of E8.5 mouse embryos (Ishii et al., 2012). We used siRNA-mediated knockdown (KD) to reduce the expression of Lats1 and Lats2 in O9-1 cells and then evaluated the expression of Snai2. Snai2 encodes a transcription factor with fundamental roles in EMT (Zhou et al., 2019) and is a known YAP target (Heallen et al., 2011; Noce et al., 2019). We found that Snai2 was upregulated in O9-1 cells treated with Lats1/2 siRNA (siL12) compared with control-treated O9-1 cells (siNC), suggesting increased EMT in the absence of Lats1/2 (Fig. 4B, Fig. S4C).
Fig. 4.
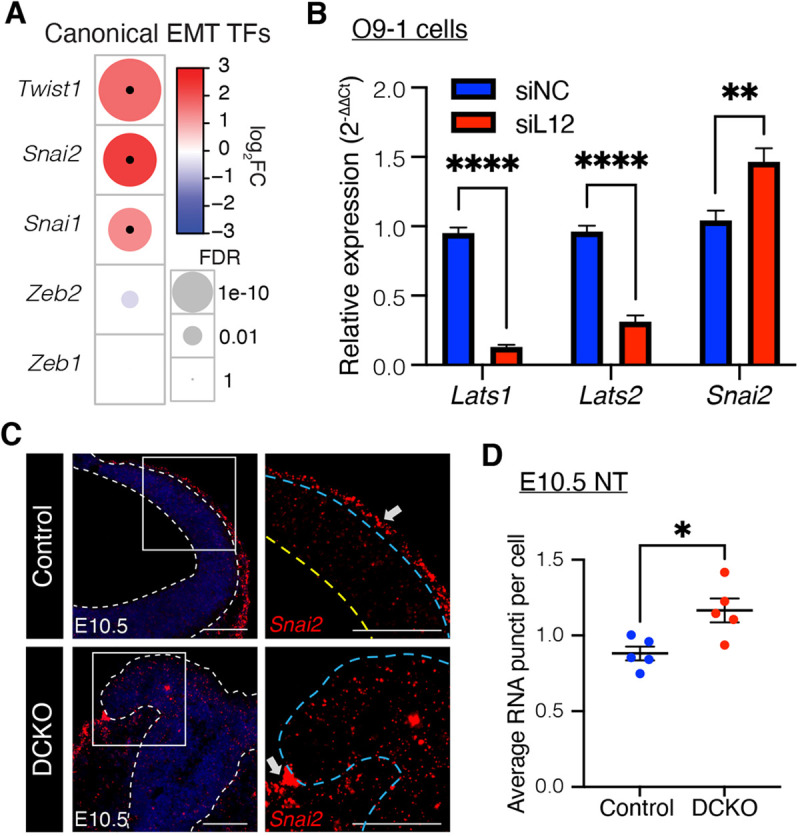
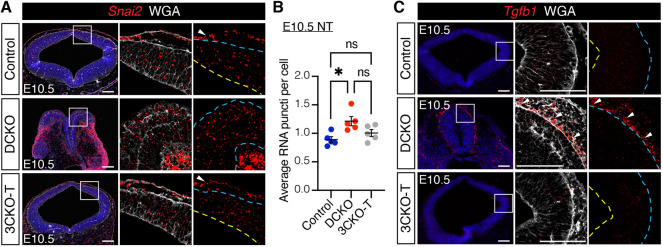
LATS1/2 regulate EMT in the neural tube. (A) Comparison of canonical epithelial-to-mesenchymal transition (EMT) transcription factors between E10.5 Lats1/2 DCKO and control NTs. Circle size represents FDR, color represents fold change, and black dots indicate significance (FDR<0.01; |log2FC|>1). (B) Relative expression of Lats1, Lats2 and Snai2 transcripts in control (siNC) and Lats1/2 knockdown (KD) (siL12) O9-1 cells. Data are from three independent experiments with two replicates each and compared using a two-tailed unpaired t-test, **P<0.01, ****P<0.0001. (C) RNAscope for Snai2 at E10.5 (red). Snai2 is detected in migrating cells between the NT and non-neural ectoderm of control NTs (top row, arrow). Snai2 transcripts are also highly expressed in the dorsal folds and neuroepithelium of DCKO NTs (bottom row, arrow). Higher magnification of boxed areas on right. Outline delineates the neuroepithelium. Yellow line indicates apical edge; blue lines indicate the basal edge. Scale bars: 100 μm. (D) Quantification of Snai2 transcript puncti per cell in NTs (control, n=5; DCKO, n=5). Data compared with a two-tailed unpaired t-test, *P<0.05. All error bars represent s.e.m.
To further investigate EMT progression in the NT, we performed fluorescence in situ hybridization (FISH) using RNAscope for Snai2. Snai2 transcript levels were significantly increased in the NT of Lats1/2 DCKO embryos compared with control embryos, at both E9.5 and E10.5 (Fig. 4C,D, Fig. S4A). Notably, Snai2 was highly expressed in migrating NC cells between the non-neural ectoderm and NT in control embryos (Fig. 4C, top row, arrow). Snai2 was also detected in migrating cells near the dorsal neural folds in Lats1/2 DCKO embryos (Fig. 4C, bottom row, arrow). Taken together, these results reveal a novel role for Lats1/2 in the progression of EMT in the cranial NT.
Lats1/2 regulates TGFB-directed EMT
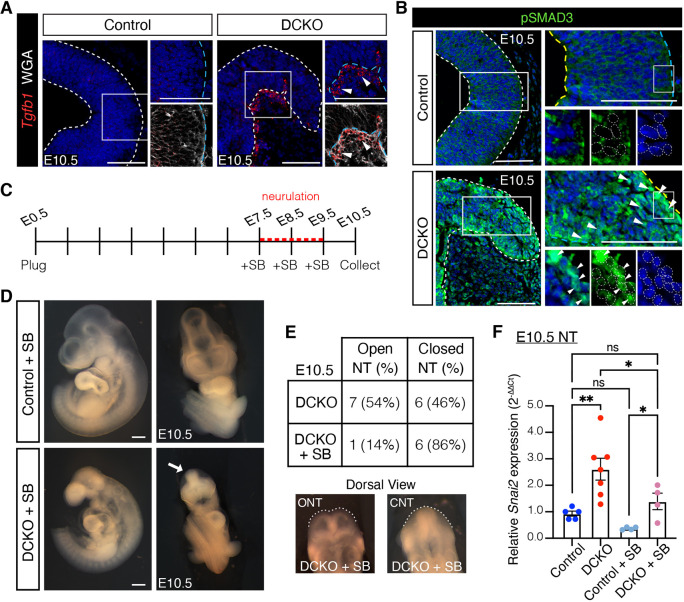
The results of transcriptional profiling predicted a significant upregulation in TGFB signaling in Lats1/2 DCKO cranial NTs (Fig. 3E). In addition, the TGFB ligand Tgfb1 was upregulated in Lats1/2 DCKO NTs. Accordingly, we found that Tfgb1 transcripts were also elevated in the NT of E9.5 Lats1/2 DCKOs (Fig. S4A). Both at E9.5 and E10.5, Lats1/2 DCKO NTs also showed increased Tgfb1 expression in basally migrating cells ventral to the neural fold area (Fig. 5A, Fig. S4A). Furthermore, we detected an increase in nuclear pSMAD3, a TGFB effector, in Lats1/2 DCKO compared with control NTs and in Lats1/2-deficient O9-1 NC cells, further supporting that TGFB signaling is upregulated in the absence of LATS1/2 regulation (Fig. 5B, Fig. S4B).
Fig. 5.
LATS1/2 regulates TGFB-directed EMT in the neural tube. (A) Tgfb1 transcripts were detected in migrating cells along the basal edge of the DCKO NT (arrowheads) by RNAscope at E10.5. Boxed areas shown at higher magnification (right). (B) Nuclear phosphorylated SMAD3 (pSMAD3) was highly expressed in DCKO neural folds compared with control neuroepithelium (arrowheads) by immunostaining at E10.5. Boxed areas shown at a higher magnification (right and bottom). Cytoplasmic signal is background. Outline delineates neuroepithelium. Nuclei are outlined in magnified panels. Yellow lines indicate apical edge; blue lines indicate basal edge. Nuclei stained with DAPI (blue). Scale bars: 100 μm. (C) TGFB inhibitor SB431542 (SB) injections were administered intraperitoneally daily for 3 days before embryo dissection at E10.5. (D) Bright-field images of treated embryos. TGFB inhibition did not alter or rescue the craniofacial region of control or DCKO embryos. Scale bars: 500 μm. (E) Top: Quantification of NT closure phenotype after TGFB inhibition. Bottom: Dorsal view of SB-treated E10.5 DCKO embryos with an open neural tube (ONT; 1) and closed neural tube (CNT; 6). (F) Relative Snai2 transcript levels in control and DCKO microdissected NT tissue in the absence or presence of SB (n=4-7 NTs). Data compared using one-way ANOVA followed by Tukey's multiple comparisons test, *P<0.05, **P<0.01. NT, neural tube; ns, not significant. All error bars represent s.e.m.
TGFB signaling induces EMT by activating the transcription of key EMT transcription factors (Moustakas and Heldin, 2016; Xu et al., 2009). To determine whether Lats1/2 deficiency results in increased TGFB-dependent EMT, we inhibited TGFB signaling using the small-molecule inhibitor SB-431542 (SB), which efficiently inhibits TGFB type I receptor (TGFBR1) and the downstream phosphorylation of SMAD2 and SMAD3 with no effects on BMP signaling (Inman et al., 2002; Vogt et al., 2011). SB treatment prevented the increased expression of pSMAD3 in Lats1/2 KD O9-1 cells (Fig. S4B). TGFB inhibition by SB also prevented Snai2 upregulation in Lats1/2 KD O9-1 cells without altering Lats1 and Lats2 transcript levels, revealing a role for TGFB signaling in EMT activation in the absence of Lats1 and Lats2 (Fig. S4C).
To determine the role of TGFB activation in Lats1/2 DCKO NTs, pregnant mice were injected intraperitoneally with 10 mg/kg of SB daily for 3 days before embryo collection (Fig. 5C). SB treatment did not disturb Lats1 and Lats2 transcript levels in the NT of SB-treated embryos (Fig. S4D). Inhibiting TGFB signaling with SB did not alter the gross morphology of Lats1/2 DCKO embryos (Fig. 5D and Fig. 1A), with the exception that most of these embryos had closed NTs at E10.5 in contrast to nontreated Lats1/2 DCKO embryos (Fig. 5E). Nevertheless, SB inhibition of TGFBR1 decreased Snai2 transcript levels in the NT of Lats1/2 DCKO embryos (Fig. 5F). Together, our data indicate that Lats1/2 regulate TGFB-directed EMT in the dorsal region of the cranial NT.
Yap/Taz deficiency rescues the neuroepithelial cell infiltration phenotype in the neural tube of Lats1/2 DCKO mutants
RNA-seq results showed that YAP/TAZ targets were significantly enriched among the genes upregulated in Lats1/2 DCKO NTs compared with control NTs (Fig. 3A). Based on this observation, we hypothesized that Lats1/2 deficiency would result in increased YAP/TAZ transcriptional activity. To test this, we generated Lats1/2 DCKO embryos with Yap/Taz heterozygosity (haplo embryos). At E10.5, Yap/Taz haploinsufficiency decreased neuroepithelial cell infiltration into the Lats1/2-deficient NT ventricle but did not rescue the craniofacial phenotype completely (Fig. 6A,B). Interestingly, the NT of haplo embryos was closed and the non-neural ectoderm layer fused (Fig. 6B). Moreover, mitotic cells in haplo embryos contoured the edge between the NT and the infiltrating cells (Fig. 6C), revealing some suppression of the neuroepithelial cell polarization defect in Lats1/2 DCKOs. Yap/Taz haploinsufficiency did not alter neuroepithelial cell proliferation (Fig. 6D).
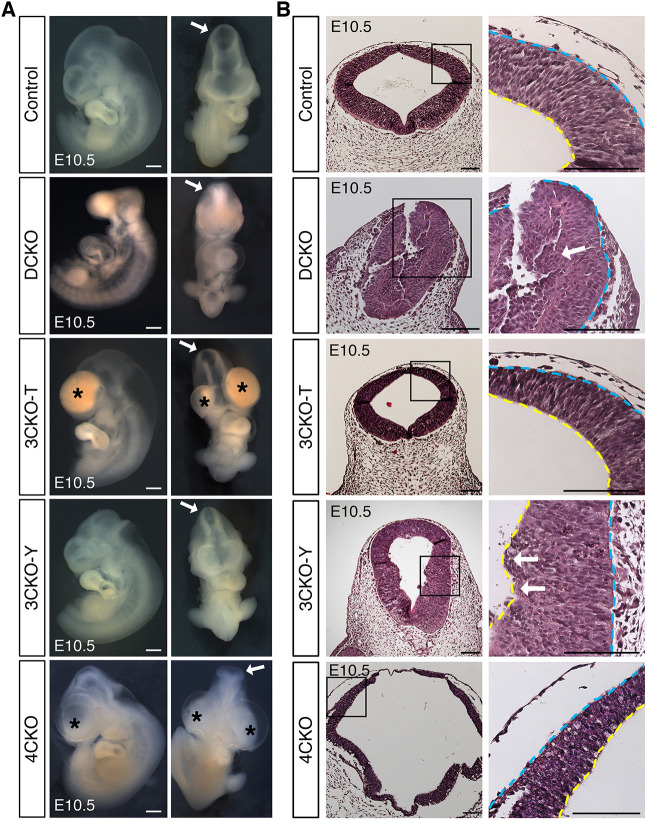
We next generated conditional quadruple mutants deficient in Lats1, Lats2, Yap and Taz (4CKO). The deletion of both Yap and Taz in addition to Lats1/2 in the embryonic NT resulted in craniofacial phenotypes that were different from those seen in the Lats1/2 DCKOs at E10.5, including enlarged and hemorrhaging forebrain and branchial arches, as well as closed but folded and misshapen NTs (Fig. 7A, Table S4). All Lats1/2 DCKOs had NT, forebrain and pharyngeal arch defects. Remarkably, Wnt1-Cre2, Lats1F/Δ, Lats2F/Δ, YapF/+, TazF/Δ mutant embryos (3CKO-Y) with Yap haploinsufficiency had a closed NT without any obvious craniofacial phenotypes compared with control embryos (Fig. 7A, Table S4). Only one 3CKO-Y embryo was found among several litters collected, although this was not significantly different from expected Mendelian ratios. Instead, Wnt1-Cre2, Lats1F/Δ, Lats2F/Δ, YapF/Δ, TazF/+ mutant embryos (3CKO-T) with Taz haploinsufficiency also showed a closed NT comparable to that of controls, but abnormalities were seen in the forebrain and mandible (Fig. 7A, Table S4). Embryonic deletion of Yap in the NC has been associated with disrupted craniofacial structures, including enlarged branchial blood vessels, hemorrhage in the forebrain and mandible, and NT abnormalities in mice (Wang et al., 2016).
Fig. 7.
Hippo signaling maintains neuroepithelial cell behavior in the dorsal neural tube. (A) Bright-field images of E10.5 rescue embryos. 3CKO-T and 3CKO-Y embryos showed comparable midbrain morphology to that of control (arrows). 3CKO-T and 4CKO embryos had forebrain and pharyngeal arch defects (asterisks). Scale bars: 500 μm. (B) H&E staining of coronal sections. Neuroepithelium integrity is recovered in 3CKO NTs. Some apical extrusion into the ventricle is detected in 3CKO-Y NTs (arrows), like the infiltration sites in DCKO NTs. Boxed areas are shown at a higher magnification (right). Scale bars: 100 μm. Yellow lines indicate apical edge; blue lines indicate basal edge.
Histological evaluation indicated that the cranial NT of 3CKO-T embryos was comparable to that of control embryos; however, in the 3CKO-Y NT, we still detected the apical edge disruption and cell infiltration phenotypes observed in the NT of Lats1/2 DCKO embryos (Fig. 7B). Although the 4CKO embryos showed NTDs, the cell infiltration phenotype was not present (Fig. 7B). Importantly, the NTs of 3CKO-T and 3CKO-Y embryos were closed, and both the neuroepithelium and the non-neural ectoderm were fused correspondingly (Fig. 7B). Neuroepithelial cells retained their pseudostratified structure, and mitotic cells were found on the apical edge of 4CKO, 3CKO-T and 3CKO-Y embryos (Fig. S5A). Yap/Taz modulation, particularly Yap deletion/Taz haploinsufficiency in 3CKO-T embryos, rescued the neuroepithelial phenotypes in response to Lats1/2 deficiency. Therefore, the regulation of YAP/TAZ by LATS1 and LATS2 is required for neuroepithelial cell integrity and architecture.
YAP activation stimulates TGFB signaling-induced EMT in the dorsal neural tube
To further determine the role of Hippo signaling in EMT progression, we compared transcript levels of Snai2 in the NT of 3CKO-T Yap/Taz rescue embryos with those of control and Lats1/2 DCKO embryos. Like control embryos, 3CKO-T rescue embryos expressed Snai2 transcripts in the neuroepithelium and in migrating NC cells between the NT and the non-neural ectoderm (Fig. 8A). Snai2 transcript levels were decreased in 3CKO-T rescue embryos compared with Lats1/2 DCKO embryos (Fig. 8A,B). Thus, these findings suggest that Hippo signaling blocks neuroepithelial cell migration into the ventricular space of the NT and aberrant EMT.
Fig. 8.
Hippo signaling controls YAP-directed activation of EMT. (A) Snai2 transcripts were detected in E10.5 neuroepithelial and migrating cells between the NT and non-neural ectoderm of control and 3CKO-T NTs (arrowheads) by RNAscope. (B) Quantification of Snai2 puncti per cell in the NT (control, n=5; DCKO, n=5; 3CKO-T, n=5). Data compared using one-way ANOVA followed by Tukey's multiple comparisons test, *P<0.05. NT, neural tube; ns, not significant. Error bars represent s.e.m. (C) At E10.5, Tgfb1 transcripts were increased in neuroepithelial and basally migrating cells of DCKO NTs (arrowheads) but not in those of control and 3CKO-T rescue NTs by RNAscope. Nuclei stained with DAPI (blue). Cell membranes stained with WGA (white). Boxed areas are shown at a higher magnification (right). Scale bars: 100 μm. Outline delineates neuroepithelium, with yellow lines indicating apical edge and blue lines indicating basal edge.
Previously, crosstalk between YAP/TAZ and the TGFB/SMAD2/3 pathway was reported, whereby YAP/TAZ promote the nuclear localization of regulatory SMADs (Alarcón et al., 2009; Narimatsu et al., 2015; Pefani et al., 2016; Varelas et al., 2008, 2010). Yap/Taz modulations in 3CKO-T rescue embryos not only decreased Snai2 expression in the NT but also reduced Tgfb1 ligand expression in the neuroepithelium to that seen in the control (Fig. 8C). Furthermore, the Tgfb1+ migrating cells detected in Lats1/2 DCKO embryos were not detected when Yap was deleted in 3CKO-T rescue embryos (Fig. 8C). Therefore, the regulation of YAP by LATS1/2 is required to prevent the upregulation of TGFB signaling and dysregulated TGFB-induced EMT in the NT and NC (Fig. 9). Our data suggest that crosstalk between the downstream Hippo signaling effectors YAP/TAZ and the TGFB signaling pathway promote EMT in the dorsal NT that is in part regulated by upstream Hippo signaling kinases.
Fig. 9.
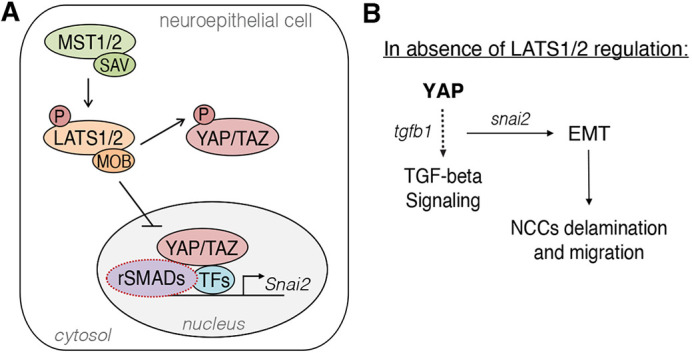
LATS1 and LATS2 phosphorylate downstream effectors YAP/TAZ to regulate TGFB-induced EMT. (A) LATS1/2 are necessary to restrict YAP/TAZ activity in the cranial neuroepithelium. When not phosphorylated, YAP/TAZ translocate into the nucleus and interact with other co-activators, and possibly receptor-regulated SMADs (rSMADs), to activate the transcription of Snai2, among other targets. P, phosphorylation. (B) Lats1/2 deficiency triggered TGFB signaling and EMT in the cranial NT. YAP hyperactivation and TGFB upregulation promoted EMT in the neuroepithelium, which led to loss of polarity and neuroepithelial cell delamination and migration. NCC, neural crest cell.
DISCUSSION
Hippo signaling is essential for craniofacial development
Deletion of both Lats1 and Lats2 in the neuroepithelium caused craniofacial and NT abnormalities with embryonic lethality; however, one copy of Lats1 or Lats2 circumvented both phenotypes. Thus, Lats1 and Lats2 likely have redundant roles during early embryonic development in the dorsal anterior NT, consistent with other developmental contexts (Lorthongpanich et al., 2013; Reginensi et al., 2016).
An interplay of signals from surrounding tissues, including the non-neural ectoderm and mesoderm, promote neural induction of the neural plate and, consequently, NC development within the dorsal folds of the closing NT (Golding et al., 2000; Grenier et al., 2009; Trainor and Krumlauf, 2000). We found that most Lats1/2 DCKOs lacked non-neural ectoderm fusion due to the NTDs. Because the non-neural ectoderm does not fuse in Lats1/2 DCKOs, the absence of its juxtaposing configuration to the NT can also affect NC behavior (Dickinson et al., 1995; Moury and Jacobson, 1990; Selleck and Bronner-Fraser, 1995).
The regulation of YAP/TAZ by LATS1/2 is required for craniofacial development. Expressed specifically in craniofacial tissues, the transcription factor FOXO6 activates Lats1/Lats2 expression, increasing YAP/TAZ phosphorylation and Hippo signaling activation to restrict proliferation and control growth and morphogenesis of the face postnatally (Sun et al., 2018). Although Hippo signaling is widely associated with proliferation control, we found that modulation of the Hippo kinases LATS1/2 in the NC did not alter neuroepithelial proliferation. Likewise, genetic deletion of Hippo effectors Yap/Taz in the NC does not affect proliferation or neuroepithelial integrity in the NT despite playing a role in cranial NC development and differentiation (Hindley et al., 2016; Manderfield et al., 2014; Wang et al., 2016). Conditional deletion of Yap/Taz in the cranial NC results in enlarged branchial arch blood vessels, vascular hemorrhaging and craniofacial defects due to impaired smooth muscle differentiation (Manderfield et al., 2015; Wang et al., 2016).
Intriguingly, Lats1/2 deficiency in NC cells resulted in only cranial NT phenotypes, with no obvious phenotypes in the posterior NT. NC cells contribute to multiple cell types and tissues throughout the whole embryo (Gandhi and Bronner, 2021). Cranial NC cells make major contributions to the skeletal, connective and neurogenic tissues of the craniofacial complex, while other NC cells give rise to neurons and glia of the peripheral nervous system, connective tissue of cardiac structures, secretory cells of the endocrine system and pigment cells of the skin (Bhatt et al., 2013; Gandhi and Bronner, 2021; George et al., 2020; Martik and Bronner, 2017; Vega-Lopez et al., 2017). Furthermore, only the cranial NC population generates ectomesenchymal derivatives (Bhatt et al., 2013; Lee et al., 2013; Soldatov et al., 2019; Zalc et al., 2021).
Recently, decreased LATS1/2 protein levels were reported in neural progenitor cells from anencephalic human fetuses with a deleterious recessive mutation in NUAK2 (Bonnard et al., 2020). NUAK2 alterations disrupt the apical actomyosin network in NT-like structures due to insufficient Hippo–YAP signaling, preventing neural plate folding and fusion (Bonnard et al., 2020). As the neural plate folds, the neural plate border specifier genes activate the expression of NC-specifier genes such as Foxd3, Sox9/10, Myc, Id2, Snai1/2 and Ets1 (Martik and Bronner, 2021; Simões-Costa and Bronner, 2015; Simões-Costa et al., 2014). Importantly, the NC gene regulatory network differs between axial levels; e.g. the transcription factor Ets1, which regulates Sox10 and Foxd3, is uniquely expressed in the cranial, but not trunk, NC (Barembaum and Bronner, 2013; Rothstein et al., 2018; Théveneau et al., 2007). We found that Ets1 was significantly upregulated in the NT of Lats1/2 DCKO embryos (log2fc=1.44; FDR=1.17×10−4), supporting our hypothesis that cranial NC expansion occurs in the absence of LATS1/2 regulation.
Hippo kinases LATS1/2 likely have essential and specific roles in cranial NT closure due to their unique ectomesenchymal potential, suggesting that Hippo signaling may be involved in the gene regulatory network of cranial NC cells. Furthermore, tissue mechanical forces are required for craniofacial morphogenesis, and the Hippo pathway has been reported to play crucial roles in mediating mechanical signals (Du et al., 2021; Dupont et al., 2011; Schroeder and Halder, 2012; Wang and Martin, 2017; Wu and Guan, 2021). Therefore, Hippo signaling regulation at the dorsal cranial NT appears to be required for NT closure, NC expansion, and NC migration and diversification.
Lats1/2 are required for neuroepithelial cell apicobasal attachment and TGFB-induced EMT regulation
LATS kinases have been associated with maintenance of epithelial cell integrity. In Caenorhabditis elegans, the Lats homolog wts-1 was found in the subapical region in the intestine, where it maintains apical and junctional protein localization through YAP regulation (Kang et al., 2009; Lee et al., 2019). Moreover, genetic ablation of Lats1/2 in the developing lung and pancreas of mice results in altered epithelial branching due to apicobasal polarity defects, aberrant EMT initiation and increased cell differentiation through YAP/TAZ (Braitsch et al., 2019; Nantie et al., 2018). Collectively, our data indicate an evolutionarily conserved function of LATS kinases in the regulation of apicobasal protein localization in NT development.
Neuroepithelial cells in the dorsal NT include NC cells that delaminate and undergo EMT before migrating across the embryo. Our RNA-seq data from Lats1/2 DCKO NTs suggested that, in addition to increased TGFB signaling, changes occurred in cellular movement, morphology and organization (Fig. 3). The loss of neuroepithelial integrity and the upregulation of EMT transcription factors in the Lats1/2 DCKO NT indicated that aberrant EMT initiation and dysregulated NC cell movement occurred in the absence of LATS1/2 regulation. Loss of polarity and EMT are required for NC delamination during development; however, the dysregulation of these processes has been associated with migratory and invasive characteristics in cancer (Bucay et al., 2017; David et al., 2016; Jung et al., 2019; Thiery et al., 2009). Moreover, the TGFB-induced overexpression of the EMT transcription factor Snai2 has been associated with tumorigenesis and cancer progression (Bucay et al., 2017; Li et al., 2014; Slabáková et al., 2011; Zhou et al., 2019). We found that Snai2 levels were upregulated in neuroepithelial tissue and infiltrating cells of Lats1/2 DCKO NTs compared with controls. The inhibition of TGFB signaling partially suppressed the NT closure defect seen in Lats1/2 DCKO embryos in addition to preventing increased Snai2 transcription, providing functional evidence that LATS1/2 regulation prevents dysregulated TGFB signaling in cranial neural folds.
LATS1/2 control neural crest EMT through YAP/TAZ
YAP/TAZ transcriptional targets were upregulated in the absence of Lats1/2 (Fig. 3A). Combining Yap and/or Taz deficiency into Lats1/2 DCKOs rescued the embryonic neuroepithelial phenotypes. Lats deficiency is associated with loss of polarity and branching morphogenesis disruption, and YAP/TAZ activity plays a central role in these phenotypes (Braitsch et al., 2019; Nantie et al., 2018; Reginensi et al., 2016). The suppression of neuroepithelial and NTDs in Lats1/2 DCKOs by reducing YAP/TAZ dosage supports the notion that a Hippo signaling-dependent mechanism is required for NC induction and delamination.
Recent studies have confirmed that Hippo signaling is essential for NC development (Bhattacharya et al., 2020; Hindley et al., 2016; Kumar et al., 2019; Wang et al., 2016). Yap/Taz deficiency in the NC leads to early lethality of mutant embryos with severe craniofacial phenotypes, including an abnormal NT, disrupted facial mesenchyme and vascular defects (Manderfield et al., 2014; Wang, 2020; Wang et al., 2016). In Yap/Taz rescue embryos, additional craniofacial phenotypes are associated with Yap deficiency. Interestingly, all mutant embryos from the genetic rescue experiments lacking both alleles of Yap, regardless of Lats1, Lats2 or Taz allele number or combination, exhibited enlarged and/or hemorrhaging forebrain and/or branchial arches (Table S4). However, in 3CKO-T rescue embryos, Yap deletion and Taz haploinsufficiency reverted the neuroepithelial phenotypes caused by the loss of Lats1/2. Instead, Yap haploinsufficiency and Taz deletion in 3CKO-Y rescue embryos partially rescued the neuroepithelial phenotypes caused by the loss of Lats1/2. Although YAP and TAZ have redundant roles during development, the variation in rescue phenotypes after Yap/Taz modulation can be attributed to the functional differences between Yap and Taz and differences in their expression at distinct times throughout development.
Hippo and TGFB pathways crosstalk to regulate EMT in the dorsal neural tube
The TGFB superfamily is essential for NT patterning and NC proliferation and differentiation during development (Chesnutt et al., 2004; Iwata et al., 2010; Tang et al., 2010; Wurdak et al., 2005). Activation of TGFB signaling results in phosphorylation of SMAD2/3 in the TGFB pathway, promoting their nuclear accumulation and transcriptional activity. Although nuclear pSMAD2/3 activity is enhanced by the nuclear translocation of YAP/TAZ, the regulation of YAP/TAZ by Hippo signaling prevents the nuclear accumulation and activation of SMADs (Nishio et al., 2016; Varelas et al., 2008). Moreover, Hippo signaling prevents YAP/TAZ transcriptional activity and restricts TGFB activation in response to cell density and changes in polarity (Narimatsu et al., 2015; Varelas et al., 2010). The NT is composed of pseudostratified neuroepithelial cells packed at high cell density. We showed that Hippo signaling inhibition in the NT resulted in abnormal neuroepithelial cell apicobasal attachment, EMT and cellular infiltration into the neural ventricle due to YAP/TAZ hyperactivation. Interestingly, cranial NC cells lost apicobasal polarity and underwent EMT, decreasing collective cell density. Our results suggest an expansion of the NC population that initiates EMT and infiltrates the NT ventricle.
In the absence of Hippo signaling regulation in the NT, TGFB signaling was increased, which was reverted by YAP deficiency. Others have suggested that a positive regulation exists between the TGFB/SMAD pathway and YAP/TAZ activation. In a lung cancer cell line, TGFB stimulates the degradation of the Hippo-signaling scaffold RASSF1A, allowing YAP to interact with SMAD2. RASSF1A regulates the TGFB-induced YAP/SMAD2 interaction and promotes SMAD2 cytoplasmic retention limiting transcriptional activity (Pefani et al., 2016). In the mouse liver, TGFB ligand production is increased by YAP/TAZ nuclear translocation, activation and interaction with SMAD2/3, whereas YAP is activated through TGFB signaling (Nishio et al., 2016). Moreover, genetic and chemical modulation of TGFB decreased YAP transcriptional activity. YAP also triggers early endothelial-to-mesenchymal transition (EndMT), an endothelial-specific form of EMT, in lung-derived endothelium (Savorani et al., 2021). Although YAP/TAZ stimulate SMAD activity to regulate TGFB-induced cellular responses during development and cancer biology, TGFB signaling can also positively regulate YAP/TAZ.
In conclusion, we have elucidated the role of the Hippo signaling kinases LATS1 and LATS2 in the neuroepithelial cell population in the dorsal cranial NT. LATS1/2 maintain neuroepithelial cell architecture in the NT by regulating YAP/TAZ activity (Fig. 9). YAP/TAZ hyperactivity promotes EMT through interaction with TGFB signaling components. Therefore, Hippo signaling is required to control NC delamination and movement in the NT.
MATERIALS AND METHODS
Mice
All animal experiments performed in this study were approved by the Baylor College of Medicine Institutional Review Board. The Wnt1-Cre2, Lats1F/F, Lats2F/F, YapF/F, TazF/F and mTmG mouse lines and alleles used in this study have been previously described (Heallen et al., 2011; Lewis et al., 2013; Muzumdar et al., 2007; Xin et al., 2011). These mouse lines are maintained by the research laboratory in the Transgenic Mouse Facility at Baylor College of Medicine. Tail genomic DNA was used for genotyping by PCR.
Wnt1-Cre2, Lats1F/+, Lats2F/+ males were crossed with Lats1F/F, Lats2F/F females to generate neuroepithelium-specific conditional Lats1/2 mutants. The Wnt1-Cre2 transgene has been reported to be active in the male germline, and therefore, the male mouse passes on recombined (Δ) alleles irrespective of Wnt1-Cre2 inheritance (Dinsmore et al., 2022). Females were checked for plugs in the morning. Noon on the day a plug was discovered was considered as E0.5. Embryos were collected at different time points in phosphate-buffered saline (PBS), and yolk sacs were used for genotyping. Embryos were somite matched when possible. For genetic rescue experiments, Wnt1-Cre2, Lats1F/+, Lats2F/+ or Wnt1-Cre2, Lats1F/+, Lats2F/+, YapF/+, TazF/+ males were crossed with Lats1F/F, Lats2F/F, YapF/F, TazF/F females to generate neural crest-specific Lats1/2 mutants with varying alleles of Yap and Taz. Rescue embryos were collected at E10.5 in PBS, and yolk sacs were used for genotyping. The mouse genotype nomenclature used is described in Table S1.
Tissue processing and histology
All embryos were collected at the chosen time points in PBS and fixed in 10% formalin or 4% paraformaldehyde (PFA). Fixed embryos were dehydrated through a series of ethanol washes, followed by xylene clearing and paraffin embedding. The embryos were embedded carefully to maintain the same orientation and were sectioned at 5-7 µm. Sections were carefully chosen to represent matching regions between control and mutant embryos. For Hematoxylin and Eosin (H&E) staining, sections were rehydrated, stained with Mayer's Hematoxylin and Alcoholic Eosin Y, dehydrated in ethanol, cleared in xylene and mounted.
Tissue immunofluorescence and FISH
Immunofluorescence and FISH were performed on formalin-fixed and paraffin-embedded sections. For immunofluorescence, sections were rehydrated and processed for antigen retrieval. Antigens were retrieved by boiling the sections in Antigen Unmasking Solution (Vector Labs, H-3300). Tissues were permeabilized by incubating sections in 0.5% Triton X-100 for 15 min and blocked in 10% donkey serum for 1 h. Primary antibodies against phospho-YAP (Cell Signaling Technology, 4911; 1:100), beta-catenin (Cell Signaling Technology, 8480; 1:200), pHH3 (Cell Signaling Technology, 9701; 1:100), GFP (Abcam, ab6673; 1:200), N-cadherin (GeneTex, GTX127345; 1:200), laminin (Novus, NB300-14; 1:200) and pSMAD3 (Novus, NBP1-77836SS; 1:200) were used at the dilutions recommended by the manufacturer and were visualized with Alexa fluorochrome-conjugated secondary antibodies. pSMAD3 staining was amplified using tyramide signal amplification (TSA). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). For FISH, RNAscope was performed according to the manufacturer's instructions with the RNAscope 2.5 HD Assay-RED protocol (ACD) using Probe-Mm-Snai2 (ACD, 451191) and Probe-Mm-Tgfb1 (ACD, 407751). Nuclei counterstaining was performed using DAPI for 10 min at room temperature. WGA conjugates were added and incubated with DAPI to outline neuroepithelial cells. Images were taken using a Zeiss LSM 780 confocal microscope.
Image analysis and quantification
The fluorescence intensity of beta-catenin in the neural tube was measured on original CZI files using the line intensity processing tool in the Zen Black microscopy software (Carl Zeiss, Germany). For each section, multiple lines were manually drawn from the basal membrane to the apical edge of the neural tube across the apicobasal axis. Beta-catenin staining intensity across each line was recorded. Owing to the difference in neural tube width between samples, line profiles were normalized to a range of 0.0 to 1.0 from basal to apical boundaries, respectively (Fig. 2F, top). The normalized distance was binned into 100 intervals (intervals of 0.01), and the fluorescence intensities within each interval were averaged. The final basal to apical intensity line profile (Fig. 2F, bottom) represents the average of all intensity profiles measured from control and DCKO embryos. The P-value for the difference in intensity was calculated for each bin along the 100 intervals of the line profile.
Neural tube width and immunofluorescence for pHH3, pSMAD3 and Snai2 were quantified manually using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Neural tube width was measured on WGA-stained sections, from the basal membrane to the apical edge. For pHH3 immunofluorescence quantification, pHH3 percentages were calculated as the number of positive-staining nuclei over the total number of nuclei in the neural tube. For Snai2 RNAscope quantification, individual dots were counted and divided by the total number of cells to obtain the average number of puncti per neuroepithelial cell in the neural tube. All analyses included data from at least three embryos of each genotype unless mentioned otherwise.
µCT
E10.5 embryos were collected in PBS and fixed in 4% PFA overnight at 4°C. The next day, samples were washed with PBS and immersed in 0.1 N iodine solution overnight at room temperature. Embryos were mounted in 1% agarose in microtubes and scanned on a Bruker SKYSCAN 1272 scanner. Images were acquired at 3 µm/pixel at 70 kV and 142 µA with a 0.5-mm aluminum filter. Tomography projection images were reconstructed using SkyScan NRecon (Bruker, Belgium) and visualized with CTVox (Bruker, Belgium) and Imaris (Bitplane, Switzerland) software.
Real-time quantitative PCR (qPCR)
Total RNA was isolated from the microdissected cranial neural tubes of E10.5 embryos using an RNeasy Plus Micro Kit (Qiagen) and processed for cDNA synthesis using Super Script II Reverse Transcriptase (Invitrogen). All real-time thermal cycling was performed with a StepOne Real-time PCR System (Thermo Fisher Scientific). SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) was used for real-time thermal cycling. Relative gene expression, calculated with the Livak–Schmittgen method 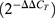 , was normalized to that of housekeeping genes Hprt or Gapdh. Graphs represent the average fold change relative to control samples, and error bars are shown, indicating s.e.m.
, was normalized to that of housekeeping genes Hprt or Gapdh. Graphs represent the average fold change relative to control samples, and error bars are shown, indicating s.e.m.
Transcriptional profiling and analysis
Bulk RNA was isolated from E10.5 microdissected cranial neural tubes (two DCKO and two controls) using an RNeasy Plus Micro Kit in accordance with the manufacturer's instructions (Qiagen). Paired-end, polyA-enriched RNA libraries were generated with a Kapa stranded RNA-seq kit (KR0960), according to the manufacturer's instructions, and were pooled in equimolar amounts. Sequencing was performed using Illumina NextSeq500 for 75 bp reads. Libraries were sequenced to a depth of 38 to 47 million paired reads.
Transcripts were aligned to the mouse genome (GRCm38/mm10, retrieved from the UCSC genome browser, genome.ucsc.edu, 23 May 2012) using TopHat2 v2.1.1 (Kim et al., 2013; Trapnell et al., 2009). Low-quality reads were filtered with bamtools using the following settings: -isMapped true -isDuplicate false -mapQuality “>10” (Barnett et al., 2011). Reads were mapped and counted after alignment using StringTie v1.3.3 (Pertea et al., 2015, 2016). Differential expression testing was performed using edgeR v3.18.1 (McCarthy et al., 2012; Robinson and Smyth, 2007, 2008; Robinson et al., 2010; Zhou et al., 2014) and limma v3.30.13 (Ritchie et al., 2015) packages in R v3.4.0. Genes with low expression were removed within each condition using median log2-transformed counts per gene per million mapped reads of 1 and a union generated from each condition. Differential expression analysis was performed using a general linear model framework in edgeR. Downstream analysis was performed using IPA (Qiagen; content build 16 June 2019; Krämer et al., 2014). Data were visualized using the gplots v3.0.1.1 (https://github.com/talgalili/gplots), RColorBrewer v1.1-2 (https://CRAN.R-project.org/package=RColorBrewer), ggplot2 v3.1.0 (Wickham, 2016), scales v1.0.0 (https://CRAN.R-project.org/package=scales) and stringr v1.4.0 (https://CRAN.R-project.org/package=stringr) packages in R. The data were deposited in Gene Expression Omnibus (GEO) with the accession number GSE182721.
O9-1 cell culture
For in vitro experiments, the cranial neural crest cell line O9-1 was used (Ishii et al., 2012; Nguyen et al., 2018). Cells were obtained from Sigma-Aldrich (SCC049), and the laboratory stock was examined for mycoplasma contamination on arrival and prior to experimentation. O9-1 cells were maintained undifferentiated in basal medium containing Dulbecco's modified Eagle medium supplemented with 15% fetal bovine serum, 0.1 mM MEM nonessential amino acids, 1 mM sodium pyruvate, 55 µM β-mercaptoethanol, 100 units/ml penicillin/streptomycin, 2 mM L-glutamine, 25 ng/ml basic fibroblast growth factor (R&D Systems, 233-FB) and 1000 U leukemia inhibitory factor (Millipore, ESG1106). For the siRNA KD experiments in O9-1 cells, siRNA SMARTpools targeting Lats1 and Lats2 were purchased from Dharmacon, and the transfections were performed according to a typical RNAiMAX transfection procedure (Thermo Fisher Scientific). RNA was extracted 48 h after transfection to determine Snai2 transcript levels. For immunofluorescence, O9-1 cells were fixed in 10% formalin for 10 min, permeabilized in 0.5% Triton X-100 for 15 min and blocked in 10% donkey serum for 1 h. A primary antibody for pSMAD3 (Santa Cruz Biotechnology, sc-517575) was used at the dilution recommended by the manufacturer and was visualized with Alexa fluorochrome-conjugated secondary antibodies. Nuclei were stained with DAPI. Experiments with O9-1 cells were performed in duplicate, and all experiments were repeated at least three times.
TGFB inhibition
To inhibit TGFB signaling both in vivo and in vitro, we used the small-molecule inhibitor SB-431542 (Tocris, 1614). Stock solutions of SB-431542 were prepared in dimethyl sulfoxide (DMSO) according to the manufacturer's recommendations. For in vivo experiments, SB-431542 (10 mg/kg in 5% DMSO) was administered intraperitoneally daily to pregnant mice at E7.5, E8.5 and E9.5, and embryos were collected at E10.5. Embryos were imaged and processed, or neural tubes were microdissected for RNA extraction and analyzed with qPCR. For in vitro experiments, O9-1 cells were treated with SB-431542 (10 µM) at the time of siRNA KD. After 48 h, untreated and treated cells were analyzed with immunofluorescence and qPCR to determine the effect of TGFB inhibition.
Statistical analyses
Statistical analyses were performed using GraphPad Prism version 9.0.0 for macOS (GraphPad Software, San Diego CA, USA). A chi-squared test was used for analyzing collected embryos numbers. A two-sample Z-test was used to determine the significance of the proportion of embryos with open neural tubes between control and Lats1/2 DCKO embryos and between SB treated and nontreated Lats1/2 DCKO embryos. Two-tailed unpaired t-test and one-way ANOVA were used to determine significance for subsequent experiments. P-values <0.05 were considered significant. The results are expressed as the mean±s.e.m. The number of samples and the statistical tests used are described in the figure legends.
Supplementary Material
Acknowledgements
We thank Jason Kirk and the Optical Imaging and Vital Microscopy Core at Baylor College of Medicine for support on imaging and imaging processing. We also thank Dr Nicole Stancel for editorial assistance in the revision of the manuscript.
Footnotes
Author contributions
Conceptualization: I.M.M.T., J.W., J.F.M.; Methodology: I.M.M.T.; Software: J.D.S.; Validation: I.M.M.T.; Formal analysis: I.M.M.T., J.D.S.; Investigation: I.M.M.T., X.Z.; Resources: I.M.M.T., J.D.S., X.Z., J.W., J.F.M.; Data curation: J.D.S.; Writing - original draft: I.M.M.T., J.D.S., J.W., J.F.M.; Writing - review & editing: I.M.M.T., J.D.S., J.W., J.F.M.; Visualization: I.M.M.T., J.D.S.; Supervision: J.W., J.F.M.; Project administration: J.F.M.; Funding acquisition: J.W., J.F.M.
Funding
This work was supported by grants from the National Institutes of Health (R01HL127717, R01HL130804 and R01HL118761 to J.F.M.; K01DE026561, R03DE025873, R01DE029014, R56HL142704 and R01HL142704 to J.W.; T32GM088129 to I.M.M.T.; T32HL139430 and F32HL156465 to J.D.S.), the Vivian L. Smith Foundation (J.F.M.), the LeDucq Foundation Transatlantic Networks of Excellence in Cardiovascular Research (14CVD01 to J.F.M.), the MacDonald Research Fund Award (16RDM001 to J.F.M.) and the Saving Tiny Hearts Society (J.F.M.). Deposited in PMC for release after 12 months.
Data availability
Data were deposited in the GEO database (accession number GSE182721).
Contributor Information
Jun Wang, Email: jun.wang@uth.tmc.edu.
James F. Martin, Email: jfmartin@bcm.edu.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200860.reviewer-comments.pdf
References
- Adzick, N. S., Thom, E. A., Spong, C. Y., Brock, J. W., Burrows, P. K., Johnson, M. P., Howell, L. J., Farrell, J. A., Dabrowiak, M. E., Sutton, L. N.et al. (2011). A randomized trial of prenatal versus postnatal repair of myelomeningocele. N. Engl. J. Med. 364, 993-1004. 10.1056/NEJMoa1014379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón, C., Zaromytidou, A.-I., Xi, Q., Gao, S., Yu, J., Fujisawa, S., Barlas, A., Miller, A. N., Manova-Todorova, K., Macias, M. J.et al. (2009). Nuclear CDKs drive smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757-769. 10.1016/j.cell.2009.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barembaum, M. and Bronner, M. E. (2013). Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Dev. Biol. 382, 567-575. 10.1016/j.ydbio.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett, D. W., Garrison, E. K., Quinlan, A. R., Strömberg, M. P. and Marth, G. T. (2011). BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691-1692. 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye, L. M. and Link, B. A. (2007). Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J. Neurosci. 27, 10143-10152. 10.1523/JNEUROSCI.2754-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, S., Diaz, R. and Trainor, P. A. (2013). Signals and switches in mammalian neural crest cell differentiation. Cold Spring Harb. Perspect. Biol. 5, a008326. 10.1101/cshperspect.a008326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, D., Azambuja, A. P. and Simoes-Costa, M. (2020). Metabolic reprogramming promotes neural crest migration via Yap/Tead signaling. Dev. Cell 53, 199-211.e6. 10.1016/j.devcel.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, H., Kancherla, V., Moorthie, S., Darlison, M. W. and Modell, B. (2018). Estimates of global and regional prevalence of neural tube defects for 2015: a systematic analysis. Ann. N. Y. Acad. Sci. 1414, 31-46. 10.1111/nyas.13548 [DOI] [PubMed] [Google Scholar]
- Bonnard, C., Navaratnam, N., Ghosh, K., Chan, P. W., Tan, T. T., Pomp, O., Ng, A. Y. J., Tohari, S., Changede, R., Carling, D.et al. (2020). A loss-of-function NUAK2 mutation in humans causes anencephaly due to impaired Hippo-YAP signaling. J. Exp. Med. 217, 1-5. 10.1084/jem.20191561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, L. D., Moore, C. A., Khoury, M. J. and Erickson, J. D. (1999). Neural-tube defects. N. Engl. J. Med. 341, 1509-1519. 10.1056/NEJM199911113412006 [DOI] [PubMed] [Google Scholar]
- Braitsch, C. M., Azizoglu, D. B., Htike, Y., Barlow, H. R., Schnell, U., Chaney, C. P., Carroll, T. J., Stanger, B. Z. and Cleaver, O. (2019). LATS1/2 suppress NFκB and aberrant EMT initiation to permit pancreatic progenitor differentiation. PLoS Biol. 17, e3000382. 10.1371/journal.pbio.3000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner, M. E. and Simões-Costa, M. (2016). The neural crest migrating into the twenty-first century. Curr. Top. Dev. Biol. 116, 115-134. 10.1016/bs.ctdb.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucay, N., Bhagirath, D., Sekhon, K., Yang, T., Fukuhara, S., Majid, S., Shahryari, V., Laura Tabatabai, Z., Greene, K. L., Hashimoto, Y.et al. (2017). A novel microRNA regulator of prostate cancer epithelial-mesenchymal transition. Cell Death Differ. 24, 1263-1274. 10.1038/cdd.2017.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cearns, M. D., Escuin, S., Alexandre, P., Greene, N. D. E. and Copp, A. J. (2016). Microtubules, polarity and vertebrate neural tube morphogenesis. J. Anat. 229, 63-74. 10.1111/joa.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S., Njah, K., Pobbati, A. V., Lim, Y. B., Raju, A., Lakshmanan, M., Tergaonkar, V., Lim, C. T. and Hong, W. (2017). Agrin as a mechanotransduction signal regulating YAP through the Hippo Pathway. Cell Rep. 18, 2464-2479. 10.1016/j.celrep.2017.02.041 [DOI] [PubMed] [Google Scholar]
- Chenn, A., Zhang, Y. A., Chang, B. T. and McConnell, S. K. (1998). Intrinsic polarity of mammalian neuroepithelial cells. Mol. Cell. Neurosci. 11, 183-193. 10.1006/mcne.1998.0680 [DOI] [PubMed] [Google Scholar]
- Chesnutt, C., Burrus, L. W., Brown, A. M. C. and Niswander, L. (2004). Coordinate regulation of neural tube patterning and proliferation by TGFβ and WNT activity. Dev. Biol. 274, 334-347. 10.1016/j.ydbio.2004.07.019 [DOI] [PubMed] [Google Scholar]
- Das, R. M. and Storey, K. G. (2014). Apical abscission alters cell polarity and dismantles the primary cilium during neurogenesis. Science 343, 200-204. 10.1126/science.1247521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, C. J., Huang, Y.-H., Chen, M., Su, J., Zou, Y., Bardeesy, N., Iacobuzio-Donahue, C. A. and Massagué, J. (2016). TGF-β tumor suppression through a lethal EMT. Cell 164, 1015-1030. 10.1016/j.cell.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, M. E., Selleck, M. A., McMahon, A. P. and Bronner-Fraser, M. (1995). Dorsalization of the neural tube by the non-neural ectoderm. Development 121, 2099-2106. 10.1242/dev.121.7.2099 [DOI] [PubMed] [Google Scholar]
- Diepenbruck, M., Waldmeier, L., Ivanek, R., Berninger, P., Arnold, P., van Nimwegen, E. and Christofori, G. (2014). Tead2 expression levels control the subcellular distribution of yap and taz, zyxin expression and epithelial-mesenchymal transition. J. Cell Sci. 127, 1523-1536. 10.1242/jcs.139865 [DOI] [PubMed] [Google Scholar]
- Dinsmore, C. J., Ke, C.-Y. and Soriano, P. (2022). The Wnt1-Cre2 transgene is active in the male germline. Genesis 60, e23468. 10.1002/dvg.23468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, W., Bhojwani, A. and Hu, J. K. (2021). FACEts of mechanical regulation in the morphogenesis of craniofacial structures. Int. J. Oral Sci. 13, 4. 10.1038/s41368-020-00110-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, S., Morsut, L., Aragona, M., Enzo, E., Giulitti, S., Cordenonsi, M., Zanconato, F., Le Digabel, J., Forcato, M., Bicciato, S.et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179-183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Gandhi, S. and Bronner, M. E. (2021). Seq your destiny: neural crest fate determination in the genomic era. Annu. Rev. Genet. 55, 349-376. 10.1146/annurev-genet-071719-020954 [DOI] [PubMed] [Google Scholar]
- George, R. M., Maldonado-Velez, G. and Firulli, A. B. (2020). The heart of the neural crest: cardiac neural crest cells in development and regeneration. Development 147, dev188706. 10.1242/dev.188706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding, J. P., Trainor, P., Krumlauf, R. and Gassmann, M. (2000). Defects in pathfinding by cranial neural crest cells in mice lacking the neuregulin receptor erbB4. Nat. Cell Biol. 2, 103-109. 10.1038/35000058 [DOI] [PubMed] [Google Scholar]
- Gonzalez, D. M. and Medici, D. (2014). Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 7, re8. 10.1126/scisignal.2005189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, N. D. E. and Copp, A. J. (2014). Neural tube defects. Annu. Rev. Neurosci. 37, 221-242. 10.1146/annurev-neuro-062012-170354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier, J., Teillet, M.-A., Grifone, R., Kelly, R. G. and Duprez, D. (2009). Relationship between Neural Crest Cells and Cranial Mesoderm during Head Muscle Development. PLoS One 4, e4381. 10.1371/journal.pone.0004381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse, S. D., Berry, R. J., Mick Tilford, J., Kucik, J. E. and Waitzman, N. J. (2016). Retrospective assessment of cost savings from prevention: folic acid fortification and Spina Bifida in the U.S. Am. J. Prev. Med. 50, S74-S80. 10.1016/j.amepre.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen, T., Zhang, M., Wang, J., Bonilla-Claudio, M., Klysik, E., Johnson, R. L. and Martin, J. F. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458-461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, A., Saade, M., Menendez, A., Marti, E. and Pons, S. (2014). Sustained Wnt/β-catenin signalling causes neuroepithelial aberrations through the accumulation of aPKC at the apical pole. Nat. Commun. 5, 1-13. 10.1038/ncomms5168 [DOI] [PubMed] [Google Scholar]
- Hindley, C. J., Condurat, A. L., Menon, V., Thomas, R., Azmitia, L. M., Davis, J. A. and Pruszak, J. (2016). The Hippo pathway member YAP enhances human neural crest cell fate and migration. Sci. Rep. 6, 1-9. 10.1038/srep23208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, F., Hirai, S.-I., Akimoto, K., Koyama, H., Miyata, T., Ogawa, M., Noguchi, S., Sasaoka, T., Noda, T. and Ohno, S. (2006). Inactivation of aPKCλ results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development 133, 1735-1744. 10.1242/dev.02330 [DOI] [PubMed] [Google Scholar]
- Inman, G. J., Nicolás, F. J., Callahan, J. F., Harling, J. D., Gaster, L. M., Reith, A. D., Laping, N. J. and Hill, C. S. (2002). SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65-74. 10.1124/mol.62.1.65 [DOI] [PubMed] [Google Scholar]
- Ishii, M., Arias, A. C., Liu, L., Chen, Y.-B., Bronner, M. E. and Maxson, R. E. (2012). A stable cranial neural crest cell line from mouse. Stem Cells Dev. 21, 3069-3080. 10.1089/scd.2012.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, J.-I., Hosokawa, R., Sanchez-Lara, P. A., Urata, M., Slavkin, H. and Chai, Y. (2010). Transforming growth factor-β regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J. Biol. Chem. 285, 4975-4982. 10.1074/jbc.M109.035105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H.-Y., Fattet, L., Tsai, J. H., Kajimoto, T., Chang, Q., Newton, A. C. and Yang, J. (2019). Apical–basal polarity inhibits epithelial–mesenchymal transition and tumour metastasis by PAR-complex-mediated SNAI1 degradation. Nat. Cell Biol. 21, 359-371. 10.1038/s41556-019-0291-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., Shin, D., Yu, J.-R. and Lee, J. (2009). Lats kinase is involved in the intestinal apical membrane integrity in the nematode Caenorhabditis elegans. Development 136, 2705-2715. 10.1242/dev.035485 [DOI] [PubMed] [Google Scholar]
- Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R. and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo, Y., Suetsugu, T., Suda, M., Mimori-Kiyosue, Y., Toida, K., Baba, S. A., Kimura, A. and Matsuzaki, F. (2011). Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 30, 1690-1704. 10.1038/emboj.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer, A., Green, J., Pollard, J., Jr and Tugendreich, S. (2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523-530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., Nitzan, E. and Kalcheim, C. (2019). YAP promotes neural crest emigration through interactions with BMP and Wnt activities. Cell Commun. Signal. 17, 69. 10.1186/s12964-019-0383-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. T. H., Nagai, H., Nakaya, Y., Sheng, G., Trainor, P. A., Weston, J. A. and Thiery, J. P. (2013). Cell delamination in the mesencephalic neural fold and its implication for the origin of ectomesenchyme. Development 140, 4890-4902. 10.1242/dev.094680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Kang, J., Ahn, S. and Lee, J. (2019). The hippo pathway is essential for maintenance of apicobasal polarity in the growing intestine of caenorhabditis elegans. Genetics 213, 501-515. 10.1534/genetics.119.302477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Q.-Y., Zhang, H., Zhao, B., Zha, Z.-Y., Bai, F., Pei, X.-H., Zhao, S., Xiong, Y. and Guan, K.-L. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo Pathway. Mol. Cell. Biol. 28, 2426-2436. 10.1128/MCB.01874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, A. E., Vasudevan, H. N., O'Neill, A. K., Soriano, P. and Bush, J. O. (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 379, 229-234. 10.1016/j.ydbio.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Zhao, Z., Xu, C., Zhou, Z., Zhu, Z. and You, T. (2014). HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett. 355, 130-140. 10.1016/j.canlet.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Ling, H.-H., Kuo, C.-C., Lin, B.-X., Huang, Y.-H. and Lin, C.-W. (2017). Elevation of YAP promotes the epithelial-mesenchymal transition and tumor aggressiveness in colorectal cancer. Exp. Cell Res. 350, 218-225. 10.1016/j.yexcr.2016.11.024 [DOI] [PubMed] [Google Scholar]
- Lorthongpanich, C., Messerschmidt, D. M., Chan, S. W., Hong, W., Knowles, B. B. and Solter, D. (2013). Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 27, 1441-1446. 10.1101/gad.219618.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X., Wang, H., Ji, J., Xu, W., Sun, Y., Li, W., Zhang, X., Chen, J. and Xue, L. (2017). Hippo signaling promotes JNK-dependent cell migration. Proc. Natl. Acad. Sci. USA 114, 1934-1939. 10.1073/pnas.1621359114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderfield, L. J., Engleka, K. A., Aghajanian, H., Gupta, M., Yang, S., Li, L., Baggs, J. E., Hogenesch, J. B., Olson, E. N. and Epstein, J. A. (2014). Pax3 and Hippo signaling coordinate melanocyte gene expression in neural crest. Cell Rep. 9, 1885-1895. 10.1016/j.celrep.2014.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderfield, L. J., Aghajanian, H., Engleka, K. A., Lim, L. Y., Lui, F., Jain, R., Li, L., Olson, E. N. and Epstein, J. A. (2015). Hippo signaling is required for Notch-dependent smooth muscle differentiation of neural crest. Development 142, 2962-2971. 10.1242/dev.125807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik, M. L. and Bronner, M. E. (2017). Regulatory logic underlying diversification of the neural crest. Trends Genet. 33, 715-727. 10.1016/j.tig.2017.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik, M. L. and Bronner, M. E. (2021). Riding the crest to get a head: neural crest evolution in vertebrates. Nat. Rev. Neurosci. 22, 616-626. 10.1038/s41583-021-00503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, D. J., Chen, Y. and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288-4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, J. P., Tamblyn, L., Elia, A., Migon, E., Shehabeldin, A., Matysiak-Zablocki, E., Lemmers, B., Salmena, L., Hakem, A., Fish, J.et al. (2004). Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 23, 3677-3688. 10.1038/sj.emboj.7600371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z., Qiu, Y., Lin, K. C., Kumar, A., Placone, J. K., Fang, C., Wang, K.-C., Lu, S., Pan, M., Hong, A. W.et al. (2018). RAP2 mediates mechanoresponses of the Hippo pathway. Nature 560, 655-660. 10.1038/s41586-018-0444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, J. R. and Irvine, K. D. (2018). The hippo signaling network and its biological functions. Annu. Rev. Genet. 52, 65-87. 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, Y., Sakane, F. and Hashimoto, K. (2015). N-cadherin-based adherens junction regulates the maintenance, proliferation, And differentiation of neural progenitor cells during development. Cell Adhes. Migr. 9, 183-192. 10.1080/19336918.2015.1005466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, J. D. and Jacobson, A. G. (1990). The origins of neural crest cells in the axolotl. Dev. Biol. 141, 243-253. 10.1016/0012-1606(90)90380-2 [DOI] [PubMed] [Google Scholar]
- Moustakas, A. and Heldin, C.-H. (2016). Mechanisms of TGFβ-induced epithelial–mesenchymal transition. J. Clin. Med. 5, 63. 10.3390/jcm5070063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. and Luo, L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nantie, L. B., Young, R. E., Paltzer, W. G., Zhang, Y., Johnson, R. L., Verheyden, J. M. and Sun, X. (2018). Lats1/2 inactivation reveals Hippo function in alveolar type I cell differentiation during lung transition to air breathing. Development 145, dev163105. 10.1242/dev.163105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu, M., Samavarchi-Tehrani, P., Varelas, X. and Wrana, J. L. (2015). Distinct polarity cues direct Taz/Yap and TGFβ receptor localization to differentially control TGFβ-induced Smad signaling. Dev. Cell 32, 652-656. 10.1016/j.devcel.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Nguyen, B. H., Ishii, M., Maxson, R. E. and Wang, J. (2018). Culturing and Manipulation of O9-1 Neural Crest Cells. J. Vis. Exp. 140, 1-10. 10.3791/58346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio, M., Sugimachi, K., Goto, H., Wang, J., Morikawa, T., Miyachi, Y., Takano, Y., Hikasa, H., Itoh, T., Suzuki, S. O.et al. (2016). Dysregulated YAP1/TAZ and TGF-β signaling mediate hepatocarcinogenesis in Mob1a/1b -deficient mice. Proc. Natl. Acad. Sci. 113, E71-E80. 10.1073/pnas.1517188113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noce, V., Battistelli, C., Cozzolino, A. M., Consalvi, V., Cicchini, C., Strippoli, R., Tripodi, M., Marchetti, A. and Amicone, L. (2019). YAP integrates the regulatory Snail/HNF4α circuitry controlling epithelial/hepatocyte differentiation. Cell Death Dis. 10, 768. 10.1038/s41419-019-2000-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden, C. (2017). Pseudostratified epithelia – cell biology, diversity and roles in organ formation at a glance. J. Cell Sci. 130, 1859-1863. 10.1242/jcs.192997 [DOI] [PubMed] [Google Scholar]
- Norden, C., Young, S., Link, B. A. and Harris, W. A. (2009). Actomyosin is the main driver of Interkinetic nuclear migration in the retina. Cell 138, 1195-1208. 10.1016/j.cell.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefani, D.-E., Pankova, D., Abraham, A. G., Grawenda, A. M., Vlahov, N., Scrace, S. and O’ Neill, E. (2016). TGF-β targets the Hippo pathway scaffold RASSF1A to facilitate YAP/SMAD2 nuclear translocation. Mol. Cell 63, 156-166. 10.1016/j.molcel.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T. and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290-295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650-1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi, A., Enderle, L., Gregorieff, A., Johnson, R. L., Wrana, J. L. and McNeill, H. (2016). A critical role for NF2 and the Hippo pathway in branching morphogenesis. Nat. Commun. 7, 12309. 10.1038/ncomms12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W. and Smyth, G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D. and Smyth, G. K. (2007). Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23, 2881-2887. 10.1093/bioinformatics/btm453 [DOI] [PubMed] [Google Scholar]
- Robinson, M. D. and Smyth, G. K. (2008). Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321-332. 10.1093/biostatistics/kxm030 [DOI] [PubMed] [Google Scholar]
- Robinson, M. D., McCarthy, D. J. and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rofail, D., Maguire, L., Kissner, M., Colligs, A. and Abetz-Webb, L. (2013). A review of the social, psychological, and economic burdens experienced by people with spina bifida and their caregivers. Neurol. Ther. 2, 1-12. 10.1007/s40120-013-0007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, M., Bhattacharya, D. and Simoes-Costa, M. (2018). The molecular basis of neural crest axial identity. Dev. Biol. 444, S170-S180. 10.1016/j.ydbio.2018.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso, D. L., Pearson, C. A., Gaber, Z. B., Miquelajauregui, A., Li, S., Portera-Cailliau, C., Morrisey, E. E. and Novitch, B. G. (2012). Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron 74, 314-330. 10.1016/j.neuron.2012.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati, F. and Rijli, F. M. (2003). Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci. 4, 806-818. 10.1038/nrn1221 [DOI] [PubMed] [Google Scholar]
- Sauer, F. C. (1935). Mitosis in the neural tube. J. Comp. Neurol. 62, 377-405. 10.1002/cne.900620207 [DOI] [Google Scholar]
- Savorani, C., Malinverno, M., Seccia, R., Maderna, C., Giannotta, M., Terreran, L., Mastrapasqua, E., Campaner, S., Dejana, E. and Giampietro, C. (2021). A dual role of YAP in driving TGFβ-mediated endothelial-to-mesenchymal transition. J. Cell Sci. 134, jcs251371. 10.1242/jcs.251371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, M. C. and Halder, G. (2012). Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin. Cell Dev. Biol. 23, 803-811. 10.1016/j.semcdb.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Selleck, M. A. J. and Bronner-Fraser, M. (1995). Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development 121, 525-538. 10.1242/dev.121.2.525 [DOI] [PubMed] [Google Scholar]
- Simões-Costa, M. and Bronner, M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242-257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa, M., Tan-Cabugao, J., Antoshechkin, I., Sauka-Spengler, T. and Bronner, M. E. (2014). Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome Res. 24, 281-290. 10.1101/gr.161182.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabáková, E., Pernicová, Z., Slavíčková, E., Staršíchová, A., Kozubík, A. and Souček, K. (2011). TGF-β1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate 71, 1332-1343. 10.1002/pros.21350 [DOI] [PubMed] [Google Scholar]
- Soldatov, R., Kaucka, M., Kastriti, M. E., Petersen, J., Chontorotzea, T., Englmaier, L., Akkuratova, N., Yang, Y., Häring, M., Dyachuk, V.et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, 1-14. 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Spear, P. C. and Erickson, C. A. (2012). Apical movement during interkinetic nuclear migration is a two-step process. Dev. Biol. 370, 33-41. 10.1016/j.ydbio.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John, M. A. R., Tao, W., Fei, X., Fukumoto, R., Carcangiu, M. L., Brownstein, D. G., Parlow, A. F., McGrath, J. and Xu, T. (1999). Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21, 182-186. 10.1038/5965 [DOI] [PubMed] [Google Scholar]
- Sun, S. and Irvine, K. D. (2016). Cellular organization and cytoskeletal regulation of the Hippo signaling network. Trends Cell Biol. 26, 694-704. 10.1016/j.tcb.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., da Fontoura, C. S. G., Moreno, M., Holton, N. E., Sweat, M., Sweat, Y., Lee, M. K., Arbon, J., Bidlack, F. B., Thedens, D. R.et al. (2018). FoxO6 regulates Hippo signaling and growth of the craniofacial complex. PLoS Genet. 14, e1007675. 10.1371/journal.pgen.1007675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., Snider, P., Firulli, A. B. and Conway, S. J. (2010). Trigenic neural crest-restricted Smad7 over-expression results in congenital craniofacial and cardiovascular defects. Dev. Biol. 344, 233-247. 10.1016/j.ydbio.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau, E. and Mayor, R. (2012). Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 366, 34-54. 10.1016/j.ydbio.2011.12.041 [DOI] [PubMed] [Google Scholar]
- Théveneau, E., Duband, J.-L. and Altabef, M. (2007). Ets-1 confers cranial features on neural crest delamination. PLoS ONE 2, e1142. 10.1371/journal.pone.0001142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery, J. P., Acloque, H., Huang, R. Y. J. and Nieto, M. A. (2009). Epithelial-mesenchymal transitions in development and disease. Cell 139, 871-890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Trainor, P. and Krumlauf, R. (2000). Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat. Cell Biol. 2, 96-102. 10.1038/35000051 [DOI] [PubMed] [Google Scholar]
- Trapnell, C., Pachter, L. and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas, X., Sakuma, R., Samavarchi-Tehrani, P., Peerani, R., Rao, B. M., Dembowy, J., Yaffe, M. B., Zandstra, P. W. and Wrana, J. L. (2008). TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 10, 837-848. 10.1038/ncb1748 [DOI] [PubMed] [Google Scholar]
- Varelas, X., Samavarchi-Tehrani, P., Narimatsu, M., Weiss, A., Cockburn, K., Larsen, B. G., Rossant, J. and Wrana, J. L. (2010). The crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev. Cell 19, 831-844. 10.1016/j.devcel.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Vega-Lopez, G. A., Cerrizuela, S. and Aybar, M. J. (2017). Trunk neural crest cells: formation, migration and beyond. Int. J. Dev. Biol. 61, 5-15. 10.1387/ijdb.160408gv [DOI] [PubMed] [Google Scholar]
- Vogt, J., Traynor, R. and Sapkota, G. P. (2011). The specificities of small molecule inhibitors of the TGFß and BMP pathways. Cell. Signal. 23, 1831-1842. 10.1016/j.cellsig.2011.06.019 [DOI] [PubMed] [Google Scholar]
- Wallingford, J. B., Niswander, L. A., Shaw, G. M. and Finnell, R. H. (2013). The continuing challenge of understanding, preventing, and treating neural tube defects. Science 339, 1222002. 10.1126/science.1222002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. (2020). Hippo pathway interacts with Wnt pathway to regulate the neural crest cells. FASEB J. 34, 1-10. 10.1096/fsb2.21134 [DOI] [Google Scholar]
- Wang, J. and Martin, J. F. (2017). Hippo pathway: an emerging regulator of craniofacial and dental development. J. Dent. Res. 96, 1229-1237. 10.1177/0022034517719886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Xiao, Y., Hsu, C. W., Martinez-Traverso, I. M., Zhang, M., Bai, Y., Ishii, M., Maxson, R. E., Olson, E. N., Dickinson, M. E.et al. (2016). Yap and taz play a crucial role in neural crest-derived craniofacial development. Development 143, 504-515. 10.1242/dev.127647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Liu, S., Heallen, T. and Martin, J. F. (2018). The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 15, 672-684. 10.1038/s41569-018-0063-3 [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer. [Google Scholar]
- Williams, J., Mai, C. T., Mulinare, J., Isenburg, J., Flood, T. J., Ethen, M., Frohnert, B. and Kirby, R. S. (2016). Updated estimates of neural tube defects prevented by mandatory folic acid fortification — United States, 1995–2011. Morb. Mortal. Wkly. Rep. 64, 1-5. [PMC free article] [PubMed] [Google Scholar]
- Wong, G. K. W., Baudet, M.-L., Norden, C., Leung, L. and Harris, W. A. (2012). Slit1b-Robo3 signaling and N-cadherin regulate apical process retraction in developing retinal ganglion cells. J. Neurosci. 32, 223-228. 10.1523/JNEUROSCI.2596-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z. and Guan, K.-L. (2021). Hippo signaling in embryogenesis and development. Trends Biochem. Sci. 46, 51-63. 10.1016/j.tibs.2020.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdak, H., Ittner, L. M., Lang, K. S., Leveen, P., Suter, U., Fischer, J. A., Karlsson, S., Born, W. and Sommer, L. (2005). Inactivation of TGFβ signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 19, 530-535. 10.1101/gad.317405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y., Hill, M. C., Zhang, M., Martin, T. J., Morikawa, Y., Wang, S., Moise, A. R., Wythe, J. D. and Martin, J. F. (2018). Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev. Cell 45, 153-169.e6. 10.1016/j.devcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, M., Kim, Y., Sutherland, L. B., Qi, X., McAnally, J., Schwartz, R. J., Richardson, J. A., Bassel-Duby, R. and Olson, E. N. (2011). Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 4, ra70. 10.1126/scisignal.2002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Lamouille, S. and Derynck, R. (2009). TGF-Β-induced epithelial to mesenchymal transition. Cell Res. 19, 156-172. 10.1038/cr.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Zheng, L., Yuan, Q., Zhen, G., Crane, J. L., Zhou, X. and Cao, X. (2018). Transforming growth factor-β in stem cells and tissue homeostasis. Bone Res. 6, 2. 10.1038/s41413-017-0005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. and Miura, M. (2013). How to form and close the brain: Insight into the mechanism of cranial neural tube closure in mammals. Cell. Mol. Life Sci. 70, 3171-3186. 10.1007/s00018-012-1227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, Y., Lindemann, M., Colligs, A. and Snowball, C. (2011). Economic burden of neural tube defects and impact of prevention with folic acid: A literature review. Eur. J. Pediatr. 170, 1391-1400. 10.1007/s00431-011-1492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Mu, J., Guo, Q., Yang, L., Zhang, J., Liu, Z., Yu, B., Zhang, T. and Xie, J. (2017). Transcriptomic profile analysis of mouse neural tube development by RNA-Seq. IUBMB Life 69, 706-719. 10.1002/iub.1653 [DOI] [PubMed] [Google Scholar]
- Zaganjor, I., Sekkarie, A., Tsang, B. L., Williams, J., Razzaghi, H., Mulinare, J., Sniezek, J. E., Cannon, M. J. and Rosenthal, J. (2016). Describing the prevalence of neural tube defects worldwide: A systematic literature review. PLoS One 11, e0151586. 10.1371/journal.pone.0151586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc, A., Sinha, R., Gulati, G. S., Wesche, D. J., Daszczuk, P., Swigut, T., Weissman, I. L. and Wysocka, J. (2021). Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 371, eabb4776. 10.1126/science.abb4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Lindsay, H. and Robinson, M. D. (2014). Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 42, e91. 10.1093/nar/gku310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., Gross, K. M. and Kuperwasser, C. (2019). Molecular regulation of Snai2 in development and disease. J. Cell Sci. 132, jcs235127. 10.1242/jcs.235127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.