Abstract
The effects of the MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms on bladder cancer risk have been evaluated in some studies. However, the results were conflicting and ambiguous. Therefore, we aimed to perform a comprehensive meta-analysis to investigate the association of these polymorphisms with risk of bladder cancer from all eligible case-control studies. PubMed, Web of science, Scopus, SID, CNKI and SciELO databases were searched to identify all relevant studies published up to 1 January, 2021. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to estimate the strength of associations. A total of 20 case-control studies including 11 studies with 3463 cases and 3927 controls on MTHFR rs1801133 (677C>T) and 9 studies with 3177 cases and 3502 controls on rs180113 (1298A>C) polymorphism were selected. Pooled data revealed that the MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms were not associated with risk bladder cancer in overall. Stratified analysis by ethnicity revealed that the MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms were associated with bladder cancer risk in Asians, but not in Caucasians. There was no publication bias. The current meta-analysis revealed that the MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms were not risk factor for development of bladder cancer globally. However, large sample size, well-designed, and population-based studies should be performed to verify the association of the MTHFR polymorphisms with bladder cancer risk.
Key Words: Bladder cancer, urinary neoplasms, folate, MTHFR, polymorphism, meta-analysis
Introduction
Bladder cancer is the second most common genitourinary malignancy, after prostate cancer in human (Oeyen et al., 2019; Lenis et al., 2020). It is the sixth most prevalent malignancy in the United States and causes more than 16,000 deaths annually (Degeorge et al., 2017), which represents 4.4% of all new cancer diagnoses in the USA (Wong et al., 2018). More than 60% of all bladder cancer cases and half of all the 165,000 bladder cancer deaths occur in the less developed regions of the world (Antoni et al., 2017). Bladder cancer is more common in men than women, with a respective incidence of 9.6 among men and 2.4 among women per 100,000 person-year globally, respectively (Bouffioux, 1984). Bladder cancer is one of the most expensive cancer to care for from diagnosis to death due to the frequent procedures required for this malignancy monitoring and treatment (Andreassen et al., 2016).
The leading risk factor for development of bladder cancer is tobacco use, which actually accounts for more 50 percent of the cases and increases the chance of development of the disease by three times compared to not smoking individuals (Freedman et al., 2011; Mobley and Baum, 2015). Moreover, studies have found that occupational exposures (such as paint, textiles, rubber, leather, and dyes) and pollutants in drinking water (such as arsenic and chlorinated byproducts) constitute the second most important risk factor for development of bladder cancer (Gu and Wu, 2011; Aminian et al., 2014). Therefore, bladder cancer is an excellent model for studying genetic susceptibility and gene-environment interaction in cancer etiology (Gu and Wu, 2011). Since the major environmental risk factors for development of bladder cancer have been identified (Letaiová et al., 2012; Al-Zalabani et al., 2016) some efforts were made in the last few years to identify genetic variations in the pathways involved in the carcinogenesis processes, including metabolism of carcinogens, DNA repair, cell cycle checkpoints, apoptosis and inflammatory response (Grotenhuis et al., 2010; Gu and Wu, 2011). It is well-known that folate metabolism may be has an important role in development of several tumours through its involvement in both DNA methylation and nucleotide synthesis (Li et al., 2013). It has been shown that cigarette smoke exposure is associated with decreased serum levels of folate and vitamin B12 antioxidants (Tungtrongchitr et al., 2003; Wu et al., 2007). Folate and other B vitamins play important roles in the one-carbon metabolism pathway, which is associated with DNA methylation, synthesis and impaired DNA repair (Pan et al., 2019). The enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR) is involved in the circulation form of folate as it catalyzes the irreversible reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate (Maruti et al., 2009; Soleimani-Jadidi et al., 2020; Tabatabaei et al., 2020).
The candidate gene approaches revealed that functional polymorphisms at the MTHFR gene may be play an important role in development of bladder cancer (Mannino et al., 2003; Tungtrongchitr et al., 2003; Baghestani et al., 2018; Ahmadi et al., 2021). The human MTHFR gene is located on chromosome 1 at 1p36.3, composed of 11 exons and consists of 17 kb (Rosenberg et al., 2002). It was found that the rs1801133 (677C>T) in exon 4 and rs180113 (1298A>C) in exon 7 of the MTHFR gene resulted in amino acid substitution and a reduction of MTHFR activity ( Liu et al., 2020). To date, several epidemiological studies have evaluated the association of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with susceptibility to bladder cancer (You et al., 2013; Xu and Zuo, 2020). However, these associations were still inconclusive. Although two meta-analyses have reported the association of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms and bladder cancer risk (Wang et al., 2009; Xu et al., 2013; Akbari et al., 2015), they did not perform subgroup analysis by country of origin and source of controls. Thus, to comprehensively estimate the association of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with susceptibility to bladder cancer, we carried out this systematic review and meta-analysis.
Materials and Methods
Search Strategy
We carried out a comprehensive online literature search on electronic databases including PubMed, Scopus, EMBASE, Web of Knowledge, Cochrane Library, Google Scholar, Scientific Information Database (SID), WanFang, VIP, Chinese Biomedical Database (CBD), Scientific Electronic Library Online (SciELO) and China National Knowledge Infrastructure (CNKI) database to identify all relevant studies on the association of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with susceptibility to bladder cancer up to 1 January, 2021. We used the combination of following keywords and terms: (‘’Bladder Cancer’’ OR ‘’Urinary Cancer’’ OR ‘’Urinary Bladder Neoplasm’’) AND (“Methylenetetrahydrofolate Reductase” OR ‘’MTFR’’ OR “Methionine Synthase Reductase” OR “Folate Pathway’’) and (‘’MTHFR 677C>T’’ OR ‘’C677T’’ OR ‘’rs1801133’’ OR ‘’p.Ala222Val’’ OR ‘’A222V’’ OR ‘’g.11796321G>A’’) AND (‘‘MTHFR 1298A>C’’ OR ‘‘MTHFR Glu222Val’’ OR ‘‘rs1801131’’) AND (‘’Gene’’ OR ‘’Genotype’’ OR ‘’Allele’’ OR ‘’Polymorphism’’ OR ‘’ Single nucleotide polymorphisms’’ OR ‘’SNP’’ OR ‘’Variation’’ OR ‘’Mutation’’). Languages were limited to English, Portuguese, Farsi and Chinese. Moreover, the reference lists of retrieved studies including case-control studies, previous meta-analyses and reviews were manually searched to find other relevant publications.
Selection Criteria
Studies meeting the following criteria were included: 1) studies with case-control or cohort design; 2) studies evaluated the association of the MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with bladder cancer risk; 3) genotype distributions in cases and healthy controls were available for calculating an odds ratio (OR) with 95% confidence interval (CI). The following were exclusion criteria: 1) animal studies or in vitro studies; 2) studies evaluated other polymorphisms at MTHFR gene; 3) case only studies; 4) linkage studies and family based studies (sibling, twins and trios-parents studies); 5) studies did not report genotype frequencies; 6) abstracts, posters, case reports, reviews, meta-analyses, commentaries, editorials, conference articles, and proceedings; 7) duplicates of previous published studies or studies with overlapping data. If more than one study was published by the same author(s) using repeated or overlapped data, the most complete one or more recently published study was selected.
Data Extraction
Two authors carefully reviewed and extracted data from all eligible studies according to the inclusion criteria. If any disagreement appeared, a third author was consulted to resolve the dispute and the final consensus was made by the majority of the votes. The following data were extracted from each study: the name of first author, year of publication, country of origin, ethnicity (Caucasian, Asian, African, Mixed populations), source of controls (hospital based or population based), genotyping methods, sample size, alleles and genotypes frequencies for MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms in cases and controls, Minor Allele Frequency (MAFs) and Hardy-Weinberg equilibrium (HWE) in healthy controls. The ‘‘mixed’’ group means mixed or unknown populations.
Quality score assessment
The Newcastle-Ottawa Score (NOS) were performed to assess the quality of included studies in the meta-analysis and to assess the various aspects of the methodology used by the observational research, which are relevant to the quality of the study. This standard assessed 3 sections (selection of cases, comparability of groups, and determination of exposure) and 8 items. In the selection and exposure categories, a quality research item received 1 star, and a comparable category could receive at most 2 stars. The quality assessment values ranged from 0 stars (worst) to 9 stars (best), and studies with a score ≥7 were defined as high quality. Generally, the study which scored at least 5 points was considered to be included in meta-analysis and any discrepant opinions were resolved by discussion and consensus.
Statistical Analysis
Crude odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated to evaluate the strength of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with susceptibility to bladder cancer. The statistical significance of pooled ORs was assessed by the Z test, in which p-value less than 0.05 was considered as statistically significant. The associations was estimated under all five genetic models, i.e., allele (B vs. A), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB+BA vs. AA), and the recessive (BB vs. BA+AA). A test of between-studies heterogeneity was conducted using Cochran’s Q test, in which P ≤ 0.01 indicated a significant heterogeneity. In addition, I2 statistic was used to quantify the proportion of the between-study heterogeneity (range of 0 to 100%: I2≤50%, no heterogeneity; I2≥50%, presence of heterogeneity). Thus, when the heterogeneity was absent the fixed-effect model (Mantel-Haenszel method) was used to calculate the overall or pooled OR; otherwise, the random-effects model (DerSimonian and Laird method) was applied. To explore sources of between-study heterogeneity, we have performed subgroup analysis by ethnicity, country, source of controls and HWE status. We used the Chi-squared test to evaluate Hardy-Weinberg equilibrium (HWE) in controls, and we considered p < 0.05 as a significant deviation from HWE (HWE-violating) (Bahrami et al., 2020). Sensitivity analyses were performed to assess the stability of the results by sequential removing of each study. The Begg’s visual inspection of funnel plot and the Egger’s regression tests were used to evaluate publication bias in the literature, in which P<0.05 was considered statistically significant. All of the statistical calculations were performed using Comprehensive Meta-Analysis (CMA) software version 2.0 (Biostat, USA). Two-sided P-values < 0.05 were considered statistically significant.
Results
Characteristics of Selected Studies
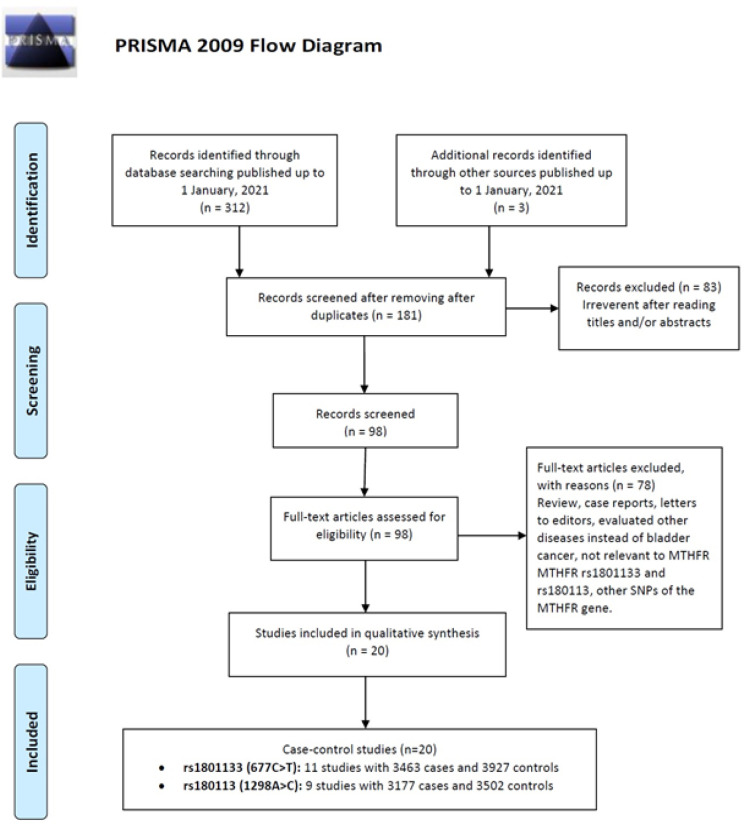
As shown in Figure 1, our initial search yielded 315 studies on MTHFR polymorphisms and bladder cancer, with duplicate studies removed resulting in 181 studies remaining. Among them, 83 studies were excluded based on titles and abstracts. Following the inclusion exclusion criteria 78 studies were excluded to case reports, review, previous meta-analyses, and other polymorphisms of MTHFR gene or lack of the relevant data. Finally, a total of 20 case-control studies from eleven independent papers (Kimura et al., 2001; Sanyal et al., 2004; Moore et al., 2004, 2007; Lin et al., 2004; Karagas et al., 2005; Cai et al., 2009; Wang et al., 2009; Rouissi et al., 2009; Chung et al., 2010; Safarinejad et al., 2011; Amooee et al., 2019) were selected. The main characteristics of each study identified are listed in Table 1. Of them, eleven case-control studies with 3,463 cases and 3,927 controls were on MTHFR rs1801133 (677C>T) and nine case-control studies with 3,177 cases and 3,502 controls were on MTHFR rs180113 (1298A>C). For the MTHFR rs1801133 (677C>T), five studies were conducted on Caucasians, four on Asians and one study on African and mixed population. For the MTHFR rs180113 (1298A>C), four studies were conducted on Caucasians, three on Asians and one study on African and mixed population. The eligible studies were published between 2001 and 2011. In term of study design, there were 14 hospital-based (HB) and six population-based (PB). Genotyping methods were conducted using restrictive fragment length polymorphism (PCR-RFLP) and TaqMan. The alleles, genotypes and minor allele frequencies (MAFs) distributions for both MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms in the cases and controls are present in Table 1. The distribution of genotypes in the controls was in agreement with Hardy-Weinberg equilibrium (HWE) for all selected studies (Table 1). The NOS score of eligible articles ranged from 7 to 9, which indicated that all included studies were of high quality (Table 1).
Figure 1.
Flow Chart for the Process of Selecting Eligible Studies
Table 1.
Main Characteristics of Studies Included in the Meta-Analysis
| First Author/Year | Country | Genotyping | SOC | Case/Control | Cases | Controls | MAFs | HWE | NOS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Ethnicity) | Method | Genotypes | Allele | Genotypes | Allele | ||||||||||||
| rs1801133 | CC | CT | TT | C | T | CC | CT | TT | C | T | |||||||
| Kimura 2001 | Germany(Caucasian) | PCR-RFLP | HB | 165/150 | 70 | 80 | 15 | 220 | 110 | 65 | 73 | 12 | 203 | 97 | 0.323 | 0.169 | 7 |
| Moore 2004 | Argentina(Mixed) | PCR-RFLP | PB | 106/109 | 45 | 42 | 19 | 132 | 80 | 32 | 59 | 18 | 123 | 95 | 0.436 | 0.292 | 7 |
| Lin 2004 | USA(Caucasian) | PCR-RFLP | HB | 448/448 | 199 | 197 | 52 | 595 | 301 | 218 | 177 | 53 | 613 | 283 | 0.316 | 0.069 | 9 |
| Sanyal 2004 | Sweden(Caucasian) | PCR-RFLP | HB | 309/246 | 173 | 113 | 23 | 459 | 159 | 121 | 102 | 23 | 344 | 148 | 0.301 | 0.822 | 7 |
| Karagas 2005 | USA(Caucasian) | PCR-RFLP | PB | 350/543 | 140 | 171 | 39 | 451 | 249 | 227 | 245 | 71 | 699 | 387 | 0.356 | 0.701 | 9 |
| Moore 2007 | Spain(Caucasian) | TaqMan | HB | 1041/1049 | 418 | 478 | 145 | 1314 | 768 | 402 | 486 | 161 | 1290 | 808 | 0.385 | 0.48 | 7 |
| Cai 2009 | China(Asian) | PCR-RFLP | HB | 312/325 | 82 | 169 | 61 | 333 | 291 | 113 | 170 | 42 | 396 | 254 | 0.391 | 0.075 | 7 |
| Rouissi 2009 | Tunisia(African) | PCR-RFLP | PB | 185/191 | 87 | 86 | 12 | 260 | 110 | 81 | 90 | 20 | 252 | 130 | 0.34 | 0.494 | 7 |
| Wang 2009 | China(Asian) | PCR-RFLP | HB | 239/250 | 66 | 128 | 45 | 260 | 218 | 88 | 132 | 30 | 308 | 192 | 0.384 | 0.066 | 9 |
| Chung 2010 | China(Asian) | PCR-RFLP | HB | 150/300 | 80 | 57 | 13 | 217 | 83 | 141 | 123 | 36 | 405 | 195 | 0.325 | 0.256 | 7 |
| Safarinejad 2011 | Iran(Asian) | PCR-RFLP | HB | 158/316 | 67 | 74 | 17 | 208 | 108 | 144 | 142 | 30 | 430 | 202 | 0.32 | 0.555 | 7 |
| Total | 3463/3927 | 1427 | 1595 | 625 | 5044 | 2477 | 1632 | 1799 | 562 | 5063 | 2791 | 0.366 | 0.064 | 8 | |||
| rs180113 | AA | AC | CC | A | C | AA | AC | CC | A | C | |||||||
| Moore 2004 | Argentina(Mixed) | PCR-RFLP | PB | 106/108 | 52 | 45 | 9 | 149 | 63 | 55 | 45 | 8 | 155 | 61 | 0.282 | 0.77 | 7 |
| Lin 2004 | USA(Caucasian) | PCR-RFLP | HB | 448/447 | 219 | 199 | 30 | 637 | 259 | 213 | 197 | 37 | 623 | 271 | 0.303 | 0.361 | 9 |
| Sanyal 2004 | Sweden(Caucasian) | PCR-RFLP | HB | 311/245 | 145 | 133 | 33 | 423 | 199 | 110 | 111 | 24 | 331 | 159 | 0.324 | 0.6 | 7 |
| Karagas 2005 | USA(Caucasian) | PCR-RFLP | PB | 350/542 | 173 | 146 | 31 | 492 | 208 | 267 | 220 | 55 | 754 | 330 | 0.304 | 0.333 | 9 |
| Moore 2007 | Spain(Caucasian) | TaqMan | HB | 1068/1078 | 537 | 457 | 74 | 1531 | 605 | 557 | 429 | 92 | 1543 | 613 | 0.284 | 0.467 | 7 |
| Cai 2009 | China(Asian) | PCR-RFLP | HB | 312/325 | 215 | 91 | 6 | 521 | 103 | 226 | 92 | 7 | 544 | 106 | 0.163 | 0.504 | 7 |
| Rouissi 2009 | Tunisia(African) | PCR-RFLP | PB | 185/191 | 97 | 78 | 10 | 272 | 98 | 121 | 60 | 10 | 302 | 80 | 0.309 | 0.478 | 7 |
| Wang 2009 | China(Asian) | PCR-RFLP | HB | 239/250 | 169 | 67 | 3 | 405 | 73 | 171 | 75 | 4 | 417 | 83 | 0.166 | 0.186 | 9 |
| Safarinejad 2011 | Iran(Asian) | PCR-RFLP | HB | 158/316 | 48 | 85 | 25 | 181 | 135 | 178 | 115 | 23 | 471 | 161 | 0.255 | 0.46 | 8 |
| Total | 3177/3502 | 1655 | 1301 | 221 | 4611 | 1743 | 1898 | 1344 | 260 | 5140 | 1864 | 0.266 | 0.3 | ||||
PCR-RFLP, Polymerase Chain Reaction Restriction Fragment Length Polymorphism; SOC, source of controls; HB, Hospital Based; PB, Population Based; MAFs, Minor Allele Frequencies; HWE, Hardy-Weinberg Equilibrium; NOS, Newcastle-Ottawa Score.
Quantitative Data Synthesis
MTHFR rs1801133 (677C>T) Polymorphism
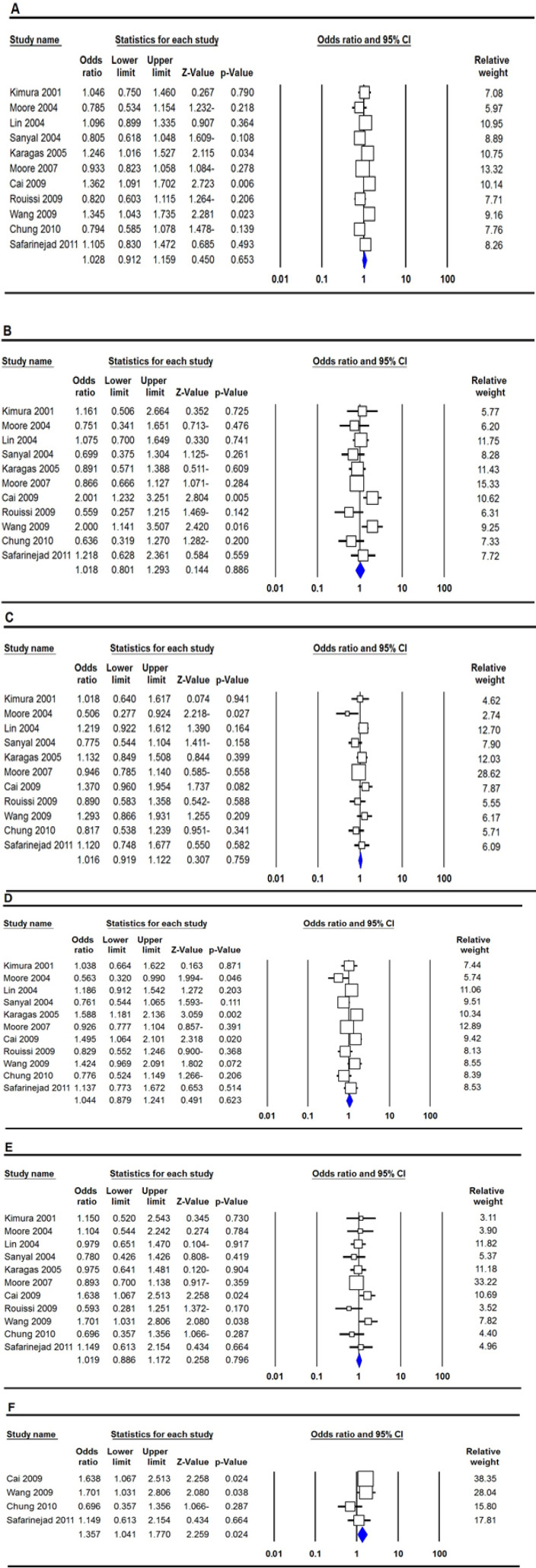
The summary of association between the MTHFR rs1801133 (677C>T) polymorphism and bladder cancer risk are shown in Table 2. Overall, the combined data did not show a significant association between MTHFR rs1801133 (677C>T) polymorphism and increased risk of bladder cancer globally under all five genetic models, i.e., allele (T vs. C: OR = 1.028, 95% CI 0.912-1.159, p=0.653, Fig 2A), homozygote (TT vs. CC: OR = 1.018, 95% CI 0.801-1.293, p=0.886, Fig 2B), heterozygote (TC vs. CC: OR = 1.016, 95% CI 0.919-1.122, p=759, Figure 2C), dominant (TT+TC vs. CC: OR = 1.044, 95% CI 0.879-1.241, p=0.623, Fig 2D), and recessive (TT vs. TC+CC: OR = 1.019, 95% CI 0.886-1.172, p=0.796, Fig 2E). Moreover, we have performed subgroup analysis by ethnicity, country of origin and source of controls. Stratified analysis by ethnicity showed that MTHFR rs1801133 (677C>T) polymorphism was associated with an increased risk of bladder cancer in Asians under the recessive genetic model (TT vs. TC+CC: OR = 1.357, 95% CI 1.041-1.770, p=0.024, Fig 2F), but not in Caucasians. There was no significant association between MTHFR rs1801133 (677C>T) polymorphism and risk of bladder cancer by source of controls and in Chinese population.
Table 2.
Summary Risk Estimates for Association between MTHFR rs1801133 (677C>T) Polymorphism and Bladder Cancer Risk
| Subgroup | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Overall | T vs. C | Random | 62.67 | 0.003 | 1.028 | 0.912-1.159 | 0.450 | 0.653 | 0.640 | 0.846 |
| TT vs. CC | Random | 52.97 | 0.019 | 1.018 | 0.801-1.293 | 0.144 | 0.886 | 0.755 | 0.960 | |
| TC vs. CC | Fixed | 37.10 | 0.103 | 1.016 | 0.919-1.122 | 0.307 | 0.759 | 0.212 | 0.557 | |
| TT+TC vs. CC | Random | 65.24 | 0.001 | 1.044 | 0.879-1.241 | 0.491 | 0.623 | 0.275 | 0.841 | |
| TT vs. TC+CC | Fixed | 29.94 | 0.161 | 1.019 | 0.886-1.172 | 0.258 | 0.796 | 0.876 | 0.895 | |
| Ethnicity | ||||||||||
| Caucasian | T vs. C | Fixed | 56.16 | 0.058 | 1.004 | 0.922-1.093 | 0.087 | 0.931 | 1.000 | 0.761 |
| TT vs. CC | Fixed | 0.00 | 0.785 | 0.902 | 0.750-1.087 | -1.084 | 0.278 | 1.000 | 0.734 | |
| TC vs. CC | Fixed | 19.51 | 0.290 | 1.007 | 0.891-1.139 | 0.116 | 0.907 | 0.806 | 0.945 | |
| TT+TC vs. CC | Random | 70.99 | 0.008 | 1.068 | 0.843-1.353 | 0.542 | 0.588 | 1.000 | 0.719 | |
| TT vs. TC+CC | Fixed | 0.00 | 0.937 | 0.923 | 0.775-1.098 | -0.909 | 0.364 | 0.806 | 0.555 | |
| Asian | T vs. C | Random | 67.64 | 0.026 | 1.143 | 0.906-1.443 | 1.126 | 0.260 | 0.089 | 0.119 |
| TT vs. CC | Random | 65.33 | 0.034 | 1.381 | 0.834-2.285 | 1.255 | 0.209 | 0.089 | 0.126 | |
| TC vs. CC | Fixed | 22.33 | 0.277 | 1.149 | 0.945-1.398 | 1.392 | 0.164 | 0.089 | 0.239 | |
| TT+TC vs. CC | Fixed | 58.16 | 0.067 | 1.195 | 0.991-1.440 | 1.864 | 0.062 | 0.089 | 0.316 | |
| TT vs. TC+CC | Fixed | 46.83 | 0.130 | 1.357 | 1.041-1.770 | 2.259 | 0.024 | 0.089 | 0.120 | |
| Country | ||||||||||
| Chinese | T vs. C | Random | 77.86 | 0.011 | 1.149 | 0.836-1.579 | 0.856 | 0.392 | 0.292 | 0.269 |
| TT vs. CC | Random | 75.70 | 0.016 | 1.416 | 0.723-2.775 | 1.014 | 0.311 | 0.296 | 0.276 | |
| TC vs. CC | Fixed | 47.94 | 0.146 | 1.159 | 0.926-1.450 | 1.286 | 0.198 | 0.296 | 0.437 | |
| TT+TC vs. CC | Random | 71.78 | 0.029 | 1.190 | 0.795-1.784 | 0.845 | 0.398 | 0.296 | 0.517 | |
| TT vs. TC+CC | Fixed | 62.37 | 0.070 | 1.316 | 0.804-2.154 | 1.091 | 0.275 | 0.296 | 0.269 | |
| Source of Controls | ||||||||||
| HB | T vs. C | Random | 63.94 | 0.007 | 1.046 | 0.911-1.199 | 0.636 | 0.525 | 0.901 | 0.647 |
| TT vs. CC | Random | 60.66 | 0.013 | 1.120 | 0.834-1.503 | 0.752 | 0.452 | 1.000 | 0.560 | |
| TC vs. CC | Fixed | 28.16 | 0.203 | 1.033 | 0.924-1.155 | 0.570 | 0.569 | 0.901 | 0.647 | |
| TT+TC vs. CC | Random | 54.00 | 0.033 | 1.057 | 0.893-1.249 | 0.643 | 0.520 | 1.000 | 0.527 | |
| TT vs. TC+CC | Fixed | 41.95 | 0.099 | 1.045 | 0.895-1.220 | 0.556 | 0.578 | 1.000 | 0.620 | |
| PB | T vs. C | Random | 72.83 | 0.025 | 0.954 | 0.690-1.320 | -0.282 | 0.778 | 1.000 | 0.186 |
| TT vs. CC | Fixed | 0.00 | 0.589 | 0.786 | 0.556-1.110 | -1.367 | 0.172 | 1.000 | 0.352 | |
| TC vs. CC | Fixed | 65.05 | 0.067 | 0.951 | 0.762-1.186 | -0.448 | 0.654 | 0.296 | 0.107 | |
| TT+TC vs. CC | Random | 84.75 | 0.001 | 0.937 | 0.510-1.721 | -0.210 | 0.834 | 0.296 | 0.153 | |
| TT vs. TC+CC | Fixed | 0.00 | 0.437 | 0.911 | 0.658-1.260 | -0.564 | 0.573 | 1.000 | 0.694 | |
HB, Hospital Based; PB, Population Based
Figure 2.
Forest Plot for Association between MTHFR rs1801133 (677C>T) polymorphism and Bladder Cancer Risk. A: allele model (T vs. C); B: homozygote model (TT vs. CC); C: heterozygote model (TC vs. CC); D: dominant model (TT+TC vs. CC); E: recessive model (TT vs. TC+CC); and F: Asians (recessive model: TT vs. TC+CC)
MTHFR rs180113 (1298A>C) Polymorphism
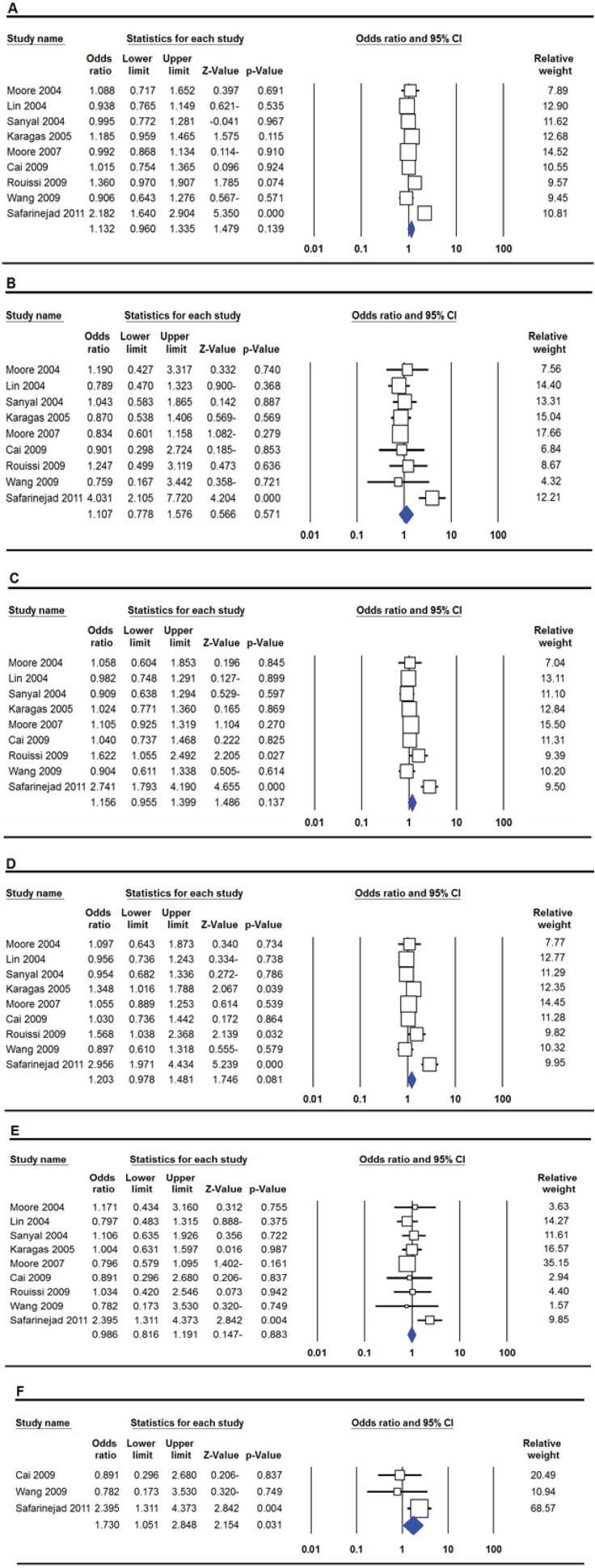
The summary of association between the MTHFR rs180113 (1298A>C) polymorphism and risk of bladder cancer are presented in Table 3. Pooled ORs demonstrated that MTHFR rs180113 (1298A>C) polymorphism was not significantly associated with bladder cancer risk globally under all five genetic models, i.e., allele (C vs. A: OR = 1.132, 95% CI 0.960-1.335, p=0.139, Fig 3A), homozygote (CC vs. AA: OR = 1.107, 95% CI 0.778-1.576, p=0.571, Fig 3B), heterozygote (CA vs. AA: OR = 1.158, 95% CI 0.955-1.399, p=0.137, Fig 3C), dominant (CC+CA vs. AA: OR = 1.203, 95% CI 0.978-1.481, p=0.081, Fig 3D), and recessive (CC vs. CA+AA: OR = 0.986, 95% CI 0.816-1.191, p=0.883, Fig 3E). Moreover, we carried out subgroup analyses by ethnicity and source of controls. Stratified analysis by ethnicity revealed that there was a significant association between the MTHFR rs180113 (1298A>C) polymorphism and increased risk of bladder cancer in Asians under the recessive genetic model (CC vs. CA+AA: OR = 1.730, 95% CI 1.051-2.848, p=0.031, Fig 3F), but not in Caucasians. Moreover, subgroup analysis by source of controls showed a significant association between MTHFR rs180113 (1298A>C) polymorphism and risk of bladder cancer in population based (PB) group of studies under two genetic models, i.e., allele (C vs. A: OR = 1.209, 95% CI 1.025-1.425, p=0.024) and dominant (CC+CA vs. AA: OR = 1.358, 95% CI 1.097-1.682, p=0.005), but not in hospital based (HB) studies (Table 3).
Table 3.
Summary Risk Estimates for Association between MTHFR rs180113 (1298A>C) Polymorphism and Bladder Cancer Risk
| Subgroup | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Overall | A vs. C | Random | 73.99 | ≤0.001 | 1.132 | 0.960-1.335 | 1.479 | 0.139 | 0.251 | 0.368 |
| AA vs. CC | Random | 60.9 | 0.009 | 1.107 | 0.778-1.576 | 0.566 | 0.571 | 0.348 | 0.467 | |
| AC vs. CC | Random | 66.55 | 0.002 | 1.158 | 0.955-1.399 | 1.486 | 0.137 | 0.602 | 0.456 | |
| AA+AC vs. CC | Random | 73.59 | ≤0.001 | 1.203 | 0.978-1.481 | 1.746 | 0.081 | 0.175 | 0.353 | |
| AA vs. AC+CC | Fixed | 28.46 | 0.192 | 0.986 | 0.816-1.191 | -0.147 | 0.883 | 0.754 | 0.457 | |
| Ethnicity | ||||||||||
| Caucasian | A vs. C | Fixed | 0 | 0.426 | 1.015 | 0.926-1.112 | 0.31 | 0.757 | 0.734 | 0.757 |
| AA vs. CC | Fixed | 0 | 0.904 | 0.861 | 0.689-1.074 | -1.326 | 0.185 | 0.734 | 0.484 | |
| AC vs. CC | Fixed | 0 | 0.757 | 1.038 | 0.918-1.175 | 0.598 | 0.55 | 0.308 | 0.033 | |
| AA+AC vs. CC | Fixed | 19.49 | 0.293 | 1.066 | 0.946-1.202 | 1.053 | 0.292 | 1 | 0.959 | |
| AA vs. AC+CC | Fixed | 0 | 0.684 | 0.879 | 0.709-1.089 | -1.179 | 0.238 | 0.308 | 0.289 | |
| Asian | A vs. C | Random | 89.76 | ≤0.001 | 1.267 | 0.727-2.207 | 0.836 | 0.403 | 0.296 | 0.49 |
| AA vs. CC | Random | 73.95 | 0.021 | 1.58 | 0.479-5.210 | 0.751 | 0.452 | 1 | 0.2 | |
| AC vs. CC | Random | 88.07 | ≤0.001 | 1.361 | 0.714-2.595 | 0.935 | 0.35 | 1 | 0.496 | |
| AA+AC vs. CC | Random | 90.54 | ≤0.001 | 1.39 | 0.688-2.811 | 0.918 | 0.359 | 1 | 0.561 | |
| AA vs. AC+CC | Fixed | 44.14 | 0.167 | 1.73 | 1.051-2.848 | 2.154 | 0.031 | 1 | 0.176 | |
| Source of Controls | ||||||||||
| HB | A vs. C | Random | 82.12 | ≤0.001 | 1.102 | 0.879-1.383 | 0.842 | 0.4 | 0.452 | 0.533 |
| AA vs. CC | Random | 74.69 | 0.001 | 1.146 | 0.681-1.927 | 0.513 | 0.608 | 0.452 | 0.626 | |
| AC vs. CC | Random | 75.73 | 0.001 | 1.145 | 0.886-1.478 | 1.036 | 0.3 | 1 | 0.636 | |
| AA+AC vs. CC | Random | 80.8 | ≤0.001 | 1.153 | 0.874-1.521 | 1.007 | 0.314 | 0.452 | 0.512 | |
| AA vs. AC+CC | Fixed | 54.68 | 0.051 | 0.971 | 0.781-1.207 | -0.264 | 0.792 | 0.452 | 0.594 | |
| PB | A vs. C | Fixed | 0 | 0.689 | 1.209 | 1.025-1.425 | 2.254 | 0.024 | 1 | 0.98 |
| AA vs. CC | Fixed | 0 | 0.726 | 0.973 | 0.657-1.441 | -0.136 | 0.892 | 1 | 0.139 | |
| AC vs. CC | Fixed | 37.1 | 0.204 | 1.158 | 0.932-1.441 | 1.322 | 0.186 | 1 | 0.717 | |
| AA+AC vs. CC | Fixed | 0 | 0.582 | 1.358 | 1.097-1.682 | 2.808 | 0.005 | 1 | 0.783 | |
| AA vs. AC+CC | Fixed | 0 | 0.963 | 1.032 | 0.705-1.511 | 0.164 | 0.869 | 0.296 | 0.384 | |
HB, Hospital Based; PB, Population Based
Figure 3.
Forest Plot for Association between MTHFR rs180113 (1298A>C) Polymorphism and Bladder Cancer Risk. A: allele model (A vs. C); B: homozygote model (AA vs. CC); C: heterozygote model (AC vs. CC); D: dominant model (AA+AC vs. CC); E: recessive model (AA vs. AC+CC) and F: Asians (recessive model: TT vs. TC+CC)
Minor Allele Frequencies (MAFs)
The minor allele frequencies (MAFs) for MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms in healthy controls is shown in Table 1. There were ethnic variations in the allele and genotype distributions for these polymorphisms. MAFs in controls for MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms were 36.6% and 26.6%, respectively. Moreover, the mutant allele frequency for MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms were 39.0% and 27.4%, respectively. Thus, the mutant and wild allele frequency of MTHFR rs180113 (1298A>C) polymorphism were less than MTHFR rs1801133 (677C>T) polymorphism.
Between-Study Heterogeneity
As shown in Tables 2 and 3, there was a significant between-study heterogeneity in overall population under most genetic models for both MTHFR rs1801133 (677C>T) and rs180113 (1298A>C). Thus, we utilized a random-effects model (DerSimonian and Laird method) for those genetic models. To explore the potential sources of between-study heterogeneity, we conducted subgroup analyses by ethnicity, country of origin and source of controls. Results revealed a significant heterogeneity in Asians studies for both MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms. However, the heterogeneity was reduced or disappeared in Caucasian studies. Subgroup analysis revealed that ethnicity and source of controls (for rs180113) might be source of heterogeneity in the current meta-analysis.
Sensitivity Analysis
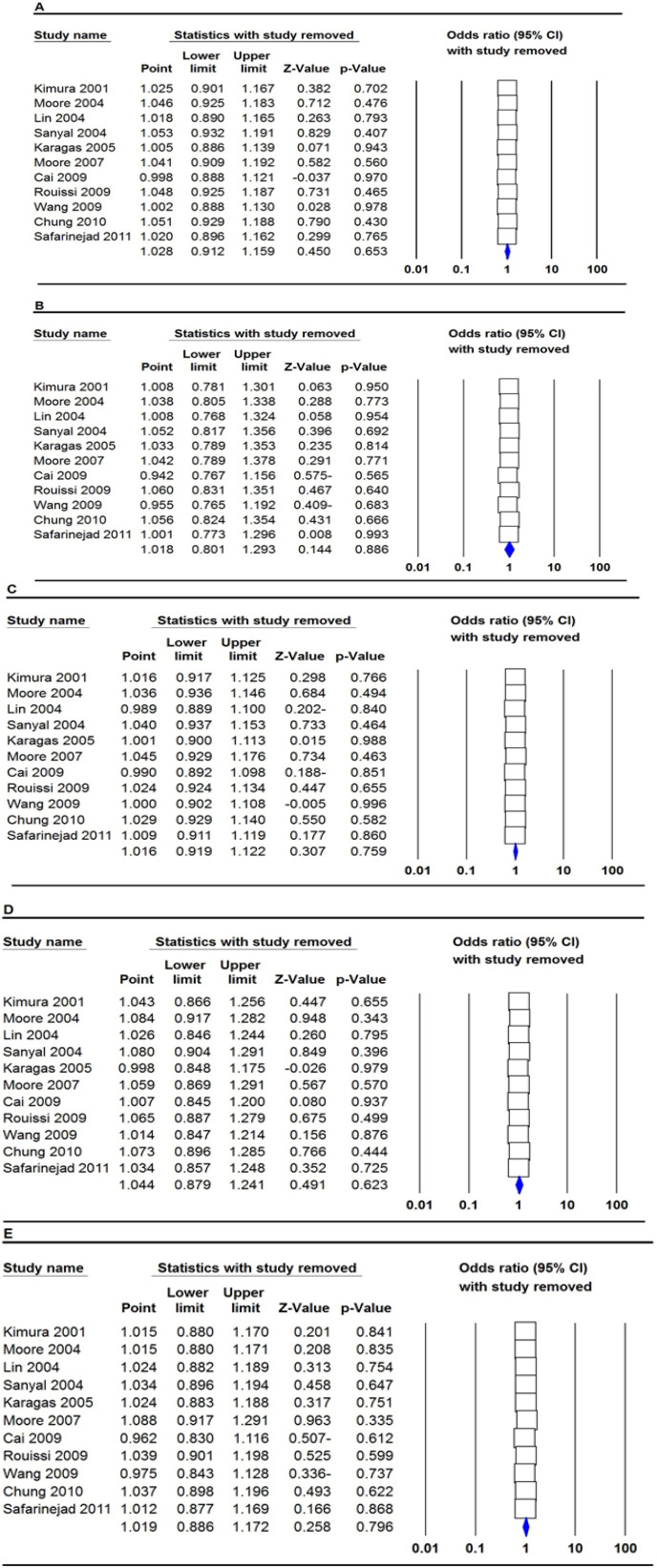
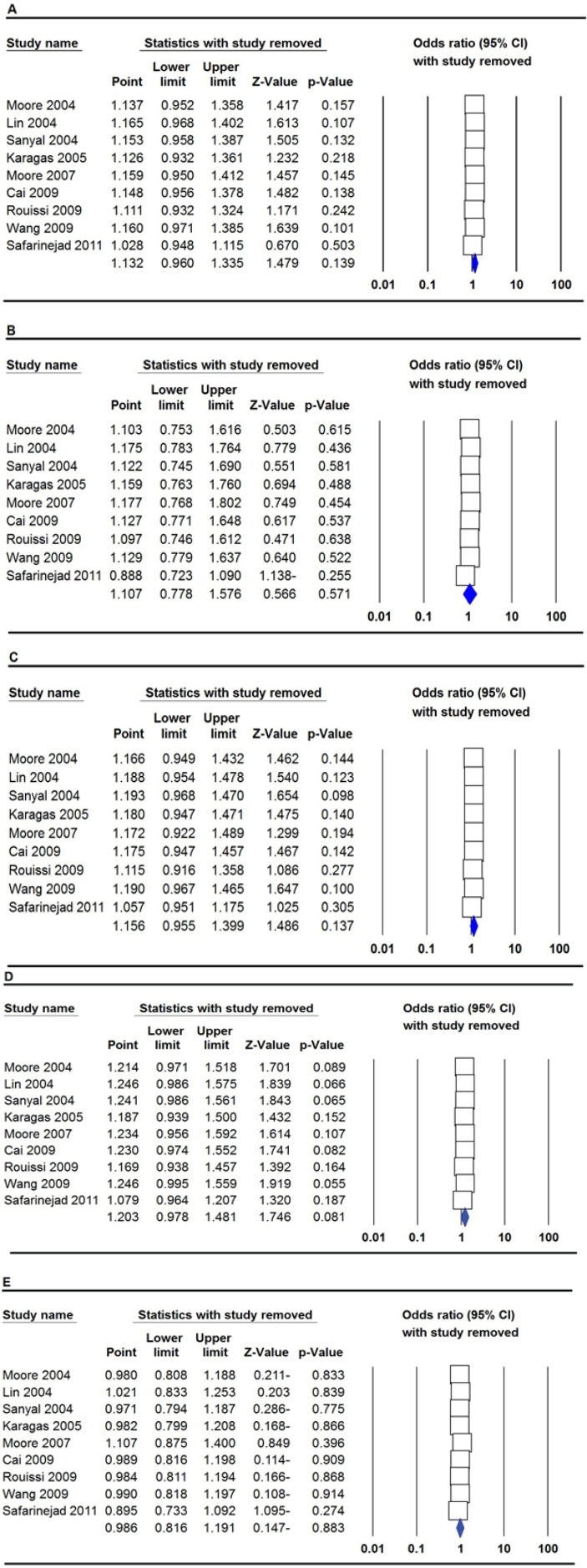
We conducted a sensitivity analysis to evaluate the stability of the results by sequentially removing each study from our meta-analysis. As shown in Figures 4 and 5, the data from sensitivity analysis revealed that none of the studies changed the pooled OR for MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) under all five genetic models, and it shows that the meta-analysis is stable.
Figure 4.
Sensitivity Analysis of Each Study Included in This Meta-Analysis for MTHFR rs1801133 (677C>T) Polymorphism by Omitting each Data Set in the Meta-Analysis. A: allele model (T vs. C); B: homozygote model (TT vs. CC); C: heterozygote model (TC vs. CC); D: dominant model (TT+TC vs. CC); E: recessive model (TT vs. TC+CC)
Figure 5.
Sensitivity Analysis of each Study Included in This Meta-Analysis for MTHFR rs180113 (1298A>C) by Omitting each Data Set in the Meta-Analysis. A: allele model (A vs. C); B: homozygote model (AA vs. CC); C: heterozygote model (AC vs. CC); D: dominant model (AA+AC vs. CC); E: recessive model (AA vs. AC+CC)
Publication Bias
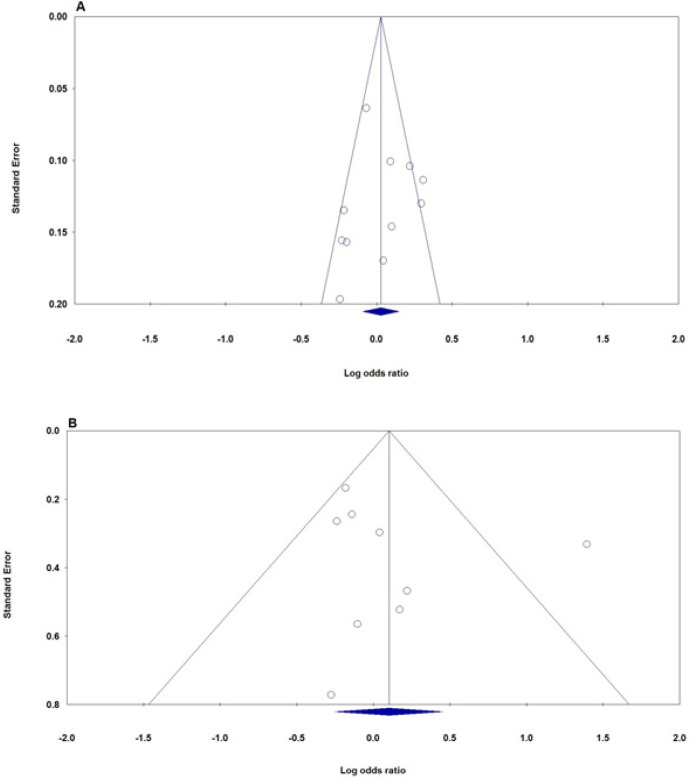
We performed potential publication bias with the Begg’s test and the Egger’s test. As Figure 6 indicated, the symmetrical funnel plot indicated that there is no significant publication bias for both for MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms under all five genetic models. Moreover, the Egger’s test was performed to provide the statistical evidence of funnel plot (Tables 2 and 3).
Figure 6.
The Funnel Plots of Publication Bias for Association between MTHFR Polymorphisms and Bladder Cancer Risk. A: MTHFR rs1801133 (677C>T) (allele model: T vs. C); B: MTHFR rs180113 (1298A>C) (homozygote model: AA vs. CC)
Discussion
MTHFR is a key enzyme in folate metabolism, which converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate that is required for the remethylation of homocysteine to methionine (Gohari et al., 2019; Karimi-Zarchi et al., 2019; Sadeghiyeh et al., 2020; Bahrami et al., 2021). The rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms are responsible for the synthesis of a thermolabile form of MTHFR enzyme with decreased enzymatic activity (Kiseljaković et al., 2008). It is suggested that the homozygote mutant genotypes of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms are linked with higher plasma homocysteine level (Azarpira et al., 2018; Niktabar et al., 2021). Some published molecular epidemiological studies have demonstrated an association between MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms and an increased risk of bladder cancer. However, Safarinejad et al., reported that MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms did not risk factor for development of bladder cancer among Iranian population (Safarinejad et al., 2011). This trend inconsistent results between the rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms at MTHFR gene and risk of bladder cancer may be caused for limited number of related studies and small sample sizes. In the current meta-analysis, we evaluated the associations between rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms at MTHFR gene and susceptibility to bladder cancer based on 20 eligible studies. Our pooled OR indicated that both rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms at MTHFR gene were not associated with susceptibility to bladder cancer globally.
In the current meta-analysis, pooled ORs showed that MTHFR rs1801133 (677C>T) polymorphism was not associated with an increased risk of bladder cancer in overall population. Stratified analysis by showed that both polymorphisms were associated with bladder cancer in Asians, but not in Caucasian. Similarly, Shi et al., (2014) in a meta-analysis of 3,463 cases and 3,927 controls revealed that the MTHFR rs1801133 (677C>T) polymorphism was not associated with risk of bladder cancer in overall population. However, their stratified analysis by ethnicity showed that the MTHFR rs1801133 (677C>T) polymorphism was significantly associated with susceptibility to bladder cancer in Middle Eastern populations. Li et al., (2013) in a meta-analysis based on 15 case-control studies with 3,570 cases and 3,926 healthy subjects demonstrated that the MTHFR rs1801133 (677C>T) polymorphism did not associate with risk of bladder cancer. Their subgroup analysis still revealed that the rs1801133 (677C>T) polymorphism was not risk factor for bladder cancer by ethnicity and sources of controls. The previous meta-analyses were not performed subgroup analyses by country of origin for MTHFR rs1801133 (677C>T) polymorphism. Thus, this polymorphism association with bladder cancer needs to be evaluated. Moreover, their conclusions reliability is considerably smaller than that needed to achieve the robust conclusions.
Our pooled data failed to show a significant association between the MTHFR rs180113 (1298A>C) and an increased risk of bladder risk in the global population. Our subgroup analysis by ethnicity showed a significant association between MTHFR rs180113 (1298A>C) and bladder cancer risk in Asians, but not in Caucasians. Moreover, some epidemiological studies revealed that the MTHFR rs180113 (1298A>C) polymorphism was associated with susceptibility to bladder risk in Chinese (Lin et al., 2004; Cai et al., 2009). However, our subgroup analysis by country of origin did not show an association between the MTHFR rs180113 (1298A>C) polymorphism and susceptibility to bladder cancer in Chinese patients. Similarly, Safarinejad et al., in case-control study revealed that the MTHFR rs180113 (1298A>C) was not risk factor for development of bladder cancer in Iranian population (Ghaemmaghami et al., 2008; Safarinejad et al., 2011; Binesh et al., 2012). According to our findings, the association of the MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with bladder cancer may be due to differences in ethnicity, genetic background, life style, smoking habits, and etc. in a meta-analysis, Zhang et al., (2018) verified the relationship between TP53 codon 72 and bladder cancer risk in Asians, but not Caucasians. These results indicated that genetic variants might be an ethnicity related factor of susceptibility to bladder cancer. In another way, it seemed that different populations with multiple genetic backgrounds have different genetic variants risk in development of bladder cancer (McConkey et al., 2010; Meng et al., 2017). Still, further studies are needed to explore this difference between Asians and Caucasians.
Between-study heterogeneity may have affected a meta-analysis result when interpreting of the pooled ORs (Edraki et al., 2019; Sayad, Ahmadi, Nekouian, et al., 2020; Sayad, Ahmadi, Moradi, et al., 2020). In the current meta-analysis, significant between study heterogeneity existed for both MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms under most genetic models. Subgroup analysis revealed that ethnicity might be the potential source of heterogeneity in the meta-analysis (Karimi-Zarchi et al., 2013; Mojtaba Sohrevardi et al., 2016). However, heterogeneity may be due to many factors, such as differences in the characteristics of controls, life style, diverse genotyping methods, small sample size, and a mixed population from different geographic regions (Yang et al., 2014). Moreover, certain HWE deviations were revealed in the distributions of controls in some included studies, which may be due to the small sample size or other experimental technique errors in the study. However, we have selected all eligible studies even HWE-violating studies.
There are potential limitations in our meta-analysis should be considered. First, the numbers of studies as well as sample sizes for each ethnicity and some subgroup analyses were relatively limited, which Type-II error might not be dismissed. Second, the research subjects of the included studies were mostly from Asian and Caucasian origins. Thus, the bias of racial diversity could not be avoided and the results are not applicable to all populations. Third, the reliability and authenticity of our results may be influenced by the limited number of studies and small sample sizes. Fourth, moderate heterogeneity existed in some genetic models for both MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms. And the subsequent meta-regression could not identify any interfering factors contributing to heterogeneity. Selection bias, although no publication bias was observed, might be a possible major source of between-study heterogeneity in this meta-analysis. Fifth, the detailed individual data in some studies was not available; leading to failure to adjust risk of MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms with bladder cancer based on potential risk factors, such as age, gender, smoking, occupation, environmental factors, lifestyle habits, and other covariates of this disease. Finally, bladder cancer as other malignancies is a multi-factorial disease that results from complex interactions between various genetic and environmental factors. However, due to the lack of the individual original data in the selected studies, we were unable to evaluate the effect of gene-environment interactions, gene-gene interactions and also different polymorphisms within MTHFR gene on development of bladder cancer.
In conclusion, our pooled data revealed that MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms might be not risk factor for development of bladder cancer. However, stratified analysis by ethnicity revealed that both MTHFR rs1801133 (677C>T) and rs180113 (1298A>C) polymorphisms were significantly associated with an increased risk of bladder cancer in Asians. Further studies with larger sample size, well-designed, and population-based studies among different ethnicities required to validate our findings.
Abbreviations
MTHFR: Methylenetetrahydrofolate Reductase
CBD: Biomedical Database
CNKI: Chinese National Knowledge Infrastructure
NOS: Newcastle-Ottawa Score
OR: Odds Ratio
CI: Confidence Interval
HWE: Hardy-Weinberg Equilibrium
CMA: Comprehensive Meta-Analysis
HB: Hospital-Based
PB: Population-Based
SOC: source of controls
MAFs: Minor Allele Frequencies
PCR-RFLP: Polymerase Chain Reaction Restriction Fragment Length Polymorphism
Author Contribution Statement
Conceived and designed the study and experiments: SAD and FA. Performed the experiments: HM, JSY and SHS. Analyzed the data: SAD and HN. Contributed reagents/materials/analysis tools: FA, SK and MZS. Wrote the paper: HN, JSY and EA. All authors reviewed the manuscript.
Acknowledgements
I would like to express my sincere gratitude to Dr. Elahe Akbarian for his motivation, knowledge and support during the course of this research.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable for this manuscript.
Vailability of data and material
The datasets generated during and/or analyzed during this study are the corresponding author on reasonable request.
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- Ahmadi SAY, Sayad S, Shahsavar F, et al. Expression of Angiogenesis-related Genes in a Group of Iranian Cases of Breast Cancer. Curr Pharmacogenomics Person Med. 2021;17:197–205. [Google Scholar]
- Akbari MT, Naderi A, Saremi L, et al. Methionine synthase A2756G variation is associated with the risk of retinoblastoma in Iranian children. Cancer Epidemiol. 2015;39:1023–5. doi: 10.1016/j.canep.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Al-Zalabani AH, Stewart KFJ, Wesselius A, Schols AMWJ, Zeegers MP. Modifiable risk factors for the prevention of bladder cancer: a systematic review of meta-analyses. Eur J Epidemiol. 2016;31:811–51. doi: 10.1007/s10654-016-0138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminian O, Saburi A, Mohseni H, et al. Occupational risk of bladder cancer among Iranian male workers. Urol Ann. 2014;6:135–8. doi: 10.4103/0974-7796.130643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amooee A, Lookzadeh MH, Mirjalili SR, et al. Association of rs2435357 and rs1800858 polymorphisms in Ret Proto-Oncogene with hirschsprung disease: systematic review and meta-analysis. Arq Bras Cir Dig. 2019;32:e1448. doi: 10.1590/0102-672020190001e1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen BK, Aagnes B, Gislefoss R, Andreassen M, Wahlqvist R. Incidence and Survival of urothelial carcinoma of the urinary bladder in Norway 1981-2014. BMC Cancer. 2016;16:799. doi: 10.1186/s12885-016-2832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Azarpira MR, Ghilian MM, Sobhan MR, et al. Association of MTHFR and TNF-α genes polymorphisms with susceptibility to Legg-Calve-Perthes disease in Iranian children: A case-control study. J Orthop. 2018;15:984–7. doi: 10.1016/j.jor.2018.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghestani AR, Shahmirzalou P, Sayad S, Akbari ME, Zayeri F. Comparison cure rate models by DIC criteria in Breast Cancer data. Asian Pac J Cancer Prev. 2018;19:1601–6. doi: 10.22034/APJCP.2018.19.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami R, Schwartz DA, Asadian F, et al. Association of MTHFR 677C>T polymorphism with IUGR and placental abruption risk: A systematic review and meta-analysis. Eur J Obstet Gynecol Reprod Biol. 2021;256:130–9. doi: 10.1016/j.ejogrb.2020.11.016. [DOI] [PubMed] [Google Scholar]
- Bahrami R, Shajari A, Aflatoonian M, et al. Association of REarranged during Transfection (RET) c 73 + 9277T > C and c 135G > a Polymorphisms with Susceptibility to Hirschsprung Disease: A Systematic Review and Meta-Analysis. Fetal Pediatr Pathol. 2020;39:476–90. doi: 10.1080/15513815.2019.1672225. [DOI] [PubMed] [Google Scholar]
- Binesh F, Zarchi MK, Vahedian H, Rajabzadeh Y. Primary malignant lymphoma of the uterine cervix. BMJ Case Rep. 2012;2012:bcr2012006675. doi: 10.1136/bcr-2012-006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffioux CR. Epidemiology of bladder cancer. Prog Clin Biol Res. 1984;162:11–25. [PubMed] [Google Scholar]
- Cai DW, Liu XF, Bu RG, et al. Genetic polymorphisms of MTHFR and aberrant promoter hypermethylation of the RASSF1A gene in bladder cancer risk in a Chinese population. Int J Appl Basic Med Res. 2009;37:1882–9. doi: 10.1177/147323000903700625. [DOI] [PubMed] [Google Scholar]
- Chung C-J, Pu Y-S, Su C-T, et al. Polymorphisms in one-carbon metabolism pathway genes, urinary arsenic profile, and urothelial carcinoma. Cancer Causes Control. 2010;21:1605–13. doi: 10.1007/s10552-010-9589-3. [DOI] [PubMed] [Google Scholar]
- Degeorge KC, Holt HR, Hodges SC. Bladder Cancer: Diagnosis and Treatment. Am Fam Physician. 2017;96:507–14. [PubMed] [Google Scholar]
- Edraki M, Noeezad Z, Bahrami R, Pourahmad S, Shirazi ZH. Effect of Spiritual Care Based on “Ghalbe Salim” Model on Anxiety among Mothers with Premature Newborns Admitted to Neonatal Intensive Care Units. Iran J Neonatol. 2019;10:50–7. [Google Scholar]
- Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. JAMA. 2011;306:737–45. doi: 10.1001/jama.2011.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami F, Zarchi MK, Mousavi A. Surgical Management of Primary Vulvar Lymphangioma Circumscriptum and Postradiation: Case Series and Review of Literature. J Minim Invasive Gynecol. 2008;15:205–8. doi: 10.1016/j.jmig.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Gohari M, Dastgheib AS, Jafari-Nedooshan J, et al. Association of MTHFR 677C>T, 1298A>C and MTR 2756A>G Polymorphisms with Risk of Retinoblastoma. Klin Onkol. 2019;32:375–379. doi: 10.14735/amko2019375. [DOI] [PubMed] [Google Scholar]
- Grotenhuis AJ, Vermeulen SH, Kiemeney LA. Germline genetic markers for urinary bladder cancer risk, prognosis and treatment response. Future Oncol. 2010;6:1433–60. doi: 10.2217/fon.10.109. [DOI] [PubMed] [Google Scholar]
- Gu J, Wu X. Genetic susceptibility to bladder cancer risk and outcome. Personalized Med. 2011;8:365–74. doi: 10.2217/PME.11.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarahzadeh MH, Asadian F, Farbod M, et al. Cancer and Coronavirus Disease (COVID-19): Comorbidity, Mechanical Ventilation, and Death Risk. J Gastrointestinal Cancer. 2021;52:80–4. doi: 10.1007/s12029-020-00529-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagas MR, Park S, Nelson HH, et al. Methylenetetrahydrofolate reductase (MTHFR) variants and bladder cancer: a population-based case-control study. Int J Hyg Environ Health. 2005;208:321–7. doi: 10.1016/j.ijheh.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Karimi-Zarchi M, Dehghani-Firoozabadi R, Tabatabaie A, et al. A comparison of the effect of levonorgestrel IUD with oral medroxyprogesterone acetate on abnormal uterine bleeding with simple endometrial hyperplasia and fertility preservation. Clin Exp Obstet Gynecol. 2013;40:421–4. [PubMed] [Google Scholar]
- Karimi-Zarchi M, Moghimi M, Abbasi H, et al. Association of MTHFR 677C > T polymorphism with susceptibility to ovarian and cervical cancers: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2019;20:2569–77. doi: 10.31557/APJCP.2019.20.9.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura F, Florl AR, Steinhoff C, et al. Polymorphic methyl group metabolism genes in patients with transitional cell carcinoma of the urinary bladder. Mutat Res. 2001;458:49–54. doi: 10.1016/s1383-5726(01)00010-3. [DOI] [PubMed] [Google Scholar]
- Kiseljaković E, Jadrić R, Hasić S, Skenderi F, Resić H, Winterhalter-Jadrić M. Polymorphism in methylentetra-hydrofolate reductase gene: Important role in diseases. Bosn J Basic Med Sci. 2008;8:165–9. doi: 10.17305/bjbms.2008.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenis AT, Lec PM, Chamie K. Bladder cancer a review. JAMA. 2020;324:1980–91. doi: 10.1001/jama.2020.17598. [DOI] [PubMed] [Google Scholar]
- Letaiová S, Medveová A, Ovíková A, et al. Bladder cancer, a review of the environmental risk factors. Environ Health. 2012;11:S11. doi: 10.1186/1476-069X-11-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Hu YP, Yang Z, Sun T. Association between MTHFR Ala222Val (rs1801133) polymorphism and bladder cancer susceptibility: A systematic review and meta-analysis. Tumour Biol. 2013;34:2565–72. doi: 10.1007/s13277-013-0802-3. [DOI] [PubMed] [Google Scholar]
- Lin J, Spitz MR, Wang Y, et al. Polymorphisms of folate metabolic genes and susceptibility to bladder cancer: a case-control study. Carcinogenesis. 2004;25:1639–47. doi: 10.1093/carcin/bgh175. [DOI] [PubMed] [Google Scholar]
- Liu JH, Zhu C, Zheng K, et al. MTHFR Ala222Val polymorphism and clinical characteristics confer susceptibility to suicide attempt in chronic patients with schizophrenia. Sci Rep. 2020;10:1–9. doi: 10.1038/s41598-020-57411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannino DM, Mulinare J, Ford ES, Schwartz J. Tobacco smoke exposure and decreased serum and red blood cell folate levels: Data from the third National Health and Nutrition Examination Survey. Nicotine Tob Res. 2003;5:357–62. doi: 10.1080/1462220031000094330. [DOI] [PubMed] [Google Scholar]
- Maruti SS, Ulrich CM, Jupe ER, White E. MTHFR C677T and postmenopausal breast cancer risk by intakes of one-carbon metabolism nutrients: A nested case-control study. Breast Cancer Res. 2009;11:R91. doi: 10.1186/bcr2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey DJ, Lee S, Choi W, et al. Molecular genetics of bladder cancer: Emerging mechanisms of tumor initiation and progression. Urol Oncol. 2010;28:429–40. doi: 10.1016/j.urolonc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XY, Shi MJ, Chen JF, et al. Association between the TACC3 rs798766 Polymorphism and Risk of Urinary Bladder Cancer: A Synthesis Based on Current Evidence. Dis Markers. 2017;2017:7850708. doi: 10.1155/2017/7850708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley D, Baum N. Smoking: Its Impact on Urologic Health. Rev Urol. 2015;17:220–5. [PMC free article] [PubMed] [Google Scholar]
- Mojtaba Sohrevardi S, Nosouhi F, Hossein Khalilzade S, et al. Evaluating the effect of insulin sensitizers metformin and pioglitazone alone and in combination on women with polycystic ovary syndrome: An RCT. Int J Reprod Biomed. 2016;14:743. [PMC free article] [PubMed] [Google Scholar]
- Moore LE, Malats N, Rothman N, et al. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer. 2007;120:2452–8. doi: 10.1002/ijc.22565. [DOI] [PubMed] [Google Scholar]
- Moore LE, Wiencke JK, Bates MN, et al. Investigation of genetic polymorphisms and smoking in a bladder cancer case-control study in Argentina. Cancer Lett. 2004;211:199–207. doi: 10.1016/j.canlet.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Niktabar SM, Aarafi H, Dastgheib SA, et al. Association of MTHFR 1298A > C Polymorphism with Susceptibility to Non-Syndromic Cleft Lip with or without Palate: A Case-Control Study and Meta-Analysis. Fetal Pediatr Pathol. 2021;40:1–17. doi: 10.1080/15513815.2019.1683918. [DOI] [PubMed] [Google Scholar]
- Oeyen E, Hoekx L, De Wachter S, et al. Bladder cancer diagnosis and follow-up: The current status and possible role of extracellular vesicles. Int J Mol Sci. 2019;20:821. doi: 10.3390/ijms20040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Su M, Huang G, et al. MTHFR C677T genetic polymorphism in combination with serum Vitamin B2, B12 and aberrant DNA methylation of P16 and P53 genes in esophageal squamous cell carcinoma and esophageal precancerous lesions: A case–control study. Cancer Cell Int. 2019;19:288. doi: 10.1186/s12935-019-1012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N, Murata M, Ikeda Y, et al. The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am J Hum Genet. 2002;70:758–62. doi: 10.1086/338932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouissi K, Ouerhani S, Oliveira E, et al. Polymorphisms in one-carbon metabolism pathway genes and risk for bladder cancer in a Tunisian population. Cancer Genet Cytogenet. 2009;195:43–53. doi: 10.1016/j.cancergencyto.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Sadeghiyeh T, Dastgheib SA, Lookzadeh MH, et al. Association of MTHFR 677C > T and 1298A > C polymorphisms with susceptibility to attention deficit and hyperactivity disorder. Fetal Pediatr Pathol. 2020;39:422–9. doi: 10.1080/15513815.2019.1666330. [DOI] [PubMed] [Google Scholar]
- Safarinejad MR, Shafiei N, Safarinejad S. Genetic susceptibility of methylenetetrahydrofolate reductase (MTHFR) gene C677T, A1298C, and G1793A polymorphisms with risk for bladder transitional cell carcinoma in men. Med Oncol. 2011;28:398–412. doi: 10.1007/s12032-010-9723-9. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–34. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- Sayad S, Ahmadi SAY, Moradi M, et al. A meta-analysis on diagnostic accuracy of serum HLA-G level in breast cancer. Expert Rev Precis Med Drug Dev. 2020;5:109–14. [Google Scholar]
- Sayad S, Ahmadi SAY, Nekouian R, Panahi M, Anbari K. Epidemiological and pathological characteristics of post-surgical cases of invasive breast cancer among ethnicities of Iran in 2018: A single center cross-sectional study. Arch Oncol. 2020;26:6–9. [Google Scholar]
- Shi R, Zhao Z, Zhou H, Zhou J TW. Lack of association between MTHFR Ala222Val and Glu429Ala polymorphisms and bladder cancer risk: A meta-analysis of case-control studies. Biomed Rep. 2014;2:396–403. doi: 10.3892/br.2014.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani-Jadidi S, Meibodi B, Javaheri A, et al. Association between Fetal MTHFR A1298C (rs1801131) Polymorphism and Neural Tube Defects Risk: A Systematic Review and Meta-Analysis. Fetal Pediatr Pathol. 2020;2020:1–18. doi: 10.1080/15513815.2020.1764682. [DOI] [PubMed] [Google Scholar]
- Tabatabaei RS, Fatahi-Meibodi N, Meibodi B, et al. Association of Fetal MTHFR C677T Polymorphism with Susceptibility to Neural Tube Defects: A Systematic Review and Update Meta-Analysis. Fetal Pediatr Pathol. 2020;2020:1–17. doi: 10.1080/15513815.2020.1775734. [DOI] [PubMed] [Google Scholar]
- Tungtrongchitr R, Pongpaew P, Soonthornruengyot M, et al. Relationship of tobacco smoking with serum vitamin B12, folic acid and haematological indices in healthy adults. Public Health Nutr. 2003;6:675–81. doi: 10.1079/phn2003483. [DOI] [PubMed] [Google Scholar]
- Venkatesulu BP, Chandrasekar VT, Giridhar P, et al. A systematic review and meta-analysis of cancer patients affected by a novel coronavirus. medRxiv. 2020;5:20115303. doi: 10.1093/jncics/pkaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhu H, Fu G, et al. Polymorphisms of methylenetetrahydrofolate reductase and methionine synthase genes and bladder cancer risk: A case-control study with meta-analysis. Clin Exp Med. 2009;9:9–19. doi: 10.1007/s10238-008-0013-1. [DOI] [PubMed] [Google Scholar]
- Wong MCS, Fung FDH, Leung C, et al. The global epidemiology of bladder cancer: a joinpoint regression analysis of its incidence and mortality trends and projection. Sci Rep. 2018;8:1129. doi: 10.1038/s41598-018-19199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lin X, Dinney CP, Gu J, Grossman HB. Genetic polymorphism in bladder cancer. Front Biosci. 2007;12:192–213. doi: 10.2741/2058. [DOI] [PubMed] [Google Scholar]
- Xu W, Zhang H, Wang F, Wang H. Quantitative assessment of the association between MHTFR C677T (rs1801133, Ala222Val) polymorphism and susceptibility to bladder cancer. Diagn Pathol. 2013;8:95. doi: 10.1186/1746-1596-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zuo L. Association between methylenetetrahydrofolate reductase gene rs1801131 A/C polymorphism and urinary tumors’ susceptibility. Hereditas. 2020;157:16. doi: 10.1186/s41065-020-00129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao Y, Feng T, et al. Meta-analysis of the rs4779584 polymorphism and colorectal cancer risk. PLoS One. 2014;9:e89736. doi: 10.1371/journal.pone.0089736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You W, Li Z, Jing C, et al. MTHFR C677T and A1298C Polymorphisms were associated with bladder cancer risk and disease progression: A meta-analysis. DNA Cell Biol. 2013;32:260–7. doi: 10.1089/dna.2012.1931. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang Y, Qin Z, et al. TP53 codon 72 Polymorphism and bladder cancer risk: A meta-analysis and emphasis on the role of tumor or smoking status. J Cancer. 2018;9:3522–31. doi: 10.7150/jca.26264. [DOI] [PMC free article] [PubMed] [Google Scholar]