Abstract
Background:
Biopsy is the gold standard for oral squamous cell carcinoma (OSCC) diagnosis. Salivary biomarkers provide promising complementary alternative diagnostic adjunct for its simple non- invasive collection and technique and to screen large population.
Objective:
To summarize and compare the existing evidence on diagnostic accuracy of salivary biomarkers with their estimation method in detecting early oral squamous cell carcinoma.
Methods:
The review protocol is registered under PROSPERO(CRD42021225704). PubMed, Google Scholar, EBSCOhost were searched from 2000 to 2020 to identify the screening potential of eight salivary biomarkers: mRNA, miRNA, DUSP100, s100P, IL-8, IL-1B, TNF-a and MMP-9. True-positive, false-positive, true-negative, false-negative, sensitivity, specificity values were extracted or calculated if not present for each study. Quality of selected studies was evaluated based on QUADAS 2 tool. Meta-analysis was performed using a bivariate model parameter for the sensitivity and specificity and summary points, summary receiver operating curve (SROC), confidence region, and prediction region were calculated.
Results:
Eighteen studies were included for qualitative synthesis and out of that 13 for meta-analysis. Sensitivity and specificity were calculated with AUC. For mRNA it was 91% and 90% with 0.96 AUC, miRNA had 91% and 91% with 0.95 AUC for PCR. IL-1B had 46% and 60% with 0.61 AUC, S100p had 45% and 90% with 0.57 AUC for ELISA. IL-8 had 54% and 74% for ELISA and 89% and 90% for PCR with 0.79 AUC and DUSP1 had 32% and 87% for ELISA and 76% and 83% for PCR with 0.83 AUC respectively.
Conclusion:
Early detection of OSCC was best achieved by screening for salivary mRNA and miRNA estimated by PCR. Further investigation is required into salivary RNA as novel biomarkers and these salivary biomarkers may be potentially used for non-invasive diagnosis of early OSCC.
Key Words: Accuracy, diagnosis, meta-analysis, oral cancer, saliva, salivary biomarker
Introduction
Saliva is considered as mirror of human health. Its composition reflects levels of hormonal, immunological, toxicological and infectious disease markers. Due to its simple collection, non-invasive method, less time consuming and inexpensive saliva has been proposed as a diagnostic medium of choice (Motamayl et al., 2010).
Squamous cell carcinomas of oral cavity are the most common malignancies (Dhanuthai et al., 2018). They are usually defined as carcinoma with squamous differentiation, arising from mucosal epithelium. The disease presents as flat, scale-like forms, found lining the mouth and throat, which are easily detectable due to their superficial location (Montero at al., 2015). Compared to five years survival rates for breast cancer (89%) and prostate cancer (99%) its five-year survival rate is 62% (Siegel at al., 2013). Oral squamous cell carcinoma (OSCC) is considered accounts for 90% of all oral cancers. Each year around 5,75,000 new cases are diagnosed and 3,35,000 deaths occur worldwide. Its high frequency in Central and South East Asian countries (India, Bangladesh, Sri Lanka, Thailand, Indonesia and Pakistan) are well documented (Saxena at al., 2017). Oral cancers are predominant in both sexes accounting for one third of all cancers in South East Asian countries. The highest incidences of oral cancer in world is seen in India, with estimated incidence of 12.48 cases per 100,000 population in males and 5.52 cases per 100,000 populations in females (Kampfrath at al., 2013). The male to female ratio is 4:1 for OSCC (Bhatt at al., 2010).
Oral cancer is usually diagnosed when it gets symptomatic but by this stage around 2/3rd of patients develops advanced disease with regional metastasis (Prasad at al., 2013). Delayed detection is primary reason for high morbidity and mortality rates, and this strongly supports the need to perk up early detection of oral cancers (Jemal at al., 2011). The gold standard for oral cancer diagnosis is still a biopsy, which is not suited for screening purposes due to its invasive nature, high cost, and need for specially trained medical personal and equipment. There is constant search for biomarkers in saliva, a body fluid which can be easily collected, (Jemal at al., 2011). Salivary biomarker offers a promising diagnostic adjunct due to its simple non- invasive collection method and can be employed to screen large population (Warnakulasuriya at al., 2009).
Analysis of literature reveals that current investigative approaches for improving oral cancer detection consists of salivary proteins, salivary proteases, salivary RNA, transcriptomic and proteomic classes of biomarkers which includes mRNA, miRNA, DUSP100, s100P, IL-8, IL-1B, TNF-a, MMP-9 (Markopoulos at al., 2012).
Analysis of oral cytokine levels gives an idea regarding early detection of OSCC (Zhang at al., 2012). The saliva of patients recently diagnosed with OSCC had significantly increased levels of IL-8 (Markopoulos at al., 2010). Moreover, few articles also conclude that salivary protein IL1-B can be used as a biomarker for oral cancer (Shaw at al., 2016).
Few researchers have explained the benefits of evaluating the expression of key OSCC-associated messenger RNA (mRNA). In 2006, salivary mRNA transcript analysis was done in a validation cohort of 32 patients with OSCC and 32 controls. 7 transcripts were significantly increased in OSCC including Dual Specificity 1 Protein (DUSP-1) and small calcium Protein 100 (S100P) mRNA. These biomarkers had combined overall sensitivity and specificity of 91%, thereby positioning them amongst the most discriminatory panels of cancer biomarkers arising from human body fluids (Cheng et al., 2015; Zhang et al., 2015; Li et al 2004).
Few research has shown the utility of microRNAs (miRNAs) as a biomarker for solid tumors (Shpitzer et al., 2007). Studies have shown different expression of miRNAs within cancerous cells compared with normal cells (Elashoff et al., 2012). miRNAs were differentially expressed in saliva when comparing OSCC patients and healthy subjects, which support their use as a diagnostic tool for oral cancer detection (Shpitzer et al., 2009; Adisa et al., 2011).
Few studies have demonstrated that increased levels of salivary MMP-9 and TNF-a was seen in patients diagnosed with OSCC having sensitivity of 91% and 90.3% which could be a useful, non- invasive technique in diagnosis, prognosis, treatment and follow- up of OSCC (Adeyemi et al., 2011; Aa et al.,2018).
Understanding the diagnostic accuracy would help clinicians to reach correct diagnosis and choose most effective treatment. Diagnostic accuracy includes sensitivity, specificity and summary receiver operating characteristics (SROC) analysis.
Sensitivity and specificity explain the diagnostic ability of a test to correctly identify diseased and non- diseased respectively. They are independent of disease prevalence which refers to the probability of disease in a specific population at a given time and summary receiver operating characteristics (SROC) analysis is used to evaluate the predictive power for diagnosis.
For an accurate diagnosis, clinicians should have thorough knowledge regarding diagnostic values of each salivary biomarker before its use. It is notable that a large variation is seen regarding the diagnostic accuracy of salivary biomarkers among individual studies. For example, Brinkmann et al (Brinkmann et al., 2011) showed that sensitivity of IL-8 was 60% and specificity was 78%, whereas Li et al (Li et al., 2004) showed that sensitivity of IL-8 was 88% and specificity was 88%.
There have been already few reviews published on various salivary biomarkers in neck and head region (Sudbo et al., 2001; Li et al., 2015; Viet et al., 2008). Till date, no studies have provided a comprehensive, quantitative analysis of salivary biomarkers on which diagnostic reasoning of early oral squamous cell carcinoma can be established. Therefore, the aim of this systematic review is to compare the diagnostic accuracy of promising classes of principal salivary biomarkers estimated by polymerase chain reaction (PCR) and enzyme linked immunosorbent assay (ELISA) for early diagnosis of OSCC in adults through a meta- analysis.
Materials and Methods
Protocol and Registration
The systematic review and meta-analysis protocol was registered at the international prospective register of systematic reviews (PROSPERO- CRD42021225704) and performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis – Diagnostic Test Accuracy (PRISMA- DTA) checklist (Salameh et al., 2020).
Study Design
The following focused research question in the Participants (P), Index test (I), reference standard (R) and target condition (T) format was proposed “Is there a difference in the diagnostic accuracy of salivary biomarkers (Index Test) compared to biopsy (gold standard) for the early detection of oral squamous cell carcinoma (OSCC) in adults? Studies evaluating salivary biomarkers along with their method of assessment as compared to biopsy and reporting measures of diagnostic test accuracy such as sensitivity and specificity in detecting OSCC in adults suspected of having OSCC were eligible for inclusion. Studies evaluating the performance of only salivary biomarkers (index test) for the diagnosis of OSCC were also included for summarizing the evidence. Studies investigating combined salivary biomarkers were included.
Eligibility Criteria
Inclusion Criteria. The inclusion criteria were as follows:
(1) Study Design: In-vivo studies- Observational studies or Clinical trials comparing the diagnostic accuracy of salivary biomarkers with biopsy.
(2) Participant characteristics: patients diagnosed with oral squamous cell carcinoma aged 18 years and older
(3) Outcome measurements: Diagnostic accuracy including sensitivity, specificity, accuracy, determined using different methods irrespective of the methods of quantifying the outcomes.
(4) Articles written in English language
(5) Articles from 2000 – 2020 and available as free full text
Exclusion Criteria. The exclusion criteria were as follows:
(1) Non-clinical studies, in-vitro studies, and animal studies. Studies reporting about a single intervention were also excluded.
(2) Studies done on individuals less than 18 years of age.
(3) Studies not fully available in the database.
(4) Article reporting only abstracts were also excluded.
(5) Studies not reporting primary outcomes of accuracy, sensitivity, and specificity as well as where primary outcomes are not possible to calculate from the given raw data.
Search protocol and study selection
A comprehensive electronic search was performed till 31st December 2020 for the studies published within the last 20 years (from 2000 to 2020) using the following databases: PubMed and EBSCOhost to retrieve articles in the English language. The searches in the clinical trials database, cross-referencing and grey literature were conducted using Google Scholar, Greylist, and OpenGrey. In addition to the electronic search, a hand search was also made, and reference lists of the selected articles were screened.
Search Strategy
Appropriate key words and Medical Subject Heading (MeSH) terms were selected and combined with Boolean operators like AND. The search strategy used was as follows: (salivary biomarkers AND sensitivity AND specificity AND oral cancer), (saliva AND biomarkers AND diagnosis).
The search and screening, according to the previously established protocol were conducted by two review authors. A two-phase selection of articles was conducted. In phase one, two reviewers reviewed titles and abstracts of all articles. Articles that did meet inclusion criteria were excluded. In phase-two, selected full articles were independently reviewed and screened by same reviewers. Any disagreement was resolved by discussion. When mutual agreement between two reviewers was not reached, a third reviewer was involved to make final decision. The final selection was based on consensus among all three authors.
Data extraction
For all included studies, following descriptive study details were extracted by two independent reviewing authors (and) using pilot-tested customized data extraction forms: authors, study year, mean age of participants, sample size, type of salivary biomarker (transcriptomic, proteomic, salivary proteases, salivary proteins and salivary RNA), method of salivary biomarker detection (ELISA and PCR), main study results like sensitivity, specificity, true positive, true negative, false positive, false negative and conclusion. Quantitative data of sensitivity and specificity were compiled from each study and using these quantitative data, values like true positive, true negative, false positive and false negatives were calculated manually for the studies using the below formula’s where the data was not provided by authors. The corresponding authors were contacted via email where further information was needed.
a) False positive = (1-specificity) x (1- diseased cases/ total sample)
b) True negative = specificity x (1- diseased cases/total sample)
c) True positive = sensitivity x diseased cases/ total sample
d) False negative = (1- sensitivity) x diseased cases/total sample
Assessment of methodological quality
The methodological quality or the risk of bias was evaluated using Quality Assessment for Diagnostic Accuracy Studies -2 (QUADAS-2) tool (Whiting et al., 2011). The QUADAS-2 is a revised tool developed to assess quality of diagnostic studies through its four domains: patient selection, index test, reference standard, flow and timing of participants. Each domain had signalling questions with options of “Yes”, “No” or “Unclear”. The overall risk of bias was assessed as high: if answered ‘No’ to any question, Low: if answered ‘Yes’ to all questions and Unclear: if answered ‘Unclear’ to all questions or accompanied by any ‘Yes’. Risk of bias summary and applicability concern was graphically plotted using Review Manager (RevMan) software version 5.3.
Statistical analysis and data synthesis
Raw data was used to calculate sensitivity and specificity for each biomarker with their estimation method. For overall accuracy, we calculated pooled sensitivity, pooled specificity with 95% confidence interval, area under summary receiver operating characteristic. (Interpretation of AUC values were as follows: value above 80% were considered as excellent, between 70% and 80% as good, between 60% and 69% as fair and below 60% as poor outcomes for a diagnostic test (Jones et al., 2005). To assess the impact of heterogeneity, Higgins I2 test was used. This test represents the proportion of variability due to heterogeneity rather than due to sampling error (Lijmer et al., 2002). According to I2 test statistic the heterogeneity could be low (I2 <50%) or high (I2 >50%). Subgroup analysis was also carried out. Results were presented graphically as coupled forest plot for each salivary biomarker with their estimation method using Meta-Disc 1.4 software.
Additional analysis
Additional analysis was performed with positive likelihood ratio (PLR) and negative likelihood ratio (NLR) using DerSimonian-Laird’s estimator considering random effect model. Positive likelihood ratio (PLR) in range of 2-5, 5-10 and >10 represents small, moderate and large increase in probability of disease when test is positive while Negative likelihood ratio (NLR) in range of 0.2-0.5, 0.2-0.1 and <0.1 represents small, moderate and large decrease in probability of disease when test is negative (Grimes et al., 2005).
Results
Study Selection
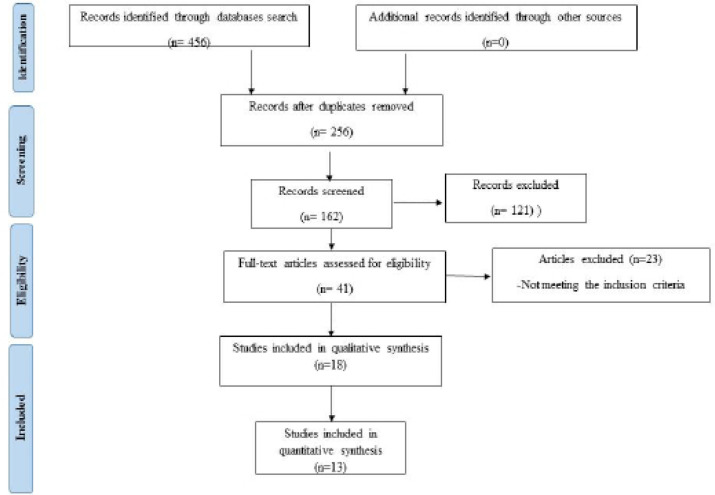
A flowchart of identification, inclusion and exclusion of studies is shown in Figure 1. After duplicates removal, reference list of all included studies was screened. Of which 121 studies were excluded. After this full text articles were assessed for eligibility and articles that did not meet inclusion criteria were excluded. Only eighteen studies fulfilled eligibility criteria and were included in qualitative synthesis. Of those, only thirteen were adequate to use for meta-analysis.
Figure 1.
PRISMA Flowchart of Literature Search and Selection Criteria
Study Characteristics
A summary of descriptive characteristics of all included 18 studies is provided in (Supplemental Table S1). Data was evaluated from aggregate of 1436 patients with mean age of 53.75 years. The articles were published between 2000 to 2020 and conducted in eight countries: seven studies (li et al., 2004; Hu et al., 2008; John et al., 2004; Park et al., 2009; Tanaka et al., 2016; Bonne et al., 2012; Zimmermann et al., 2008) in USA, four studies (Deepthi et al., 2020; Panta et al., 2014; Singh et al., 2020; Smriti et al., 2021) in India, two studies (Chu et al., 2019; Lee et al., 2018) in Taiwan, one study (Feng et al., 2019) in China, one study (Prestiyanti et al., 2020) in Indonesia, one study (Cristaldi et al., 2019) in Italy, one study (Brinkmann et al., 2011) in Serbia and one study (Young et al., 2020) in South Korea. According to saliva-omics classification: six were salivary RNA studies (Cristaldi et al., 2019; Hu et al., 2008; Park et al., 2009; Tanaka et al., 2016; Young et al., 2020; Zimmermann et al., 2008), four were transcriptomic studies (Brinkmann et al., 2011; John et al., 2009; Lee et al., 2018; Panta et al., 2014), two were salivary proteases studies (Feng et al., 2019; Smriti et al., 2021), one salivary proteomic study (Chu et al., 2019) one salivary protein study (Deepthi et al., 2020). Four of the studies (Lee et al., 2018; Prestiyanti et al., 2020; Singh et al., 2020; Bonne et al., 2012) included two complementary approaches (both transcriptomic and proteomic). Salivary biomarkers were estimated by ELISA in 10 studies (Brinkmann et al., 2011; Deepthi et al., 2020; Feng et al., 2019; Hu et al., 2008; Lee et al., 2018; Prestiyanti et al., 2020; Singh et al., 2020; Smriti et al., 2021; Bonne et al., 2012; Chu et al., 2019) and by PCR in eight studies (Li et al., 2004; Cristaldi et al., 2019; John et al., 2004; Panta et al., 2014; Park et al., 2009; Tanaka et al., 2016; Young et al., 2020; Zimmermann et al., 2008). All studies utilized same reference standard: tissue biopsy.
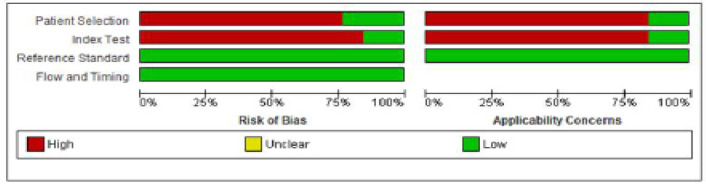
Risk of Bias within Studies
Although none of included studies were classified as low risk of bias for all four domains. Patient selection was considered as high risk of bias in all studies, which was mainly due to method of patient enrollment, nature of study design and implementing inappropriate exclusion. Only three studies (Brinkmann et al., 2011; Panta et al., 2014; Zimmermann et al., 2008), reported low risk of bias with respect to patient selection domain.
The index test was considered to be at low risk of bias only in two studies (Panta et al., 2014; Yakob et al., 2014). High risk of bias was reported with respect to index test domain due to insufficient details reported as to whether results of index test was interpreted without prior knowledge of reference standard results, lack of pre-specification of a test-positive threshold and statement of conflict of interest.
Similarly, the reference standard and flow and timing domain was considered at low risk in all studies.
The risk of bias and applicability concern summary and graph is depicted in Figures 2 and 3.
Figure 2.
Risk of Bias and Applicability Concerns Summary: Review Authors' Judgements about Each Domain for Each Included Study
Figure 3.
Risk of Bias and Applicability Concerns Graph: Review Authors' Judgements about Each Domain Presented as Percentages Across Included Studies
Synthesis of Results
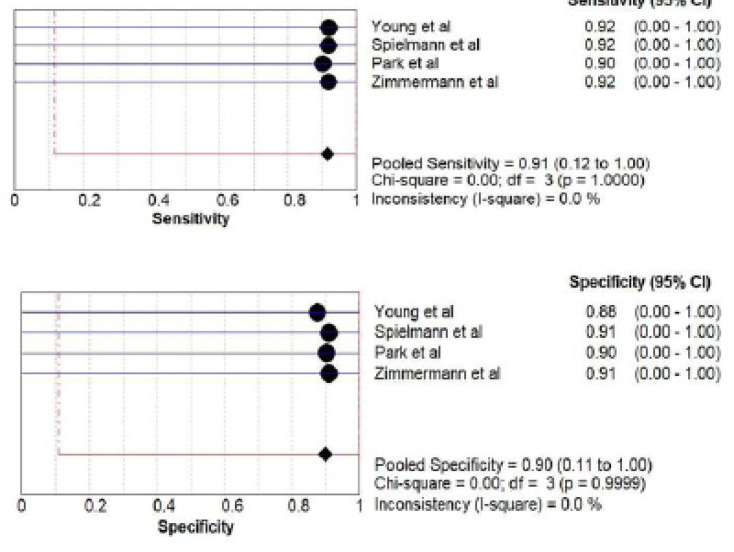
A) mRNA salivary biomarker estimation by PCR: a total of 295 patients from four studies (Young et al., 2020; Spielmann et al., 2011; Park et al., 2009; Zimmermann et al., 2008) investigated accuracy of mRNA. The pooled sensitivity was 0.91 (CI 0.12 -1.00) and pooled specificity was 0.90(CI 0.11 -1.00) as shown in Figure 4.
Figure 4.
Pooled Sensitivity and Specificity for mRNA Salivary Biomarker Estimated by PCR
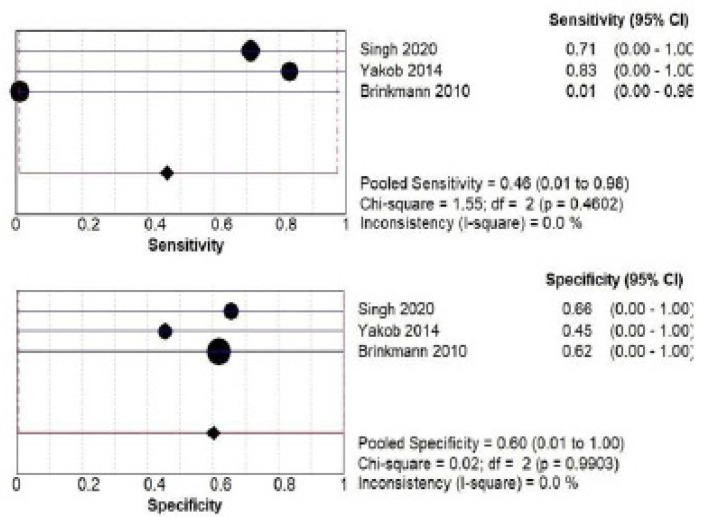
B) IL-1B salivary biomarker estimation by ELISA: a total of 345 patients from three studies (Singh., 2020; Yakob et al., 2014; Brinkmann 2010) investigated accuracy of IL-1B estimated by ELISA. The pooled sensitivity was 0.46 (CI 0.01- 0.98) and pooled specificity was 0.60 (CI 0.01- 1.00) as shown in Figure 5.
Figure 5.
Pooled Sensitivity and Specificity for IL-1B Salivary Biomarker Estimated by ELISA
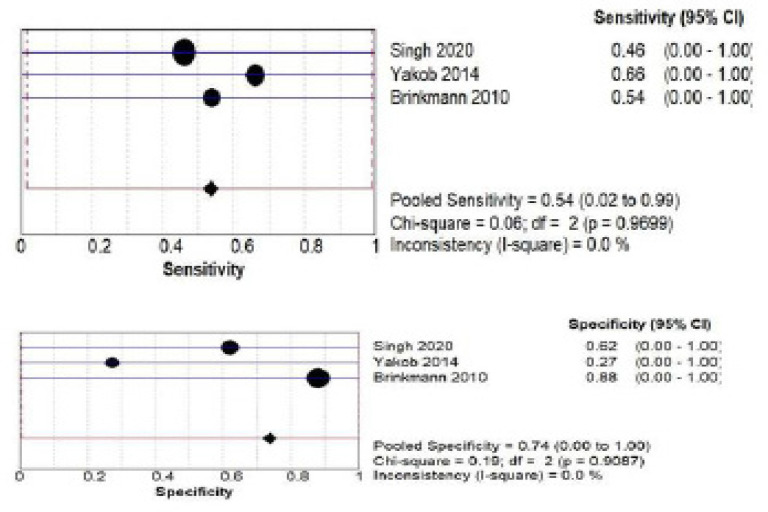
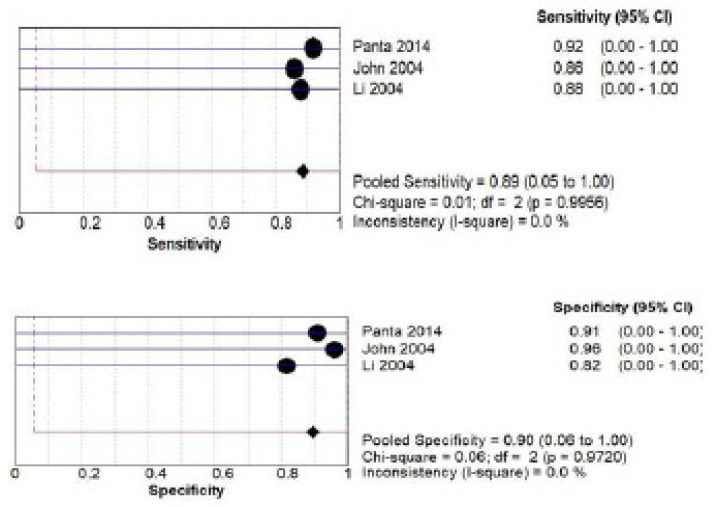
C) IL-8 salivary biomarker estimation Both by ELISA and PCR: a total of 345 patients from three studies (Singh., 2020; Yakob et al., 2014; Brinkmann et al., 2010) investigated accuracy of IL-8 biomarker estimated by ELISA. The pooled sensitivity was 0.54 (CI 0.02 – 0.99) and pooled specificity was 0.74 (CI 0.00 – 1.00) as shown in Figure 6 (a) while for PCR method a total of 192 patients from three studies (Panta et al., 2014; John et al., 2004; Li et al., 2004) investigated accuracy of IL-8. The pooled sensitivity was 0.89 (CI 0.05- 1.00) and pooled specificity was 0.90 (CI 0.06- 1.00) as shown in Figure 6 (b).
Figure 6. a.
Pooled Sensitivity and Specificity for IL-8 Salivary Biomarker Estimated by ELISA
Figure 6.b.
Pooled Sensitivity and Specificity for IL-8 Salivary Biomarker Estimated by PCR
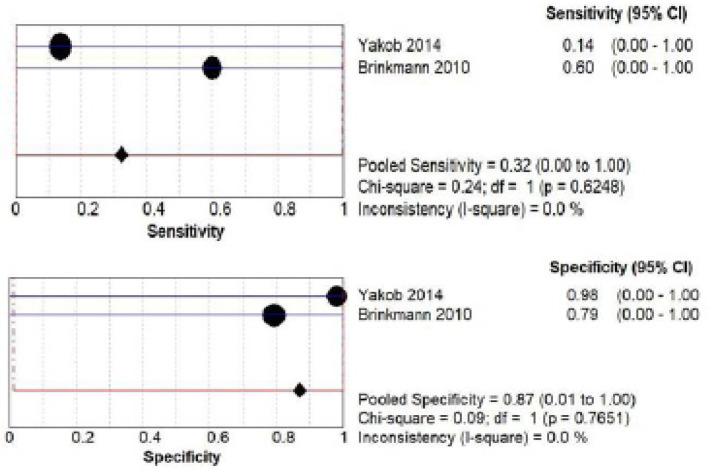
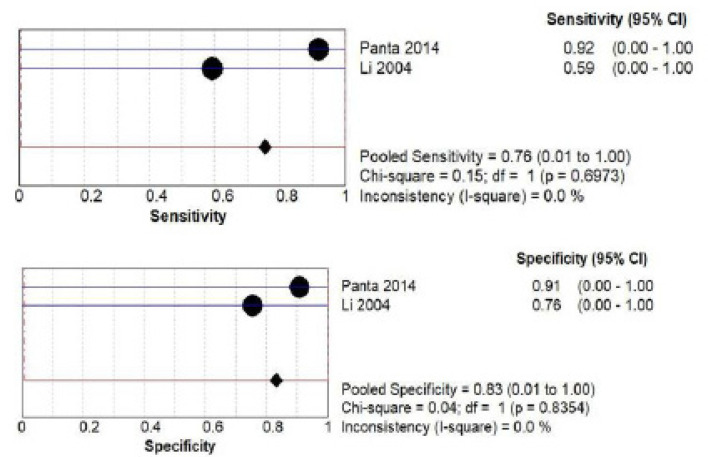
D) DUSP1 salivary biomarker estimation Both by ELISA and PCR: a total of 186 patients from two studies (Yakob et al., 2014; Brinkmann et al., 2010) investigated accuracy of DUSP1 by ELISA. The pooled sensitivity was 0.32 (CI 0.00- 1.00) and pooled specificity was 0.87 (CI 0.00- 1.00) as shown in Figure 7 (a) while a total of 128 patients from two studies (Panta et al., 2014; Li et al., 2004) investigated accuracy of DUSP1 by PCR. The pooled sensitivity was 0.76 (CI 0.00- 1.00) and pooled specificity was 0.83 (CI 0.01- 1.00) as shown in Figure 7 (b).
Figure 7.a.
Pooled Sensitivity and Specificity for DUSP1 Salivary Biomarker Estimated by ELISA
Figure 7. b.
Pooled Sensitivity and Specificity for DUSP1 Salivary Biomarker Estimated by PCR
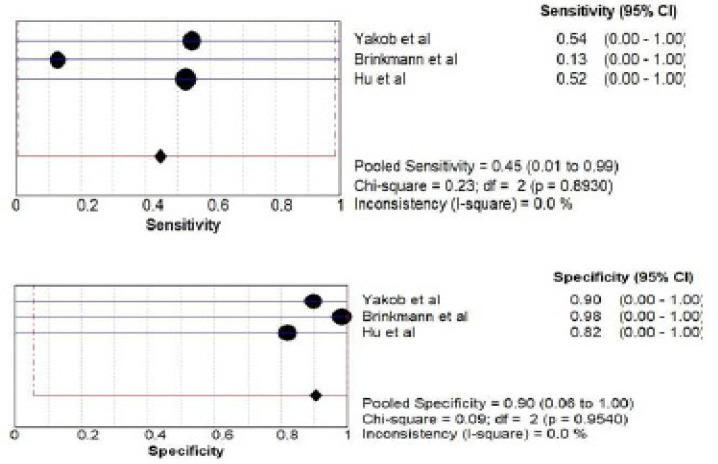
E) s100P biomarker estimation by ELISA: a total of 314 patients from three studies (Yakob et al., 2014; Brinkmann et al., 2011; Hu et al., 2008) investigated accuracy of s100P biomarker. The pooled sensitivity was 0.45 (CI 0.00- 0.99) and pooled specificity was 0.90 (CI 0.01- 1.00) as shown in Figure 8.
Figure 8.
Pooled Sensitivity and Specificity for s100P Salivary Biomarker Estimated by PCR
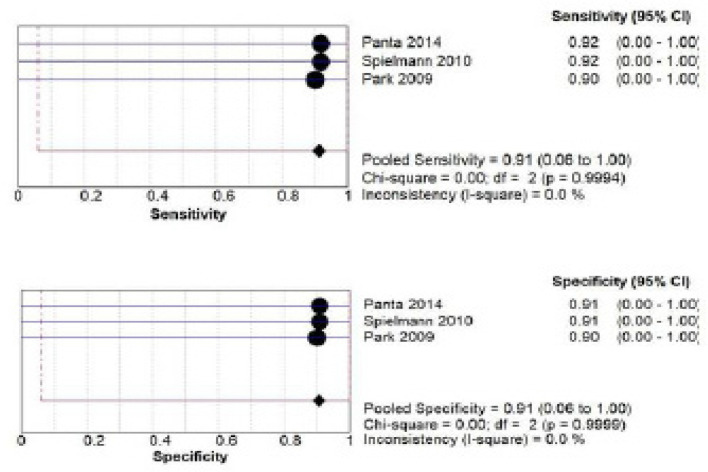
F) miRNA salivary biomarker estimation by PCR: a total of 228 patients from three studies (Panta et al., 2014; Spielmann et al., 2010; Park et al., 2009) investigated accuracy of miRNA biomarker. The pooled sensitivity was 0.91 (CI 0.06- 1.00) and pooled specificity was 0.91 (CI 0.06- 1.00) as shown in Figure 9.
Figure 9.
Pooled Sensitivity and Specificity for miRNA Salivary Biomarker Estimated by PCR
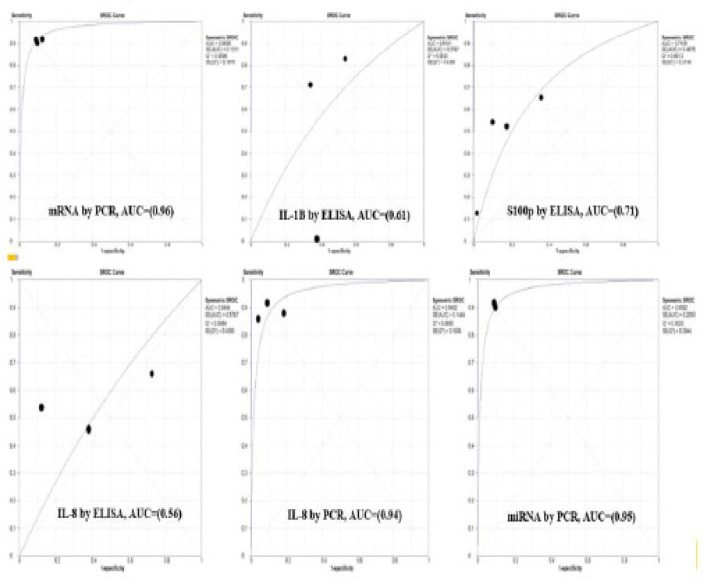
The sensitivities and specificities for studies ranged from 13% to 92% and from 27% to 91%. The area under the curve (AUC) with summary receiver operating characteristics (SROC) curve was plotted for all the biomarkers with their estimation method as shown in Figure 10.
Figure 10.
The Area under the Curve (AUC) with Summary Receiver Operating Characteristics (SROC) Curve was Plotted for all the Biomarkers with Their Estimation Method as Shown in Figure 10. Note: The highest AUC was seen for mRNA (0.96) and miRNA (0.95) estimated by PCR which was considered excellent and the lowest AUC was seen for IL-8 (0.56) estimated by ELISA which was considered poor
Additional analysis
A diagnostic table was constructed for each salivary biomarker with their estimation method (Supplemental Table S2). In this table, all measurements (true positive, true negative, false positive, false negative, sensitivity, specificity of each individual study, pooled sensitivity, pooled specificity, positive likelihood ratio, negative likelihood ratio of each individual study, pooled positive and pooled negative likelihood ratio) are shown.
Table 1.
Showing Measure of Accuracy of Salivary Biomarkers in Early OSCC Detection: Area under Curve (AUC) and Standard Error
| Salivary Biomarkers with their Method of Detection | Area Under Curve (AUC) Value | Standard Error (SE) |
|---|---|---|
| 1. mRNA (PCR) | 0.96 | 0.13 |
| 2. IL-1B (ELISA) | 0.61 | 0.57 |
| 3. IL-8 | ||
| a) IL-8 (ELISA) | 0.56 | 0.57 |
| b) IL-8 (PCR) | 0.94 | 0.14 |
| 4. S100p | ||
| a) S100p (ELISA) | 0.75 | 0.79 |
| 5. miRNA (PCR) | 0.95 | 0.2 |
Discussion
The aim of this systematic review and meta-analysis is to summarize existing evidence on principal salivary biomarkers and to compare their accuracy in diagnosing early oral squamous cell carcinoma in adults. To the best of our knowledge, this is the first systematic review and meta-analysis which provides a comprehensive quantitative analysis of salivary biomarkers in early oral squamous cell carcinoma diagnosis. A total of 1048 patients from thirteen eligible studies were included in meta-analysis. Most of the salivary biomarkers overall had good diagnostic accuracy. To further evaluate their diagnostic accuracy, we calculated positive and negative likelihood ratio. Furthermore, we also conducted a subgroup analysis. While method of estimation may be a potential source of heterogeneity, our subgroup analysis clearly indicates that salivary biomarkers had high diagnostic accuracy.
Although our study represents the first work which provides a comprehensive quantitative analysis on diagnostic accuracy of salivary biomarkers, three meta-analysis (Guerra et al., 2015; Ding et al., 2016; AlAli et al., 2020) addresses potentiality of salivary biomarkers as a clinical tool for head and neck carcinoma as well as oral, oesophageal and pancreatic cancer. (Guerra et al., 2015) analysed 15 studies which evaluated different type of biomarkers like mRNA, proteins, metabolites and DNA. Sensitivity and specificity of biomarkers varied from 14% to 100% and 38% to 100% respectively. However, (Ding et al., 2016) evaluated only overall diagnostic values of salivary miRNA for cancer detection, being the sensitivity and specificity reported as 77%. This data added the interest of salivary miRNAs as a diagnostic tool for cancer detection. While, (Al Alli et al., 2020) evaluated diagnostic accuracy of CYFRA 21-1 and MMP-9 for oral squamous cell carcinoma. The author reported pooled sensitivity and specificity for CYFRA-21 ranging from 84% to 94% and from 84% to 96% while MMP-9 had pooled sensitivity and specificity ranging from 76% to 100% and from 27% to 100%. The study was limited by poor overall quality of included studies.
Most of included studies were at high risk of selection bias arising from use of a ‘case-control’ study design. In addition, patient sampling and/or recruitment into studies were insufficiently reported. Among the included studies, only three studies (Brinkmann et al., 2011; Panta et al., 2014; Zimmermann et al., 2008) had sufficiently reported patient selection process. All studies used biopsy as reference standard and salivary biomarkers as index test. However, insufficient detail and lack of clarity in reporting studies made it difficult to assess risk of bias. Therefore, use of STARD (Cohen et al., 2016) checklist in reporting primary studies could have facilitated the quality appraisal. Reporting guidelines for primary diagnostic studies should be followed strictly and studies should address all potential source of bias and applicability concern as indicated in QUADAS-2 tool (Whiting et al., 2011).
Among the included studies for analysis, four studies (Panta et al., 2014; Singh et al., 2020; Smriti et al., 2021; Deepthi et al., 2020) were from India. It is important to keep in mind that India itself accounts for fifth of all oral cancer cases worldwide, and all oral cancer cases developed from potentially malignant disorders seen in patients including betel quid users (Wong et al., 2008; Gupta et al., 1980). Studies have shown that chemicals in betel quid have cytotoxic and genotoxic effects on mucosal epithelial cells due to the generation of reactive oxygen species (ROS), genetic damage and micronuclei formation (Gonzalez et al., 2020). Subsequently similar studies should be performed in other ethnic populations. To overcome these challenges, research efforts should be addressed to validate salivary biomarkers for cancer diagnosis, characterization and monitoring (Gonzalez et al., 2020).
Saliva as a diagnostic fluid has shown to express altered levels of biomarkers not only in OSCC but also in various oral and other systemic disease (Hilden et al., 2005; Zhang et al., 2010; Streckfus et al., 2008; Li et al., 2012; Lee et al., 2012). Salivary biomarkers offer to be a promising tool for oral cancer screening and diagnosis with high sensitivity and specificity. A number of molecular screening tests have been established to detect cancer in early stages (Baron et al., 2012). However, current clinical blood-based screening/diagnostic tests are inaccurate and not specific enough and has many limitations (Nair et al., 2018).
This study provides information on the accuracy and applicability of salivary biomarkers in improving cancer detection through dynamic and non-invasive method. Highest sensitivity and specificity were observed for mRNA 90% and 91% and miRNA 91% and 91% estimated by PCR. While lowest sensitivity and specificity was observed for IL-1B estimated by ELISA. Also, the pooled positive likelihood ratio (PLR) of 9.77 was highest for miRNA estimated by PCR, indicating that patients with oral cancer have a 9.77 folds higher chance of having a positive test result compared to cancer free patients. By contrary the pooled negative likelihood ratio (NLR) was 0.10, indicating the probability of a patient having cancer is 10% if the test shows negative result. The individual sensitivity and specificity make the mRNA and miRNA as better biomarkers, while an overall holistic AUC value of 0.96 for mRNA and 0.95 for miRNA highlights these biomarkers as more accurate overall. Analysis of other biomarkers revealed comparatively low sensitivity and specificity value compared to mRNA and miRNA biomarkers. The higher AUC value for these biomarkers suggests a more easily interpretable and meaningful measure of performance in correctly diagnosing the target condition.
This study is limited by overall quality of included studies. Further standardised diagnostic test accuracy studies that minimises potential sources of bias through rigorous design, conduct and reporting are needed. Future research must focus on the accuracy of current potential principal salivary biomarkers in detection of OSCC with clear and robust methodology.
In conclusion, among all biomarkers mRNA and miRNA biomarker estimation by PCR had overall excellent accuracy while other biomarkers had low to poor overall accuracy, which is mostly due to their high dependence on expert technical ability for their execution and interpretation. Our findings provide evidence on ability of salivary biomarkers for early cancer screening and diagnosis. Despite of some limitations, our study findings demonstrate that salivary biomarkers overall have high sensitivity and specificity to be used as a non-invasive method for early oral squamous cell carcinoma diagnosis. Thus, we can conclude salivary biomarkers for secondary level of prevention for early OSCC under early diagnosis and prompt treatment.
Appendix
1. Supplementary data (Table S1) related to the descriptive data characteristics of all the included studies
2. Supplementary data (Table S2) related to the overall diagnostic accuracy of salivary biomarker along with their method of estimation.
Author Contribution Statement
Dr. Amar Kumar Shaw; Contribution towards the manuscript: Concepts, design, definition of intellectual content, literature search, data acquisition, data analysis, statistical analysis, manuscript preparation, manuscript editing. Dr. Vikram Garcha; Contribution towards the manuscript: Manuscript preparation, manuscript editing, manuscript review. Dr. Vittaldas Shetty; Contribution towards the manuscript: Manuscript editing, manuscript review, manuscript proof reading. Dr. Vineet Vinay; Contribution towards the manuscript: Design, data analysis, statistical analysis, manuscript preparation, manuscript editing, manuscript review. Dr. Ketaki Bhor; Contribution towards the manuscript: Definition of intellectual content, manuscript preparation, manuscript editing, manuscript review. Dr. Kadambari Ambildhok; Contribution towards the manuscript: Manuscript preparation, manuscript editing, manuscript review. Dr. Purnima Karande; Contribution towards the manuscript: Literature search, data acquisition, manuscript preparation.
Acknowledgements
The study was approved by the Institutional Ethical Committee, Sinhgad Technical Education Society’s Sinhgad Dental College and Hospital Pune, bearing Approval No. SDCH/IEC/2020-21/30
The study was approved by Scientific Advisory Committee, Sinhgad Technical Education Society, Sinhgad Dental College and Hospital Pune, bearing approval number: SDCH/SAC/2020-21/30
Conflict of interest
The authors declare NO conflicts of interests
References
- Aa O, Ao A, To A, et al. Orofacial Cancers. Med J Zambia. 2018;45:179–88. [Google Scholar]
- Adeyemi BF, Olusanya AA, Lawoyin JO. Oral Squamous Cell Carcinoma, Socioeconomic Status and History of Exposure to Alcohol and Tobacco. J Natl Med Assoc. 2011;103:498–502. doi: 10.1016/s0027-9684(15)30364-3. [DOI] [PubMed] [Google Scholar]
- Adisa AO, Adeyemi BF, Oluwasola AO, et al. Clinico-pathological profile of head and neck malignancies at University College Hospital, Ibadan, Nigeria. Head Face Med. 2011;7:1–9. doi: 10.1186/1746-160X-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlAli AM, Walsh T, Maranzano M. CYFRA 21-1 and MMP-9 as salivary biomarkers for the detection of oral squamous cell carcinoma: a systematic review of diagnostic test accuracy. Int J Oral and Maxillofac Surg. 2020;49:973–83. doi: 10.1016/j.ijom.2020.01.020. [DOI] [PubMed] [Google Scholar]
- Baron JA. Screening for cancer with molecular markers: progress comes with potential problems. Nat Rev Cancer. 2012;12:368–71. doi: 10.1038/nrc3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AN, Mathur R, Farooque A, Verma A, Dwarakanath BS. Cancer biomarkers - Current perspectives. Indian J Med Res. 2010;143 [PubMed] [Google Scholar]
- Bonne NJ, Wong DTW. Salivary biomarker development using genomic, proteomic and metabolic approaches. Genome Med. 2012;4:1–12. doi: 10.1186/gm383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann O, Kastratovic DA, Dimitrijevic MV, et al. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol. 2011;47:51–5. doi: 10.1016/j.oraloncology.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YL, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clin Transl Med. 2014;3:1–10. doi: 10.1186/2001-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6:1–17. doi: 10.1136/bmjopen-2016-012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristaldi M, Mauceri R, Di Fede O, et al. Salivary Biomarkers for Oral Squamous Cell Carcinoma Diagnosis and Follow-Up: Current Status and Perspectives. Front Physiol. 2019;10:1–12. doi: 10.3389/fphys.2019.01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepthi G, Nanadan SRK, Kulkarni P. Salivary Tumour Necrosis Factor-a as a Biomarker in Oral Leukoplakia and Oral Squamous Cell Carcinoma. Asian Pac J Cancer Prev. 2020;20:87–93. doi: 10.31557/APJCP.2019.20.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanuthai K, Rojanawatsirivej S, Thosaporn W, et al. Oral cancer: A multicenter study. Med Oral. 2018;23:23–9. doi: 10.4317/medoral.21999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ma Q, Liu F, Zhao L, Wei W. The Potential Use of Salivary miRNAs as Promising Biomarkers for Detection of Cancer: A Meta-Analysis. Chalmers J. 2016;editor. PLoS One: 1–12. doi: 10.1371/journal.pone.0166303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elashoff D, Zhou H, Reiss J, et al. Prevalidation of Salivary Biomarkers for Oral Cancer Detection. Cancer Epidemiol Biomark Prev. 2012;21:664–72. doi: 10.1158/1055-9965.EPI-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Li Q, Chen J, et al. Salivary protease spectrum biomarkers of oral cancer. Int J Oral Sci. 2019;11:1–12. doi: 10.1038/s41368-018-0032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes DA, Schulz KF. Refining clinical diagnosis with likelihood ratios. Lancet. 2005;365:1500–5. doi: 10.1016/S0140-6736(05)66422-7. [DOI] [PubMed] [Google Scholar]
- Guerra ENS, Acevedo AC, Leite AF, et al. Diagnostic capability of salivary biomarkers in the assessment of head and neck cancer: A systematic review and meta-analysis. Oral Oncol. 2015;51:805–18. doi: 10.1016/j.oraloncology.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Gupta PC, Mehta FS, Daftary DK, et al. Incidence rates of oral cancer and natural history of oral precancerous lesions in a 10-year follow-up study of Indian villagers. Commun Dent Oral Epidemiol. 1980;8:287–333. doi: 10.1111/j.1600-0528.1980.tb01302.x. [DOI] [PubMed] [Google Scholar]
- Hilden J. What properties should an overall measure of test performance possess? Clin Chem. 2005;51:471–72. doi: 10.1373/clinchem.2004.041376. [DOI] [PubMed] [Google Scholar]
- Hu S, Arellano M, Boontheung P, et al. Salivary Proteomics for Oral Cancer Biomarker Discovery. Clin Cancer Res. 2008;14:6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA A Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jones CM, Athanasiou T. Summary Receiver Operating Characteristic Curve Analysis Techniques in the Evaluation of Diagnostic Tests. Ann Thorac Surg. 2005;79:16–20. doi: 10.1016/j.athoracsur.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Kampfrath T. Statistical assessment of biomarker performance. Clin Chimica Acta. 2013;419:102–7. doi: 10.1016/j.cca.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Lee LT, Wong YK, Hsiao HY, W et al. Evaluation of saliva and plasma cytokine biomarkers in patients with oral squamous cell carcinoma. Int J Oral and Maxillofac Surg. 2018;47:699–707. doi: 10.1016/j.ijom.2017.09.016. [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Kim JH, Zhou H, Kim BW, Wong DT. Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J Mol Med. 2012;90:427–34. doi: 10.1007/s00109-011-0829-0. [DOI] [PubMed] [Google Scholar]
- Lijmer JG, Bossuyt PMM, Heisterkamp SH. Exploring sources of heterogeneity in systematic reviews of diagnostic tests. Stat Med. 2002;21:1525–37. doi: 10.1002/sim.1185. [DOI] [PubMed] [Google Scholar]
- Li X. Spectral analysis of human saliva for detection of lung cancer using surface-enhanced Raman spectroscopy. J Biomed Opt. 2012;17:1–5. doi: 10.1117/1.JBO.17.3.037003. [DOI] [PubMed] [Google Scholar]
- Li Y-F, Hsiao Y-H, Lai Y-H, et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics. 2015;10:229–36. doi: 10.1080/15592294.2015.1006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, St. John MAR, Zhou X, et al. Salivary Transcriptome Diagnostics for Oral Cancer Detection. Clin Cancer Res. 2004;10:8442–50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- Markopoulos AK, Michailidou EZ, Tzimagiorgis G. Salivary Markers for Oral Cancer Detection. TODENT J. 2010;4:172–8. doi: 10.2174/1874210601004010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markopoulos AK. Current Aspects on Oral Squamous Cell Carcinoma. TODENT J. 2012;6:126–30. doi: 10.2174/1874210601206010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero PH, Patel SG. Cancer of the Oral Cavity. Surg Oncol Clin N Am. 2015;24:491–508. doi: 10.1016/j.soc.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamayl F, Davodi P, Dalband M, Hendi S. Saliva as a mirror of the body health. Avicenna J Dent Rs. 2010;1:41–55. [Google Scholar]
- Nair M, Sandhu SS, Sharma AK. Cancer molecular markers: A guide to cancer detection and management. Semin Cancer Biol. 2018;52:39–55. doi: 10.1016/j.semcancer.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Panta P, Venna VR. Salivary RNA Signatures in Oral Cancer Detection. Anal Cell Pathol. 2014;4:1–7. doi: 10.1155/2014/450629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer Detection. Clin Cancer Res. 2009;15:5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad G, McCullough M. Chemokines and Cytokines as Salivary Biomarkers for the Early Diagnosis of Oral Cancer. Int J Dent. 2013;9:1–7. doi: 10.1155/2013/813756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestiyanti NMI. The role of salivary biomarker as a diagnostic tool in oral cancer: a literature review. Intisari Sains Medis. 2020;11:112–7. [Google Scholar]
- Rapado-González Ó, Martínez-Reglero C, Salgado-Barreira Á, et al. Salivary biomarkers for cancer diagnosis: a meta-analysis. Ann Med. 2020;52:131–44. doi: 10.1080/07853890.2020.1730431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salameh J-P, Bossuyt PM, McGrath TA, et al. Preferred reporting items for systematic review and meta-analysis of diagnostic test accuracy studies (PRISMA-DTA): explanation, elaboration, and checklist. BMJ. 2020;10:1–17. doi: 10.1136/bmj.m2632. [DOI] [PubMed] [Google Scholar]
- Saxena S, Sankhla B, Sundaragiri K, Bhargava A. A Review of Salivary Biomarker: A Tool for Early Oral Cancer Diagnosis. Adv Biomed Res. 2017;6:1–7. doi: 10.4103/2277-9175.211801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics. Cancer Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Shaw R, Beasley N. Aetiology, risk factors for head, neck cancer. United Kingdom National Multidisciplinary Guidelines. J Laryngol Otol. 2016;130:9–12. doi: 10.1017/S0022215116000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpitzer T, Bahar G, Feinmesser R, Nagler RM. A comprehensive salivary analysis for oral cancer diagnosis. J Cancer Res Clin Oncol. 2007;133:613–7. doi: 10.1007/s00432-007-0207-z. [DOI] [PubMed] [Google Scholar]
- Singh P, Verma JK, Singh JK. Validation of Salivary Markers, IL-1β, IL-8 and Lgals3bp for Detection of Oral Squamous Cell Carcinoma in an Indian Population. Sci Rep. 2020;10:1–9. doi: 10.1038/s41598-020-64494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smriti K, Ray M, Chatterjee T, et al. Salivary MMp-9 as a Biomarker for the Diagnosis of Oral Potentially Malignant Disorders and Oral Squamous Cell Carcinoma. Asian Pac J Cancer Prev. 2020;21:233–8. doi: 10.31557/APJCP.2020.21.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis. 2011;14:345–54. doi: 10.1111/j.1601-0825.2010.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. John MAR, Li Y, Zhou X, et al. Interleukin 6 and Interleukin 8 as Potential Biomarkers for Oral Cavity and Oropharyngeal Squamous Cell Carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:929–35. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- Streckfus CF, Mayorga-Wark O, Arreola D, et al. Breast Cancer Related Proteins Are Present in Saliva and Are Modulated Secondary to Ductal Carcinoma In Situ of the Breast. Clin Cancer Investig. 2008;26:159–67. doi: 10.1080/07357900701783883. [DOI] [PubMed] [Google Scholar]
- Sudbø J, Kildal W, Risberg B, et al. DNA Content as a Prognostic Marker in Patients with Oral Leukoplakia. N Engl J Med. 2001;344:1270–8. doi: 10.1056/NEJM200104263441702. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tanaka M, Tanaka T. Oral Carcinogenesis and Oral Cancer Chemoprevention: A Review. Pathol Res Int. 2016;16:1–10. [Google Scholar]
- Viet CT, Schmidt BL. Methylation Array Analysis of Preoperative and Postoperative Saliva DNA in Oral Cancer Patients. Cancer Epidemiol Biomark & Prev. 2008;17:3603–11. doi: 10.1158/1055-9965.EPI-08-0507. [DOI] [PubMed] [Google Scholar]
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Whiting PF. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- Wong DT, Segal A, Wong DT. enhancing disease detection and making medicine better. Eur J Dent Educ. 2008;12:22–9. doi: 10.1111/j.1600-0579.2007.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakob M, Fuentes L, Wang M, Abemayor E, Wong D. Salivary biomarkers for detection of oral squamous cell carcinoma – current state and recent advances. Curr Oral Health Rep. 2014;1:133–41. doi: 10.1007/s40496-014-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young OH, Kang S-M, Kang SH, et al. Potential Salivary mRNA Biomarkers for Early Detection of Oral Cancer. J Clin Med. 2020;9:1–12. doi: 10.3390/jcm9010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Sun H, Wang X. Saliva Metabolomics Opens Door to Biomarker Discovery, Disease Diagnosis, and Treatment. Appl Biochem Biotechnol. 2012;168:1718–27. doi: 10.1007/s12010-012-9891-5. [DOI] [PubMed] [Google Scholar]
- Zhang A, Sun H, Yan G, Wang P, Wang X. Metabolomics for Biomarker Discovery: Moving to the Clinic. Biomed Res Int. 2015;2:1–6. doi: 10.1155/2015/354671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Farrell JJ, Zhou H, Elashoff D, et al. Salivary Transcriptomic Biomarkers for Detection of Resectable Pancreatic Cancer. Gastroenterology. 2010;138:949–57. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann BG, Wong DT. Salivary mRNA targets for cancer diagnostics. Oral Oncol. 2008;44:425–9. doi: 10.1016/j.oraloncology.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]