Abstract
Objective:
To evaluate the frequency distribution of viral infections in Peruvian Breast Cancer (BC) lesions and its association with clinicopathological features. Additionally, a prospective evaluation of p16 and Tumor-infiltrating lymphocytes (TIL) levels were performed for developing a comprehensive analysis.
Methods:
Detection of high risk- human papillomavirus (HR- HPV) through qPCR was performed in 447 BC and 79 non-cancer frozen samples. Paired paraffin samples from 238 BC were stained with Human cytomegalovirus (HCMV) and p16 immunohistochemistry. TIL was calculated in 397 BC cases.
Results:
HCMV was positive in 72.5%. HR- HPV was detected in 2.9% of BC and 1.3% of non-malignant samples. P16+ was found in 28.15% and median TIL percentage was 30. HR- HPV infection was associated with non-ductal histology (p=0.003) and p16+ (p=0.017). Positive P16+ was associated with higher T stage (p=0.022), grade (p=0.009), TIL level (p=0.002), and triple-negative phenotype (p=0.021).
Conclusion:
HCMV is frequent, but HR- HPV infection is unusual in Peruvian BC. P16+ is associated with HR- PVH infection, high TIL and aggressive features.
Key Words: Human papillomavirus, cytomegalovirus, p16 protein, polymerase chain reaction, immunohistochemistry
Introduction
Human papillomaviruses (HPV) are small double-stranded DNA viruses that infect squamous epithelia. HPV are responsible for almost all cervical carcinomas, that are the second most frequent malignancy in Peruvian women(Sullcahuaman-Allende et al., 2015; Walboomers et al., 1999), as well as other malignancies (Hausen, 2000; Wong et al., 2002). Malignancies related to HPV infection have different clinical features than those not related, and recent clinical trial results have demonstrated that they have high response to immunotherapy (Seiwert et al., 2016).
Breast cancer is a heterogeneous disease that has important differences in clinicopathological features and prognosis according to ethnicity like high rate of Triple-Negative (TNBC) phenotype in Latin American women (Castaneda et al., 2018). Tumor-infiltrating lymphocytes (TILs) is a pathological feature that evaluates the host immune system activity against a BC tumor, is higher in TNBC, and predicts response to chemotherapy and immunotherapy as well as survival (Galvez et al., 2018).
It has been proven though, that human mammary epithelial cell immortalization in in-vitro model is efficiently obtained with the expression of high-risk (HR)-HPV oncogenes E6 and E7 (Dimri et al., 2005). Some studies have described the presence of HPV DNA in breast cancer specimens with molecular techniques, however, most studies evaluate Caucasian women and include small number of cases. They report that HPV-harboring breast tumors are associated with aggressive features like younger age, grade-3, higher Ki67 index and, estrogen receptor (ER)-negative status (de Villiers et al., 2004; Wrede et al., 1992). Viral oncoprotein E7 inactivates the phosphorylated Retinoblastoma protein, and it up-regulates the p16 protein expression. And, the association of HPV DNA presence with p16 staining has been strongly described in different malignancies. Overexpression of p16 in breast tumors from Caucasian women has similarly been associated with aggressiveness features like higher grades, TNBC phenotype, and advanced stages (Lebok et al., 2016).
Human cytomegalovirus (HCMV) is a double-stranded DNA virus that infects nearly 50–90% of the population worldwide, and produces either asymptomatic or mild discomfort. HCMV gene products have been reported to be involved in cell cycle dysfunction, genome instability and cell immortalization. Several HCMV proteins have demonstrated to have the ability to induce cell transformation in in-vitro and in-vivo experiments (Fortunato et al., 2000; Soroceanu & Cobbs, 2011). Presence of HCMV antigens has been described in breast cancer samples evaluated in small series from Caucasian women (Taher et al., 2013).
There is little information about the prevalence of HR-HPV and HCMV infections as well as p16 expression in breast tumors of South American women. Few of them have evaluated their relationship with tumor features. In this study, we detected HR-HPV in frozen tissue from BC and non-malignant breast tissue, as well as in swabs from oral cavity of cases with positive results in breast tumors. Expression of p16 and presence of HCMV protein in breast cancer tissues was determined in paired paraffin breast cancer samples. This viral infection information was contrasted with clinicopathological features including TNBC phenotype and TIL level.
Materials and Methods
Patients and Samples
Four hundred forty-seven female breast cancer and 79 non-cancer (screening) frozen samples were collected from routine biopsies or surgeries and saved at the biobank of the Instituto Nacional de Enfermedades Neoplasicas (INEN) in Lima-Peru. Breast cancer cases that tested positive for HPV were followed up, contacted when possible and oral samples were taken through brushing with a long cotton swab. Paraffin samples from breast cancer cases with available large specimens were obtained from pathology archives. Clinicopathological features were collected from patient files. A special focus was applied to determine the story of HPV-related malignancies (squamous cervical, vulvar, vaginal, penile, anal, and oropharyngeal cancers). Tumor-infiltrating lymphocytes (TIL) were prospectively evaluated in available H&E slides (n=397) by a pathologist (JS). This study was approved by the Institutional Review Board and the Ethics Board of the INEN. Written informed consent was obtained from every patient.
DNA extraction and HR-HPV detection
Cellular DNA was prepared using Maxwell® RSC Tissue DNA kit (Promega Corporation, Madison, WI, USA) for the freshly breast tissues and Maxwell® RSC Buccal Swab DNA kit (Promega Corporation, Madison, WI, USA) for the mouth swab samples, the quantification by Fluorometry using the QUANTUS fluorometer (Promega Corporation, Madison, WI, USA). DNA of HR-HPV was detected by quantitative PCR (qPCR) using probe directed to the HPV types 16 and 18 were detected with The Primer design genesig Kit for human papillomavirus 16 y 18 (HPV16 and HPV18), the primer and probe mix used exploits the so-called TaqMan® principle. A total volume of 20 μl PCR reaction containing 1 μl of each primer and probe mix, 10 μl of Precision PLUS2X qPCR Master Mix, 4 μl of RNase/DNase free water, and 5 μl extracted DNA (100 ng). The amplification was performed under the cyclic program; enzyme activation step of 95ºC, 2 min, followed by 50 cycles of 95ºC, 10 sec; 60ºC, 60 sec (data collection) by using LightCycler ® 96 Instrument (Roche, USA).
Immunohistochemistry staining
Immunohistochemistry (IHC) stainings for HCMV and p16 status were performed using the DAKO EnVision™ FLEX+ detection system together with the Autostainer Link instrument (DAKO, Carpentaria, California) on paraffin embedded tissue. Antigen was retrieved using EnVision™ FLEX Target Retrieval Solution, High pH. The mouse monoclonal antibodies used were antihuman p16 antibody (clone 16p04, JC2, BSB 5828, prediluted, Bio SB, Santa Barbara, California) and antihuman CMV (clone 8B1.2, 2882686-MAB810R, 1:100, Merck Millipore, Temecula, CA, USA). The EnVision™ FLEX+, Mouse High pH Kit (Dako, K8002, Glostrup, Denmark) was used to perform the assay according to the manufacturer’s instructions. It contains the substrate chromogen 3-3′-diaminobenzidine (DAB), which, on staining results in a brown-colored precipitate at the antigen site. In negative control sections, the specificity of the antisera was tested by replacing the primary antibodies with normal serum. For p16 staining, cervical cancer tissue sections were used as positive controls. A colon and regional node with pathology image and serological test-positive for HCMV infection were selected for positive control.
Staining and lecture were performed only in tumorectomy or mastectomy material with available residual material at pathology archive. Cases were considered positive for p16 immunohistochemical expression if strong nuclear and cytoplasmic staining was detected in more than 50% of malignant cells as previously described (block staining) (Doxtader and Katzenstein, 2012), while HCMV was based on the presence of labeling in tumor cell nucleus or cytoplasm, or stromal cells (Taher et al., 2013); and both were performed by a pathologist (JS).
Statistics analysis
The statistical analysis of the data was carried out using SPSS. For the nominal or ordinal categorical variables frequencies and percentages were used. In the categorical variables, the association with the Pearson Chi-Square test or Fisher’s exact test was evaluated. The statistical tests were evaluated at a level of significance of 5%.
Results
Median age in our series of 447 breast cancer lesions was 51 years, most were in stage III (35%), T4 (35.8), ductal histology (89.5%), grade III (63%) and Luminal- B phenotype (53.8%). Median TIL level was 30% (Table 1).
Table 1.
Clinicopathological Features of Breast Cancer Cases Evaluated for HPV and p16 Staining
| Features | Total | HPV (n=447) | p16 staining (n=238) | ||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | P | Negative | Positive | P | ||
| n (%) | n (%) | n (%) | n (%) | n (%) | |||
| Age (year) | 0.577 | 0.456 | |||||
| ≤ 51 year | 206 (46.5) | 201 (97.5) | 5 (2.5) | 70 (69.3) | 31 (30.7) | ||
| > 51 year | 241 (53.5) | 233 (96.6) | 8 (3.4) | 101 (73.7) | 36 (26.3) | ||
| Tumor size | 0.82 | 0.022 | |||||
| I | 28 (6.2) | 28 (100.0) | 0 (0.0) | 14 (93.3) | 1 (6.6) | ||
| II | 195 (43.6) | 189 (96.0)) | 6 (4.0) | 92 (76.6) | 28 (23.4) | ||
| III | 54 (12.4) | 51 (94.4) | 3 (5.6) | 20 (55.5) | 16 (44.5) | ||
| IV | 160 (35.8) | 156 (97.5) | 4 (2.5) | 44 (68.8) | 20 (31.2) | ||
| Stage | 0.549 | 0.076 | |||||
| I | 17 (4.6) | 17 (100.0) | 0 (0.0) | 8 (80.0) | 2 (20.0) | ||
| II | 146 (45.4) | 141 (96.6) | 5 (3.4) | 99 (76.7) | 30 (23.3) | ||
| III | 225 (36.9) | 217 (96.4) | 8 | 57 (65.5) | 30 (34.5) | ||
| IV | 59 (13.1) | 59 (100.0) | 0 (0.0) | 7 (58.3) | 5 (41.7) | ||
| Grade | 0.097 | 0.009 | |||||
| I | 10 (2.5) | 10 (100.0) | 0 (0.0) | 5 (62.5) | 3 (37.5) | ||
| II | 151 (33.9) | 143 (95.0) | 8 (5.0) | 69 (82.2) | 13 (17.8) | ||
| III | 284 (63.3) | 279 (98.2) | 5 (1.8) | 97 (65.5) | 51 (34.5) | ||
| TIL (%) | 0.809 | 0.002 | |||||
| <30 | 218 (54.9) | 211 (96.8) | 7 (3.2) | 96 (81.4) | 22 (18.6) | ||
| ≥30 | 179 (45.1) | 174 (97.2) | 5 (2.8) | 73 (62.9) | 43 (37.1) | ||
| Histology | 0.003 | 0.145 | |||||
| Ductal | 400 (89.5) | 392 (98.0) | 8 (2.0) | 71.8 | 58 (28.3) | ||
| Lobulillar | 25 (5.6) | 22 (88.0) | 3 (12.0) | 14 (87.5) | 2 (12.5) | ||
| Other | 22 (4.9) | 20 (90.9) | 2 (9.1) | 9 (56.3) | 7 (41.2) | ||
| Phenotype | 0.181 | 0.021 | |||||
| Luminal-A | 69 (15.7) | 65 (15.0) | 4 (30.8) | 37 (82.2) | 8 (17.8) | ||
| Luminal-B | 236 (53.8) | 230 (97.5) | 6 (2.5) | 95 (71.9) | 37 (28.1) | ||
| HER-2 | 66 (15.0) | 64 (97.0) | 2 (3.0) | 20 (80.0) | 5 (20) | ||
| TNBC | 68 (15.5) | 67 (98.5) | 1 (1.5) | 18 (51.4) | 17 (48.6) | ||
| p16 | 0.017 | ||||||
| Negative | 171 (71.8) | 166 (97.1) | 5 (2.9) | - | - | ||
| Positive | 67 (28.2) | 60 (89.5) | 7 (10.5) | - | - | ||
TNBC, triple negative breast cancer; TIL, Tumor Infiltrating Lymphocytes; P.value<0.05
HCMV detection
There were 169 (72.5%) cases with HCMV-positive staining in malignant cells from 233 evaluated cases. Positive staining was found in tumor cell cytoplasm and nucleus staining in 48 tumors. Intensity of tumor staining was moderate in 28 (16.6%) and mild in 141 (83.4%) cases. They were not associated with age (p=0.22), clinical-stage (p=0.95), grade (p=0.2), TIL (p=0.989), histology (p=0.45), phenotype (p=0.2) nor p16 (p=0.48). Staining for HCMV was associated with PVH (p=0.04) (all 10 PVH positive with evaluated paired paraffin sample for HCMV were positive for co-infection). Additionally, stromal cells were positively stained in 214 cases (91.8%).
HPV detection
HPV was detected in 13 of 447 (2.9%) tumor samples of breast cancer. Infection by HPV-16 was found in 11 and HPV-18 in 2 cancer cases. HR-HPV breast cancer infection was associated with non- ductal histology (p=0.003), but not associated with age (p=0.577), T stage (p=0.82), clinical-stage (p=0.549), grade (p=0.07), TIL (p=0.5) nor molecular subtype (p=0.181) (Table 1). There were six papillary breast cancer cases in our series, and only one had HPV-16 infection (this case was also p16-positive).
Eleven cases with positive breast tumor samples for HPV underwent the qPCR analysis also in oral mucosa. Only one oral mucosa was an HPV positive result, however, by a different subtype (HPV-18 in oral mucosa and HPV-16 in breast tissue). We did not find medical information on the prior diagnosis of HPV-related malignancy in any of the 13 positive cases.
Additionally, HPV was detected in 1 (HPV16) of the 79 benign lesion samples of the breast, and this rate was not different than rates in the malignant samples (p=0.404). We could not obtain paired oral mucosa samples for any of the non-malignant cases.
Determination of p16 expression
Positive p16 expression of the protein was found in 28.15% of the cases (67/238), and was associated with HR-HPV+ (p=0.017), higher T stage (p=0.022), higher grade (p=0.009), higher TIL level (p=0.002), and TNBC phenotype (p=0.021) (Table 1) (Figure 1). There were only three available paraffin samples of papillary lesions for p16 staining, and 1 of them was positive.
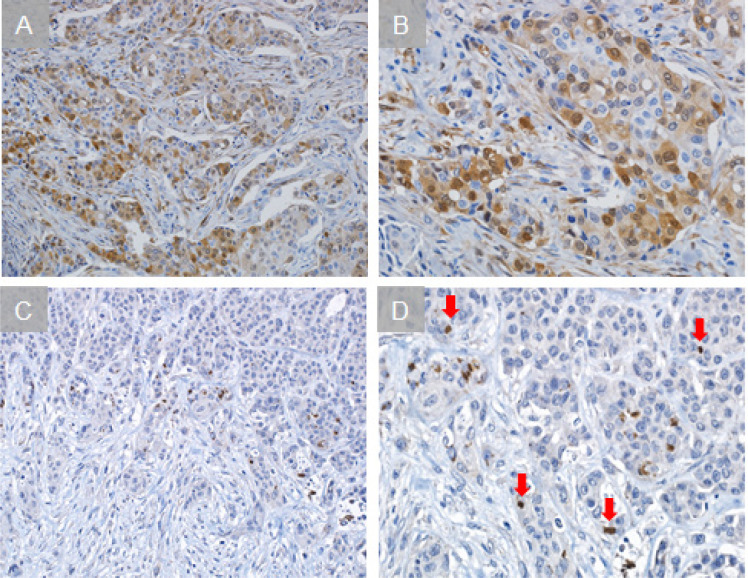
Figure 1.
Immunohistochemical Staining of P16 and HCMV. Representative sections with positive staining for p16 (A), and CMV (C) are shown alongside higher magnification insets from the same tissue sections (B and D, respectively). Red arrows show positive cells
Discussion
In the present study, we detected HCMV in 72% of breast cancer samples, which is similar to the previously published (7.4%- 100%) (Eghbali et al., 2012; Harkins et al., 2010; Taher et al., 2013; Tsai et al., 2005). Presence of HR-HPV DNA sequences was found only in 13 (2.87%) out of 447 frozen breast cancer specimens. All available cases with detection of HR-HPV also stained positive for HCMV.
The prevalence of HR-HPV infection in the cancer group was lower than the reported range in previous studies: 25–86%. The majority (84.6%) of the identified types were HPV-16 stain, similar to the described in other series, and were associated with positive p16 expression (Table 1) (Damin et al., 2004; de Villiers et al., 2004; Hennig et al., 1999; Lebok et al., 2016).
Since this sexually transmitted virus affects mostly young people and produces skin papilloma in HPV-related malignancies, it was expected that infected patients would be younger, had papillary morphology or would have larger skin involvement. However, not differences for age, T-stage, or papillary histology were associated with the infection status in our series as well as in previously published series (Damin et al., 2004; de Villiers et al., 2004; Hennig et al., 1999).
Some previous reports find that cases with HPV infection in breast tissue have higher possibilities of developing cervical cancer with a similar HPV genotype (de Villiers et al., 2004; Hennig et al., 1999; Widschwendter et al., 2004). However, we did not find coinfection of HR-HPV in breast tumors and paired oral mucosa. Positive cases did not have a previous story of cervical cancer or other HPV-related malignancies, which indicates that breast cancer infection would be isolated to the chest and do not behave as a systemic infection. Similarly, other South-American series found less than 10% infection rates and no predisposition for cervical infection nor cancer (Aguayo et al., 2011). TSimilarly, other reports describe the finding of HPV in other unexpected sites such as Hodgkin’s lymphoma and bronchopulmonary cancer (Cheng et al., 2001).
In addition to the previously described association between p16 expression and TNBC phenotype, we for the first time found its association with high TIL levels. This relationship could be explained by the fact that TNBC has both accelerated cell proliferation that involve the role of p16 for cell cycle regulation as well as high level of mutations, epigenetic alterations and neoantigens including genes of the Retinoblastoma pathway. And both features attract and increase intratumoral level of lymphocytes (Castaneda et al., 2019; Kozomara et al., 2018; Lebok et al., 2016; Zhang et al., 2021).
We analyzed only the HR-HPV genotypes 16 and 18 as they are the most frequently described in cervical carcinomas. However, other highly prevalent genotypes described in Peruvian cervical cancer like HR-HPV genotypes: HPV 31 and HPV 33, and the low-risk genotypes: HPV6 and HPV11 have not been evaluated.
This information indicates that prevalence of HVMV infection is high but not associated with clinicopathological features in our Peruvian cohort. HPV infection in breast tumors of Peruvian women is extremely low. However, the alteration of its associated pathway, Retinoblastoma (detected by positive p16 expression), was found in around a quarter of our breast cancer cases and was associated with high TIL levels as well as with tumor aggressiveness features like TNBC phenotype.
Author Contribution Statement
GC conceived the study, CAC and MC designed, and supervised the study, analyzed and interpreted the participants’ data, then wrote the manuscript, JMC, JD, MDLC, JA, GC, HLG, JM and HG, collected and interpreted the participants’ data, JS, NS and LAB collected and analyzed the participants’ data, FA interpreted the participants’ data, then revised and edited the manuscript; All authors read, edited and approved the final manuscript.
Acknowledgments
We would like to extend our appreciation to Dr. Sandro Casavilca and Dr. Carolina Belmar for their administrative support and scientific advice.
Informed consent
Written informed consent was obtained from all participants.
Compliance with Ethical Standards
All procedures conducted in the study were under the ethical standards of the institutional review board of the of Instituto Nacional de Enfermedades Neoplasicas (approval no. INEN-15-12). The study was processed under the ethical standards of the Helsinki declaration.
Funding
This study was supported by CONCYTEC, under contracts #204-2015-FONDECYT and #198-2015-FONDECYT.
Disclosure of potential conflicts of interest
No authors declare that there are no conflicts of interest.
References
- Aguayo F, Khan N, Koriyama C, et al. Human papillomavirus and Epstein-Barr virus infections in breast cancer from chile. Infect Agent Cancer. 2011;6:7. doi: 10.1186/1750-9378-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda CA, Castillo M, Torres-Cabala C, et al. Relationship between tumor-associated immune infiltrate and p16 staining over clinicopathological features in acral lentiginous melanoma. Clin Transl Oncol. 2019;21:1127–34. doi: 10.1007/s12094-019-02033-x. [DOI] [PubMed] [Google Scholar]
- Castaneda CA, Castillo M, Villarreal-Garza C, et al. Genetics, tumor features and treatment response of breast cancer in Latinas. Breast Cancer Manag. 2018;7:571–80. [Google Scholar]
- Cheng Y-W, Chiou H-L, Sheu G-T, et al. The association of human papillomavirus 16/18 infection with lung cancer among nonsmoking Taiwanese women. Cancer Res. 2001;61:2799–2803. [PubMed] [Google Scholar]
- Damin APS, Karam R, Zettler CG, et al. Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Res Treat. 2004;84:131–7. doi: 10.1023/B:BREA.0000018411.89667.0d. [DOI] [PubMed] [Google Scholar]
- de Villiers EM, Sandstrom RE, zur Hausen H, et al. Presence of papillomavirus sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast. Breast Cancer Res. 2004;7:1–11. doi: 10.1186/bcr940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G, Band H, Band V. Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Res. 2005;7:171–9. doi: 10.1186/bcr1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxtader EE, Katzenstein ALA. The relationship between p16 expression and high-risk human papillomavirus infection in squamous cell carcinomas from sites other than uterine cervix: a study of 137 cases. Hum Pathol. 2012;43:327–32. doi: 10.1016/j.humpath.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Eghbali M, Ghane M, Mirinargesi M. Frequency of cytomegalovirus (CMV) in benign and malignant tumors. Int J Mol Clin Microbiol. 2012;2:175–9. [Google Scholar]
- Fortunato EA, Dell’Aquila ML, Spector DH. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2000;97:853–8. doi: 10.1073/pnas.97.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez M, Castaneda CA, Sanchez J, et al. Clinicopathological predictors of long-term benefit in breast cancer treated with neoadjuvant chemotherapy. World J Clin Oncol. 2018;9:33–41. doi: 10.5306/wjco.v9.i2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins LE, Matlaf LA, Soroceanu L, et al. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausen H zur. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–8. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- Hennig EM, Suo Z, Thoresen S, et al. Human papillomavirus 16 in breast cancer of women treated for high grade cervical intraepithelial neoplasia (CIN III) Breast Cancer Res Treat. 1999;53:121–35. doi: 10.1023/a:1006162609420. [DOI] [PubMed] [Google Scholar]
- Kozomara Z, Supic G, Krivokuca A, Magic , et al. Promoter hypermethylation of p16, BRCA1 and RASSF1A genes in triple-negative breast cancer patients from Serbia. J BUON. 2018;23:684–91. [PubMed] [Google Scholar]
- Lebok P, Roming M, Kluth M, et al. p16 overexpression and 9p21 deletion are linked to unfavorable tumor phenotype in breast cancer. Oncotarget. 2016;7:81322. doi: 10.18632/oncotarget.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–65. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullcahuaman-Allende Y, Castro-Mujica MC, Farro RM, et al. Demographic characteristics of human papillomavirus detected by PCR-RFLP in peruvian women. Rev Peru Med Exp Salud Publica. 2015;32:509–14. [PubMed] [Google Scholar]
- Taher C, de Boniface J, Mohammad AA, et al. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One. 2013;8:e56795. doi: 10.1371/journal.pone.0056795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Tsai C, Cheng M, et al. Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma, and thyroid tumor tissues. J Med Virol. 2005;75:276–81. doi: 10.1002/jmv.20267. [DOI] [PubMed] [Google Scholar]
- Walboomers JMM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:9–12. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Widschwendter A, Brunhuber T, Wiedemair A, et al. Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J Clin Virol. 2004;31:292–7. doi: 10.1016/j.jcv.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Wong M, Pagano , JS , Schiller , JT , et al. New associations of human papillomavirus, Simian virus 40, and Epstein-Barr virus with human cancer. J Natl Cancer Inst. 2002;94:1832–6. doi: 10.1093/jnci/94.24.1832. [DOI] [PubMed] [Google Scholar]
- Wrede D, Luqmani YA, Coombes RC, Vousden KH. Absence of HPV 16 and 18 DNA in breast cancer. Br J Cancer. 1992;65:891–4. doi: 10.1038/bjc.1992.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Wang YQ, Zhang JH, et al. Methylated p16 gene is associated with negative expression of estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 in breast cancer. Eur J Gynaecol Oncol. 2021;42:530–6. [Google Scholar]