Abstract
Background
Irritable bowel syndrome (IBS) is the most prevalent functional bowel disorder, but its pathophysiology is still unknown. Although a microbial signature associated with IBS severity has been suggested, its association with IBS severity still remains largely unknown.
Aims
This study aims to assess longitudinal dynamics of fecal microbiota and short-chain fatty acids (SCFAs) in different IBS severity groups and study the association with stool pattern, diet, depression, anxiety, and quality of life (QoL).
Methods
A longitudinal study was performed, including n = 91 IBS patients and n = 28 matched controls. All participants collected fecal samples for microbiota composition and SCFA analysis and completed validated questionnaires regarding IBS severity, stool pattern, depression, anxiety, and IBS-QoL at two timepoints with four weeks in-between. Diet was assessed at the first timepoint.
Results
Over time, 36% of IBS patients changed in severity group, and 53% changed in predominant stool pattern. The largest proportion of microbiota variation was explained by the individual (R2 = 70.07%). Microbiota alpha diversity and composition, and SCFAs did not differ between IBS severity groups, nor between IBS and controls. Relative abundances of Bifidobacterium, Terrisporobacter, and Turicibacter consistently differed between IBS and controls, but not between IBS severity groups. Large dynamics over time were observed in the association of microbiota composition with questionnaire data where IBS symptom severity was associated at T1 but not at T2.
Conclusions
Fecal microbiota and SCFA signatures were not consistently associated with IBS severity over time, indicating the importance of repeated sampling in IBS research.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-022-07543-3.
Keywords: Irritable bowel syndrome, Fecal microbiota composition, Short-chain fatty acids, Severity
Introduction
Irritable bowel syndrome (IBS) is the most commonly diagnosed functional gastrointestinal disorder, with a global prevalence around 11% [1]. IBS reduces quality of life (QoL) and increases healthcare costs [2]. Several factors have been associated with IBS, such as alterations in the gut-brain axis [3], visceral hypersensitivity [4], increased intestinal permeability [5], and altered fecal microbiota composition [6, 7]. Different studies have been performed to identify microbial signatures in IBS patients, but a general consensus in IBS-related profiles is lacking [8]. This inconsistency could be due to the large individual variation in microbiota composition and cohort-specific characteristics [9], as well as cross-sectional study designs. Moreover, there is large variation in symptom severity and stool pattern within and between individuals with IBS [5], and instability in fecal microbiota over time [10, 11]. Furthermore, studies often do not include other covariates like diet and psychological state, which can be different in IBS and are associated with fecal microbiota [8, 12, 13].
Recently, Tap et al. (2017) were the first to explore IBS symptom severity related to the microbiota, and they cross-sectionally identified a microbial signature of 90 operational taxonomic units (OTUs) associated with IBS severity [14], which provides a new research direction to investigate microbial signatures with taking IBS severity into consideration. Although a recent study by Mars and colleagues longitudinally identified species-level taxa associated with severity of symptoms in diarrhea-predominant IBS [15], the consistency of microbial signatures associated with IBS symptom severity over time still remains unknown. Moreover, alterations in fecal short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate, have also been observed between IBS and controls [16]. These alterations were associated with bloating, abdominal pain, and QoL in IBS [17]. However, the association of SCFA with IBS severity and its consistency over time are unknown.
Therefore, we investigated the dynamics of fecal microbiota composition and SCFA levels in different IBS severity groups compared to controls, and the association with stool pattern, diet, depression, anxiety, and QoL over a period of four weeks. These dynamics and associations were also investigated between IBS and controls. We hypothesized that over time, IBS severity would demonstrate a regression to the mean, while fecal microbiota signatures associated with severity would remain stable.
Methods
This was an observational longitudinal study with two timepoints (T1 and T2) with four weeks in-between and included IBS patients and controls who were matched for age, gender, and body mass index (BMI) at T1 (no significance at group level). All participants signed an informed consent. The study was approved by the medical ethical committee of Wageningen and was registered at Clinicaltrials.gov (NCT03720314, https://clinicaltrials.gov/ct2/show/NCT03720314).
Study Participants
Participants were recruited using the Wageningen University subject database, and recruitment calls on websites and social media. Participants were aged 18–65 years, lived near Wageningen, and had a BMI between 18.5 and 30.0 kg/m2. IBS patients had to fulfill the Rome IV criteria or had to be diagnosed with IBS by a physician. Exclusion criteria were presence of any other gastrointestinal or systemic diseases, antibiotics use < 3 months before study start, pregnancy, or breastfeeding. We aimed to include 100 IBS patients and 30 matched controls at T1, to be able to detect a difference of 3.6 ± 4.9% in similarity of microbiota over time [10]. After T1, the 30 IBS patients with the least symptoms and 30 with the most severe symptoms were selected for T2, to assess the regression to the mean hypothesis and fecal microbiota dynamics. Controls completed both timepoints.
Fecal Microbiota Profiling
Participants collected a fecal sample at both timepoints. After collection, the fecal material was immediately stored in the participants’ home freezer. Fecal samples were transported on dry ice by research staff to the laboratory on average within 1.1 ± 1.2 days, where it was stored immediately at − 80 °C until further analysis.
Fecal microbiota composition was determined by sequencing the V4 region of the 16S ribosomal RNA (rRNA) gene (Illumina Hiseq2500, 150 bp paired end). As previously described, 0.25 g feces (wet weight) was used for DNA isolation with the repeated bead beating method [18]. Subsequently, DNA was purified using the Maxwell® 16 Total RNA system (Promega, Madison, WI, USA) with the 16 Tissue LEV Total RNA purification Kit Cartridge (XAS1220). Amplification was performed in duplicate with uniquely barcoded primers [19] 515F (5’-GTGYCAGCMGCCGCGGTAA-3’) [20] and 806R (5’-GGACTACNVGGGTWTCTAAT-3’) [21]. Reaction conditions and library preparation were performed as described previously [19]. Afterward, the libraries were purified with the CleanPCR kit (CleanNA, The Netherlands) and sent to Eurofins Genomics Germany GmbH (Konstanz, Germany) for sequencing. NG-Tax 2.0 was used to process the raw sequencing data for Amplicon Sequencing Variant (ASV) picking with default settings and for taxonomic assignments using the SILVA database (version 128) [22, 23]. Sequencing data was submitted to the European Nucleotide Archive with accession number PRJEB44533.
SCFA Profiling
SCFAs were measured as described previously with minor modifications [24]. In total, 0.4 g of feces (wet weight) were mixed thoroughly with 1.6 mL ultrapure water to extract SCFAs (acetate, propionate, and butyrate). Subsequently, the mixture was centrifuged (21,130 × g, 10 min) to get the supernatant. Afterward, 0.4 mL supernatant was added to 0.6 mL 10 mM DMSO as the internal standard in 0.1 N H2SO4 solution, and analyzed by High-Performance Liquid Chromatography (HPLC, LC-2030C, Shimazu, Kyoto, Japan) with a Shodex SH1821 column (Showa Denko K.K., Tokyo, Japan).
Questionnaires
Both IBS and controls completed all questionnaires at T1 and T2 for comparison. IBS severity was assessed using the validated IBS symptom severity score (IBS-SSS), which was used to classify severity (no symptoms ≤ 75; mild = 76–175; moderate = 176–300, and severe > 300 IBS) [25]. Both of the continuous severity score and severity grouping were used in analysis. QoL was assessed with the 34-item IBS-QoL, which gave a score for total IBS-QoL, and subscales dysphoria, interference with activity, body Image, health worry, food avoidance, social reaction, sexual life, and relationship [26]. Participants completed the hospital anxiety and depression score (HADS) [27]. A score ≥ 8 indicated substantial depressive or anxious symptoms [28]. Furthermore, the predominant stool pattern of the previous week was assessed by letting participants rank their stools of the week before sampling from most to least frequent, using the seven types of the validated Bristol Stool chart [29]. Participants also indicated the Bristol stool scale of the fecal sample. Habitual dietary intake of the previous month was assessed at T1 using a semi-quantitative 83-item Food Frequency Questionnaire [30, 31]. Dietary intake was calculated using the Dutch Food Composition table [32]. Furthermore, IBS patients were asked if they were currently following the Fermentable Oligo, Disaccharides, Monosaccharides, and Polyols (FODMAP) diet. Participants were instructed to keep their diet similar during the study period.
Statistical Analyses
Microbiota data were analyzed in R version 4.0.0 [33] and questionnaire data in SPSS version 25 (Armonk, NY, USA: IBM Corp.). Continuous data are presented as mean ± standard deviation, or median and interquartile range when skewed. Categorical data are presented as counts and percentages. Differences in the questionnaire data between IBS and controls were tested with an independent sample T test or Mann–Whitney U test when not normally distributed. Differences in questionnaire data between IBS subgroups and controls were tested with a one-way Analysis of Variance (ANOVA), or a Wilcoxon test when not normally distributed, with Bonferroni corrected post hoc testing. An unpaired Wilcoxon test was used to test differences in SCFAs between IBS subgroups and controls, or between IBS and controls. Differences for categorical data were assessed using chi-square tests. Associations of acetate, propionate, and butyrate (dependent variable) with questionnaire data (independent variables) were determined using linear mixed models.
Alpha diversity (within sample diversity) and beta diversity (between sample diversity) were calculated at ASV level using Phyloseq [34]. Alpha diversity metrics, ASV richness, and Shannon diversity were calculated. To visualize beta diversity, principal coordinate analysis (PCoA) based on unweighted (considering presence/absence of ASVs) and weighted (considering ASVs and their relative abundance) Unifrac [35] distances was performed. An unpaired Wilcoxon test was used to compare genus-level taxa between IBS and controls, or between IBS subgroups and controls at both timepoints. The p values for multiple pairwise tests were corrected for multiple testing using Benjamini–Hochberg false-discovery rate (FDR). The Vegan package [36] was used to assess the association of microbiota composition with questionnaire and dietary variables, using permutational multivariate analysis of variance (PERMANOVA).
A (corrected) p value ≤ 0.05 was considered statistically significant, and 0.05 ≤ (corrected) p value < 0.1 was considered a trend.
Results
A total of n = 91 IBS and n = 30 controls participated, with n = 55 IBS and n = 28 controls included for longitudinal analyses (Fig. 1). Baseline characteristics differed between IBS and controls for IBS-SSS (p < 0.001), IBS-QoL (p < 0.001), anxiety (p = 0.001), and depression (p = 0.004, Table 1). When data were stratified for IBS severity and compared with controls, IBS-QoL, anxiety, and depression remained different (p’s < 0.05). Distribution of the predominant stool patterns was not different between IBS severity groups. There was no significant difference in dietary intake of energy, fat, carbohydrates, polysaccharides, dietary fiber, alcohol, or water between IBS and controls (Supplementary Table 1). However, IBS patients had a lower intake of protein, maltose, and lactose. Moreover, 22% of IBS patients followed a FODMAP for whom lactose intake was significantly lower to those not following a FODMAP diet (p = 0.007).
Fig. 1.
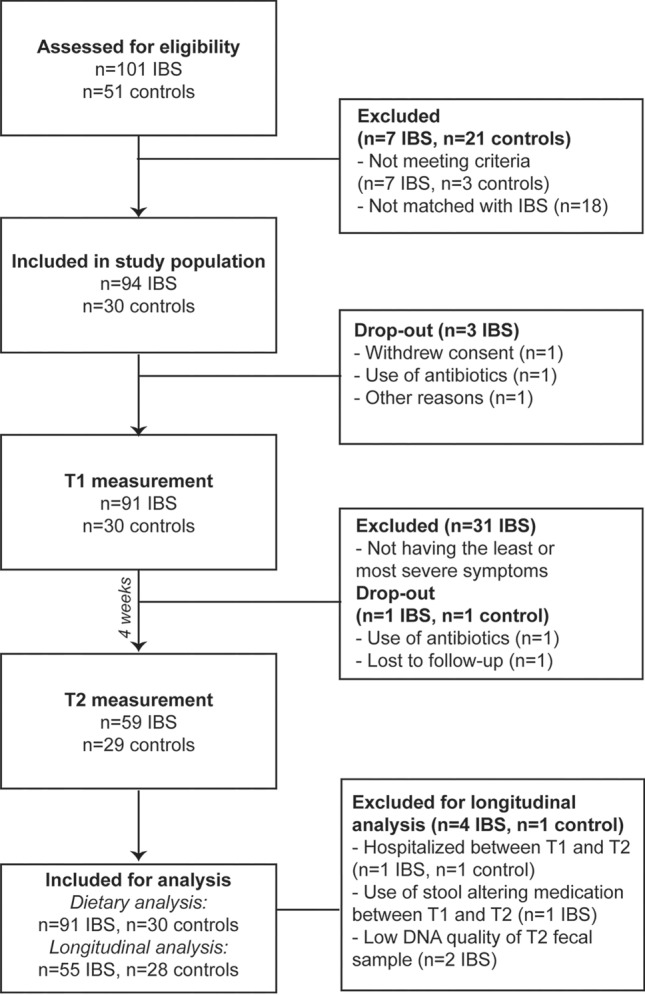
Flowchart of participant recruitment
Table 1.
Baseline characteristics of the study population
| IBS (n = 91) | Control (n = 30) | p value | |
|---|---|---|---|
| Age, years | 41.7 ± 14.4 | 39.4 ± 16.9 | .500 |
| Gender n (%) male | 19 (21) | 4 (13) | .361 |
| BMI, kg/m2 | 22.9 ± 2.9 | 23.2 ± 3.1 | .566 |
| IBS-SSS | 150 (110–230) | 55 (27–90) | .000 |
| Severity groups n (%) | .000 | ||
| No symptoms (≤ 75) | 7 (8) | 19 (63) | |
| Mild (76–175) | 48 (53) | 11 (37) | |
| Moderate (176–300) | 24 (26) | 0 (0) | |
| Severe (> 300) | 12 (13) | 0 (0) | |
| Predominant stool pattern n (%) | .183 | ||
| Constipation | 26 (29) | 14 (47) | |
| Diarrhea | 32 (35) | 6 (20) | |
| Mixed | 19 (21) | 4 (13) | |
| Unspecified | 14 (15) | 6 (20) | |
| IBS-QoL | 72.2 ± 16.8 | 98.6 ± 3.6 | .000 |
| Anxiety | |||
| Score | 6.0 (3.0–10.0) | 4.0 (2.8–6.0) | .001 |
| Substantial anxious symptoms n (%)† | 35 (38) | 3 (10) | .004 |
| Depression | |||
| Score | 2.0 (1.0–5.0) | 1.0 (0.0–2.2) | .004 |
| Substantial depressive symptoms n (%)† | 9 (10) | 2.0 1 (3) | .258 |
Data present mean ± standard deviation or median (interquartile range) when skewed
BMI body mass index; IBS-SSS IBS symptom severity score; IBS-QoL IBS quality of life
†Based on the hospital anxiety and depression score (HADS) cutoff > 8
Instability of IBS Symptom Severity Over Time
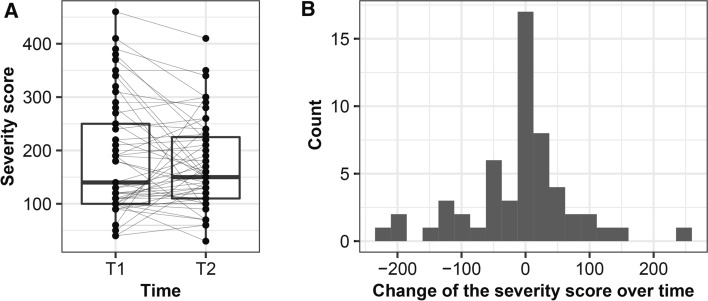
Instability of the IBS severity score was observed over time (Fig. 2A). The severity score decreased by ≥ 50 in 14 IBS patients and increased by ≥ 50 in 9 IBS patients (Fig. 2B). Furthermore, over time, 20 (36%) of the IBS patients changed in severity groups (Supplementary Fig. 1A). For other IBS symptoms, a large variation over time was also observed: 29 (53%) of the IBS patients changed in their predominant stool patterns (Supplementary Fig. 1B). Total IBS-QoL (Δ = 2.4, p = 0.028), sub-scores dysphoria (Δ = 3.1, p = 0.032), body image (Δ = 3.9, p = 0.004), and impact on relationship (Δ = 4.1, p = 0.010) increased, while other IBS-QoL sub-scores, anxiety, and depression remained stable over time.
Fig. 2.
Instability of severity score over time. Dynamics of IBS symptom severity score over time (A), and values were presented as interquartile with a boxplot. Samples taken at different timepoints are connected by solid lines per subject. Distribution of change of the severity score over time with a histogram (B)
No Difference in Microbiota Alpha and Beta Diversity Between IBS Severity Groups and Controls
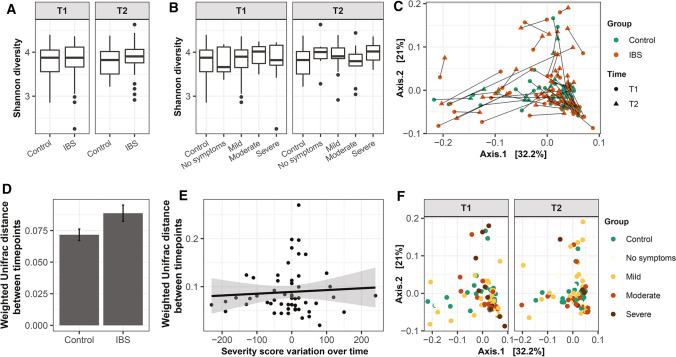
ASV richness and Shannon diversity were not different between IBS and controls, and between IBS with different predominant stool patterns and controls nor between IBS severity groups and controls (Fig. 3A, B, Supplementary Fig. 2) at both timepoints. This indicates the number and distribution of microbial ASVs are similar between these groups. PERMANOVA based on unweighted and weighted Unifrac distance (Fig. 3F) revealed no significant difference between IBS severity groups and controls as well as between IBS with different predominant stool patterns and controls. Between IBS and controls, the unweighted Unifrac-based observation was similar, while based on weighted Unifrac, a trend at T1 (p = 0.073) and T2 (p = 0.064) was observed (Fig. 3C). This indicates that the relative abundance of microbial taxa plays a role in the differences of microbiota composition between IBS and controls, rather than the presence or absence of the microbial taxa. Longitudinally, the change in fecal microbiota composition in IBS and controls was not different (Fig. 3D, p = 0.27), indicating that the temporal stability of the microbiota of IBS patients was similar to that of controls. Moreover, microbiota variation over time was not associated with a change in severity score (Fig. 3E).
Fig. 3.
Analysis of fecal microbiota alpha and beta diversity for IBS patients and controls and severity groups over time. Shannon diversity displayed as interquartile with boxplot, stratified for IBS and controls (A), symptom severity groups and controls (B). PCoA of microbiota composition based on weighted Unifrac distances, stratified for IBS and controls. Samples taken at different timepoints are connected by solid lines per subject (C). Comparison of microbiota composition stability based on weighted Unifrac distances over time between IBS patients and controls Values are presented as mean ± standard error (D). Linear model indicated no association between the changed severity score and weighted Unifrac distance of microbiota composition over time (E). PCoA of microbiota composition based on weighted Unifrac distances, stratified for symptom severity groups and controls (F)
Differences in Genus-Level Taxa Between IBS and Controls Were Not Associated with IBS Symptom Severity
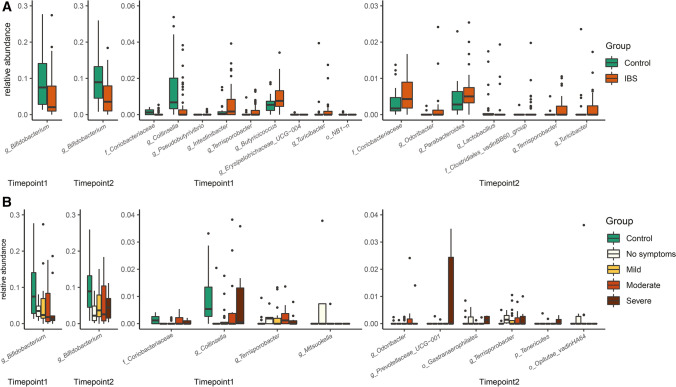
Relative abundances were different for ten genus-level taxa at T1 and eight at T2 between IBS and controls (Fig. 4A, Supplementary Table 2). However, of those only Bifidobacterium, Terrisporobacter, and Turicibacter were consistently different over time. The relative abundance of Bifidobacterium was lower in IBS compared to controls (pT1 = 0.0003; pT2 = 0.0003). In contrast, the relative abundances of Terrisporobacter (pT1 = 0.010; pT2 = 0.004) and Turicibacter (pT1 = 0.042; pT2 = 0.0013) were consistently higher in IBS patients over time. However, these taxa were not associated with IBS severity (Fig. 4B, Supplementary Table 3). The relative abundances of five genus-level taxa at T1 and seven at T2 were significantly different within the severity groups or between severity groups and controls. Remarkably, one uncultured taxon within the Tenericutes phylum (p = 0.021) and Prevotellaceae_UCG-001 (p = 0.021) was significantly higher in severe IBS patients at T2 compared to controls. However, none of these taxa were consistently different between IBS severity groups. In addition, the change of the genus-level taxa over time was not correlated with the change of the severity score over time (Supplementary Table 4). Next to IBS severity, we also assessed associations between fecal microbiota composition and predominant stool patterns in IBS over time. Some genus-level taxa were associated with predominant stool patterns, such as Alistipes with constipation. However, in line with the IBS severity observations these associations were only observed at a single timepoint, and not consistent over time (Supplementary Table 5).
Fig. 4.
Genus-level taxa that significantly differed in relative abundance between IBS patients and controls (A), or between severity groups and controls (B). Data are presented as interquartile with boxplot
Dynamics of the Association Between Questionnaire Data and Microbiota Composition Over Time
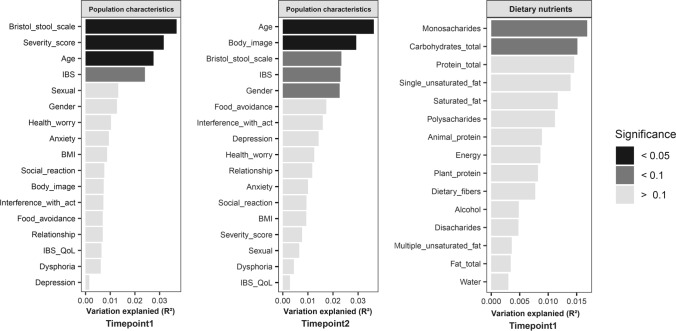
The largest proportion of microbiota variation was explained by the individual (R2 = 70.07%), when data of both timepoints was included. Age was significantly and consistently associated with microbiota composition at T1 (R2 = 2.75%) and T2 (R2 = 3.63%, Fig. 5). IBS explained a stable proportion of microbiota variation over time (R2T1 = 2.41%, R2T2 = 2.30%). At T1, Bristol stool scale and symptom severity score were significantly associated with microbiota composition, and explained, respectively, the first and second largest proportion of microbiota variation out of all participant characteristics (R2Bristol stool = 3.69%, R2Severity = 3.17%). However, at T2 the proportion of variation explained by Bristol stool scale and symptom severity score decreased, and was not significant anymore. This indicates large dynamics of explained microbiota variation by participant characteristics over time. We did not observe an association between dietary intake and microbiota variation. Moreover, no correlations were found between macronutrient, lactose, or maltose intake and relative abundance of Bifidobacterium, Terrisporobacter, and Turicibacter.
Fig. 5.
Large dynamics in explained variation of microbiota composition based on weighted Unifrac distances by population characteristics and dietary nutrients
No Difference in SCFAs Between IBS, Controls, and Severity Groups
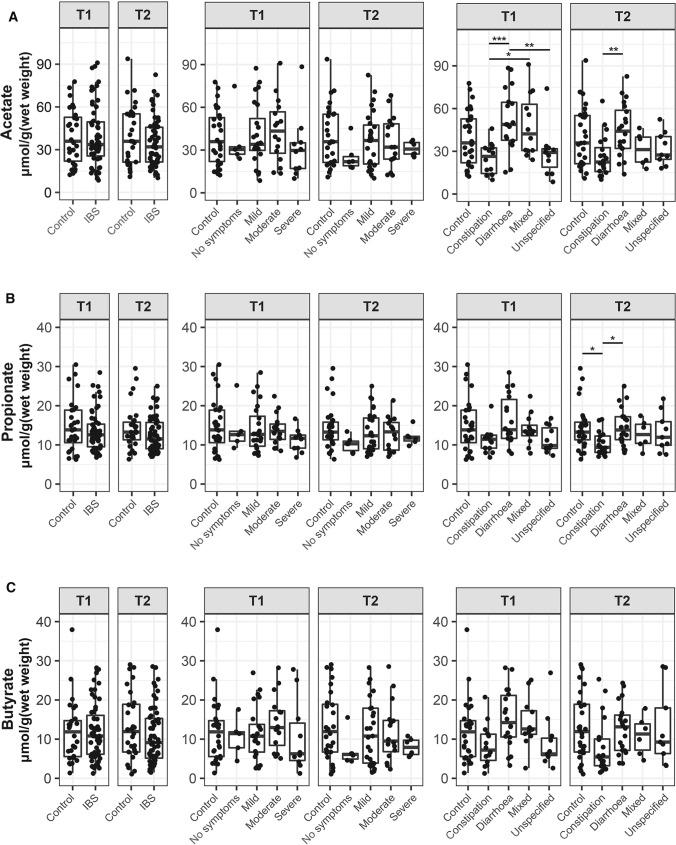
Large within and between person variation in SCFA levels was observed at both timepoints (Fig. 6), but no differences were found between IBS severity groups or IBS and controls. When subjects were stratified based on the predominant stool pattern, acetate was consistently higher in diarrhea-predominant IBS compared to patients with constipation (pT1 = 0.000; pT2 = 0.008). Linear mixed model analysis revealed that acetate, propionate, and butyrate were not associated with psychological factors over time (Supplementary Table 6). No correlations were observed between diet and SCFAs, except for lactose intake and butyrate (r = -0.182, p = 0.046, Supplementary Table 7).
Fig. 6.
No differences in fecal acetate (A), propionate (B), and butyrate (C) between controls and IBS, and severity groups over time, but lower level acetate in constipation-predominant IBS patients. Values were presented as interquartile with boxplot. Significance between groups was test by Wilcoxon. * p < 0.05; ** p < 0.01
Discussion
We performed a longitudinal study to examine the dynamics of fecal microbiota and SCFA levels between IBS severity and controls over time. Moreover, we assessed the association between fecal microbiota and SCFAs with stool pattern, IBS-QoL, anxiety, depression, and diet. We did not find any difference in microbial alpha diversity, composition, or SCFAs between the control and IBS severity groups. Multiple taxa were significantly different in relative abundance between IBS and controls were found at different timepoints, but consistent differences were only observed for Bifidobacterium, Terrisporobacter, and Turicibacter. Moreover, consistently lower acetate levels were found in only constipation-predominant IBS patients. Longitudinal analysis showed a large within and between subject variation in IBS severity, predominant stool pattern, and their association with microbiota composition.
Our results showed no significant difference of fecal microbial alpha diversity and composition in IBS, indicating that the number, distribution, and phylogenetic identity of microbial ASVs were similar between IBS and controls. This is in line with some studies [14, 37], but not all [38–41]. These conflicting results could be due to different methods used for the analysis, variations in discriminative power of different 16S rRNA gene regions, variations in inclusion and exclusion criteria, as well as heterogeneity of IBS cohorts [8]. Although the individuality explained the largest proportion of microbiota variation when looking at genus-level taxa, we observed several consistent microbial signatures associated with IBS over time. Lower relative abundance of Bifidobacterium in IBS patients has been reported in previous studies with a cross-sectional study design [8], which we found to be consistent over time. Remarkably, we found both Bifidobacterium and lactose consumption significantly lower in IBS patients compared to controls. Lactose has been suggested as one of the carbon sources of Bifidobacterium [42, 43]. However, the relative abundance of Bifidobacterium and lactose were not correlated in our study. Interestingly, Bifidobacterium supplements have been reported to improve IBS symptoms [44, 45], however, relief of symptoms was not always associated with an increase in relative abundance of Bifidobacterium [46]. This supports our observation that Bifidobacterium was not associated with symptom severity, but with IBS itself. Remarkably, we observed consistently higher relative abundance of Terrisporobacter and Turicibacter in IBS. Both taxa are thought to regulate the biosynthesis and release of serotonin and may play a role in IBS pathophysiology [47–50]. As these taxa have not been associated with IBS before, this finding may provide potential targets for future research.
Due to the accessibility without invasive procedures, fecal samples were the most commonly used to explore the role of gut microbiota in IBS [8]. It is evident that fecal samples only represent the end of the colon and previous studies have shown that comparing samples from small intestine and colon provides relevant insights into the microbiota at other locations in the intestine of IBS patients [37, 51]. Hence, our study cannot exclude that potential key microbes at other locations in relation to IBS are overlooked. Nevertheless, longitudinal studies require repeated sampling without disturbing the intestine, taking samples from other locations nearly impossible without invasive procedures.
In our study, we found some taxa to be associated with IBS severity at one of the two timepoints. However, differences were not consistent over time. IBS symptom severity itself changed drastically within four weeks, and the explained variance of microbiota composition by severity also indicated large dynamics over time. Moreover, we did not find any correlation between the change of the severity score over time with the change of the genus-level taxa in relative abundance over time, which indicates that IBS symptom severity seems not the reason causing changes in the relative abundance of fecal microbial taxa. A cross-sectional study by Tap et al. (2017) [14] and a longitudinal study by Mars et al. (2020) [15] have suggested that microbial signatures are associated with IBS symptom severity. However, the dynamics of the microbial signatures over time has not been determined in these two studies. Our study shows that the microbial signature associated with symptom severity is not stable longitudinally. Therefore, caution is needed in identifying signatures based on cross-sectional comparisons, which may change over time.
Increasing evidence indicates that IBS symptoms and microbiota composition are associated with carbohydrate intake [52, 53], and especially the FODMAPs [54, 55]. In our study, we did not find an association between microbiota composition and carbohydrate intake. Furthermore, after comparing the microbiota composition between IBS patients following the FODMAP diet or not, no difference was found. This might be due to the large dietary differences within the FODMAP diet and between studies. Further studies assessing the effects of carbohydrates on IBS symptoms and microbiota are needed.
The main microbial metabolites, SCFAs, have been suggested as a biomarker of IBS [16, 56]. However, approximately 80% of SCFAs produced in the gut are absorbed, and therefore not found in fecal samples [57], which may limit the effectiveness of fecal SCFAs as a biomarker. In our study, we did not find differences in SCFA levels between IBS and controls, while we confirmed that acetate was consistently lower in constipation-predominant IBS compared to diarrhea-predominant IBS [16]. This might be explained by the shorter transit time in diarrhea, leaving less time for absorption of SCFA in the gut, as shown in people with slow colonic transit [58].
Our study is strengthened by the longitudinal design, which enabled us to assess dynamics of fecal microbiota and SCFAs associated with IBS severity. Moreover, we assessed diet and psychological factors, which are altered in IBS, thus giving a more complete overview of the IBS patient. However, due to our observational study design we cannot determine causality. In addition, given the large variability over a short period of time in symptom severity scores, maintaining consistently equal-sized groups of severity was challenging.
In conclusion, consistent fecal microbiota and SCFA signature associated with IBS severity were not found. Interestingly, the relative abundances of the genera Bifidobacterium, Terrisporobacter, and Turicibacter were consistently different between IBS and controls over time, giving directions for future explorations. The importance of inclusion of multiple timepoints was demonstrated by the large within and between person variation of observed IBS severity, stool pattern, and their association with fecal microbiota composition over time. Hence, conclusion of single-timepoint studies in the past should be reconsidered, and future studies are highly recommended to take time-dynamics into account.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Leander Dijk for his help in laboratory analysis. Moreover, we thank Jennifer Klein Gunnewiek, Janyke Geluk, Zoe Verdaasdonk, Leonie Horne, and Ilse Schilderinck for their help with data collection. The research by Taojun Wang is financially supported by the China Scholarship Council (File No. 201600090211).
Abbreviations
- IBS
Irritable bowel syndrome
- SCFAs
Short-chain fatty acids
- QoL
Quality of life
- OTUs
Operational taxonomic units
- T1
Timepoint 1
- T2
Timepoint 2
- BMI
Body mass index
- rRNA
Ribosomal ribonucleic acid
- ASV
Amplicon sequencing variant
- HPLC
High-performance liquid chromatography
- SSS
Symptom severity score
- HADS
Hospital anxiety and depression score
- FODMAPs
Fermentable oligo, disaccharides, monosaccharides, and polyols
- ANOVA
Analysis of variance
- PCoA
Principal coordinate analysis
- FDR
False-discovery rate
- PERMANOVA
Permutational multivariate analysis of variance
- CI
Confidence interval
- SD
Standard deviation
- NA
Not available
Author's contributions
TW contributed to laboratory work, statistical analysis and interpretation of data, and drafting manuscript. IR contributed to study concept and design, statistical analysis and interpretation of data, and drafting manuscript. GH assists in study concept and design and interpretation of data and critically revised the manuscript. NR contributed to study supervision and critically revised the manuscript. BW contributed to study supervision and critically revised the manuscript. NW, project leader, contributed to study supervision and critically revised the manuscript. CG, project leader, contributed to study supervision and critically revised the manuscript. HS contributed to study supervision and critically revised the manuscript. EZ drafted study design, assisted in interpretation of data, led the study supervision, and critically revised the manuscript.
Funding
This research was supported by a Public Private Partnership grant from the Top consortium for Knowledge and Innovation Agri & Food by the Dutch Ministry of Economic Affairs (TKI-AF-16012) and associated companies. The funding agencies had no role in the collection, analysis, and interpretation of data; or in the preparation, review, or approval of the manuscript. Taojun Wang was financially supported by the China Scholarship Council (File No. 201600090211). Gerben Hermes was supported by the project MASTER (Microbiome Applications for Sustainable food systems through Technologies and Enterprise) funded by the European Union’s H2020 research and innovation program under Grant Agreement No. 818368.
Declarations
Conflict of interest
The authors report no conflict of interest.
Ethical approval
The study was approved by the medical ethics committee of Wageningen (18/06) and conducted according to the declaration of Helsinki. All participants signed a written informed consent. The study was registered at Clinicaltrials.gov under number NCT03720314 on October 25, 2018.
Footnotes
An editorial commenting on this article is available at 10.1007/s10620-022-07551-3
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Taojun Wang and Iris Rijnaarts contributed equally.
Contributor Information
Taojun Wang, Email: taojun.wang@wur.nl.
Iris Rijnaarts, Email: iris.rijnaarts@wur.nl.
Gerben D. A. Hermes, Email: gerben.hermes@wur.nl
Nicole M. de Roos, Email: nicole.deroos@wur.nl
Ben J. M. Witteman, Email: ben.witteman@wur.nl
Nicole J. W. de Wit, Email: nicole.dewit@wur.nl
Coen Govers, Email: coen.govers@wur.nl.
Hauke Smidt, Email: hauke.smidt@wur.nl.
Erwin G. Zoetendal, Email: erwin.zoetendal@wur.nl
References
- 1.Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin Epidemiol. 2014;6:71. doi: 10.2147/CLEP.S40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA. 2015;313:949–958. doi: 10.1001/jama.2015.0954. [DOI] [PubMed] [Google Scholar]
- 3.Koloski NA, Jones M, Kalantar J, et al. The brain–gut pathway in functional gastrointestinal disorders is bidirectional: a 12-year prospective population-based study. Gut. 2012;61:1284–1290. doi: 10.1136/gutjnl-2011-300474. [DOI] [PubMed] [Google Scholar]
- 4.Caldarella MP, Milano A, Laterza F, et al. Visceral sensitivity and symptoms in patients with constipation-or diarrhea-predominant irritable bowel syndrome (IBS): effect of a low-fat intraduodenal infusion. Am J Gastroenterol. 2005;100:383–389. doi: 10.1111/j.1572-0241.2005.40100.x. [DOI] [PubMed] [Google Scholar]
- 5.Ford AC, Lacy BE, Talley NJ. Irritable bowel syndrome. N Engl J Med. 2017;376:2566–2578. doi: 10.1056/NEJMra1607547. [DOI] [PubMed] [Google Scholar]
- 6.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 7.Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol-Gastrointest Liver Physiol 2017;312:G52-G62. [DOI] [PMC free article] [PubMed]
- 8.Pittayanon R, Lau JT, Yuan Y, et al. Gut microbiota in patients with irritable bowel syndrome—a systematic review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 9.Hermes GD, Reijnders D, Kootte RS, et al. Individual and cohort-specific gut microbiota patterns associated with tissue-specific insulin sensitivity in overweight and obese males. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-64574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mättö J, Maunuksela L, Kajander K, et al. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome—a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Maukonen J, Satokari R, Mättö J, et al. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–633. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- 12.Luna RA, Foster JA. Gut brain axis: diet microbiota interactions and implications for modulation of anxiety and depression. Curr Opin Biotechnol. 2015;32:35–41. doi: 10.1016/j.copbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Rajilić-Stojanović M, Jonkers DM, Salonen A, et al. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am J Gastroenterol. 2015;110:278. doi: 10.1038/ajg.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tap J, Derrien M, Törnblom H, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152(111–123):e8. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 15.Mars RA, Yang Y, Ward T, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182(1460–1473):e17. doi: 10.1016/j.cell.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q, Jia Q, Song L, et al. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: a systematic review and meta-analysis. Medicine 2019;98. [DOI] [PMC free article] [PubMed]
- 17.Tana C, Umesaki Y, Imaoka A, et al. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–e115. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 18.Salonen A, Nikkilä J, Jalanka-Tuovinen J, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Ramiro-Garcia J, Hermes GD, Giatsis C, et al. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Research 2016;5. [DOI] [PMC free article] [PubMed]
- 20.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 21.Apprill A, McNally S, Parsons R, et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 22.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yilmaz P, Parfrey LW, Yarza P, et al. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An R, Wilms E, Smolinska A, et al. Sugar beet pectin supplementation did not alter profiles of fecal microbiota and exhaled breath in healthy young adults and healthy elderly. Nutrients. 2019;11:2193. doi: 10.3390/nu11092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis CY, Morris J, Whorwell PJ. The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment Pharmacol Therap. 1997;11:395–402. doi: 10.1046/j.1365-2036.1997.142318000.x. [DOI] [PubMed] [Google Scholar]
- 26.Patrick DL, Drossman DA, Frederick IO, et al. Quality of life in persons with irritable bowel syndrome (development and validation of a new measure) Dig Dis Sci. 1998;43:400–411. doi: 10.1023/A:1018831127942. [DOI] [PubMed] [Google Scholar]
- 27.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67:361–370. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- 28.Bjelland I, Dahl AA, Haug TT, et al. The validity of the Hospital Anxiety and Depression Scale: an updated literature review. J Psychosom Res. 2002;52:69–77. doi: 10.1016/S0022-3999(01)00296-3. [DOI] [PubMed] [Google Scholar]
- 29.Blake M, Raker J, Whelan K. Validity and reliability of the Bristol Stool Form Scale in healthy adults and patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Therap. 2016;44:693–703. doi: 10.1111/apt.13746. [DOI] [PubMed] [Google Scholar]
- 30.Siebelink E, Geelen A, de Vries JH. Self-reported energy intake by FFQ compared with actual energy intake to maintain body weight in 516 adults. Br J Nutr. 2011;106:274–281. doi: 10.1017/S0007114511000067. [DOI] [PubMed] [Google Scholar]
- 31.Streppel MT, de Vries JH, Meijboom S, et al. Relative validity of the food frequency questionnaire used to assess dietary intake in the Leiden Longevity Study. Nutr J. 2013;12:75. doi: 10.1186/1475-2891-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NEVO-online versie. Bilthoven: RIVM, 2019/6.0.
- 33.Team RC. R: A language and environment for statistical computing. 2013.
- 34.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oksanen J, Blanchet F, Friendly M, et al. vegan: Community ecology package. R package version 2.5-4. R Foundation for Statistical Computing, 2019.
- 37.Hugerth LW, Andreasson A, Talley NJ, et al. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 2020;69:1076–1084. doi: 10.1136/gutjnl-2019-318717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll IM, Ringel-Kulka T, Keku TO, et al. Molecular analysis of the luminal-and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol-Gastroint Liver Physiol 2011;301:G799-G807. [DOI] [PMC free article] [PubMed]
- 39.Carroll IM, Ringel-Kulka T, Siddle JP, et al. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–e248. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundin J, Rangel I, Fuentes S, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Therap. 2015;41:342–351. doi: 10.1111/apt.13055. [DOI] [PubMed] [Google Scholar]
- 41.Jeffery IB, Das A, O’Herlihy E, et al. Differences in fecal microbiomes and metabolomes of people with vs without irritable bowel syndrome and bile acid malabsorption. Gastroenterology. 2020;158:1016–1028. doi: 10.1053/j.gastro.2019.11.301. [DOI] [PubMed] [Google Scholar]
- 42.Chia LW, Mank M, Blijenberg B, et al. Cross-feeding between Bifidobacterium infantis and Anaerostipes caccae on lactose and human milk oligosaccharides. Beneficial Microbes. 2021;12:69–83. doi: 10.3920/BM2020.0005. [DOI] [PubMed] [Google Scholar]
- 43.Parche S, Beleut M, Rezzonico E, et al. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J Bacteriol. 2006;188:1260–1265. doi: 10.1128/JB.188.4.1260-1265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guglielmetti S, Mora D, Gschwender M, et al. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life––a double-blind, placebo-controlled study. Aliment Pharmacol Therap. 2011;33:1123–1132. doi: 10.1111/j.1365-2036.2011.04633.x. [DOI] [PubMed] [Google Scholar]
- 45.Andresen V, Gschossmann J, Layer P. Heat-inactivated Bifidobacterium bifidum MIMBb75 (SYN-HI-001) in the treatment of irritable bowel syndrome: a multicentre, randomised, double-blind, placebo-controlled clinical trial. Lancet Gastroenterol Hepatol. 2020;5:658–666. doi: 10.1016/S2468-1253(20)30056-X. [DOI] [PubMed] [Google Scholar]
- 46.Wilson B, Rossi M, Kanno T, et al. β-Galactooligosaccharide in conjunction with low FODMAP diet improves irritable bowel syndrome symptoms but reduces fecal Bifidobacteria. Am J Gastroenterol. 2020;115:906–915. doi: 10.14309/ajg.0000000000000641. [DOI] [PubMed] [Google Scholar]
- 47.Stasi C, Bellini M, Bassotti G, et al. Serotonin receptors and their role in the pathophysiology and therapy of irritable bowel syndrome. Tech Coloproctol. 2014;18:613–621. doi: 10.1007/s10151-013-1106-8. [DOI] [PubMed] [Google Scholar]
- 48.Yano JM, Yu K, Donaldson GP, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fung TC, Vuong HE, Luna CD, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4:2064–2073. doi: 10.1038/s41564-019-0540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labus JS, Osadchiy V, Hsiao EY, et al. Evidence for an association of gut microbial Clostridia with brain functional connectivity and gastrointestinal sensorimotor function in patients with irritable bowel syndrome, based on tripartite network analysis. Microbiome. 2019;7:1–15. doi: 10.1186/s40168-019-0656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dlugosz A, Winckler B, Lundin E, et al. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci Rep. 2015;5:1–9. doi: 10.1038/srep08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamal A, Pimentel M. Influence of dietary restriction on irritable bowel syndrome. Am J Gastroenterol. 2019;114:212–220. doi: 10.1038/s41395-018-0241-2. [DOI] [PubMed] [Google Scholar]
- 53.Collins SM. A role for the gut microbiota in IBS. Nat Rev Gastroenterol Hepatol. 2014;11:497–505. doi: 10.1038/nrgastro.2014.40. [DOI] [PubMed] [Google Scholar]
- 54.Halmos EP, Christophersen CT, Bird AR, et al. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93–100. doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- 55.Staudacher HM, Lomer MC, Farquharson FM, et al. A diet low in FODMAPs reduces symptoms in patients with irritable bowel syndrome and a probiotic restores bifidobacterium species: a randomized controlled trial. Gastroenterology. 2017;153:936–947. doi: 10.1053/j.gastro.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 56.Farup PG, Rudi K, Hestad K. Faecal short-chain fatty acids-a diagnostic biomarker for irritable bowel syndrome? BMC Gastroenterol. 2016;16:51. doi: 10.1186/s12876-016-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNeil NI, Cummings J, James W. Short chain fatty acid absorption by the human large intestine. Gut. 1978;19:819–822. doi: 10.1136/gut.19.9.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Müller M, Hermes GD, Canfora EE, et al. Distal colonic transit is linked to gut microbiota diversity and microbial fermentation in humans with slow colonic transit. Am J Physiol-Gastrointest Liver Physiol 2020;318:G361–G369. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.