Abstract
Background: The autophagy machinery is reported to be employed by Coronaviruses during their replication. Beclin-1 (BECN1) and protein 1 light chain 3 (LC3) are two key elements in the autophagy process, and their inhibition can prevent the replication of some coronaviruses in vitro. Here, we aimed to investigate the expression levels of Beclin-1 and LC3 in COVID-19 patients and healthy controls, hoping to find new therapeutic targets.
Methods: This cross-sectional study was conducted in Imam Reza and Ghaem University Hospitals, Mashhad, Iran. Nasopharyngeal samples of 68 consecutive Covid-19 patients and 61 healthy controls, who have been referred to the laboratories for COVID-19 PCR testing between 21 March to 21 September 2021, were used in order to evaluate the expression of BECN1 and LC3 genes using the Real-time quantitative PCR method. Demographic and other laboratory findings of patients were extracted from the hospital electronic system. SPSS Statistics 16.0 and Graph Pad Prism 8.4.2 soft wares were used for statistical analysis. Non-parametric tests were used.
Results: BECN1 expression was significantly higher in COVID-19 patients compared to the controls (14.37±18.84 vs. 4.26±7.39, p=0.001). The expression of LC3 gene was significantly lower in patients compared to the controls (1.01±1.06 vs. 1.49±1.12, p=0.007). There was no significant correlation between the expression levels of BECN1 and LC3. Patients with lower BECN1 expression showed significantly higher RBC counts, higher Urea and lower HCO3 levels. The patients in LC3Low group showed significantly lower MCH, MCHC and PH levels compared to the others.
Conclusion: Regarding the significant difference in the expression of BECN1 and LC3 in COVID-19 patients compared to the controls, these molecules may have a role in the pathogenesis of this disease. In case of further confirmation of this role, these molecules may be used as possible therapeutic targets.
Keywords: Autophagy, Beclin 1, BECN1, Protein 1 Light Chain 3, LC3, RT-PCR, COVID-19, SARS-CoV2
↑What is “already known” in this topic:
Coronaviruses use the autophagy machinery during their replication process. It has been shown that some autophagy inhibitor drugs, especially chloroquine and hydroxychloroquine, inhibit the cytopathic effects of SARS-CoV-2.
→What this article adds:
Evaluating the expression of autophagy-related genes in COVID-19 patients may help to identify specific targets for the development of more specific and targeted therapies. The expression of the two key elements of autophagy; Beclin1 and Light Chain 3(LC3), is evaluated in this study and the expression of both of them were significantly different from controls. So, these molecules may be used as possible therapeutic targets.
Introduction
SARS-CoV-2 is a member of the Beta coronavirus genus, enveloped viruses with a Positive-strand RNA, and is responsible for the COVID-19 pandemic, which is considered a global threat (1). To date, 242,348,657 Infections and 4,927,723 death from SARS-CoV-2 has been reported worldwide (2). There is no effective specialized therapy which can be used for all patients at this time. So, revealing the pathogenesis of the disease is of urgent need.
Autophagy is an intracellular process involved in the degradation of damaged organelles, long-lived proteins, cell evolution, the pathogenesis of some cancers as well as innate immunity; through the degradation of intracellular pathogens (including viruses) and their presentation to the adaptive immune system (3). Some viruses have mechanisms to evade autophagy.In contrast, some viruses use the autophagy process during their replication (4).
The usage of the autophagy machinery during the replication process of some viruses, including HIV 1,2, hepatitis B virus, hepatitis C virus, Picornavirus and Coxsackie virus, has been reported (5-8). There are some evidence that coronaviruses use autophagy machinery for their replication. For example, Coronavirus infection leads to the formation of vesicles with double-membrane (DMVs) in the cell (9). The virus replication happens in these vesicles. In addition, the formation of DMVs is a characteristic finding in the autophagy process. Furthermore, a coronavirus protein; nsp6 has been shown to induce the formation of DMV (10). In addition, Co-localization of the coronavirus protein nsp6 with LC3, which is a stable component of autophagy vesicles, has been observed, which indicates the possible interaction of these two molecules. Based on the reports, coronaviruses may use parts of the autophagy process, not the whole process, to reproduce themselves (11, 12). Another evidence that highlights the importance of autophagy in the pathogenesis of SARA-COV2 comes from studies that showed that inhibitors of autophagy are able to prevent coronavirus replication in cells. For example, in a study by Gorshkov K et al., six autophagic inhibitor drugs especially chloroquine and hydroxychloroquine, were studied regarding their ability in inhibition of the cytopathic effects of SARS-CoV-2 and most of them showed significant inhibitory effects (13). BECN1 protein is part of the phosphatidylinositol-3 kinase class III (PIK3C3) complex. Together with ATG14 and UVRAG (radiation resistance gene), it plays an essential role in nucleation and autophagosome formation in the early stages of autophagy (14). Furthermore, inhibitors of the nucleation stage (ATG14/Beclin1/ VPS34) of autophagy were able to inhibit SARS-CoV-2 replication in human lung cells (15). LC3 protein is located in the autophagosome membrane in the early stages and is preserved until the end of the process so that it can be used as a marker to identify the autophagosome (11). LC3 downregulation has been shown to protect cells from infection by some coronaviruses including SARS and mouse hepatitis virus (16).
Excess production of inflammatory cytokines has been reported in COVID-19 patients with severe disease. It has been reported that BECN1 induces the production of inflammatory cytokines such as IL6 and IL-1 and directly increases TNFα (17). Furthermore, recent studies have suggested that COVID-19 increases apoptosis via the accumulation of autophagosomes in the cells (15). So, it may be possible to decrease apoptosis and cell death by inhibiting the autophagy process.
As seen from the mentioned studies, autophagy is probably a fundamental part of SARS-CoV-2 replication in cells and BECN1 and LC3 are key elements in this process. In this study, we investigated the expression of BECN1 and LC3 in the nasopharyngeal samples of COVID-19 patients and healthy controls hoping to find possible targets for the development of new and more targeted therapies. As it has been reported, some autophagy inhibitors, such as chloroquine and hydroxychloroquine, are broad-acting, and have considerable side effects, so finding more targeted medications will be of invaluable help for the treatment of patients.
Methods
Study design and participants
This cross-sectional study was conducted in Imam Reza as well as Ghaem University Hospitals, Mashhad University of Medical Sciences, Mashhad, Iran. The study was done on nasopharyngeal samples of 68 consecutive Covid-19 patients and 61 healthy controls who had been referred to the laboratory for COVID-19 PCR testing between 21 March to 21 September 2021. All patients had positive PCR results for COVID-19. The control group was asymptomatic individuals who needed a COVID-19 test for traveling, entering dormitories or so on and their Covid-19 PCR results were negative. The demographic and other laboratory test results of patients were also extracted. These tests were including hematological parameters (white blood cells [WBC], lymphocyte count [LYM], neutrophil count [neut,], red blood cells [RBC], hemoglobin [HGB], hematocrit [HCT], mean corpuscular volume [MCV], Platelet, mean platelet volume [MPV], platelet distribution width [PWD]), coagulation parameters (D-dimer, PT, PTT), inflammatory markers (Erythrocyte sedimentation rate [ESR] and C-reactive protein [CRP]) and biochemical markers (alanine aminotransferase [ALT], aspartate aminotransferase [AST], creatinine), Urea, lactate dehydrogenase [LDH], PH, HCO3, cTnI [cardiac troponin I] and BS [blood sugar] test.
RNA isolation and complementary DNA synthesis
Immediately after sampling, RNA was extracted using RNeasy Kit (add bio, Korea Cat. No. 10119) according to the instruction protocol. Following the extraction, the quality of RNA was evaluated by measuring the absorption at 260 & 280 & 230 nm by the Nano Drop 2000 Spectrophotometer (Thermo Scientific, USA). A ratio of A260/A280 = 1.8 - 2.1 was considered high purity. cDNA synthesis was done by means of cDNA Synthesis Kit (add bio, Korea22701). 5µl of RNA were mixed with 10 µl of 2x Reaction Buffer, 2 µl dNTP, 2 µl oligo dt (10x random hexamer), and 1 µl Enzyme solution. Thermo cycler temperatures and time conditions are as follows:
The reaction mixture (20 µl) was incubated at 50 ºC for 60 minutes; then the temperature was raised to 80 ºC for 5 minutes to stop the reaction.
After RNA isolation and complementary DNA preparation, Real-Time quantitative RT-PCR (RQ-PCR) reactions were performed by The Applied Bio-system Step One Plus Real-Time PCR Systems (Applied Bio-systems). The Relative expression of BECN1 and LC3 genes was evaluated by 2 -ΔΔCt method using GAPDH gene as an internal control gene in the same samples. Primers for LC3 and Beclin1 genes and GAPDH gene (as an internal control) were designed by PubMed blast software. Table 1 shows the primer sequences used in this study. Each reaction contained 5 μL of master mix (SYBR Green), 2 μL of CDNA, 0.15 μL of each primer and probe, and DEPC water to a final volume of 10.5 μL. 40 cycles of 95°C for 5 min, denaturation at 95°C for 30 s followed by extension at 60°C for 1.5 min were applied. All samples were analyzed in duplicate wells.
Table 1. Primer sequences used in real-time PCR.
| Primer | Sequence 5'-3' | Ampliconsize (b.p.) | |
|---|---|---|---|
| Primer sequences used in real-time PCR | |||
| BECN1 | F1 | 5′-CAA GAT CCT GGA CCG TGT CA-3′ | 191 bp |
| R1 | 5′-TGG CAC TTT CTG TGG ACA TCA-3′ | ||
| LC3 | F1 | 5′-ATG CCG TCG GAC AAG ACC TT-3′ | 360 bp |
| R1 | 5′-TTA CAC TGA CAA TTT CAT CCC G-3′ | ||
| GAPDH | F1 | 5′-TGC ACC ACC AAC TGC TTA-3′ | 87 bp |
| R1 | 5′-GAG GGC ATG GAC TGT GGT CAT-3′ | ||
Statistical analysis
The Kolmogorov-Smirnov test was used to evaluate the normal distribution of the data. As the LC3 and BECN1 expressions did not show normal distribution, non-parametric tests were used to compare the expression levels of LC3 and BECN1 between the patients and controls. The changes in the expression of the studied genes compared to the control group were evaluated using fold change expression. Fold change values of 2 or more than 2 were considered overexpression. The values equal to or less than 0.5 were interpreted as down-regulation and fold change values between 0.5 to 2 were considered to have no change in the expression. Based on the median of the expression level of each gene, patients were divided into two groups: low expression and high expression for each gene of interest. All data analyses were performed using SPSS Statistics 16.0 and Graph Pad Prism 8.4.2 software. p values less than 0.05 were considered statistically significant.
Results
Expressions level of LC3 and Beclin1 gens
Detailed demographic and laboratory information from 68 patients with covid-19 are listed in Table 2. The mean (SD) age of the patient was 60.72±16.8 years, ranging between 27 and 87 years. 44 (62.9%) of patients were male. The control group included 61 participants with approximately same sex distribution (M/F ratio: 1.5) and mean (SD) age of 58.7 ±17.54 years, ranging between 20 to 88 years. Thirteen (19%) patients died during hospitalization. The BECN1 gene expression was significantly higher in Covid-19 patients compared to controls (p=0.001, Figure 1). The mean expression of BECN1 (2 –ΔΔCt ± SD) for patients and the control group was 14.37 ±18.84 and 4.26± 7.39, respectively. The Beclin1 gene showed upper expression in 36 (57%) patients. Fifteen (24%) patients showed underexpression and the others showed no change in the expression of Beclin1 gene. The expression of LC3 gene was significantly lower in patients compared to controls (p=0.007, Figure 1). The mean expression of LC3 (2 –ΔΔCt ± SD) for patients with covid-19 and the control group were 1.007 ±1.06 and 1.489± 1.12, respectively. The LC3 gene was down-regulated in 27 (44%) patients, while 13 (21%) showed up-regulation and the others showed no changes in LC3 expression. The correlation between LC3 and BECN1 expressions was evaluated using Spearman's test. There was no significant correlation between LC3 and Beclin1 expressions (p=0.703, r=- 0.036).
Table 2. Clinical characteristics of COVID 19 patients based on BECLIN1 and LC3 expression level.
| Characteristic | All patients | LC3low | LC3high | P | Beline1 low | Beline1high | P |
|---|---|---|---|---|---|---|---|
| gender (male/ female), N=68 | (41/26) | 15/15 | 21/10 | 0.159 | 19/13 | 19/12 | 0.877 |
| Outcome (survived/ death), N=68 | (47/13) | 20/7 | 20/6 | 0.810 | 19/7 | 23/4 | 0.327 |
| Age, mean, N= 68 | 60.72 (16.80) | 61.13 (14.9) | 59.87 (18.93) | 0.773 | 63.56 (16.71) | 58.74 (16.82) | 0.258 |
| MCV (fl), mean (SD), N= 68 | 85.29 (5.28) | 84.72 (3.58) | 86.01 (5.21) | 0.512 | 84.40 (5.39) | 85.79 (5.01) | 0.470 |
| MCH (pg), mean (SD), N= 68 | 27.91 (2.31) | 27.27 (1.71) | 28.53 (1.97) | 0.026 | 27.60 (2.48) | 28.08 (2.16) | 0.858 |
| MCHC (g/dl), mean (SD), N= 68 | 32.71 (1.5) | 32.18 (1.39) | 33.17 (1.31) | 0.008 | 32.67 (1.71) | 32.72 (1.39) | 0.747 |
| RBC (×10^6/µL), mean (SD), N= 68 | 4.67 (0.7) | 4.61 (0.59) | 4.67 (0.77) | 0.337 | 4.84 (0.64) | 4.57 (0.58) | 0.036 |
| HGB (g/l), mean (SD), N= 68 | 131.6 (19.3) | 126.4 (18.9) | 135.8 (19.6) | 0.040 | 134.0 (20.0) | 128.4 (17.9) | 0.151 |
| HCT (%), mean (SD), N= 68 | 40.18 (5.13) | 39.26 (5.12) | 40.87 (5.24) | 0.105 | 40.94 (5.02) | 39.20 (4.98) | 0.072 |
| Platelet (x 10^3/µL), mean (SD), N= 68 | 209.78 (82.19) | 208.10 (86.67) | 211.87 (84.07) | 0.655 | 221.59 (81.09) | 195.71 (84.20) | 0.173 |
| Neut Absolute (× 10^3/µL), mean (SD), N= 64 | 7.41 (4.37) | 7.42 (4.45) | 7.15 (3.83) | 0.750 | 7.84 (4.747) | 7.17 (4.23) | 0.626 |
| Lymphocytes (×10^3/µL), mean (SD), N= 66 | 0.98 (0.54) | 0.90 (0.58) | 0.99 (0.47) | 0.387 | 0.93 (0.46) | 1.07 (0.58) | 0.419 |
| RDW-CV (%), mean (SD), N=68 | 14.23 (1.48) | 14.31 (1.68) | 14.22 (1.17) | 0.681 | 14.17 (1.39) | 14.23 (1.57) | 0.989 |
| WBC (×10^9/L), mean (SD), N= 68 | 8.67 (4.38) | 8.78 (4.43) | 8.24 (4.07) | 0.530 | 9.21 (4.73) | 8.35 (4.29) | 0.509 |
| PDW (fl), mean (SD), N= 64 | 13.45 (2.41) | 13.55 (2.79) | 13.19 (1.91) | 0.854 | 13.08 (1.93) | 13.57 (2.72) | 0.600 |
| MPV (fl), mean (SD), N= 68 | 10.11 (1.17) | 10.13 (1.31) | 10.06 (1.09) | 0.817 | 9.84 (1.13) | 10.30 (1.18) | 0.236 |
| INR, mean (SD), N= 50 | 1.05 (0.17) | 1.05 (0.11) | 1.05 (0.22) | 0.462 | 1.03 (0.06) | 1.07 (0.24) | 0.931 |
| PT (s), mean (SD), N= 50 | 10.79 (1.85) | 10.78 (1.33) | 10.88 (2.39) | 0.554 | 10.69 (1.19) | 11.02 (2.39) | 0.800 |
| PTT (s), mean (SD), N= 49 | 31.08 (22.37) | 28.27 (28.27) | 34.38 (32.16) | 0.991 | 34.65 (32.98) | 28.37 (5.20) | 0.741 |
| ESR (mm/h), mean (SD), N= 31 | 41.06 (28.96) | 36.20 (22.71) | 46.60 (34.87) | 0.595 | 40.37 (25.65) | 42.17 (34.76) | 0.826 |
| Total-Billirubin (mg/dL), mean (SD), N= 43 | 0.79 (0.31) | 0.81 (0.33) | 0.77 (0.31) | 0.637 | 0.78 (0.27) | 0.79 (0.36) | 0.865 |
| Direct-Billirubin (mg/dL), mean (SD), N= 43 | 0.25 (0.14) | 0.26 (0.14) | 0.24 (0.15) | 0.469 | 0.26 (0.17) | 0.24 (0.12) | 0.680 |
| AST (U/L), mean (SD), N= 53 | 63.53 (48.44) | 66.34 (57.23) | 56.73 (35.69) | 0.967 | 62.04 (41.63) | 69.38 (57.16) | 0.936 |
| ALT (U/L), mean (SD), N= 54 | 70.25 (92.82) | 80.84 (125.81) | 56.92 (45.13) | 0.719 | 78.83 (121.94) | 67.04 (58.00) | 0.846 |
| ALP (U/L), mean (SD), N= 48 | 212.26 (1.58) | 237.18 (222.7) | 184.85 (69.21) | 0.511 | 238.95 (224.21) | 196.78 (80.69) | 0.760 |
| LDH (U/L), mean (SD), N= 51 | 729.49 (430.23) | 801.98 (533.98) | 686.17 (311.92) | 0.967 | 689.64 (324.52) | 807.45 (590.14) | 0.880 |
| CRP (mg/L), mean (SD), N= 61 | 108.2 (90.16) | 110.81 (104.62) | 105.42 (75.65) | 0.736 | 96.75 (76.01) | 111.92 (98.98) | 0.756 |
| Na (mEq/L), mean (SD), N=55 | 135.13 (4.27) | 134.46 (4.21) | 135.29 (4.61) | 0.375 | 134.56 (4.06) | 135.28 (4.44) | 0.176 |
| k (mEq/L), mean (SD), N=55 | 4.32 (0.55) | 4.41 (0.61) | 4.23 (0.53) | 0.344 | 4.42 (0.57) | 4.26 (0.56) | 0.270 |
| BS (mg/dL), mean (SD), N= 42 | 147.55 (66.79) | 149.4 (67.1) | 149.3 (70.52) | 0.871 | 156.55 (64.07) | 135.37 (60.25) | 0.286 |
| urea (mg/dL), mean (SD), N= 53 | 51.40 (29.6) | 54.46 (28.05) | 49.26 (33.40) | 0.218 | 59.81 (36.47) | 40.79 (12.90) | 0.034 |
| Creatinine (mg/dL), mean (SD), N= 53 | 1.04 (0.45) | 1.08 (0.54) | 1.02 (0.39) | 0.832 | 1.07 (0.43) | 0.92 (0.28) | 0.216 |
| cTnI (pg/ml), mean (SD), N= 26 | 23.88 (54.46) | 36.79 (75.80) | 15.25 (26.33) | 1.000 | 17.05 (29.87) | 38.58 (84.82) | 0.559 |
| Ferritin, mean (SD), N=17 | 540.32 (3.33) | 623.15 (400.38) | 397.77 (242.14) | 0.281 | 448.52 (409.11) | 599.67 (250.93) | 0.328 |
| PH, mean (SD), N=49 | 7.38 (0.05) | 7.37 (0.05) | 7.40 (0.05) | 0.039 | 7.38 (0.05) | 7.39 (0.04) | 0.295 |
| HCO3, mean (SD), N=49 | 25.26 (3.62) | 25.5 (4.39) | 24.95 (2.93) | 0.233 | 24.62 (3.81) | 26.64 (2.62) | 0.033 |
| D-dimer ng/ml, mean (SD), N= 45 | 1535.44 (1435.48) | 1691.7 (1743.04) | 1367.8 (1159.92) | 0.751 | 1872.78 (1665.25) | 1026.60 (821.31) | 0.147 |
1. Table 2. Data are mean (SD) or n/N (%), where N is the number of patients with available data. p values were calculated by non-parametric tests. Abbreviations: RBC : red blood cells; HCT : hematocrit; MCV : mean corpuscular volume; MCH : mean corpuscular hemoglobin; MCHC :mean corpuscular hemoglobin concentration; PLT: platelets; WBC : white blood cells ; RDW-CV- Red Cell Distribution Width; PDW: Platelet Distribution Width; APTT: activated partial thromboplastin time; PT: prothrombin time; ESR: erythrocyte sedimentation rate; CRP: C-reactive protein; ALT: alanine aminotransferase; AST: aspartate aminotransferase; LDH: lactate dehydrogenase; BS: blood sugar; cTnI: cardiac troponin.
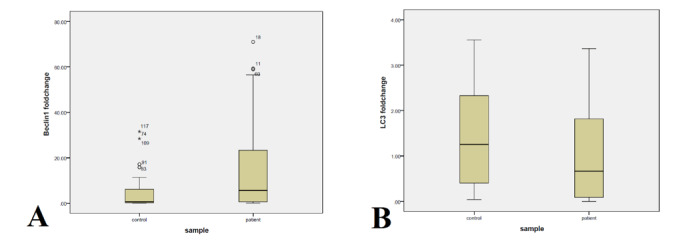
Figure 1.
Boxplot graphs of Beclin1 and LC3 expression in COVID-19 patients and controls. Beclin1 expression (A) is significantly higher in patients comparing to controls (p=0.001), while the expression of LC3 (B) shows significantly lower levels in COVID-19 patients (p=0.007).
LC3 and Beclin1 expression correlation with patient characteristics and outcome
The comparison between LC3 and BECN1 expression and patient characteristics is shown in Table 2. No significant relationship was seen between the expression level of LC3 or BECN1 and patients' outcomes. The Becline1Low group showed a higher RBC count than BECN1High group (p=0.036). In addition, these patients showed significantly higher Urea and lowered HCO3 levels (p=0.034 and 0.033, respectively). Furthermore, the BECN1Low group indicated higher mean levels of PLT, PTT, BS, D-dimer and ALT and lower mean levels of LDH and Lymphocytes. However, the differences were not statistically significant. The patients in LC3Low group showed significantly lower MCH, MCHC and PH levels compared to the others (p= 0.026, p= 0.008, p=0.039, respectively) (Table 2). This group of patients also indicated higher levels of Neutrophils, ALT, AST, LDH, CRP, Urea, D-dimer and Troponin and lower mean levels of PTT than those in LC3High group, but the differences were not statistically significant.
Discussion
SARS-CoV-2, the etiology of the current pandemic, with no concurrent specific and effective therapy, deserves urgent attention in order to elucidate its replication mechanisms and pathogenesis. It has been claimed that coronaviruses might replicate their genomes by hijacking the specific component(s) of autophagy in host cells (15, 18). Autophagy is an important mechanism in physiological homeostasis, which is also encountered in cancer pathogenesis as well as defense against intracellular pathogens, including viruses. In addition, it has been shown that the autophagy pathway is employed by some viruses, including coronaviruses, during replication (19). In this study, we assessed the expressions of LC3 and BECN1, two key elements of the autophagy pathway, in the nasopharyngeal samples of COVID-19 patients and healthy controls and found significant differences in their expressions between the patient group and controls.
BECN1 is an essential regulatory component of autophagy that controls the autophagy process at the nucleation stage, and its deregulation is associated with some diseases, including cancer, diabetes, and kidney diseases (20, 21). We found that BECN1 gene expression was significantly higher in COVID-19 patients compared to the control group and upper expression of BECN1 was seen in 57% of patients. In line with our results, Okuyan K et al. found that the Serum BECN1 levels in COVID-19 patients were higher than the healthy controls (22). As the coronaviruses use autophagy machinery during their replication (23), it is expected that BECN1 an autophagy-related molecule, is expressed in virus-infected tissues, as found in our study; and its serum level is expectedly derived from its diffusion from involved tissues. So, the results of Okuyan K et al. study confirm our results, and regarding the findings of these two studies, it is speculated that BECN1 may have a role in the pathogenesis of COVID-19. One familiar method to identify the role of a molecule in a cellular process is studying the effects of its inhibition or deletion on the cells. The next two studies evaluated the effects of inhibition and deletion of BECN1 on virus-infected cells. In a study by Yuen CK et al., human lung cells were cultured in vitro and were infected with COVID-19. Then the researchers inhibited four stages of autophagy with specific inhibitory drugs. They showed that the second group of inhibitors (class III PI3-kinase inhibitor VPS34-IN1), which target the nucleation stage of autophagy (ATG14/Beclin1/ VPS34) prevent the formation of autophagolysosome and were able to effectively inhibit the viral replication. Therefore, they suggested that these inhibitors, which target Beclin1, could be a potential target for COVID‐19 therapy (15). Furthermore, Chen X et al. showed that membrane-associated papain-like protease PLP2 (PLP2-TM) of coronaviruses induces the incomplete autophagy process. They found that PLP2-TM interacts with BECN1 and LC3. The researchers also showed that knockdown of Beclin1 results in decreased coronavirus replication (24). These studies were performed in vitro condition and emphasize the important role of BECN1 in the replication of coronaviruses including SARS-COV2. In the current study, using patient samples we showed that the expression of BECN1 was significantly higher in COVID-19 patients, which further accentuates the in vivo role of BECN1 in the pathogenesis of COVID-19. So, the results of these studies are confirmatory and complementary to each other. Regarding the findings of mentioned studies, the use of BECN1 inhibitors for the treatment of COVID-19 patients may be beneficial. Further studies as well as clinical trials, are needed to confirm this claim.
In contrast to our findings, Gassen NC et al. identified that S-phase kinase-associated protein 2 (SKP2), has an essential role in BECN1degradation, and inhibition of SKP2 reduces the MERS-COV infection. They observed that inhibition of SKP2, decreases the ubiquitination and degradation of BECN1 and improves autophagy flux which leads to a 28,000-fold decrease in the proliferation of MERS-COV (25). In addition, the researchers suggested that SKP2 and BECN1 also may be used as appropriate therapies. The results of Gassen NC et al.'s study are in contrast to the previously mentioned studies, including our study that suggest the inhibition of BECN1 may be a therapeutic approach for SARS-COV2 infection. As the Gassen NC et al. study was done on MERS Coronavirus, these differences may be explained. As reported by the last-mentioned study, activation of the autophagy decreases the MERS coronavirus proliferation. So, the antiviral effect of autophagy may be more pronounced during the MERS coronavirus infection. In contrast, the replication of SARS-CoV2 is decreased by autophagy inhibition, which emphasizes the usage of autophagy machinery by the virus during its replication. So, the two viruses may differ in using autophagy machinery during their replication.
LC3 is another major element of autophagy. LC3 is needed for the replication of some coronaviruses and its down-regulation has been shown to protect cells from infection by some coronaviruses including the mouse hepatitis virus process (16, 26). Regarding to this, we had expected that LC3 would be increased in the nasopharyngeal samples of the patients, where the virus is actively replicated. Our findings in the current study showed that the expression of LC3 gene was significantly lower in COVID-19 patients compared to healthy controls and down-regulation of LC3 was observed in 27 (44%) of patients. To the best of our knowledge LC3 expression has not been evaluated in SARS-COV2 previously and further studies are needed to confirm our results and elucidate the role of LC3 in the pathogenesis of COVID-19. Regarding another coronaviruses, some studies have evaluated the expression or role of LC3 in their pathogenesis. In a study by Regiori F et al., which was done on a Coronaviruses named mouse hepatitis virus, it was demonstrated that LC3-I-Positive EDEMosomes are hijacked by this coronavirus for its replication. They reported that down-regulation of LC3, protects cells from Coronavirus infection (16). These results are in contrast with decreased expression of LC3 in COVID-19 patients in our study. The Regiori F et al. study was done on mouse hepatitis virus which is a murine coronavirus. So, the differences may be attributable to the fact that murine coronavirus differently uses autophagy machinery, in contrast to human coronaviruses. Further studies are needed to elucidate the role of LC3 in the replication process of SARS-CoV2.
One indirect way to evaluate the effect of these molecules on the disease process maybe by assessing their correlation with disease severity. If the changes in the expression of these molecules were more pronounced in patients with more severe diseases, it would be another proof of their important role in the pathogenesis of COVID-19. Several studies have investigated different clinical and laboratory indicators regarding the severity of disease in COVID-19 patients (27-29). According to the previous studies, the blood levels of some hematological and biochemical markers such as RBC (30), Urea (31), D-dimer (32) and so on, were associated with the severity of COVID-19 disease and now are used in clinical practice for assessing the severity and prognosis of COVID-19 Patients. We assume that if a significant relationship is seen between changes in the expression of BECN1 and LC3 and blood factors, the expression of these genes may be indirectly related to the severity of the COVID-19 disease. Okuyan K et al. investigated the relationship between BECN1 blood levels with laboratory tests and the clinical severity in 108 patients with COVID-19. They found that Serum BECN1 levels were higher in COVID-19 patients compared to healthy controls. In addition, dividing the patients into the severe and non-severe groups, showed that serum BECN1 was significantly higher in the severe group (p<0.001). Furthermore, they reported a significant positive correlation between BECN1 and D-dimer, CRP, urea and LDH (p<0.001) (22). In the current study laboratory findings and demographic information of the patients showed no association with the expression levels of BECN1or LC3 in most cases. However, we found that patients with lower BECN1 expression had significantly higher RBC, higher urea levels and lower HCO3 levels (p=0.036, p=0.034. and p=0.033, respectively). The differences between the two groups in RBC counts and HCO3 seem to be clinically insignificant because the mean blood levels of both groups fall within normal limits. Although these findings are not in line with Okuyan K et al. study, regarding the usage of nasopharyngeal samples in our study, these differences can be explained. As the virus replication at the nasopharyngeal mucosa occurs early in the course of the disease, the patients who are admitted later after the onset of their symptoms may show lower virus replication, and so lower BECN1, in their nasopharynx, while they may have advanced disease progression in other body areas including lungs. This may explain the differences between BECN1 nasopharyngeal expression and BECN1 serum level regarding the severity of the disease and laboratory indicators of disease severity. Further studies with larger sample sizes are needed to elucidate the association between BECN1 as well as LC3 expressions and the severity of COVID-19.
Conclusion
To the best of our knowledge, this is the first study to examine the expression of BECN1 and LC3 in the nasopharyngeal samples of COVID-19 patients. The results indicated a significant difference in the expression of these autophagy-related molecules between the COVID-19 patients and healthy controls. According to these findings, BECN1 and LC3 may be potential therapeutic targets for the development of new drugs for the treatment of COVID-19 patients. Further studies are needed to confirm this claim.
Conflict of Interests
The authors declare that they have no competing interests.
Acknowledgment
We gratefully acknowledge our colleagues from the central laboratory of Imam Reza as well as Ghaem University Hospital, Mashhad, Iran for their help and support. The results described in this article were part of an MSc dissertation. This study was supported by a grant from Mashhad University of Medical Sciences (Pazhoohan ID: 991820).
Ethical Approval
The study protocol was approved by the Research Ethics Committee at Mashhad University of Medical Sciences (IR.MUMS.REC.1399.648).
Cite this article as : Boroumand-Noughabi S, Khoshnegah Z Amel Jamehdar S, Ayatollahi H, Sheikhi M, Rostami M, Keramati MR. Deregulation of the Expression of Beclin1 and Light Chain 3(LC3), Autophagy-Related Genes, in COVID-19 Patients. Med J Islam Repub Iran. 2022 (31 Aug);36:99. https://doi.org/10.47176/mjiri.36.99
References
- 1.Wang L, Wang Y, Ye D, Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents. 2020:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed]
- 2.WHO. Weekly Epidemiological and Operational. 2021 2021 [updated 22,Oct,2021. Available from: https://covid19
- 3.Ahmad L, Mostowy S, Sancho-Shimizu V. Autophagy-virus interplay: from cell biology to human disease. Front Cell Dev Biol. 2018:155 doi: 10.3389/fcell.2018.00155. [DOI] [PMC free article] [PubMed]
- 4.Mao J, Lin E, He L, Yu J, Tan P, Zhou Y. Autophagy and Viral Infection. Adv Exp Med Biol. 2019 doi: 10.1007/978-981-15-0606-2_5. [DOI] [PMC free article] [PubMed]
- 5.Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J. et al. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011:6319. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed]
- 6.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008:286. doi: 10.4161/auto.5377. [DOI] [PubMed]
- 7.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ. et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science (New York, NY) 2008:921. doi: 10.1126/science.1152725. [DOI] [PubMed]
- 8.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009:14046. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed]
- 9.Benvenuto D, Angeletti S, Giovanetti M, Bianchi M, Pascarella S, Cauda R. et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J Infect. 2020:e24. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed]
- 10.Guo L, Yu H, Gu W, Luo X, Li R, Zhang J. et al. Autophagy Negatively Regulates Transmissible Gastroenteritis Virus Replication. Sci Rep. 2016 doi: 10.1038/srep23864. [DOI] [PMC free article] [PubMed]
- 11.Maier HJ, Britton P. Involvement of autophagy in coronavirus replication. Viruses. 2012:3440. doi: 10.3390/v4123440. [DOI] [PMC free article] [PubMed]
- 12.Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol Biol (Clifton, NJ) 2008 doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed]
- 13.Gorshkov K, Chen CZ, Bostwick R, Rasmussen L, Xu M, Pradhan M. et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv: the preprint server for biology. 2020
- 14.Wirawan E, Vanden Berghe, Lippens S, Agostinis P, Vandenabeele P. Autophagy: for better or for worse. Cell Res. 2012:43. doi: 10.1038/cr.2011.152. [DOI] [PMC free article] [PubMed]
- 15.Yuen CK, Wong WM, Mak LF, Wang X, Chu H, Yuen KY. et al. Suppression of SARS-CoV-2 infection in ex-vivo human lung tissues by targeting class III phosphoinositide 3-kinase. J Med Virol. 2021:2076. doi: 10.1002/jmv.26583. [DOI] [PMC free article] [PubMed]
- 16.Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S. et al. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010:500. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed]
- 17.García-Pérez BE, González-Rojas JA, Salazar MI, Torres-Torres C, Castrejón-Jiménez NS. Taming the Autophagy as a Strategy for Treating COVID-19. Cells. 2020 doi: 10.3390/cells9122679. [DOI] [PMC free article] [PubMed]
- 18.Proikas-Cezanne T, Takacs Z, Dönnes P, Kohlbacher O. WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci. 2015:207. doi: 10.1242/jcs.146258. [DOI] [PubMed]
- 19.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008:27. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed]
- 20.Naguib M, Rashed LA. Serum level of the autophagy biomarker Beclin-1 in patients with diabetic kidney disease. Diabet Res Clin Pract. 2018 doi: 10.1016/j.diabres.2018.06.022. [DOI] [PubMed]
- 21.Fu LL, Cheng Y, Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int J Biochem Cell Biol. 2013:921. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed]
- 22.Okuyan HM, Dogan S, Bal T, Çabalak M. Beclin-1, an autophagy-related protein, is associated with the disease severity of COVID-19. Life Sci. 2021 doi: 10.1016/j.lfs.2021.119596. [DOI] [PMC free article] [PubMed]
- 23.Kliche J, Kuss H, Ali M, Ivarsson Y. Cytoplasmic short linear motifs in ACE2 and integrin β(3) link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci Signal. 2021 doi: 10.1126/scisignal.abf1117. [DOI] [PMC free article] [PubMed]
- 24.Chen X, Wang K, Xing Y, Tu J, Yang X, Zhao Q. et al. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014:912. doi: 10.1007/s13238-014-0104-6. [DOI] [PMC free article] [PubMed]
- 25.Gassen NC, Niemeyer D, Muth D, Corman VM, Martinelli S, Gassen A. et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat Commun. 2019:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed]
- 26.Monastyrska I, Ulasli M, Rottier PJ, Guan JL, Reggiori F, de Haan. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013:164. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed]
- 27.Hu X, Hu C, Yang Y, Chen J, Zhong P, Wen Y. et al. Clinical characteristics and risk factors for severity of COVID-19 outside Wuhan: a double-center retrospective cohort study of 213 cases in Hunan, China. Ther Adv Respir Dis. 2020 doi: 10.1177/1753466620963035. [DOI] [PMC free article] [PubMed]
- 28.Ghahramani S, Tabrizi R, Lankarani KB, Kashani SMA, Rezaei S, Zeidi N. et al. Laboratory features of severe vs. non-severe COVID-19 patients in Asian populations: a systematic review and meta-analysis. Eur J Med Res doi: 10.1186/s40001-020-00432-3. [DOI] [PMC free article] [PubMed]
- 29.Marcos M, Belhassen-García M, Sánchez-Puente A, Sampedro-Gomez J, Azibeiro R, Dorado-Díaz PI. et al. Development of a severity of disease score and classification model by machine learning for hospitalized COVID-19 patients. PloS One. 2021:e0240200. doi: 10.1371/journal.pone.0240200. [DOI] [PMC free article] [PubMed]
- 30.Bergamaschi G, Borrelli de, Aronico N, Lenti MV, Barteselli C, Merli S. et al. Anemia in patients with Covid-19: pathogenesis and clinical significance. Clin Exp Med. 2021:239. doi: 10.1007/s10238-020-00679-4. [DOI] [PMC free article] [PubMed]
- 31.Izcovich A, Ragusa MA, Tortosa F, Lavena Marzio, Agnoletti C, Bengolea A. et al. Prognostic factors for severity and mortality in patients infected with COVID-19: A systematic review. PloS One. 2020:e0241955. doi: 10.1371/journal.pone.0241955. [DOI] [PMC free article] [PubMed]
- 32.Li Y, Zhao K, Wei H, Chen W, Wang W, Jia L. et al. Dynamic relationship between D-dimer and COVID-19 severity. Br J Haematol. 2020:e24. doi: 10.1111/bjh.16811. [DOI] [PMC free article] [PubMed]


