Abstract
Background
Circular RNAs (circRNAs) appear to be important modulators in ovarian cancer. We aimed to explore the role and mechanism of circ_0025033 in ovarian cancer.
Methods
qRT-PCR was conducted to determine circ_0025033, hsa_miR-370-3p, and SLC1A5 mRNA expression. Functional experiments were conducted, including Cell Counting Kit-8 (CCK-8), 5-ethynyl-2′-deoxyuridine (EdU), flow cytometry, transwell, tube formation, xenograft tumor model assay, western blot analysis of protein levels, and analysis of glutamine metabolism using commercial kits. Their predicted interaction was confirmed using dual-luciferase reporter and RNA pull-down.
Results
circ_0025033 was upregulated in ovarian cancer; its knockdown induced proliferation, invasion, angiogenesis, glutamine metabolism, and apoptosis in vitro, and blocked tumor growth in vivo. circ_0025033 regulated ovarian cancer cellular behaviors via sponging hsa_miR-370-3p. In parallel, SLC1A5 might abolish the anti-ovarian cancer role of hsa_miR-370-3p. Furthermore, circ_0025033 affected SLC1A5 via regulating hsa_miR-370-3p.
Conclusion
circ_0025033 might promote ovarian cancer progression via hsa_miR-370-3p/SLC1A5, providing an interesting insight into ovarian cancer tumorigenesis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s11658-022-00364-2.
Keywords: Ovarian cancer, circ_0025033, hsa_miR-370-3p, SLC1A5
Highlights
circ_0025033 knockdown inhibited ovarian cancer malignant behaviors.
circ_0025033 served as a hsa_miR-370-3p sponge.
hsa_miR-370-3p targeted SLC1A5.
Supplementary Information
The online version contains supplementary material available at 10.1186/s11658-022-00364-2.
Introduction
Ovarian cancer, a common gynecological malignancy, is considered to be a global health issue correlated with increased morbidity and mortality [1]. Many patients with ovarian cancer are not diagnosed until they reach an advanced stage because early lesions are not easy to detect [2]. Although tremendous efforts have been made in ovarian cancer treatment, the 5-year overall survival rate of patients with ovarian cancer is between 35% and 40% [3]. Hence, elucidating the molecular mechanism involved in ovarian cancer is crucial for discovering effective therapeutic targets.
Unlike linear RNAs, circular RNAs (circRNAs) have special covalently closed loop structures [4]. Widely expressed in the cytoplasm of eukaryotic cells, they often exert a role in specific patterns of tissue and developmental stages [5]. circRNAs are becoming attractive biomarkers of human diseases owing to their abundance and stability [6, 7]. Emerging evidence has revealed that dysregulated circRNAs are implicated in cancer initiation and development in a wide range of tumors [8–10]. Apart from that, some circRNAs participate in ovarian cancer processes by acting as tumor suppressors or promoters [11–13]. circ_0025033 is produced by the back-splicing of its parental forkhead box M1 (FOXM1) gene (located at chr12: 2966846–2983691), whose spliced mature sequence length is 3410 bp. FOXM1 is an essential transcription regulator that might modulate multiple aspects of tumor progression [14, 15]. It has been confirmed that the downregulation of FOXM1 could effectively hinder the proliferation, migration, and invasion of ovarian cancer cells in vitro [16, 17]. A previous report indicated that circ_0025033 upregulation might boost ovarian cancer development [18]. Yet, its function and mechanism remain largely unknown in ovarian cancer.
Research in the past decades has shown that circRNAs exert their functions via competitive endogenous RNAs (ceRNAs) through binding with miRNA response elements (MREs), thereby de-repressing target mRNA expression [19, 20]. As another type of ncRNA, miRNAs might achieve the regulation of target gene via binding to their 3′ untranslated region (UTR) [21]. miRNAs as anti-oncogenes or oncogenes regulate cellular biological activities in cancer progression [22–24]. A previous report showed that has_hsa_miR-370-3p could inhibit metastatic ability in ovarian cancer cells [25]. Moreover, a recent study indicated that SLC1A5 (also called ASCT2) plays a promoter role in ovarian cancer [26]. Here, by applying bioinformatics tools, we revealed that hsa_miR-370-3p possesses binding sites with circ_0025033 and SLC1A5. Hence, we further explored whether the regulatory impact of circ_0025033 ovarian cancer development is mediated via hsa_miR-370-3p–SLC1A5.
Materials and methods
Specimen collection
After obtaining informed consent, ovarian cancer tissue samples (n = 29) along with matched adjacent normal samples were harvested from sufferers of ovarian cancer at First Affiliated Hospital of Xi'an Jiaotong University. This research had acquired approval from the ethics committee of First Affiliated Hospital of Xi'an Jiaotong University.
Cell culture and transfection
Stored under standard conditions (37 ℃; 5% CO2) in RPMI-1640 medium, human ovarian surface epithelial cells (HOSEPiC) cells (cat. no. #7310) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA). Two ovarian cancer cell lines (HEY; cat. no. CL-0671, OVCAR3; cat. no. CL-0178) were supplied by Procell (Wuhan, China), and two other cell lines (SKOV3; cat. no. BNCC338639, A2780; cat. no. BNCC351906) were obtained from BeNa Culture Collection (Beijing, China). Human umbilical vein endothelial cells (HUVECs; Procell) were grown in HUVEC-specific complete medium (Procell).
RiboBio (Guangzhou, China) provided circ_0025033 small interfering RNA (si-circ_0025033), circ_0025033-overexpressing RNA (circ_0025033), hsa_miR-370-3p mimic/inhibitor (hsa_miR-370-3p/anti-hsa_miR-370-3p), siRNA against SLC1A5 (si-SLC1A5), SLC1A5-overexpressing RNA (SLC1A5), and controls (si-NC, pCD5-ciR, miR-NC, anti-miR-NC, si-con, and pcDNA), followed by Lipofectamine 3000 reagent treatment.
Immunohistochemistry (IHC) analysis
After being fixed and embedded, tumor samples were cut into slices of 5 μm thickness. Then, Ki67 (ab15580; 1:200), SLC1A5 (ab237704; 1:500), c-Myc (ab32072; 1:200), or MMP9 (ab283575; 1:1000) at 4 ℃ were reacted with these sections overnight, which were further incubated with secondary antibody (ab205718; 1:2000). Finally, immunostaining images were obtained by microscope (Leica, Wetzlar, Germany). All antibodies were provided by Abcam (Cambridge, UK).
qRT-PCR
Using TRIzol reagent (Invitrogen), the generated total RNA was reverse transcribed according to PrimeScript RT Reagent Kit. An miRNA reverse-transcription PCR kit was used to reverse transcribe has_mR-370-3p. Subsequently, cDNA amplification was implemented according to SYBR Green Master Mix (Roche, Shanghai, China) on CFX96 PCR equipment. After GAPDH or U6 normalization, the gene levels were evaluated via the 2−ΔΔCt method. The primer sequences are listed in Table 1.
Table 1.
The sequences of primers for RT-qPCR used in this study
| Name | Sequence (5′–3′) |
|---|---|
| circ_0025033: forward | GGTGTGAGCCAGCTTGAGA |
| circ_0025033: reverse | GACGGGGGCTAGTTTTCATT |
| FOXM1: forward | TCTGCCAATGGCAAGGTCTCCT |
| FOXM1: reverse | CTGGATTCGGTCGTTTCTGCTG |
| hsa_miR-370-3p: forward | GTATGAGCCTGCTGGGGTGG |
| hsa_miR-370-3p: reverse | CAGTGCGTGTCGTGGAGT |
| SLC1A5: forward | TCCTCTTCACCCGCAAAAACCC |
| SLC1A5: reverse | CCACGCCATTATTCTCCTCCAC |
| U6: forward | CTCGCTTCGGCAGCACATATACT |
| U6: reverse | ACGCTTCACGAATTTGCGTGTC |
| GAPDH: forward | CTGACTTCAACAGCGACACC |
| GAPDH: reverse | TGCTGTAGCCAAATTCGTTGT |
In addition, to validate the circular structure of this circRNA, the RNAs generated at 37 ℃ were reacted with RNase R (Seebio, Shanghai, China). Finally, RNA expression levels were assessed with qRT-PCR. Meanwhile, to check the distribution of circ_0025033 in ovarian cancer cells, the RNA from the nuclear and cytoplasmic fractions was distinguished using PARIS Kit (Invitrogen), followed by qRT-PCR analysis.
Cell proliferation assays
After 48 h of transfection, we seeded SKOV3 and A2780 cells (5 × 103 cells per well) into 96-well plates. After incubation for 24 h, Cell Counting Kit-8 (CCK-8) solution (10 μL; Beyotime, Jiangsu, China) was added to each well, followed by analysis via microplate reader.
After 48 h of transfection, 5-ethynyl-2′-deoxyuridine (EdU) assay was conducted, where tumor cells were cultured at 2 × 104 cells per well. At 24 h post-incubation, EdU solution and paraformaldehyde (4%) were mixed with the cells into each well, which were next incubated with DAPI and analyzed using a microscope.
Flow cytometry analysis
Annexin V-FITC and PI apoptosis detection kit purchased from Yeasen (Shanghai, China) detected apoptotic cells. After 48 h of transfection, we seeded SKOV3 and A2780 cells (2 × 105 cells per well) into a six-well plate. After labeling with annexin V-FITC and PI, the solution was placed in a flow cytometer for analysis.
Transwell assay
After 48 h of transfection, SKOV3 and A2780 cell suspension was introduced into the top chamber (24-well; Costar, Corning, NY, USA) precoated with Matrigel, while the bottom counterpart contained complete medium. Cells remaining bottom were fixed and stained after 24 h, and invasion pictures were obtained using a microscope (×100; Leica).
Tube formation assay
Angiogenesis capability was assessed by tube formation assay. In brief, when transfected cells (SKOV3 and A2780) reached 80% confluence, the supernatant was collected as the conditioned medium. Twenty-four-well dishes were coated with Matrigel in each well at 37 ℃ to polymerize. Next, HUVECs were seeded into Matrigel-coated wells under different conditioned media for 6 h. Finally, results were analyzed under a microscope and using ImageJ.
Western blot assay
Total protein was extracted using RIPA lysis buffer (Solarbio, Beijing, China). After quantification of total protein using BCA protein assay kit (Solarbio), protein samples were loaded onto SDS–PAGE prior to being immunoblotted onto PVDF membranes (Millipore, Billerica, MA, USA). After incubation with primary antibodies, these membranes were incubated for 2 h with a corresponding secondary antibody (ab205718; 1:5000; Abcam). The combined signals were analyzed using enhanced chemiluminescence (ECL) (Vazyme, Nanjing, China). The primary antibodies were purchased from Abcam: SLC1A5 (ab237704; 1:1000), c-Myc (ab32072; 1:200), MMP9 (ab76003; 1:1000), and β-actin (1:2500; ab8227).
Measurement of glutamine metabolism
According to the manufacturer’s protocols, glutamine consumption, α-ketoglutarate production, and glutamate production were determined according to glutamine assay, α-ketoglutarate assay, and glutamate assay kits (Abcam), respectively.
Dual-luciferase reporter assay
These fragments of circ_0025033 or 3′ UTR of SLC1A5 with or without putative binding sites of hsa_miR-370-3p were introduced via pmirGLO vector (YouBia, Changsha, China), generating WT/MUT-circ_0025033 and WT/MUT-SLC1A5 3′ UTR. Then, SKOV3 and A2780 cells were transfected with hsa_miR-370-3p/miR-NC and reporter vectors, followed by analysis using dual-luciferase reporter gene assay kit.
RNA pull-down assay
After being transfected with biotinylated (bio)-hsa_miR-370-3p or miR-NC (GenePharma, Shanghai, China), harvested cells were lysed, followed by reaction with M-280 streptavidin (Invitrogen). Subsequently, beads were mixed with the biotinylated hsa_miR-370-3p for 10 min and analyzed via qRT-PCR.
Tumor formation assay in vivo
Twelve 5-week-old BALB/c nude mice (female; Vital River, Beijing, China) were separated into two groups, followed by subcutaneous inoculation with A2780 cells with sh-circ_0025033 or sh-NC (RiboBio). Tumor volume was measured. After inoculation for 23 days, the excised tumors from these sacrificed mice were weighed and studied. Permission to perform this experiment was provided by the Animal Care and Use Committee of First Affiliated Hospital of Xi'an Jiaotong University.
Statistical analysis
GraphPad Prism 7.0 software was used to process all data in this work, presented as mean ± standard deviation. P-value below 0.05 was considered statistically significant. Student’s t-test or one-way analysis of variance (ANOVA) was adopted for comparisons. Survival curve was analyzed by Kaplan–Meier method. Pearson’s correlation coefficient was used to determine correlations in expression.
Results
circ_0025033 was enhanced in ovarian cancer
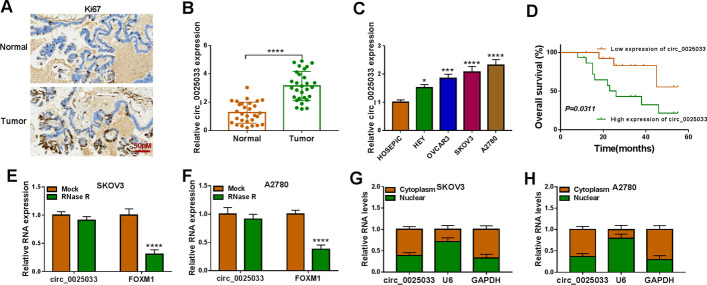
IHC analysis revealed higher Ki67 content in tumor tissue (Fig. 1A). circ_0025033 content was increased in ovarian cancer tissue and cells (HEY, OVCAR3, SKOV3, and A2780) (Fig. 1B and C). Among these ovarian cancer cells, circ_0025033 content was higher in SKOV3 and A2780 cells, so these two cell lines were selected for further analysis. Moreover, high level of circ_0025033 was predictive of poor prognosis in sufferers of ovarian cancer (Fig. 1D). In addition, linear FOXM1 mRNA was degraded by RNase R, but there was no change in circ_0025033 level (Fig. 1E and F). Localization of circ_0025033 in tumor cells was determined. Figure 1G and H shows that circ_0011298 was prominently located in tumor cell cytoplasm. Taken together, the findings show that circ_0025033 was upregulated in ovarian cancer.
Fig. 1.
Overexpression of circ_0025033 in ovarian cancer. A IHC analysis detecting Ki67 content in tumor tissue. B and C qRT-PCR analysis of circ_0025033 content in tissue samples, HOSEPiC cells, and ovarian cancer cells (HEY, OVCAR3, SKOV3, and A2780). D Kaplan–Meier curves exhibiting survival rate of sufferers of ovarian cancer with high or low level of circ_0025033. E and F Expression of circ_0025033 and FOXM1 determined via qRT-PCR after treatment of RNase R. G and H qRT-PCR analysis of circ_0025033 localization in tumor cells. *P < 0.05, ***P < 0.001, ****P < 0.0001
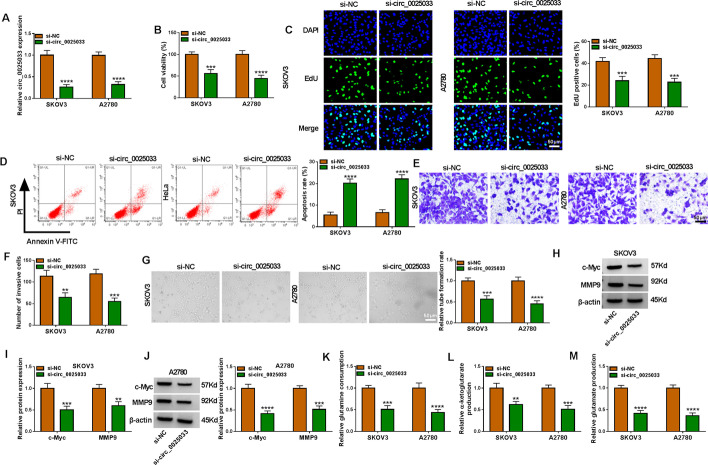
circ_0025033 absence inhibits ovarian cancer cell development
As expected, circ_0025033 content was diminished in tumor cells via si-circ_0025033 (Fig. 2A). Functionally, circ_0025033 silencing reduced cell viability and DNA synthesis in SKOV3 and A2780 cells (Fig. 2B and C). As shown in Fig. 2D, SKOV3 and A2780 cell apoptosis was increased after circ_0025033 downregulation. Meanwhile, circ_0025033 silencing blocked tumor cell invasion (Fig. 2E and F). Angiogenesis is required for tumor growth and metastasis. Tube formation assay showed that circ_0025033 interference decreased angiogenesis (Fig. 2G). Next, circ_0025033 deficiency reduced levels of proliferation/metastasis-related proteins (c-Myc and MMP9) (Fig. 2H–J). Glutamine, a non-essential amino acid, can be converted into glutamate and then transformed into α-ketoglutarate, which is involved in the tricarboxylic acid cycle to provide energy for cells [27, 28]. Glutamine metabolism is indispensable for tumor development [29]. We found that circ_0025033 silencing reduced glutamine consumption, α-ketoglutarate production, and glutamate production (Fig. 2K–M), suggesting that circ_0025033 downregulation repressed glutamine metabolism. Together, circ_0025033 absence alleviated tumor cell malignancy glutamine metabolism.
Fig. 2.
circ_0025033 knockdown hampered the malignant behavior of ovarian cancer cells in glutamine metabolism. A–M SKOV3 and A2780 cells were transfected with si-NC or si-circ_0025033. A qRT-PCR analysis of circ_0025033. B and C CCK-8 and EdU analysis of cell viability and DNA synthesis. D Apoptosis rate analyzed by flow cytometry analysis. E and F Cell invasion measured by transwell assay. G Tube formation assay for evaluating angiogenesis activity. H–J Western blot analysis of c-Myc and MMP9 levels. K–M Glutamine consumption, α-ketoglutarate production, and glutamate production measured via glutamine, α-ketoglutarate, and glutamate assay kits, respectively. **P < 0.01, ***P < 0.001, ****P < 0.0001
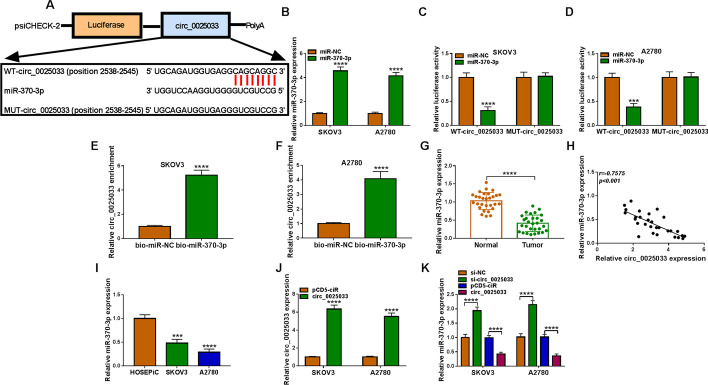
circ_0025033 directly interacted with hsa_miR-370-3p
It has been confirmed that circRNAs could exert their role by interacting with miRNAs [30]. Circinteractome software revealed that circ_0025033 shares binding sites with hsa_miR-370-3p (Fig. 3A), indicating their interaction. Figure 3B shows the overexpression efficiency of hsa_miR-370-3p (Fig. 3B), which exhibited an evident suppression in luciferase activity of WT-circ_0025033, instead of MUT-circ_0025033 (Fig. 3C and D). circ_0025033 was pulled down when using bio-hsa_miR-370-3p rather than bio-miR-NC (Fig. 3E and F). In addition, hsa_miR-370-3p content was downregulated (Fig. 3G), and its level was inversely correlated with circ_0025033 in ovarian cancer tissue (Fig. 3H). Similarly, an obvious decrease of hsa_miR-370-3p in tumor cells was found (Fig. 3I). The significant increase of hsa_miR-370-3p indicated the significant transfection efficiency of pCD-circ_0025033 (Fig. 3J). Next, hsa_miR-370-3p was upregulated via si-circ_0025033, and reduced via circ_0025033 (Fig. 3K), suggesting that circ_0025033 negatively regulates hsa_miR-370-3p expression. Overall, circ_0025033 sequestered hsa_miR-370-3p.
Fig. 3.
hsa_miR-370-3p was a target of circ_0025033. A Circinteractome was used to predict the complementary sequence between hsa_miR-370-3p and circ_0025033. B Transfection efficiency of hsa_miR-370-3p in tumor cells. C and D Their binding was confirmed using dual-luciferase reporter assay in SKOV3 and A2780 cells cotransfected with WT-circ_0025033 or MUT-circ_0025033 and hsa_miR-370-3p or miR-NC. E and F Their interaction was examined using RNA pull-down. G qRT-PCR analysis of hsa_miR-370-3p in tumor tissue. H Their correlation was determined via Pearson’s correlation analysis in tumor samples. I hsa_miR-370-3p content in HOSEPiC, SKOV3, and A2780 cells. J circ_0025033 in cells transfected with pcD5-ciR or circ_0025033 was detected using qRT-PCR. K Effect of circ_0025033 deficiency or overexpression on hsa_miR-370-3p content was examined using qRT-PCR. ***P < 0.001, ****P < 0.0001
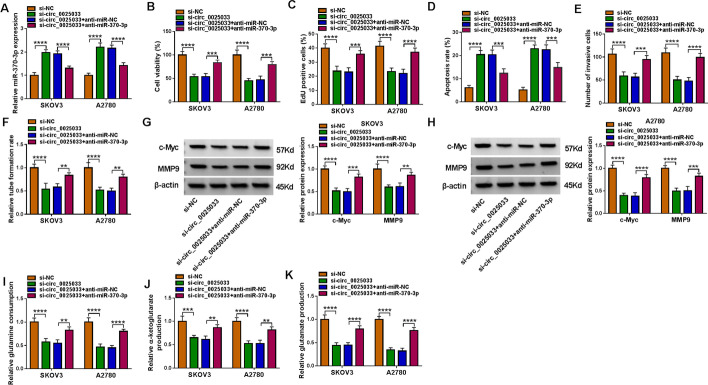
circ_0025033 knockdown restrained tumor cell malignant phenotypes via regulating hsa_miR-370-3p
We found that circ_0025033 deletion promoted hsa_miR-370-3p expression, while anti-hsa_miR-370-3p abated the effect (Fig. 4A). hsa_miR-370-3p absence mitigated circ_0025033 deficiency-mediated tumor cell viability and DNA synthesis inhibition (Fig. 4B and C). Moreover, circ_0025033 knockdown-induced apoptosis was prevented via hsa_miR-370-3p downregulation (Fig. 4D). In addition, circ_0025033 silencing constrained cell invasion and angiogenesis, and hsa_miR-370-3p inhibition reversed the phenomenon (Fig. 4E and F). Meanwhile, hsa_miR-370-3p reduction might abolish downregulation of c-Myc and MMP9 protein levels caused via circ_0025033 absence (Fig. 4G and H). Further, hsa_miR-370-3p downregulation counteracted the si-circ_0025033-caused reduction in glutamine consumption, α-ketoglutarate production, and glutamate production (Fig. 4I–K). Together, circ_0025033 regulated ovarian cancer cell behaviors by targeting hsa_miR-370-3p.
Fig. 4.
circ_0025033 silencing inhibited ovarian cancer cell malignant behaviors via targeting hsa_miR-370-3p. A–K SKOV3 and A2780 cells were transfected with si-NC, si-circ_0025033, si-circ_0025033 + anti-miR-NC, or si-circ_0025033 + anti-hsa_miR-370-3p. A Expression of hsa_miR-370-3p measured by qRT-PCR. B and C Cell proliferation detected using CCK-8 assay and EdU assay. D–F Apoptosis, invasion, and angiogenesis ability assessed using flow cytometry, transwell, and tube formation assays. G and H Western blot analysis of c-Myc and MMP9. I–K Glutamine assay kit, α-ketoglutarate assay kit, and glutamate assay kit were utilized to detect glutamine consumption, α-ketoglutarate production, and glutamate production, respectively. **P < 0.01, ***P < 0.001, ****P < 0.0001
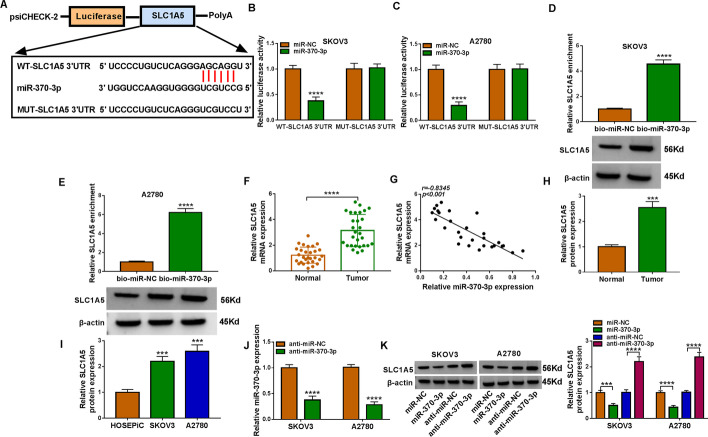
SLC1A5 acted as a direct target of hsa_miR-370-3p
starBase software revealed that hsa_miR-370-3p harbored some complementary binding sites with SLC1A5 3′ UTR (Fig. 5A). hsa_miR-370-3p overexpression strikingly reduced the luciferase activity of WT-SLC1A5 3′ UTR (Fig. 5B and C). A higher enrichment of SLC1A5 was observed in the captured fraction of bio-hsa_miR-370-3p (Fig. 5D and E). Additionally, SLC1A5 content was significantly reduced in ovarian cancer tissue (Fig. 5F), and its mRNA content was negatively correlated with the hsa_miR-370-3p level (Fig. 5G). Furthermore, SLC1A5 protein expression was notably enhanced in ovarian cancer tissue and cells (Fig. 5H and I). Transfection of anti-hsa_miR-370-3p reduced hsa_miR-370-3p expression in SKOV3 and A2780 cells (Fig. 5J). In addition, overexpression of hsa_miR-370-3p decreased SLC1A5 content in tumor cells, and hsa_miR-370-3p absence displayed the opposite effect (Fig. 5K). Taken together, the findings indicate that SLC1A5 was targeted by hsa_miR-370-3p.
Fig. 5.
SLC1A5 was a direct target of hsa_miR-370-3p. A Starbase predicted putative binding sites between hsa_miR-370-3p and SLC1A5. B–E Their interaction was confirmed via dual-luciferase reporter and RNA pull-down assays. F qRT-PCR analysis of SLC1A5 mRNA expression in tumor tissues. G Their relationship was analyzed via Pearson’s correlation coefficient. H and I Western blot analysis of SLC1A5 protein in tissue samples, HOSEPiC cells, SKOV3 cells, and A2780 cells. J Transfection efficiency of anti-hsa_miR-370-3p. K Effect of hsa_miR-370-3p upregulation or knockdown on SLC1A5 content was analyzed via western blot. ***P < 0.001, ****P < 0.0001
SLC1A5 dampened ovarian cancer cell progression
Transfection of si-SLC1A5 reduced SLC1A5 content in tumor cells (Additional file 1: Fig. S1A). Functionally, deletion of SLC1A5 notably repressed proliferation, invasion, and angiogenesis while promoting apoptosis (Additional file 1: Fig. S1B–S1F). Moreover, SLC1A5 knockdown inhibited c-Myc and MMP9 protein expression (Additional file 1: Fig. S1G and S1H). Simultaneously, glutamine consumption, α-ketoglutarate production, and glutamate production were inhibited by downregulation of SLC1A5 in tumor cells (Additional file 1: Fig. S1I-S1K). These data suggested that SLC1A5 might be an oncogene in ovarian cancer.
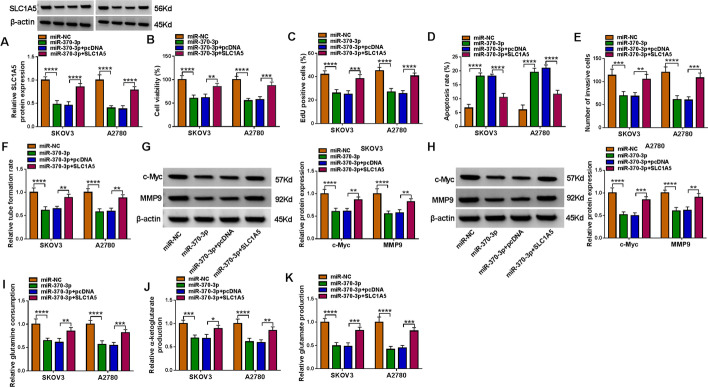
hsa_miR-370-3p targeted SLC1A5
hsa_miR-370-3p overexpression downregulated SLC1A5 protein expression, which was rescued by SLC1A5 upregulation (Fig. 6A). Apart from that, increased hsa_miR-370-3p resulted in a significant suppression in cell proliferation, while increased SLC1A5 reversed these impacts in tumor cells (Fig. 6B and C). Cell apoptosis was induced, and cell invasion and angiogenesis were inhibited, by hsa_miR-370-3p restoration, which were abated by SLC1A5 overexpression (Fig. 6D–F). Enhanced hsa_miR-370-3p reduced the protein levels of c-Myc and MMP9, while the re-introduction of SLC1A5 prevented this reduction (Fig. 6G and H). In addition, glutamine metabolism was decreased by overexpression of hsa_miR-370-3p, which was partly reversed via SLC1A5 enhancement (Fig. 6I–K). Overall, hsa_miR-370-3p inhibited ovarian cancer cell malignant behaviors via targeting SLC1A5.
Fig. 6.
hsa_miR-370-3p negatively regulated SLC1A5. A–K SKOV3 and A2780 cells were transfected with miR-NC, hsa_miR-370-3p, hsa_miR-370-3p + pcDNA, or hsa_miR-370-3p + SLC1A5. A SLC1A5 protein expression was examined using western blot. B and C CCK-8 and EdU assays assessed proliferation ability. D Apoptosis, invasion, and angiogenesis capacity was measured using flow cytometry, transwell, and tube formation assays. G and H c-Myc and MMP9 protein levels were analyzed via western blot. I–K Glutamine metabolism was evaluated via the corresponding assay kits. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
circ_0025033 regulated SLC1A5 expression through sponging hsa_miR-370-3p
As shown in Additional file 2: Fig. S2A and S2B, SLC1A5 content was dramatically downregulated via circ_0025033 absence, and hsa_miR-370-3p interference recovered the SLC1A5 content, supporting the regulatory role of the circ_0025033/hsa_miR-370-3p/SLC1A5 axis.
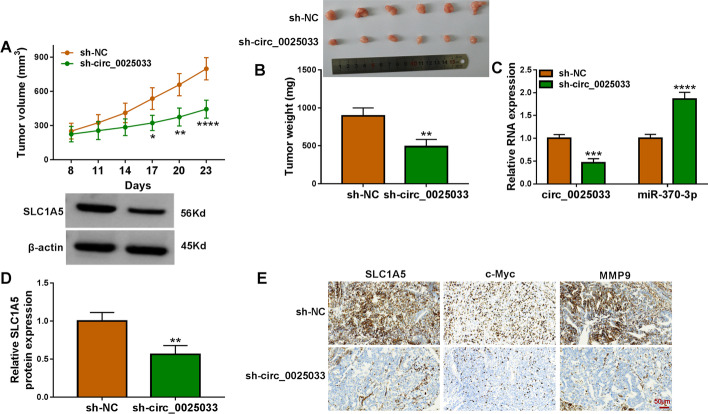
Downregulation of circ_0025033 blocked tumor growth in vivo
Mouse xenograft models of ovarian cancer were established. As shown in Fig. 7A and B, tumor growth was diminished in the sh-circ_0025033 group (Fig. 7A and B). Apart from that, we confirmed that circ_0025033 expression and SLC1A5 protein expression were remarkably reduced in the sh-circ_0025033 group, and the hsa_miR-370-3p level was increased (Fig. 7C and D). IHC analysis showed that circ_0025033 silencing suppressed SLC1A5, c-Myc, and MMP9 (Fig. 7E). Taken together, circ_0025033 knockdown repressed ovarian cancer growth in vivo.
Fig. 7.
circ_0025033 absence repressed tumor growth in vivo. A and B Effects of sh-circ_0025033 on tumor volume and weight. C circ_0025033, hsa_miR-370-3p, and SLC1A5 were examined using qRT-PCR and western blot in tumor tissue. E IHC analysis was used to detect SLC1A5, c-Myc, and MMP9 expression. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Discussion
Patients with ovarian cancer, a gynecologic malignancy, have a short survival time [31]. In this study, circ_0025033 knockdown repressed ovarian cancer cell proliferation, metastasis, angiogenesis, and glutamine metabolism and accelerated apoptosis through the hsa_miR-370-3p/SLC1A5 axis, which is expected to offer a promising treatment strategy for patients with ovarian cancer.
circRNAs have been shown to be stable in general and aberrantly expressed in various diseases [32]. These characteristics make circRNAs potential therapeutic targets or biomarkers for many diseases, especially cancers. Regarding ovarian cancer, high-throughput sequencing has identified abnormal expression of an increasing number of circRNAs [33, 34]. Nevertheless, the majority of circRNAs in ovarian cancer still need further research. circ_0025033 has been shown to promote cell invasion by targeting the miR-1304/miR-1231 axis in papillary thyroid cancer [35]. Moreover, circ_0025033 was upregulated, and its knockdown inhibited ovarian cancer cell viability and metastasis through targeting the miR-330-5p/KLK4 axis [36]. In addition, Hou and Zhang report that circ_0025033 downregulation suppressed colony formation ability, mobility, and glycolysis metabolism in ovarian cancer cells via regulation of the LSM4/miR-184 axis [18]. However, the roles of circ_0025033 in angiogenesis and glutamine metabolism have not been reported. In line with previous research, high circ_0025033 levels in tumor specimens and cells were observed. Moreover, circ_0025033 deficiency limited tumor malignant phenotypes, indicating its promoting effect in ovarian cancer.
Accumulating reports have indicated that circRNAs in the cytoplasm function as miRNA sponges, resulting in changes of target gene expression [23]. In this research, circ_0025033 was predominantly located in the cytoplasm. Hence, circ_0025033 was a hsa_miR-370-3p sponge. Cumulative evidence indicates that hsa_miR-370-3p has a strong ability to modulate tumor development. When hsa_miR-370-3p level is reduced, its increase might inhibit the development of bladder cancer [37], papillary thyroid carcinoma [38], gliomas [39], and acute myeloid leukemia [40]. However, hsa_miR-370-3p expression is enhanced and acts as a tumor-promoting miRNA in gastric carcinoma [41] and breast cancer [42]. In terms of ovarian cancer, hsa_miR-370-3p suppression abated circAGFG1 interference-mediated ovarian cancer cell growth and migration [43]. In addition, hsa_circ_0061140 absence repressed ovarian cancer cell metastasis through sponging miR-370 [44]. Herein, hsa_miR-370-3p showed a low level in ovarian cancer tissue samples and ovarian cancer cells. Rescue assays revealed that suppression of hsa_miR-370-3p counteracted circ_0025033 deficiency-triggered ovarian cancer cell proliferation, apoptosis, metastasis, angiogenesis, and glutamine metabolism inhibition, indicating that circ_0025033 promoted ovarian cancer cell progression via downregulating hsa_miR-370-3p.
Online software Starbase indicated that SLC1A5 may be an hsa_miR-370-3p target. SLC1A5, a glutamine transporter, can control glutamine uptake and is essential for tumor growth [45, 46]. SLC1A5 plays as a vital role in prostate cancer [47], gastric cancer [48], lung cancer [49], and esophageal cancer [50]. Importantly, Huang and her colleagues stated that upregulation of miR-122-5p inhibited ovarian cancer process via targeting SLC1A5 [26]. High SLC1A5 levels were associated with poor prognosis for patients with ovarian cancer [51]. In this research, SLC1A5 silencing inhibited ovarian cancer cell malignant behaviors, indicating a cancer-promoting role of SLC1A5 in ovarian cancer cells. Furthermore, SLC1A5 upregulation could abrogate hsa_miR-370-3p-triggered anti-ovarian cancer. Mechanistically, circ_0025033 could regulate SLC1A5 expression in ovarian cancer cells via binding to hsa_miR-370-3p. Consistently, tumor growth in this research was also suppressed via circ_0025033 knockdown in vivo.
In conclusion, circ_0025033 interference repressed ovarian cancer cell malignant behaviors and glutamine metabolism via the hsa_miR-370-3p/SLC1A5 axis, indicating an underlying therapeutic target for the tumor.
Supplementary Information
Additional file 1: Fig. S1. SLC1A5 and circ_0025033 had similar roles in ovarian cancer. (A-K) SKOV3 and A2780 cells were transfected with si-NC or si-SLC1A5. (A) Western blot analysis of SLC1A5 content. (B-E) Proliferation, apoptosis, and invasion were assessed using CCK-8, EdU, and flow cytometry assays, respectively. (F) Angiogenesis ability was evaluated using tube formation assay. (G and H) Western blot analysis of c-Myc and MMP9. (I-K) Glutamine metabolism was analyzed using special kits. ***P < 0.001, ****P < 0.0001.
Additional file 2: Fig. S2. Circ_0025033 sponged hsa_miR-370-3p to regulate SLC1A5 expression. (A and B) Effects of si-circ_0025033 and anti-hsa_miR-370-3p on SLC1A5 content were monitored using western blot. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Acknowledgements
None.
Abbreviations
- circRNAs
Circular RNAs
- ncRNAs
Noncoding RNAs
- FOXM1
Forkhead box M1
- miRNA
MicroRNA
- ceRNAs
Competitive endogenous RNAs
- SLC1A5/ ASCT2
Solute carrier family 1 member 5
Author contributions
All authors have been involved in the management of the patient and in the conception of the manuscript. The specific authors’ contribution is listed below. H.M.: methodology, formal analysis and investigation, writing—original draft. S.Q.: formal analysis and investigation. Y.Z.: conceptualization, formal analysis and investigation, resources. X.Y.: methodology, writing—review and editing. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
For patients: This research had acquired approval from Ethics Committee of First Affiliated Hospital of Xi'an Jiaotong University on 12 February 2020, under the number 2020ZBV13. It was conducted in accordance with the Helsinki Declaration. For animals: Animal experiments were conducted with the permission of the Animal Care and Use Committee of First Affiliated Hospital of Xi'an Jiaotong University on 5 March 2020, under the number 2020SI351.
Consent for publication
All authors have reviewed the final version of the manuscript and approve it for publication.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 3.Waldmann A, Eisemann N, Katalinic A. Epidemiology of malignant cervical, corpus uteri and ovarian tumours—current data and epidemiological trends. Geburtshilfe Frauenheilkd. 2013;73(2):123–129. doi: 10.1055/s-0032-1328266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 6.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. circRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 7.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. circRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–81 e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou XY, Yang H, Bai YQ, Li XL, Han SY, Zhou BX. hsa_circ_0006916 promotes hepatocellular carcinoma progression by activating the miR-337-3p/STAT3 axis. Cell Mol Biol Lett. 2020;25(1):47. doi: 10.1186/s11658-020-00238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang N, Jin Y, Hu Q, Cheng S, Wang C, Yang Z, et al. Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. J Ovarian Res. 2020;13(1):64. doi: 10.1186/s13048-020-00664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):1224–1233. doi: 10.1080/21691401.2019.1593999. [DOI] [PubMed] [Google Scholar]
- 13.Foruzandeh Z, Zeinali-Sehrig F, Nejati K, Rahmanpour D, Pashazadeh F, Seif F, et al. circRNAs as potent biomarkers in ovarian cancer: a systematic scoping review. Cell Mol Biol Lett. 2021;26(1):41. doi: 10.1186/s11658-021-00284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gartel AL. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 2017;77(12):3135–3139. doi: 10.1158/0008-5472.CAN-16-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochem Pharmacol. 2013;85(5):644–652. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Guo H, Wang Z, Bu H, Wang S, Wang H, et al. Cyclin F and KIF20A, FOXM1 target genes, increase proliferation and invasion of ovarian cancer cells. Exp Cell Res. 2020;395(2):112212. doi: 10.1016/j.yexcr.2020.112212. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Tu K, Liu F, Liang M, Yu K, Wang Y, et al. FoxM1 promotes the migration of ovarian cancer cell through KRT5 and KRT7. Gene. 2020;757:144947. doi: 10.1016/j.gene.2020.144947. [DOI] [PubMed] [Google Scholar]
- 18.Hou W, Zhang Y. circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract. 2021;217:153275. doi: 10.1016/j.prp.2020.153275. [DOI] [PubMed] [Google Scholar]
- 19.Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 21.Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161. [PMC free article] [PubMed] [Google Scholar]
- 22.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong ZH, Zhou F, Shi C, Xiang T, Zhou CK, Wang QQ, et al. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24:9. doi: 10.1186/s11658-018-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun N, Zhang L, Zhang C, Yuan Y. miR-144–3p inhibits cell proliferation of colorectal cancer cells by targeting BCL6 via inhibition of Wnt/beta-catenin signaling. Cell Mol Biol Lett. 2020;25:19. doi: 10.1186/s11658-020-00210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Huang Y, Deng X, Luo M, Wang X, Hu H, et al. Long noncoding RNA H19 promotes transforming growth factor-β-induced epithelial-mesenchymal transition by acting as a competing endogenous RNA of miR-370–3p in ovarian cancer cells. Onco Targets Ther. 2018;11:427–440. doi: 10.2147/OTT.S149908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Luo Y, Li X. Circ_0072995 promotes ovarian cancer progression through regulating miR-122–5p/SLC1A5 axis. Biochem Genet. 2021; [DOI] [PubMed]
- 27.Xiao D, Zeng L, Yao K, Kong X, Wu G, Yin Y. The glutamine-alpha-ketoglutarate (AKG) metabolism and its nutritional implications. Amino Acids. 2016;48(9):2067–2080. doi: 10.1007/s00726-016-2254-8. [DOI] [PubMed] [Google Scholar]
- 28.Michalak KP, Maćkowska-Kędziora A, Sobolewski B, Woźniak P. Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxid Med Cell Longev. 2015;2015:964321. doi: 10.1155/2015/964321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36(10):1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 31.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 32.Haddad G, Lorenzen JM. Biogenesis and function of circular RNAs in health and in disease. Front Pharmacol. 2019;10:428. doi: 10.3389/fphar.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 34.Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18(4):547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 35.Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun. 2019;510(2):334–338. doi: 10.1016/j.bbrc.2019.01.108. [DOI] [PubMed] [Google Scholar]
- 36.Cheng H, Wang N, Tian J, Li Y, Ren L, Shi Z. Circular RNA circ_0025033 promotes the evolvement of ovarian cancer through the regulation of miR-330–5p/KLK4 axis. Cancer Manag Res. 2020;12:2753–2765. doi: 10.2147/CMAR.S241372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Zhang R, Wang J, Jia E, Zhang J, Liu N, Chi C. lncRNA BCAR4 sponges miR-370-3p to promote bladder cancer progression via Wnt signaling. Int J Mol Med. 2020;45(2):578–588. doi: 10.3892/ijmm.2019.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, Yan C, Tao S, Wang H. circ_0058124 aggravates the progression of papillary thyroid carcinoma by activating LMO4 expression via targeting miR-370–3p. Cancer Manag Res. 2020;12:9459–9470. doi: 10.2147/CMAR.S271778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Z, Wu T, Li Y, Xu Z, Zhang S, Liu B, et al. MicroRNA-370–3p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting β-catenin. Brain Res. 2016;1644:53–61. doi: 10.1016/j.brainres.2016.04.066. [DOI] [PubMed] [Google Scholar]
- 40.Lei W, Lin J, Liu F, Chen N. Long noncoding RNA GAS6 antisense RNA1 silencing attenuates the tumorigenesis of acute myeloid leukemia cells through targeting microRNA-370-3p/Tetraspanin3 axis. Clin Hemorheol Microcirc. 2021;78(1):69–81. doi: 10.3233/CH-201039. [DOI] [PubMed] [Google Scholar]
- 41.Lo SS, Hung PS, Chen JH, Tu HF, Fang WL, Chen CY, et al. Overexpression of miR-370 and downregulation of its novel target TGFβ-RII contribute to the progression of gastric carcinoma. Oncogene. 2012;31(2):226–237. doi: 10.1038/onc.2011.226. [DOI] [PubMed] [Google Scholar]
- 42.Jin Y, Zhang M, Duan R, Yang J, Yang Y, Wang J, et al. Long noncoding RNA FGF14-AS2 inhibits breast cancer metastasis by regulating the miR-370–3p/FGF14 axis. Cell Death Discov. 2020;6:103. doi: 10.1038/s41420-020-00334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F, Zhou J. CircAGFG1 promotes cervical cancer progression via miR-370-3p/RAF1 signaling. BMC Cancer. 2019;19(1):1067. doi: 10.1186/s12885-019-6269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGivan JD, Bungard CI. The transport of glutamine into mammalian cells. Front Biosci. 2007;12:874–882. doi: 10.2741/2109. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Beaumont KA, Otte NJ, Font J, Bailey CG, van Geldermalsen M, et al. Targeting glutamine transport to suppress melanoma cell growth. Int J Cancer. 2014;135(5):1060–1071. doi: 10.1002/ijc.28749. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Hardie RA, Hoy AJ, van Geldermalsen M, Gao D, Fazli L, et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J Pathol. 2015;236(3):278–289. doi: 10.1002/path.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye J, Huang Q, Xu J, Huang J, Wang J, Zhong W, et al. Targeting of glutamine transporter ASCT2 and glutamine synthetase suppresses gastric cancer cell growth. J Cancer Res Clin Oncol. 2018;144(5):821–833. doi: 10.1007/s00432-018-2605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu K, Kaira K, Tomizawa Y, Sunaga N, Kawashima O, Oriuchi N, et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br J Cancer. 2014;110(8):2030–2039. doi: 10.1038/bjc.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J, Yang T, Peng Z, Xiao H, Jiang N, Zhang L, et al. SLC1A5 silencing inhibits esophageal cancer growth via cell cycle arrest and apoptosis. Cell Physiol Biochem. 2018;48(1):397. doi: 10.1159/000491769. [DOI] [PubMed] [Google Scholar]
- 51.Guo H, Xu Y, Wang F, Shen Z, Tuo X, Qian H, et al. Clinical associations between ASCT2 and p-mTOR in the pathogenesis and prognosis of epithelial ovarian cancer. Oncol Rep. 2018;40(6):3725–3733. doi: 10.3892/or.2018.6729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. SLC1A5 and circ_0025033 had similar roles in ovarian cancer. (A-K) SKOV3 and A2780 cells were transfected with si-NC or si-SLC1A5. (A) Western blot analysis of SLC1A5 content. (B-E) Proliferation, apoptosis, and invasion were assessed using CCK-8, EdU, and flow cytometry assays, respectively. (F) Angiogenesis ability was evaluated using tube formation assay. (G and H) Western blot analysis of c-Myc and MMP9. (I-K) Glutamine metabolism was analyzed using special kits. ***P < 0.001, ****P < 0.0001.
Additional file 2: Fig. S2. Circ_0025033 sponged hsa_miR-370-3p to regulate SLC1A5 expression. (A and B) Effects of si-circ_0025033 and anti-hsa_miR-370-3p on SLC1A5 content were monitored using western blot. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Data Availability Statement
Not applicable.